Abstract
As observed previously, tetraspanin palmitoylation promotes tetraspanin microdomain assembly. Here, we show that palmitoylated integrins (α3, α6, and β4 subunits) and tetraspanins (CD9, CD81, and CD63) coexist in substantially overlapping complexes. Removal of β4 palmitoylation sites markedly impaired cell spreading and signaling through p130Cas on laminin substrate. Also in palmitoylation-deficient β4, secondary associations with tetraspanins (CD9, CD81, and CD63) were diminished and cell surface CD9 clustering was decreased, whereas core α6β4–CD151 complex formation was unaltered. There is also a functional connection between CD9 and β4 integrins, as evidenced by anti-CD9 antibody effects on β4-dependent cell spreading. Notably, β4 palmitoylation neither increased localization into “light membrane” fractions of sucrose gradients nor decreased solubility in nonionic detergents—hence it does not promote lipid raft association. Instead, palmitoylation of β4 (and of the closely associated tetraspanin CD151) promotes CD151–α6β4 incorporation into a network of secondary tetraspanin interactions (with CD9, CD81, CD63, etc.), which provides a novel framework for functional regulation.
Introduction
The α6β4 integrin appears on epithelial and other types of cells, acts as a receptor for basement membrane laminin-5 and related laminin isoforms, and plays a key role during cell migration and tumorigenesis (Belkin and Stepp, 2000; Mercurio et al., 2001). In response to EGF receptor (EGFR) stimulation, α6β4 disconnects from the intermediate filament cytoskeleton and becomes associated with the actin cytoskeleton in lamellipodia and membrane ruffles (Mercurio et al., 2001). During this process, EGFR signaling might activate the Src family kinase fyn, leading to phosphorylation of β4 on tyrosine (Mainiero et al., 1996; Mariotti et al., 2001), or might activate conventional PKC, leading to β4 phosphorylation on serine (Rabinovitz et al., 1999). Consistent with cooperative signaling between α6β4 and growth factor receptors, α6β4 has been suggested to physically associate with fyn (Mariotti et al., 2001), EGFR (Mariotti et al., 2001), ErbB2 (Gambaletta et al., 2000; Hintermann et al., 2001), c-met (Trusolino et al., 2001), and Ron (Santoro et al., 2003).
The laminin-binding integrins (α6β4, α3β1, α6β1, and α7β1) not only form a distinct subgroup among integrins in terms of amino acid sequence similarity, but also show robust association with tetraspanin proteins (Hemler, 1998; Berditchevski, 2001). There are 32 mammalian tetraspanins, and at least a few of these are abundantly present on nearly all cell and tissue types. Tetraspanin proteins regulate cell motility, morphology, fusion, and signaling in the brain and immune system, on tumors, and elsewhere (Levy et al., 1998; Boucheix and Rubinstein, 2001; Hemler, 2001; Stipp et al., 2003b). Tetraspanins CD151, CD81, and CD9 can modulate α3β1 and α6β1 integrin–dependent neurite outgrowth, cell migration, and/or cell morphology (Yánez-Mó et al., 1998; Yauch et al., 1998; Stipp and Hemler, 2000; Kazarov et al., 2002; Zhang et al., 2002). Of particular relevance here, CD151 associates with α6β4 to regulate kidney epithelial cell morphology (Yang et al., 2002), whereas CD9–α6β4 complexes may affect primary keratinocyte cell motility (Jones et al., 1996; Baudoux et al., 2000).
Associations of tetraspanins with each other are at least partly stabilized by palmitoylation. Mutation of CD9 palmitoylation sites impaired associations with tetraspanins CD81 and CD53 (Charrin et al., 2002), and loss of CD151 palmitoylation decreased association with other tetraspanins (CD81, CD63, and CD9), without affecting integrin α3β1 association (Berditchevski et al., 2002; Yang et al., 2002). Palmitoylation of CD151 contributes to cell signaling (Berditchevski et al., 2002). In some proteins (e.g., G proteins and Src family kinases), palmitoylation leads to the diminished detergent solubility and lower protein density characteristic of lipid raft association (Dunphy and Linder, 1998; Resh, 1999). However, palmitoylation of tetraspanins CD9 and CD151 causes neither decreased protein density in sucrose gradients nor decreased detergent solubility (Berditchevski et al., 2002; Charrin et al., 2002; Yang et al., 2002).
The α6β4 integrin, like other laminin-binding integrins, associates strongly with CD151 (Sterk et al., 2000, 2002). CD151 association with laminin-binding integrins is direct, occurs early in biosynthesis, and is resistant to disruption by nonionic detergents (Yauch et al., 2000; Berditchevski et al., 2001; Kazarov et al., 2002). Removal of CD151 palmitoylation sites did not disrupt the CD151–α6β4 complex in epithelial cells, but did strongly influence α6β4 integrin–dependent cell morphology (Yang et al., 2002). In contrast to the primary (i.e., direct) associations of α3 and α6 integrins with CD151, there is an extended network of secondary (i.e., most likely indirect) associations with other tetraspanins (e.g., CD9, CD81, and CD63) that occur later in biosynthesis and are more sensitive to nonionic detergents (Berditchevski et al., 2001; Kazarov et al., 2002). These secondary-type associations are impaired upon removal of CD151 or CD9 palmitoylation sites (Berditchevski et al., 2002; Charrin et al., 2002; Yang et al., 2002).
The integrin β4 subunit was recently shown also to be palmitoylated (Gagnoux-Palacios et al., 2003). Here, we provide evidence for palmitoylation of integrins α3, α6, and β4, and show striking parallels between β4 integrin and CD151 palmitoylations, in terms of effects on (1) core complex (CD151–α6β4) formation, (2) secondary complex formation (involving CD9, CD81, and CD63), (3) integrin density, (4) integrin solubility, (5) integrin-dependent cell morphology, and (6) integrin signaling. Our results are in sharp contrast to previous suggestions that β4 palmitoylation promotes lipid raft and Src family kinase association (Mariotti et al., 2001; Gagnoux-Palacios et al., 2003). We suggest that integrin palmitoylation promotes, instead of lipid rafts, assembly of a novel type of signaling platform enriched for palmitoylated tetraspanins.
Results
A network of palmitoylated integrins and palmitoylated tetraspanins
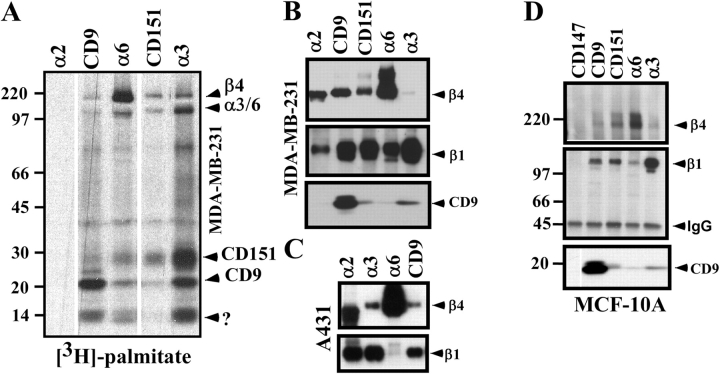
While studying palmitoylated CD151 (Yang et al., 2002), we noticed that it associates with palmitoylated proteins resembling the β4, α3, and α6 integrin subunits (Fig. 1 A, lanes 3 and 4). [3H]palmitate-labeled integrin subunits from [3H]palmitate-labeled MDA-MB-231 breast carcinoma cells were more abundant on the cell surface (Fig. 1 A, lane 3) than intracellularly (Fig. 1 A, lane 4), and were not present in tetraspanin CD82 immunoprecipitations (Fig. 1 A, lanes 1 and 2). Integrin palmitoylation was confirmed by recovery of [3H]palmitate-labeled α3 (Fig. 1 B, lane 5), α6 (Fig. 1 B, lanes 6 and 8), and β4 (Fig. 1 B, lane 6), but not α2 (Fig. 1 B, lanes 4 and 7), from stringent detergent (RIPA) lysates of A431 cells or B12 kidney epithelial cells. In A431 cells, α2, α3, and α6 integrin subunits were present at comparable levels, as indicated by cell surface labeling (Fig. 1 B, lanes 1–3), and in B12 cells, α2 and α6 were again at comparable levels (not depicted).
Figure 1.
Integrin α3, α6, and β4 subunits undergo palmitoylation. (A) MDA-MB-231 human breast carcinoma cells were pulsed with [3H]palmitate and incubated at 4°C with mAb anti-CD82 (mAb M104) or anti-CD151 (mAb 5C11). Unbound antibody was removed, cells were lysed in 1% Brij 96, and immune complexes were collected using protein G–agarose. Remaining lysate was then used for further immunoprecipitation. In, intracellular fraction; Sur, surface fraction. (B) A431 cells were pulsed with [3H]palmitate, surface labeled with biotin, and lysed in RIPA buffer, and then human α2, α3, and α6 integrins were immunoprecipitated using mAbs A2-IIE10, A3-X8, and GoH3, respectively. Proteins labeled with biotin (lanes 1–3) and [3H]palmitate (lanes 4–6) are shown. Kidney epithelial B12 cells expressing human integrin α2 and α6 were also labeled with [3H]palmitate, and then α2 and α6 were immunoprecipitated (lanes 7 and 8). Comparable cell surface α2 and α6 levels were seen by flow cytometry (not depicted). Note that, under RIPA conditions, human α6 remains associated with human (lane 6), but not mouse (lane 8) β4. White lines indicate that intervening lanes have been spliced out. (C) A431 cells were pulsed with [3H]palmitate and then chased in the presence of 100 μM unlabeled palmitic acid for up to 6 h, as indicated. Anti-α6 and anti-CD151 immunoprecipitates were analyzed for [3H] labeling of β4 and CD151 (top) or by immunoblotting (loading controls, bottom) using anti-α6 cyto tail antibody (to evaluate α6β4) and anti-α3 cyto tail antibody (to evaluate α3β1–CD151 complexes). Question mark indicates unknown protein.
