Abstract
HA95 is a chromatin-associated protein that interfaces the nuclear envelope (NE) and chromatin. We report an interaction between HA95 and the inner nuclear membrane protein lamina-associated polypeptide (LAP) 2β, and a role of this association in initiation of DNA replication. Precipitation of GST–LAP2β fusion proteins and overlays of immobilized HA95 indicate that a first HA95-binding region lies within amino acids 137–242 of LAP2β. A second domain sufficient to bind HA95 colocalizes with the lamin B–binding domain of LAP2β at residues 299–373. HA95–LAP2β interaction is not required for NE formation. However, disruption of the association of HA95 with the NH2-terminal HA95-binding domain of LAP2β abolishes the initiation, but not elongation, of DNA replication in purified G1 phase nuclei incubated in S-phase extract. Inhibition of replication initiation correlates with proteasome-mediated proteolysis of Cdc6, a component of the prereplication complex. Rescue of Cdc6 degradation with proteasome inhibitors restores replication. We propose that an interaction of LAP2β, or LAP2 proteins, with HA95 is involved in the control of initiation of DNA replication.
Keywords: chromosome; lamina-associated polypeptide; HA95; nuclear envelope; replication
Introduction
The nuclear envelope (NE)* mediates key nuclear functions by interacting with chromatin. The inner nuclear membrane (INM) harbors integral proteins that interact with the nuclear lamina and chromosomes. Among these, lamina-associated polypeptide (LAP) 2β binds chromatin in a phosphorylation-dependent manner (Foisner and Gerace, 1993; Furukawa et al., 1995) and contains a LAP2, emerin, MAN-1 (LEM) domain (Lin et al., 2000) that interacts with the DNA-bridging protein, barrier-to-autointegration factor (BAF) (Furukawa, 1999). At least six LAP2 isoforms are produced from alternative mRNA splicing in mammals (Berger et al., 1996). LAP2 proteins share an NH2-terminal 187–amino acid domain that binds DNA (Cai et al., 2001) and contains the LEM motif (residues 111–152) (Furukawa, 1999; Shumaker et al., 2001). LAP2β also interacts with the germ cell less (GCL) transcription regulator through residues 219–328 and mediates transcriptional repression alone or together with GCL (Nili et al., 2001). A lamin B–binding domain specific for LAP2β has been identified at residues 299–373 (Furukawa and Kondo, 1998). Injection of LAP2β fragments in living cells (Yang et al., 1997) and in vitro studies (Gant et al., 1999) suggests that LAP2β is involved in nuclear assembly.
We, and others, have cloned a 95-kD nuclear protein named HA95/NAKAP95/HAP95 (Orstavik et al., 2000; Seki et al., 2000; Westberg et al., 2000). HA95 displays partial homology to AKAP95, a member of the A-kinase anchoring protein family, but it lacks a protein kinase A (PKA)–binding domain. HA95 appears in photobleaching experiments as a stable protein and cofractionates with chromatin and a nuclease- and salt-resistant “matrix.” HA95 associates with itself (Orstavik et al., 2000) and interacts with RNA helicase A (RHA) to enhance expression of a constitutive transport element (CTE) involved in nuclear export of retroviral RNA (Westberg et al., 2000; Yang et al., 2001). In vitro nuclear breakdown and reassembly assays combined with antibody-blocking experiments have shown that HA95 is involved in NE breakdown and chromatin condensation, whereas a role in nuclear membrane reassembly is unlikely (Martins et al., 2000).
Initiation of DNA replication involves the assembly of prereplication complexes (preRCs) at origins of replication in G1 (Kelly and Brown, 2000; Bell and Dutta, 2002). PreRCs include the origin recognition complex (ORC), the minichromosome maintenance (MCM) complex, and the monomeric Cdc6 protein (Bell and Dutta, 2002). ORC recruits Cdc6 into the preRC in G1, and Cdc6 in turn promotes loading of MCM proteins on chromatin (Bell and Dutta, 2002). In human cells, Cdc6 levels are relatively stable during interphase (Williams et al., 1997; Saha et al., 1998). Nonetheless, a fraction of Cdc6 is exported out of the nucleus (Coleman et al., 1996) while another remains associated with chromatin during S and G2 phases (Coverley et al., 2000; Mendez and Stillman, 2000). Proteolysis of free Cdc6, not assembled into preRCs, has also been reported (Coverley et al., 2000). Thus, after origin firing at the start of S phase, preRCs are dissociated, ensuring a single round of replication per cell cycle.
We provide evidence here that HA95 interacts with LAP2β via two distinct domains. Blocking the association of HA95 with LAP2β does not affect nuclear assembly in vitro. However, disruption of the association of HA95 with the NH2-terminal HA95-binding domain of LAP2β triggers proteasome-mediated degradation of Cdc6 and inhibits initiation, but not elongation, of replication. The results suggest an implication of the association of HA95 with LAP2β in regulating the initiation phase of DNA replication.
Results
HA95 and LAP2β coprecipitate in interphase
HA95 coprecipitates with a protein complex containing LAP2β from the transformed human B cell line Bjab in interphase (Martins et al., 2000). We now show that in Bjab cells, HA95 and LAP2β coprecipitated in interphase but not at mitosis, regardless of the precipitating antibody (Fig. 1 A). Immunoblots of the nonprecipitated (S) fractions indicate that in interphase, HA95 coprecipitated all detectable LAP2β, whereas LAP2β did not precipitate all HA95 (Fig. 1 A). Immunoprecipitation of an HA95–Myc fusion protein stably expressed in Bjab cells using an anti-Myc antibody also coprecipitated LAP2β in interphase but not at mitosis (Fig. 1 B). These results suggest a cell cycle–dependent interaction of HA95 with LAP2β; however, not all HA95 resides in a complex with LAP2β.
Figure 1.
HA95 coprecipitates with LAP2β in interphase. (A) HA95 or LAP2β was immunoprecipitated (IP) from interphase and mitotic Bjab cells. Precipitates (P) and supernatants (S) were immunoblotted using anti-LAP2β or anti-HA95 antibodies. (B) Myc-tagged HA95 was immunoprecipitated from Bjab cells stably expressing HA95–Myc using anti-Myc antibodies, and precipitates were immunoblotted using indicated antibodies. Control immunoprecipitations were done with preimmune rabbit IgGs. (C) HA95-IPs were extracted with 0, 0.25, 0.5, 0.75, 1.0, or 1.25 M NaCl before sedimentation and immunoblotting of pellets and supernatants.
The strength of interaction between HA95 and LAP2β within the complex was evaluated by a 30-min extraction of anti-HA95 immune precipitates (HA95-IPs) with 0–1.25 M NaCl. Fig. 1 C shows that the interaction resisted a 0.75 M NaCl wash but was completely disrupted with 1.25 M NaCl. High salt resistance of the interaction suggests a strong association between HA95 and LAP2β, confirming our earlier cross-linking data (Martins et al., 2000).
Mapping of the HA95-binding domains of LAP2β