When A431 cells were pulse labeled with [3H]palmitate, followed by a 6-h chase, the palmitate label was retained fully on the β4 subunit (Fig. 1 C, top left), and almost completely on CD151 (Fig. 1 C, top right). Blotting of α6 and α3 (Fig. 1 C, bottom) revealed similar levels of total α6β4 and α3β1–CD151 complexes throughout the experiment. Palmitoylations of β4 and CD151 were almost completely inhibited by a protein synthesis inhibitor (cycloheximide) and by a Golgi-disrupting agent (brefeldin A) (unpublished data). Hence, CD151 and β4 palmitoylations both occur early in biosynthesis and undergo little subsequent turnover.
Results shown in Fig. 1 A suggested that additional palmitoylated proteins, including CD9, associate with palmitoylated CD151 and palmitoylated integrins. Indeed, the profiles of palmitoylated proteins associated with tetraspanins (CD9 and CD151) and integrins (α6β4 and α3β1) from MDA-MB-231 cells are strikingly similar (Fig. 2 A). No [3H]palmitate-labeled proteins associated with α2 integrin, which itself is not palmitoylated (Fig. 2 A, lane 1). Palmitoylated protein profiles for CD9, CD151, α3β1, and α6β4 were similarly congruent in two additional cell lines (MCF-10A and A431; unpublished data). Immunoprecipitation of CD9, CD151, α3, and α6 from MDA-MB-231 cells (Fig. 2 B) or MCF-10A cells (Fig. 2 D) yielded, in each case, β4, β1, and CD9, as indicated by blotting. Immunoprecipitation of α3, α6, and CD9 from A431 cells again yielded, in each case, both β4 and β1 (Fig. 2 C). Immunoprecipitation of α2 yielded β1 (as expected), but not β4 or CD9 (Fig. 2, B and C). Likewise, immunoprecipitation of CD147 (a highly expressed control cell surface protein) did not yield β1, β4, or CD9 (Fig. 2 D). Results shown in Fig. 2 indicate that complexes containing palmitoylated CD9, CD151, α6β4, and α3β1 are substantially overlapping in three different cell lines.
Figure 2.
Palmitoylated integrins associate with each other and with multiple palmitoylated tetraspanin proteins. (A) MDA-MB-231 cells were pulsed with [3H]palmitate, lysed in 1% CHAPS, and then integrins (α2, α6, and α3) and tetraspanins (CD9 and CD151) were immunoprecipitated using mAbs A2-IIE10, GoH3, A3-X8, Du-All, and 5C11, respectively. Question mark indicates unknown protein. (B) MDA-MB-231 cells were immunoprecipitated as in A, and then samples were blotted for β4 (rabbit pAb), β1 (rabbit pAb), and CD9 (C9BB), as indicated. (C) A431 cells were lysed and immunoprecipitated as in A, and then samples were blotted for β4 and β1 (as in B). Note that the band in the α2 lanes migrating slightly below the β4 band (B and C) is an Ig background band, which does not appear in the α2 lane when [3H]palmitate labeling is used (A). (D) Indicated proteins were immunoprecipitated from MCF-10A cells, as in A, and then blotted as in B. As a negative control, abundant cell surface protein CD147 was immunoprecipitated using mAb 8G6. White lines (A and D) indicate that intervening lanes have been spliced out.
Removal of integrin palmitoylation sites
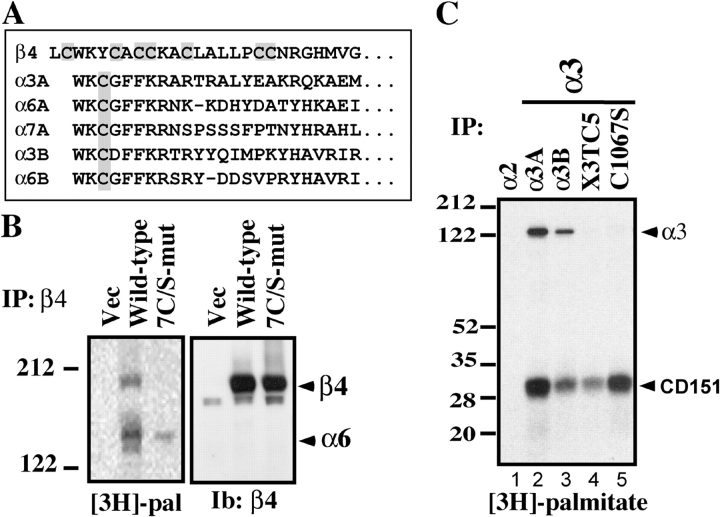
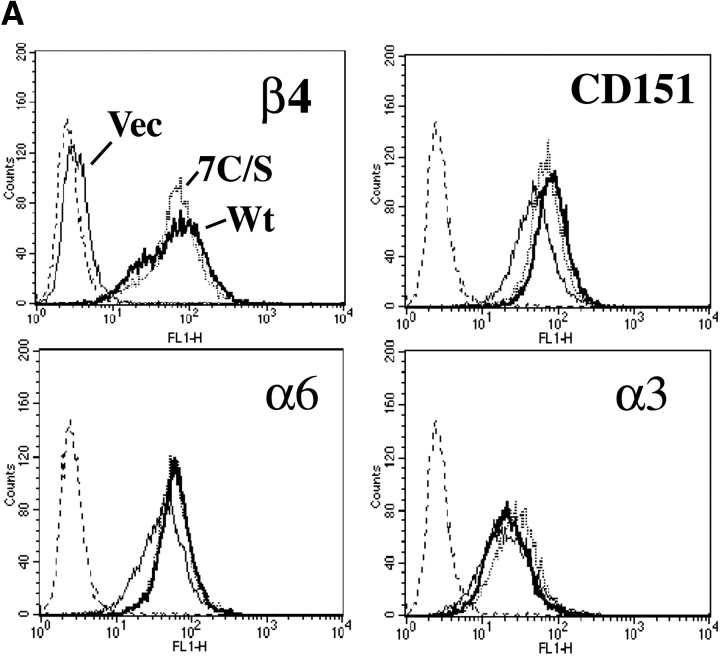
The membrane-proximal region of the β4 cytoplasmic tail contains seven potential cysteine palmitoylation sites (Fig. 3 A). Because palmitoylation often occurs on multiple clustered cysteines in the same molecule (Gundersen et al., 1994; Chapman et al., 1996; Berditchevski et al., 2002; Charrin et al., 2002; Yang et al., 2002), we prepared a “7C/S” β4 mutant, replacing all seven cysteines with serines. When stably expressed in MDA-MB-435 cells, wild-type β4, but not the 7C/S mutant, incorporated [3H]palmitate (Fig. 3 B, left). Although β4 palmitoylation was lost, the 7C/S mutant retained association with palmitoylated α6 (Fig. 3 B, left). Wild-type and mutant β4 were present at similar levels, as detected by anti-β4 immunoblotting (Fig. 3 B, right; and Fig. 4 B, middle), flow cytometry (Fig. 4 A), cell surface biotinylation (Fig. 4 B, top), and by [35S]methionine labeling (not depicted). Mutant and wild-type β4 associated similarly with their core partner, CD151 (Fig. 4 B, bottom), and brought similarly elevated amounts of CD151 and α6 to the cell surface (Fig. 4 A). Levels of cell surface α3 (Fig. 4 A) and β1 (not depicted) also remained very similar when either wild-type or mutant β4 was expressed.
Figure 3.
Identification of integrin palmitoylation sites. (A) Membrane-proximal regions containing candidate palmitoylation sites are shaded gray. The first lysine defines a putative transmembrane interface. A and B designate alternatively spliced forms of integrin cytoplasmic tails. (B) MDA-MB-435 cells stably expressing mutant or wild-type β4, or vector control, were [3H]palmitate labeled and lysed in 1% Brij 96. The α2 integrin was immunoprecipitated from vector control cells, and α6β4 was immunoprecipitated (using anti-α6 mAb GoH3) from β4-transfected cells. Shown are proteins labeled with [3H]palmitate (left) or blotted with anti-β4 antibody (right). (C) Murine B12 cells, stably expressing human integrin α subunits, were [3H]palmitate labeled and lysed in 1% RIPA. Integrins were immunoprecipitated using anti–human α2 (lane 1) and α3 (lanes 2–5) antibodies (A2-IIE10 and A3-X8) and resolved by SDS-PAGE, and [3H]palmitate was detected. The α3 subunits include α3-X3TC5 (α3 tail and transmembrane regions are replaced by those regions from α5; Yauch et al., 1998) and α3-C1067S (palmitoylation site point mutant).
Figure 4.

Comparable expression of wild-type and mutant β4 in MDA-435 cells. (A) MDA-MB-435 cells stably expressing vector or wild-type (Wt) or 7C/S β4 were analyzed using anti-β4 (mAb AA3), anti-CD151 (mAb 5C11), anti-α6 (mAb A6-ELE), and anti-α3 (mAb A3-X8). Histograms represent Wt β4 cells (thick line), 7C/S β4 cells (dashed line), background autofluorescence (dotted line), and vector control cells (thin line). Data are representative of three independent experiments. (B) MDA-MB-435 cells stably expressing vector or Wt or 7C/S β4 were surface labeled with biotin and lysed in 1% Brij 96, and then the α6β4–CD151 complex was immunoprecipitated (using anti-β4 mAb AA3) and analyzed by blotting for biotin-labeled proteins (avidin blotting; top), total β4 (rabbit pAb; middle), or CD151 (mAb 1A5; bottom).
The membrane-proximal regions of integrin α3, α6, and α7 cytoplasmic tails contain a single cysteine residue (Fig. 3 A) that is highly conserved in diverse animal species, but not in most other integrin α chains. Upon replacement of this key residue in α3 with serine (C1067S mutation), or upon replacement of the entire α3 cytoplasmic tail and transmembrane with the tail and transmembrane of α5 (X3TC5), palmitoylation was completely lost (Fig. 3 C, lanes 4 and 5). In the same experiment, palmitoylation of wild-type human α3A and α3B was readily observed (Fig. 3 C, lanes 2 and 3). Despite loss of palmitoylation in the C1067S and X3TC5 mutants, these integrins retained association with palmitoylated tetraspanin CD151 (Fig. 3 C, lanes 4 and 5).
Effects of integrin β4 palmitoylation on cell functions
We observed comparable hemidesmosome-like staining for GFP-tagged wild-type and mutant β4 in A431 cells (unpublished data). Because a high level of endogenous β4 precluded further functional studies in A431 cells, we switched to MDA-MB-435 cells, with minimal endogenous β4, for studies of stably expressed wild-type and 7C/S β4. On laminin-5, spreading of 7C/S cells was markedly impaired compared with that of cells with control vector or wild-type β4 (Fig. 5 A, top). All cells spread equally well on vitronectin (Fig. 5 A, bottom). Quantitation of multiple experiments confirmed deficient 7C/S β4 cell spreading on laminin-5 but not vitronectin substrate (Fig. 5 B). A marked defect in tyrosine phosphorylation of p130Cas was also observed for 7C/S β4 cells (Fig. 5 C, lanes 7 and 12) compared with wild-type β4 cells (Fig. 5 C, lanes 6 and 11), when plated on laminin-1 or laminin-5. No defect was seen on control ligand (vitronectin; Fig. 5 C, lanes 3 and 4), and minimal p130Cas phosphorylation was seen for cells in suspension (Fig. 5 C, lanes 1 and 2). In contrast to results with p130Cas, mutant and wild-type β4 showed little difference in phosphorylation of FAK (unpublished data). In concert with cell spreading, p130Cas is typically phosphorylated by a mechanism dependent on Src family kinases (O'Neill et al., 2000). Consistent with this, the Src family kinase inhibitor PP2 (Hanke et al., 1996) abolished both p130Cas phosphorylation (see Fig. 7 A, lanes 8–10) and wild-type β4–MDA-MB-435 cell spreading on laminin-5 (not depicted). Similar spreading and signaling defects were also seen for 7C/S β4–transfected SK-MEL-5 melanoma cells (unpublished data).
Figure 5.
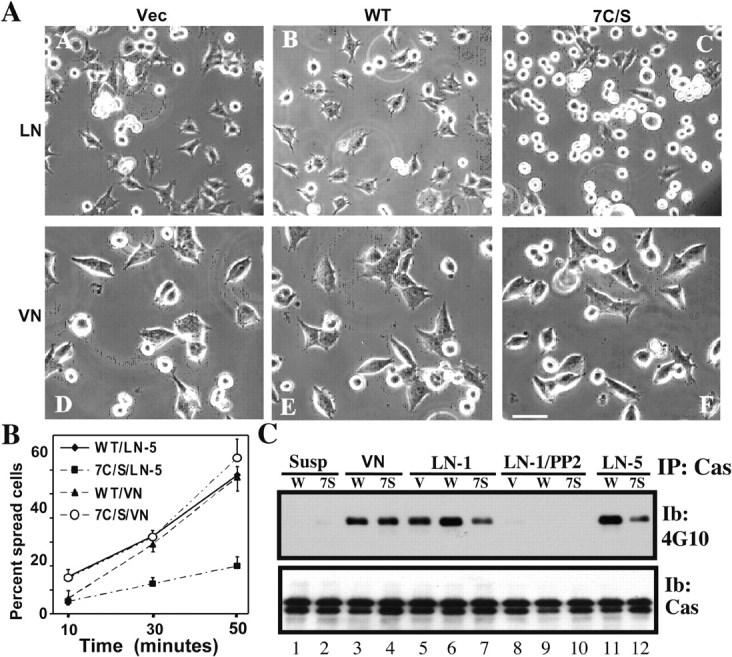
β4 integrin–mediated cell spreading. (A) MDA-MB-435 cells stably expressing vector or wild-type (WT) or 7C/S β4 were held in suspension at 37°C for 30 min, and then were plated on wells coated in PBS at 4°C overnight with either laminin-5 (LN; subpanels A–C) or vitronectin (VN; subpanels D–E), and photographed after 45 min. Cell spreading was monitored using a microscope (Axiovert 135; Carl Zeiss MicroImaging, Inc.) as described previously (Stipp and Hemler, 2000). Bar, 50 μm. (B) Cell spreading was estimated by determining the percentage of cells that were no longer round and phase-bright. The accuracy of this method was validated by quantitation of cell area using Scion Image software, which confirmed that we could readily distinguish cells in which the area had increased by ≥1.2-fold. Results (mean ± SD) are derived from at least two separate experiments, counting at least two representative fields in each experiment, with at least 50 cells/field. (C) MDA-MB-435 cells were suspended at 37°C for 30 min, and then either retained in suspension or plated on vitronectin (Vn), laminin-1, or laminin-5 for 45 min. Some samples (lanes 8–10) were preincubated with 1.0 μg/ml PP2 in DMSO for 20 min before plating. Cells were collected and lysed in RIPA, and p130Cas was immunoprecipitated. After SDS-PAGE, samples were blotted with antiphosphotyrosine mAb 4G10. To assess protein loading, the blot was stripped and reblotted with p130Cas-specific antibody. Similar results were seen in multiple experiments.
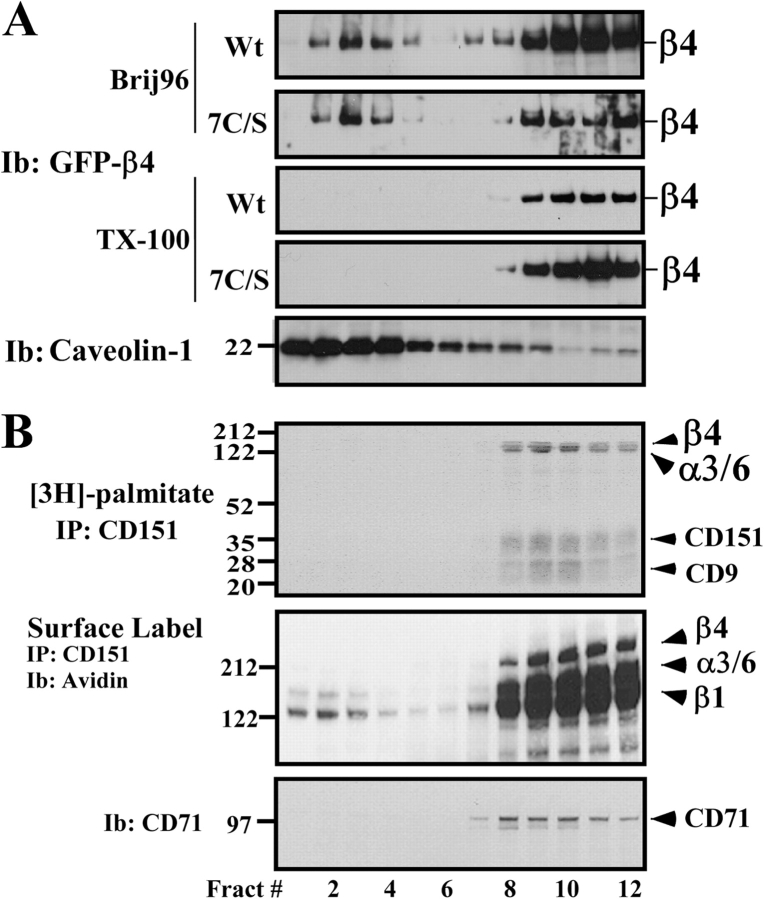
Figure 7.
Sucrose density gradient analyses of β4 integrins and integrin–tetraspanin complexes. (A) A431 cells (expressing either wild-type [Wt] or 7C/S β4–GFP) were lysed in 1% Brij 96 or 1% Triton X-100, and then fractionated on 5–35–45% discontinuous sucrose density gradients at 4°C (Claas et al., 2001). 50-μl aliquots of total lysate from each fraction were blotted for β4, using anti-GFP mAb, or for caveolin-1, using rabbit pAb. (B) Untransfected A431 cells were pulsed with [3H]palmitate, surface labeled with biotin, lysed in 1% Brij 96, and then fractionated as in A. From each of the 12 fractions, 50 μl was used for immunoprecipitation of CD151 (mAb 5C11), and [3H] labeling was detected (top), or CD151 was immunoprecipitated and proteins that were surface labeled with biotin were detected by avidin blotting (middle). Also, 50-μl aliquots of total lysate from each fraction were blotted for CD71 (transferrin receptor; bottom).
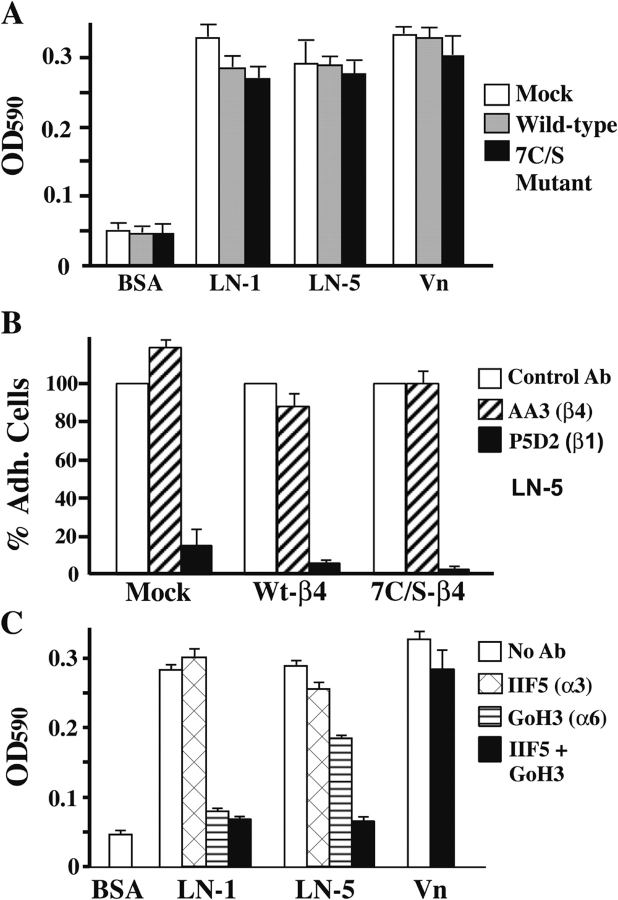
The spreading defect does not arise because of diminished cell adhesion. In a 30-min static cell adhesion assay, all cells adhered similarly to laminin-1, laminin-5, and vitronectin (Fig. 6 A). Adhesion to laminin-5 (Fig. 6 B) or laminin-1 (not depicted) was dependent on β1 rather than β4 integrins, as indicated by nearly complete blocking with an anti-β1 monoclonal antibody. On laminin-1, adhesion was almost entirely α6β1 dependent, whereas on laminin-5, α3β1 also contributed (Fig. 6 C). On vitronectin, antibodies to laminin receptors had minimal inhibitory effect. The use of α6β1, but not α6β4, for adhesion is in accord with a previous study of untransfected and β4-transfected MDA-MB-435 cells (Shaw et al., 1997). Expression levels for α3β1 and α6β1 were essentially unchanged, regardless of whether mutant or wild-type β4 was present, as seen by flow cytometry (Fig. 4 A) and immunoblotting (not depicted).
Figure 6.
β1-dependent cell adhesion is unaffected by β4 mutation. (A) To assess cell adhesion, MDA-MB-435 cells were plated on specific substrates for 30 min, and then nonadherent cells were removed by washing and adherent cells were stained using Wright-Giemsa and then quantitated at OD 590 using an ELISA 96-well plate reader. Starting with 70,000 cells/well, ∼70% cell adhesion corresponds to OD 590 = 0.3. (B) Cells were incubated with ∼10 μg/ml of the indicated mAbs at RT for 15 min before being plated on laminin-5. Adhesion was determined as in A. (C) Effects of indicated antibodies on adhesion of wild-type (Wt) β4 cells was determined as in A and B. Results shown are the means of triplicate determinations ± SD.
Lipid rafts, growth factor receptors, and Src kinases
Palmitoylation diminishes the solubility of some lipid raft–type proteins, leading to increased appearance in “light membrane” fractions of sucrose gradients and decreased extractability in nonionic detergents (Dunphy and Linder, 1998; Resh, 1999). However, CD151 palmitoylation neither decreased density in sucrose gradients nor decreased detergent solubility (Yang et al., 2002). Similarly, in A431 cells, palmitoylation of β4 (compared with 7C/S β4) did not yield decreased density in sucrose gradients in either stringent (Triton X-100) or mild (1% Brij 96) detergent conditions (Fig. 7 A). Furthermore, neither endogenous [3H]palmitate-labeled β4 nor endogenous β4 cell surface–labeled with biotin appeared in the low density fractions of a sucrose gradient, even when mild Brij 96 detergent conditions were used (Fig. 7 B). In control experiments, abundant endogenous caveolin-1 appeared in low density fractions (Fig. 7 B, Fract 1–4) and transferrin receptor (CD71) marked the dense fractions (Fig. 7 B, Fract 8–12). Furthermore, in MDA-MB-435 cells, palmitoylated β4 integrin (compared with the 7C/S mutant) showed increased solubility in nonionic detergent. Upon extraction of cells with 1% Brij 96 for 10 or 20 min, the solubility of wild-type β4 protein was 69% and 82%, respectively, relative to the total extractable β4 (in RIPA). In marked contrast, the 7C/S mutant remained at 40% solubility at the 10- and 20-min time points, although by 30 min, both mutant and wild-type β4 were completely extracted. In conclusion, palmitoylation did not endow β4 with lipid raft–like properties in either A431 or MDA-MB-435 cells.
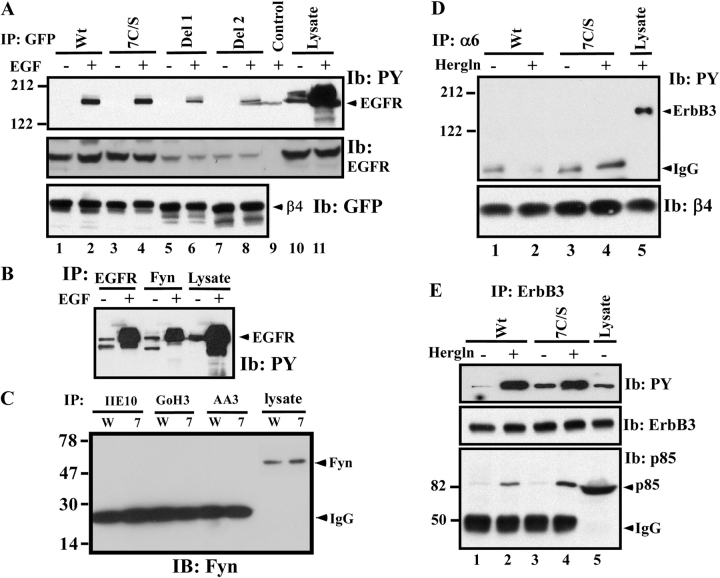
As mentioned in the Introduction, integrin β4 can associate with activated EGFR-type growth factor receptors and Src family kinases. However, in EGF-stimulated A431 cells, we observed no difference in levels of tyrosine-phosphorylated EGFR associating with GFP-tagged 7C/S and wild-type β4 (Fig. 8 A, top, lanes 2 and 4). In control experiments, deletion of portions of the β4 tail did diminish association with tyrosine-phosphorylated EGFR (Fig. 8 A, top, lanes 6 and 8) and EGFR protein (Fig. 8 A, middle, lanes 5–8). Tyrosine-phosphorylated EGFR was not detected in the absence of EGF stimulation (Fig. 8 A, top, lanes 1, 3, 5, and 7) or when β4 was not immunoprecipitated (Fig. 8 A, lane 9). Comparable amounts of β4 were recovered in each lane (Fig. 8 A, bottom, lanes 1–8). As expected (Mariotti et al., 2001), an abundance of tyrosine-phosphorylated EGFR was recovered in association with the Src family kinase fyn (Fig. 8 B), but no fyn was recovered in association with either mutant or wild-type β4 that had been immunoprecipitated with antibodies to either α6 or β4 (Fig. 8 C). In addition, there were no consistent differences in tyrosine phosphorylation of 7C/S and wild-type β4 (unpublished data). Integrin α6β4 also associates with ErbB2 (Hintermann et al., 2001), which in MDA-MB-435 cells forms dimers with ErbB3 (Adelsman et al., 1999). However, after heregulin treatment to induce ErbB phosphorylation, we observed no association of either wild-type or mutant β4 with tyrosine-phosphorylated ErbB3 (Fig. 8 D) or ErbB2 (not depicted). Also, for mutant β4 in MDA-MB-435 cells, we saw no diminution in ErbB3 tyrosine phosphorylation or ErbB3 association with the p85 subunit of PI 3-K (Fig. 8 E).
Figure 8.
β4 palmitoylation does not affect association with EGFR, fyn, or ErbB3. (A) A431 cells stably expressing wild-type (Wt) or mutant β4–GFP proteins were serum-starved for ∼12 h, stimulated with (or without) 50 ng/ml EGF for 10 min, and lysed in RIPA, and then β4 was immunoprecipitated using anti-GFP pAb. Samples were blotted for phosphotyrosine (mAb 4G10) and β4 (mAb anti-GFP). Del1, β4 deleted after the third fibronectin repeat (aa 1582); Del2, β4 deleted after the second fibronectin repeat (aa 1412). (B) Samples were prepared as in A, and then EGFR and fyn were immunoprecipitated and blotted for phosphotyrosine, using mAb 4G10. (C) A431 transfectants were stimulated and lysed as in A; α2, α6, and β4 immunoprecipitations were performed; and samples were blotted for fyn (using pAb). W, wild-type β4; 7, 7C/S β4. (D) MDA-MB-435 transfectants were treated with (or without) 100 ng/ml heregulin for 10 min and lysed in RIPA, and then α6β4 was immunoprecipitated using mAb GoH3. ErbB3 was blotted for phosphotyrosine (top) and β4 was blotted using anti-β4 pAb (bottom). (E) MDA-MB-435 cell lysate was prepared in D, and ErbB3 was immunoprecipitated. Samples were blotted for phosphotyrosine, total ErbB3, and p85 subunit of PI 3-K.
Diminished association with tetraspanins
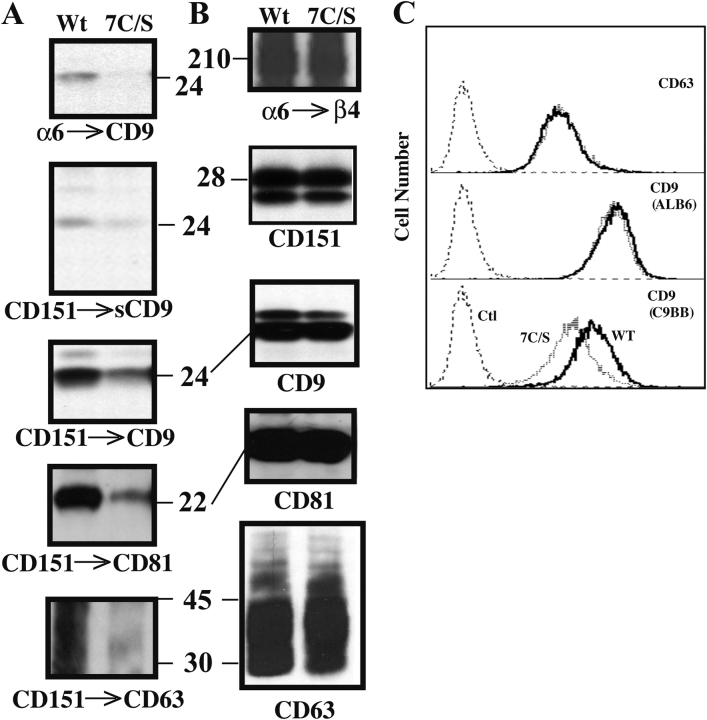
Removal of CD151 palmitoylation sites did not affect core association with α3β1 or α6β4, but did alter secondary associations with other tetraspanins (Berditchevski et al., 2002; Yang et al., 2002). Likewise, removal of β4 palmitoylation sites did not affect core association with CD151 (Fig. 4 B), but did markedly alter secondary associations with other tetraspanins (Fig. 9 A). As indicated by immunoprecipitations of α6 or CD151, association of CD9, cell surface CD9 (sCD9), CD81, and CD63 was impaired in 7C/S–MDA-MB-435 cells, in five different experiments. In control experiments, the levels of wild-type and 7C/S β4 associated with α6 were unaltered, as were the total levels of CD151, CD9, CD81, and CD63 (Fig. 9 B). Similar levels of CD63 were seen by flow cytometry (Fig. 9 C, top).
Figure 9.
Loss of β4 palmitoylation sites causes tetraspanin reorganization. (A) MDA-MB-435 cells stably expressing wild-type (Wt) or 7C/S β4 were surface labeled with biotin and lysed in 1% Brij 96, and then the α6β4–CD151 complex was immunoprecipitated using anti-α6 mAb GoH3 or anti-CD151 mAb 5C11. Panels were then blotted for CD9 (mAb C9BB; first and third panels), cell surface CD9 (avidin blotting; second panel), CD81 (mAb M38; fourth panel), or CD63 (mAb 6H1; fifth panel). (B) From an anti-α6 immunoprecipitation, surface labeled β4 was detected (avidin blotting; first panel). CD151 was immunoprecipitated (mAb 5C11) and detected (mAb 1A5; second panel), CD9 was immunoprecipitated (mAb ALB6) and blotted (mAb C9BB; third panel), CD81 was immunoprecipitated and blotted (mAb M38; fourth panel), and CD63 was immunoprecipitated and blotted (mAb 6H1; fifth panel). (C) MDA-MB-435 cells stably expressing mock, Wt, or 7C/S β4 were analyzed using anti-CD63 (mAb 6H1) and the indicated anti-CD9 antibodies. Histograms are from Wt β4 cells (thick line), 7C/S β4 cells (dashed line), and background autofluorescence (dotted line). Data are representative of three independent experiments.
Flow cytometry showed that surface levels of tetraspanin CD9 were very similar, as indicated by mAb ALB6 staining (Fig. 9 C, middle), but different as indicated by mAb C9BB staining (Fig. 9 C, bottom). Indeed, the C9BB/ALB6 staining ratio (0.59 in wild-type cells) decreased to 0.34 in 7C/S cells. After β4-transfected MDA-MB-435 cells had been cultured for a few more months, there was an even larger difference in C9BB/ALB6 ratios (0.56 in wild-type vs. 0.07 in 7C/S cells), whereas mutant and wild-type β4 levels remained equal (unpublished data). Wild-type β4–GFP and 7C/S β4–GFP were expressed in another cell line (SK-MEL-5), and again there was similar staining of CD9 by mAb ALB6, but selectively diminished staining of the mutant cells by mAb C9BB, such that the C9BB/ALB6 ratio decreased by ∼40% (unpublished data). These results indicate that cell surface tetraspanin organization is affected by β4 palmitoylation.
Functional link between CD9 and β4
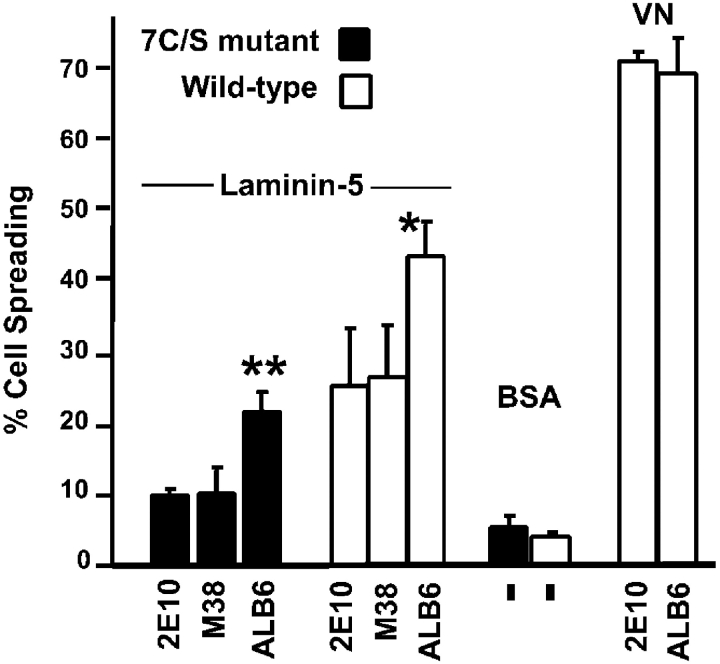
In cells where CD9 associates with α6β4, anti-CD9 antibodies can alter α6β4-dependent cell motility (Jones et al., 1996; Baudoux et al., 2000). Here, we confirm a functional connection between CD9 and laminin-binding integrins in MDA-MB-435 cells. Addition of an anti-CD9 antibody (ALB6) stimulated cell spreading in 7C/S cells, and to an even greater extent in cells expressing wild-type β4 (Fig. 10). Background spreading was observed in the presence of control anti-α2 and anti-CD81 antibodies. As expected from the results shown in Fig. 5, spreading on laminin-5 was elevated for wild-type β4 cells. The anti-CD9 mAb ALB6 did not promote spreading of either wild-type (Fig. 10) or 7C/S (not depicted) cells on vitronectin, and had no effect on static cell adhesion to laminin-1 or laminin-5.
Figure 10.
Anti-CD9 antibody stimulates cell spreading. MDA-MB-435 cells spread on the indicated substrates for 45 min, in the presence of mAb ALB6 (anti-CD9) or control antibodies (2E10, anti–integrin α2; and M38, anti-CD81). Spreading was assessed as described in the Fig. 5 legend (**, P < 0.02 vs. controls; *, P < 0.06 vs. controls). Error bars indicate SEM, and dashes indicate that no antibody was added.
Discussion
CD151 and β4 palmitoylation similarities
There are several striking similarities between β4 and tetraspanin CD151, with respect to palmitoylation. First, multiple membrane-proximal cysteines are used. Although six cysteines may contribute to CD151 palmitoylation (Berditchevski et al., 2002; Yang et al., 2002), removal of seven cysteines eliminated all detectable [3H]palmitate incorporation into β4. Removal of only one, two, three, or five membrane-proximal cysteines yielded incomplete results (Gagnoux-Palacios et al., 2003). The seven mutated cysteines are exactly conserved in β4 from multiple species (e.g., humans, rats, and mice), but do not appear in any other integrin β subunits. Second, palmitoylation of both CD151 (Berditchevski et al., 2002; Yang et al., 2002) and β4 occurs on newly synthesized protein and remains stably attached, with minimal turnover on the mature proteins. Third, removal of palmitoylation sites from CD151 (Berditchevski et al., 2002; Yang et al., 2002) or β4 increased neither protein density nor detergent solubility/extractability (see TEMs and lipid raft localization section). Fourth, removal of palmitoylation sites from CD151 (Berditchevski et al., 2002; Yang et al., 2002) or β4 did not alter static cell adhesion, but did alter α6β4-dependent cell signaling and morphology. Fifth, immunoprecipitation of either CD151 or α6β4 yielded strikingly similar patterns of palmitoylated proteins. Sixth, loss of palmitoylation from either molecule did not disrupt the core CD151–α6β4 complex. However, seventh, loss of palmitoylation from either CD151 (Berditchevski et al., 2002; Yang et al., 2002) or β4 did markedly alter secondary associations with other tetraspanins. These observations reinforce the concept that the CD151–α6β4 core complex is similarly affected by palmitoylation on either CD151 or β4.
Tetraspanin-enriched microdomains (TEMs)
It is well established that tetraspanin proteins (including CD151 and CD9) and their partners associate with each other in extended complexes known as the tetraspanin web (Trusolino et al., 2001), or TEMs (Berditchevski et al., 2002; Hemler, 2003). TEMs contain primary complexes (e.g., CD151–integrin [Serru et al., 1999; Yauch et al., 2000; Kazarov et al., 2002; Sterk et al., 2002] and tetraspanin homodimers [Kovalenko et al., 2004]) occurring through direct, protein–protein interactions. These are then brought together into extended secondary complexes, with tetraspanin palmitoylation playing a key role (Berditchevski et al., 2002; Charrin et al., 2002; Yang et al., 2002; Hemler, 2003). Given the abundance of evidence regarding CD151 palmitoylation, and the strong similarities between CD151 and β4 (see previous section), it is not surprising that β4 palmitoylation would also contribute to localization into TEMs. The evidence for this is as follows. First, in multiple cell lines, palmitoylated β4 associates with other palmitoylated proteins (CD9, CD81, CD63, and others) in a pattern that is strikingly similar whether immunoprecipitated with antibodies to CD151, to CD9, or to integrins (α6β4 or α3β1). The specificity of interaction among these particular components is emphasized by the exclusion of other abundant cell surface molecules (e.g., integrin α2β1 and tetraspanin CD82). Consistent with our results, others have also shown that CD9 can associate with α6β4 (Jones et al., 1996; Baudoux et al., 2000). Second, loss of β4 palmitoylation diminished secondary associations with other tetraspanins (CD9, CD81, and CD63), just as seen previously upon removal of CD151 and CD9 palmitoylation sites (Berditchevski et al., 2002; Charrin et al., 2002; Yang et al., 2002). Third, not only was CD9 substantially dissociated from the palmitoylation-deficient α6β4–CD151 complex, it also showed reorganization on the cell surface, which is consistent with reduced clustering. Pairs of monoclonal antibodies, diagnostic for cell surface clustering, have been described for other molecules, including CD147 (Koch et al., 1999) and CDw78 (Drbal et al., 1999). In each case, a lower affinity antibody (such as anti-CD9 mAb C9BB) showed more dependence on bivalent binding, and hence more sensitivity to cell surface clustering, compared with a higher affinity antibody (such as anti-CD9 mAb ALB6).
It is likely not a coincidence that the integrin α subunits (α3, α6, and α7) best able to interact with tetraspanins also contain palmitoylation sites. The integrin α3, α6, and α7 subunits contain single, membrane-proximal cysteine palmitoylation sites, conserved in orthologous subunits across a wide range of animal species. For α3 and α6, the key cysteine is present in both major (α3A and α6A) and minor (α3B and α6B) alternatively spliced forms. In contrast to the integrin α3, α6, and α7 subunits, the other 13 integrin α subunits (except for αE and α8) don't contain membrane-proximal cysteines.
Functional consequences
Removal of CD151 palmitoylation did not affect static cell adhesion but did alter α6β4-dependent kidney epithelial cell morphology (Yang et al., 2002) and integrin-dependent signaling in fibroblasts (Berditchevski et al., 2002). Similarly, removal of β4 palmitoylation did not alter cell surface expression levels, association with α6 or CD151, or cell adhesion to laminins, but did alter α6β4-dependent spreading and associated signaling through p130Cas in MDA-MB-435 cells. Although β4 has not previously been linked to p130Cas signaling, p130Cas is preferentially activated when cells are plated on laminin, compared with fibronectin (Gu et al., 2001). Inhibition by the Src family kinase inhibitor PP2 is consistent with cell spreading depending on Src family kinase phosphorylation of p130Cas (Nojima et al., 1996; Panetti, 2002). Presently, we cannot determine whether impaired cell spreading, due to the absence of β4 palmitoylation, is a consequence or a cause of diminished Src kinase → p130Cas signaling. Notably, Gagnoux-Palacios et al. (2003) have also reported altered Src family kinase–dependent signaling upon removal of β4 palmitoylation sites. Upon removal of palmitoylation from CD151 (Berditchevski et al., 2002) or β4, signaling through FAK was unaltered, consistent with cell adhesion being unaltered. Our results are in accord with previous evidence that FAK and p130Cas can sometimes be differentially regulated (Gu et al., 2001).
For several reasons, we suggest that β4 palmitoylation–dependent recruitment of CD151–α6β4 into TEMs plays a major role in determining cell spreading and signaling. First, diminished spreading and signaling occurred in parallel with reduced secondary associations of α6β4 with tetraspanins (CD9, CD81, and CD63) and reorganization of cell surface CD9. Second, CD9 is functionally connected to α6β4, consistent with its physical association. As seen elsewhere, anti-CD9 antibodies inhibited the motility of cultured keratinocytes under conditions where motility is at least partially α6β4-dependent (Jones et al., 1996; Baudoux et al., 2000). Consistent with this, we used an anti-CD9 antibody to stimulate MDA-MB-435 cell spreading, which is partly α6β4 dependent, as evidenced by the negative effects of β4 mutation. It was a little surprising that anti-CD9 antibody also stimulated spreading of MDA-MB-435 cells bearing mutant β4. However, we note that removal of β4 palmitoylation does not completely dissociate CD151–α6β4 from CD9 and other tetraspanins. There remain six palmitoylation sites in CD151, and one site in α6, that likely contribute to continued CD9 association. Third, an association with TEMs provides a mechanism to link CD151–α6β4 with several other molecules known to regulate cell morphology and signaling. These include other CD9-associated proteins such as EWI-2 (Stipp et al., 2003a; Kolesnikova et al., 2004), PKC (Zhang et al., 2001), and phosphatidylinositol 4-kinase (Yauch and Hemler, 2000). In this regard, CD9 has also been linked to signaling of c-kit and EGFR tyrosine kinases (Higashiyama et al., 1995; Shi et al., 2000; Anzai et al., 2002). It was suggested previously that CD9 regulation of α6β4 might only be relevant when cells are migrating and/or when the α6β4 is associated with the actin cytoskeleton rather than stably interacting with intermediate filaments within hemidesmosome-like structures (Baudoux et al., 2000). Indeed, tetraspanins CD9 and CD81 are not associated with the CD151–α6β4 complex in hemidesmosomes (Sterk et al., 2000), and mutation of β4 palmitoylation sites did not affect hemidesmosome localization, as observed here and by Gagnoux-Palacios et al. (2003).
Our β4 mutation had minimal effect on cell adhesion to laminin-1 or laminin-5. This result is consistent with previous results in which β4 in MDA-MB-435 cells modulated downstream signaling but not initial cell adhesion to laminin-1 (Shaw et al., 1997). Furthermore, our β4 mutant was deficient in spreading and signaling on both laminin-5 and laminin-1, even though only the former is a very good ligand for α6β4. Hence, a mechanism is needed to explain how α6β4 can exert substrate-dependent effects on cell morphology, signaling, and/or motility, even when it is not primarily responsible for cell adhesion. In this regard, we have demonstrated that multiple laminin-binding integrins can be linked together within overlapping complexes. Not only were the profiles of palmitoylated proteins associated with α6β4 and α3β1 strikingly similar, but also we observed α6β4 and α3β1 coimmunoprecipitations from multiple cell lines. Hence, we propose that α6β4 can physically associate with α3β1 (and likely also α6β1), in the context of TEMs. This provides a mechanism for the close coordination of initial adhesion, mediated by α3β1 (or α6β1), with subsequent α6β4-dependent spreading, signaling, or migration. Though novel, these results are not unexpected. It was previously established that α6β4, α6β1, and α3β1 all can associate with CD151 (Berditchevski, 2001), and that CD151 may appear predominantly as a homodimer (Kovalenko et al., 2004), thus having the potential to link distinct integrins. Furthermore, all of these integrins were known to associate with additional tetraspanins (CD9, CD81, CD63, etc.), which can associate with each other (Boucheix and Rubinstein, 2001; Berditchevski et al., 2002; Hemler, 2003).
TEMs and lipid raft localization
Protein palmitoylation can lead to localization into lipid rafts and decreased solubility in nonionic detergents (Dunphy and Linder, 1998; Resh, 1999). Indeed, it was suggested elsewhere that β4 palmitoylation promotes lipid raft localization, thus providing a mechanism for bringing β4 into functionally important complexes with Src family kinases such as fyn and yes (Mariotti et al., 2001; Gagnoux-Palacios et al., 2003). However, we found no evidence for palmitoylation promoting either integrin or tetraspanin association with lipid rafts. First, in β4-transfected A431 cells, the presence of β4 palmitoylation was not associated with decreased density in either Brij 96 or Triton X-100 lysates. Second, neither the palmitoylated subset nor the cell surface–labeled subset of endogenous β4 in A431 cells appeared in low density sucrose gradient fractions, even in mild Brij 96 detergent. Third, β4 palmitoylation did not decrease detergent solubility in MDA-MB-435 cells. In fact, in a short-term detergent (Brij 96) solubility assay, β4 palmitoylation increased solubility. Fourth, our β4 results are in complete agreement with prior results regarding tetraspanin palmitoylation. Protein density was not affected by removal of palmitoylation from either CD151 or CD9 (Berditchevski et al., 2002; Charrin et al., 2002; Yang et al., 2002), and CD151 palmitoylation did not decrease detergent solubility (Yang et al., 2002). Fifth, we did not see changes in association of mutant β4 with signaling proteins (EGFR, ErbB3/2, and fyn) that can be found in lipid rafts. Despite the potential of tetraspanins to associate with gangliosides and cholesterol, TEMs consistently remain distinct from lipid rafts. In contrast to lipid rafts, TEMs are resistant to cholesterol depletion, typically appear in the soluble phase upon extraction by nonionic detergents, and are not disrupted at 37°C (Hemler, 2003). An extensive proteomics analysis of lipid rafts did not reveal any tetraspanins or palmitoylated integrins (Foster et al., 2003). Conversely, we have identified, by mass spectrometry, numerous potential partners for tetraspanins and palmitoylated integrins, but this list does not contain typical raft-type proteins such as caveolins and GPI-linked proteins (unpublished data).
In conclusion, palmitoylation of β4 integrin, as seen previously for CD151, plays a key role in integrin-dependent cell spreading and signaling, and in the maintenance of secondary interactions with additional tetraspanin proteins. We suggest that CD151–α6β4, CD151–α3β1, and CD151–α6β1 assemble with other tetraspanins into novel signaling platforms (TEMs). These provide a framework for the regulation of integrin-dependent postadhesion functions (spreading, signaling, and motility) and coordination of the functions and distributions of laminin-binding integrins at the cell–substrate interface.
Materials and methods
Reagents and cells
mAbs to integrins α2 (A2-IIE10), α3 (A3-X8 and A3IVA5), α6 (A6-ELE); to tetraspanins CD9 (C9BB and DU-ALL-1), CD63 (6H1), CD81 (M38 and JS64), CD82 (M104), and CD151 (5C11 and 1A5); and to CD147 (8G6) were referenced previously (Kazarov et al., 2002; Yang et al., 2002). Other mAbs used were anti–integrin α6 (GoH3; BD Biosciences), anti-β4 (AA3; provided by V. Quaranta, Vanderbilt University, Nashville, TN) anti-CD9 (ALB6; CHEMICON International), and anti-CD71 (OKT9; American Type Culture Collection). Also used were rabbit polyclonal antibodies to β4 and β1 integrins (CHEMICON International) and to the cytoplasmic domains of integrins α3A (Dipersio et al., 1995) and α6A (Ab 6843; a gift from V. Quaranta). mAbs to phosphotyrosine (4G10) and PI 3-K p85 were purchased from Upstate Biotechnology, and antibodies to EGFR, ErbB2, and ErbB3 were purchased from Transduction Laboratories. Antibodies to FAK, fyn, Src, and p130Cas were obtained from Santa Cruz Biotechnology, Inc., and anti-GFP was obtained from CLONTECH Laboratories, Inc.
l-[35S]methionine/l-[35S]cysteine and [3H]palmitic acid were purchased from NEN Life Science Products; brefeldin A (Sigma-Aldrich) was dissolved in DMSO or ethanol at 1–2 mg/ml and stored at −80°C. EGF and heregulin-β1 were purchased from Upstate Biotechnology and R&D Systems, respectively. Sulfo-NHS-LC-Biotin and Sulfo-NHS-SS-Biotin were obtained from Pierce Chemical Co. PP2, a specific inhibitor for Src family kinases, was obtained from Calbiochem. Laminin-5 was a gift (Desmos Inc., San Diego, CA) and vitronectin was purchased from Becton Dickinson. Cell lines were grown in DME supplemented with 10% FCS (GIBCO BRL), 10 mM Hepes, and antibiotics (penicillin and streptomycin). B12 kidney epithelial cells, lacking α3 integrin, were a gift from J. Kreidberg (Children's Hospital Boston, Boston, MA; Kreidberg et al., 1996).
Integrin constructs and transfection
Point mutants (C→S) were generated in human β4 or α3 integrin subunits by PCR and subcloned into pcDNA3.1 (Invitrogen). To generate GFP-tagged β4, a 2.2-kb NH2-terminal fragment of human β4 was amplified and subcloned into EGFP-N1 vector via XhoI and EcoRI sites. Additional β4 fragments (with or without C→S point mutations) were subcloned and then were inserted via EcoRI and Kpn sites. β4 and α3 sequences were confirmed by DNA sequencing at the Dana-Farber Cancer Institute core facility. Lipofectamine (GIBCO BRL) was used for transient and stable transfections. Human integrin wild-type and mutant α subunits were stably expressed in the B12 cell line, a murine α3 integrin-null kidney epithelial cell line (Wang et al., 1999), using the Fugene 6 method (Roche Diagnostics Corporation). Integrin mutant X3TC5 is an α3 subunit with the transmembrane domain and cytoplasmic tail replaced by corresponding domains from α5 (Yauch et al., 1998).
Protein labeling, immunoprecipitation, and immunoblotting
For [3H]palmitate labeling, cell lines (at 80–90% confluence) were washed twice in PBS, serum-starved for 3–4 h, and then pulsed for 1–2 h in medium containing 0.2–0.3 mCi/ml [3H]palmitic acid plus 5% dialyzed FBS. For surface labeling with biotin, semiconfluent cells were cooled on ice, washed three times in PBS, incubated in PBS containing Sulfo-NHS-LC-Biotin at 0.1–0.2 mg/ml for 1 h at 4°C, and then washed with cold PBS containing 0.1 M glycine. Labeled cells were lysed in RIPA (1% Triton X-100, 1% deoxycholate, and 0.1% SDS) or in buffer containing 1% Brij 96 for 1 h at 4°C. Immunoprecipitation, immunoblotting, and detection of [3H]palmitate were described previously (Yang et al., 2002).
Immunofluorescence microscopy and flow cytometry
A431 cells were cultured overnight on 60-mm dishes with coverglass bottoms (MaTek Corp.). Cells were then transfected with GFP-tagged β4 constructs, and after 48 h were rinsed in PBS and photographed (Axioskop; Carl Zeiss MicroImaging, Inc.) at a magnification of 100. For flow cytometry (FACSCalibur; Becton Dickinson), stably transfected semiconfluent MDA-MB-435 cells were detached and stained on ice with either control IgG or specific mAbs, followed by FITC-conjugated secondary antibody (Biosource International).
Acknowledgments
This work was supported by National Institutes of Health grants CA42368 and CA86712.
Abbreviations used in this paper: EGFR, EGF receptor; TEM, tetraspanin-enriched microdomain.
References
- Adelsman, M.A., J.B. McCarthy, and Y. Shimizu. 1999. Stimulation of β1-integrin function by epidermal growth factor and heregulin-β has distinct requirements for erbB2 but a similar dependence on phosphoinositide 3-OH kinase. Mol. Biol. Cell. 10:2861–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai, N., Y. Lee, B.S. Youn, S. Fukuda, Y.J. Kim, C. Mantel, M. Akashi, and H.E. Broxmeyer. 2002. C-kit associated with the transmembrane 4 superfamily proteins constitutes a functionally distinct subunit in human hematopoietic progenitors. Blood. 99:4413–4421. [DOI] [PubMed] [Google Scholar]
- Baudoux, B., D. Castanares-Zapatero, M. Leclercq-Smekens, N. Berna, and Y. Poumay. 2000. The tetraspanin CD9 associates with the integrin alpha6beta4 in cultured human epidermal keratinocytes and is involved in cell motility. Eur. J. Cell Biol. 79:41–51. [DOI] [PubMed] [Google Scholar]
- Belkin, A.M., and M.A. Stepp. 2000. Integrins as receptors for laminins. Microsc. Res. Tech. 51:280–301. [DOI] [PubMed] [Google Scholar]
- Berditchevski, F. 2001. Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 114:4143–4151. [DOI] [PubMed] [Google Scholar]
- Berditchevski, F., E. Gilbert, M.R. Griffiths, S. Fitter, L. Ashman, and S.J. Jenner. 2001. Analysis of the CD151-α3β1 integrin and CD151-tetraspanin interactions by mutagenesis. J. Biol. Chem. 276:41165–41174. [DOI] [PubMed] [Google Scholar]
- Berditchevski, F., E. Odintsova, S. Sawada, and E. Gilbert. 2002. Expression of the palmitoylation-deficient CD151 weakens the association of α3β1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signalling. J. Biol. Chem. 277:36991–37000. [DOI] [PubMed] [Google Scholar]
- Boucheix, C., and E. Rubinstein. 2001. Tetraspanins. Cell. Mol. Life Sci. 58:1189–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, E.R., J. Blasi, S. An, N. Brose, P.A. Johnston, T.C. Sudhof, and R. Jahn. 1996. Fatty acylation of synaptotagmin in PC12 cells and synaptosomes. Biochem. Biophys. Res. Commun. 225:326–332. [DOI] [PubMed] [Google Scholar]
- Charrin, S., S. Manie, M. Oualid, M. Billard, C. Boucheix, and E. Rubinstein. 2002. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett. 516:139–144. [DOI] [PubMed] [Google Scholar]
- Claas, C., C.S. Stipp, and M.E. Hemler. 2001. Evaluation of prototype TM4SF protein complexes and their relation to lipid rafts. J. Biol. Chem. 276:7974–7984. [DOI] [PubMed] [Google Scholar]
- Dipersio, C.M., S. Shah, and R.O. Hynes. 1995. α3Aβ1 integrin localizes to focal contacts in response to diverse extracellular matrix proteins. J. Cell Sci. 108:2321–2336. [DOI] [PubMed] [Google Scholar]
- Drbal, K., P. Angelisova, A.M. Rasmussen, I. Hilgert, S. Funderud, and V. Horejsi. 1999. The nature of the subset of MHC class II molecules carrying the CDw78 epitopes. Int. Immunol. 11:491–498. [DOI] [PubMed] [Google Scholar]
- Dunphy, J.T., and M.E. Linder. 1998. Signalling functions of protein palmitoylation. Biochim. Biophys. Acta. 1436:245–261. [DOI] [PubMed] [Google Scholar]
- Foster, L.J., C.L. De Hoog, and M. Mann. 2003. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. USA. 100:5813–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnoux-Palacios, L., M. Dans, W. van't Hof, A. Mariotti, A. Pepe, G. Meneguzzi, M.D. Resh, and F.G. Giancotti. 2003. Compartmentalization of integrin α6β4 signaling in lipid rafts. J. Cell Biol. 162:1189–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambaletta, D., A. Marchetti, L. Benedetti, A.M. Mercurio, A. Sacchi, and R. Falcioni. 2000. Cooperative signaling between α6β4 integrin and ErbB-2 receptor is required to promote phosphatidylinositol 3-kinase-dependent invasion. J. Biol. Chem. 275:10604–10610. [DOI] [PubMed] [Google Scholar]
- Gu, J., Y. Sumida, N. Sanzen, and K. Sekiguchi. 2001. Laminin-10/11 and fibronectin differentially regulate integrin-dependent Rho and Rac activation via p130Cas-CrkII-DOCK180 pathway. J. Biol. Chem. 276:27090–27097. [DOI] [PubMed] [Google Scholar]
- Gundersen, C.B., A. Mastrogiacomo, K. Faull, and J.A. Umbach. 1994. Extensive lipidation of a Torpedo cysteine string protein. J. Biol. Chem. 269:19197–19199. [PubMed] [Google Scholar]
- Hanke, J.H., J.P. Gardner, R.L. Dow, P.S. Changelian, W.H. Brissette, E.J. Weringer, B.A. Pollok, and P.A. Connelly. 1996. Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor. Study of Lck- and FynT-dependent T cell activation. J. Biol. Chem. 271:695–701. [DOI] [PubMed] [Google Scholar]
- Hemler, M.E. 1998. Integrin-associated proteins. Curr. Opin. Cell Biol. 10:578–585. [DOI] [PubMed] [Google Scholar]
- Hemler, M.E. 2001. Specific tetraspanin functions. J. Cell Biol. 155:1103–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler, M.E. 2003. Tetraspanin proteins mediate cellular penetration, invasion and fusion events, and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19:397–422. [DOI] [PubMed] [Google Scholar]
- Higashiyama, S., R. Iwamoto, K. Goishi, G. Raab, N. Taniguchi, M. Klagsbrun, and E. Mekada. 1995. The membrane protein CD9/DRAP27 potentiates the juxtacrine growth factor activity of the membrane-anchored heparin-binding EGF-like growth factor. J. Cell Biol. 128:929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintermann, E., M. Bilban, A. Sharabi, and V. Quaranta. 2001. Inhibitory role of α6β4-associated erbB-2 and phosphoinositide 3-kinase in keratinocyte haptotactic migration dependent on α3β1 integrin. J. Cell Biol. 153:465–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, P.H., L.A. Bishop, and F.M. Watt. 1996. Functional significance of CD9 association with β1 integrins in human epidermal keratinocytes. Cell Adhes. Commun. 4:297–305. [DOI] [PubMed] [Google Scholar]
- Kazarov, A.R., X. Yang, C.S. Stipp, B. Sehgal, and M.E. Hemler. 2002. An extracellular site on tetraspanin CD151 determines α3 and α6 integrin–dependent cellular morphology. J. Cell Biol. 158:1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, C., G. Staffler, R. Huttinger, I. Hilgert, E. Prager, J. Cerny, P. Steinlein, O. Majdic, V. Horejsi, and H. Stockinger. 1999. T cell activation-associated epitopes of CD147 in regulation of the T cell response, and their definition by antibody affinity and antigen density. Int. Immunol. 11:777–786. [DOI] [PubMed] [Google Scholar]
- Kolesnikova, T.V., C.S. Stipp, R.M. Rao, W.S. Lane, F.W. Luscinskas, and M.E. Hemler. 2004. EWI-2 modulates lymphocyte integrin α4β1 functions. Blood. 103:3013–3019. [DOI] [PubMed] [Google Scholar]
- Kovalenko, O.V., X. Yang, T.V. Kolesnikova, and M.E. Hemler. 2004. Evidence for specific tetraspanin homodimers: inhibition of palmitoylation makes cysteine residues available for cross-linking. Biochem. J. 377:407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreidberg, J.A., M.J. Donovan, S.L. Goldstein, H. Rennke, K. Shepherd, R.C. Jones, and R. Jaenisch. 1996. α3β1 integrin has a crucial role in kidney and lung organogenesis. Development. 122:3537–3547. [DOI] [PubMed] [Google Scholar]
- Levy, S., S.C. Todd, and H.T. Maecker. 1998. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89–109. [DOI] [PubMed] [Google Scholar]
- Mainiero, F., A. Pepe, M. Yeon, Y. Ren, and F.G. Giancotti. 1996. The intracellular functions of α6β4 integrin are regulated by EGF. J. Cell Biol. 134:241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti, A., P.A. Kedeshian, M. Dans, A.M. Curatola, L. Gagnoux-Palacios, and F.G. Giancotti. 2001. EGF-R signaling through Fyn kinase disrupts the function of integrin α6β4 at hemidesmosomes: role in epithelial cell migration and carcinoma invasion. J. Cell Biol. 155:447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio, A.M., I. Rabinovitz, and L.M. Shaw. 2001. The α6β4 integrin and epithelial cell migration. Curr. Opin. Cell Biol. 13:541–545. [DOI] [PubMed] [Google Scholar]
- Nojima, Y., T. Mimura, N. Morino, K. Hamasaki, H. Furuya, R. Sakai, T. Nakamoto, Y. Yazaki, and H. Hirai. 1996. Tyrosine phosphorylation of p130Cas in cell adhesion and transformation. Hum. Cell. 9:169–174. [PubMed] [Google Scholar]
- O'Neill, G.M., S.J. Fashena, and E.A. Golemis. 2000. Integrin signalling: a new Cas(t) of characters enters the stage. Trends Cell Biol. 10:111–119. [DOI] [PubMed] [Google Scholar]
- Panetti, T.S. 2002. Tyrosine phosphorylation of paxillin, FAK, and p130CAS: effects on cell spreading and migration. Front. Biosci. 7:d143–d150. [DOI] [PubMed] [Google Scholar]
- Rabinovitz, I., A. Toker, and A.M. Mercurio. 1999. Protein kinase C–dependent mobilization of the α6β4 integrin from hemidesmosomes and its association with actin-rich cell protrusions drive the chemotactic migration of carcinoma cells. J. Cell Biol. 146:1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh, M.D. 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta. 1451:1–16. [DOI] [PubMed] [Google Scholar]
- Santoro, M.M., G. Gaudino, and P.C. Marchisio. 2003. The MSP receptor regulates α6β4 and α3β1 integrins via 14-3-3 proteins in keratinocyte migration. Dev. Cell. 5:257–271. [DOI] [PubMed] [Google Scholar]
- Serru, V., F.L. Naour, M. Billard, D.O. Azorsa, F. Lanza, C. Boucheix, and E. Rubinstein. 1999. Selective tetraspan–integrin complexes (CD81/α4β1, CD151/α3β1, CD151/α6β1) under conditions disrupting tetraspan interactions. Biochem. J. 340:103–111. [PMC free article] [PubMed] [Google Scholar]
- Shaw, L.M., I. Rabinovitz, H.H. Wang, A. Toker, and A.M. Mercurio. 1997. Activation of phosphoinositide 3-OH kinase by the alpha6beta4 integrin promotes carcinoma invasion. Cell. 91:949–960. [DOI] [PubMed] [Google Scholar]
- Shi, W., H. Fan, L. Shum, and R. Derynck. 2000. The tetraspanin CD9 associates with transmembrane TGF-α and regulates TGF-α–induced EGF receptor activation and cell proliferation. J. Cell Biol. 148:591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterk, L.M., C.A. Geuijen, L.C. Oomen, J. Calafat, H. Janssen, and A. Sonnenberg. 2000. The tetraspan molecule CD151, a novel constituent of hemidesmosomes, associates with the integrin α6β4 and may regulate the spatial organization of hemidesmosomes. J. Cell Biol. 149:969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterk, L.M., C.A. Geuijen, J.G. van den Berg, N. Claessen, J.J. Weening, and A. Sonnenberg. 2002. Association of the tetraspanin CD151 with the laminin-binding integrins α3β1, α6β1, α6β4 and α7β1 in cells in culture and in vivo. J. Cell Sci. 115:1161–1173. [DOI] [PubMed] [Google Scholar]
- Stipp, C.S., and M.E. Hemler. 2000. Transmembrane-4-Superfamily proteins CD151 and CD81 associate with α3β1 integrin, and selectively contribute to α3β1-dependent neurite outgrowth. J. Cell Sci. 113:1871–1882. [DOI] [PubMed] [Google Scholar]
- Stipp, C.S., T.V. Kolesnikova, and M.E. Hemler. 2003. a. EWI-2 regulates α3β1 integrin–dependent cell functions on laminin-5. J. Cell Biol. 163:1167–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp, C.S., T.V. Kolesnikova, and M.E. Hemler. 2003. b. Functional domains in tetraspanin proteins. Trends Biochem. Sci. 28:106–112. [DOI] [PubMed] [Google Scholar]
- Trusolino, L., A. Bertotti, and P.M. Comoglio. 2001. A signaling adapter function for α6β4 integrin in the control of HGF-dependent invasive growth. Cell. 107:643–654. [DOI] [PubMed] [Google Scholar]
- Wang, Z., J.M. Symons, S.L. Goldstein, A. McDonald, J.H. Miner, and J.A. Kreidberg. 1999. α3β1 integrin regulates epithelial cytoskeletal organization. J. Cell Sci. 112:2925–2935. [DOI] [PubMed] [Google Scholar]
- Yánez-Mó, M., A. Alfranca, C. Cabañas, M. Marazuela, R. Tejedor, M.A. Ursa, L.K. Ashman, M.O. De Landázuri, and F. Sánchez-Madrid. 1998. Regulation of endothelial cell motility by complexes of tetraspan molecules CD81/TAPA-1 and CD151/PETA-3 with α3β1 integrin localized at endothelial lateral junctions. J. Cell Biol. 141:791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., C. Claas, S.K. Kraeft, L.B. Chen, Z. Wang, J.A. Kreidberg, and M.E. Hemler. 2002. Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol. Biol. Cell. 13:767–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch, R.L., and M.E. Hemler. 2000. Specific interactions among transmembrane 4 superfamily (TM4SF) proteins and phosphatidylinositol 4-kinase. Biochem. J. 351:629–637. [PMC free article] [PubMed] [Google Scholar]
- Yauch, R.L., F. Berditchevski, M.B. Harler, J. Reichner, and M.E. Hemler. 1998. Highly stoichiometric, stable and specific association of integrin α3β1 with CD151 provides a major link to phosphatidylinositol 4-kinase and may regulate cell migration. Mol. Biol. Cell. 9:2751–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch, R.L., A.R. Kazarov, B. Desai, R.T. Lee, and M.E. Hemler. 2000. Direct extracellular contact between integrin α3β1 and TM4SF protein CD151. J. Biol. Chem. 275:9230–9238. [DOI] [PubMed] [Google Scholar]
- Zhang, X.A., A.L. Bontrager, and M.E. Hemler. 2001. Transmembrane-4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific β1 integrins. J. Biol. Chem. 276:25005–25013. [DOI] [PubMed] [Google Scholar]
- Zhang, X.A., A.R. Kazarov, X. Yang, A.L. Bontrager, C.S. Stipp, and M.E. Hemler. 2002. Function of the tetraspanin CD151-α6β1 integrin complex during cellular morphogenesis. Mol. Biol. Cell. 13:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]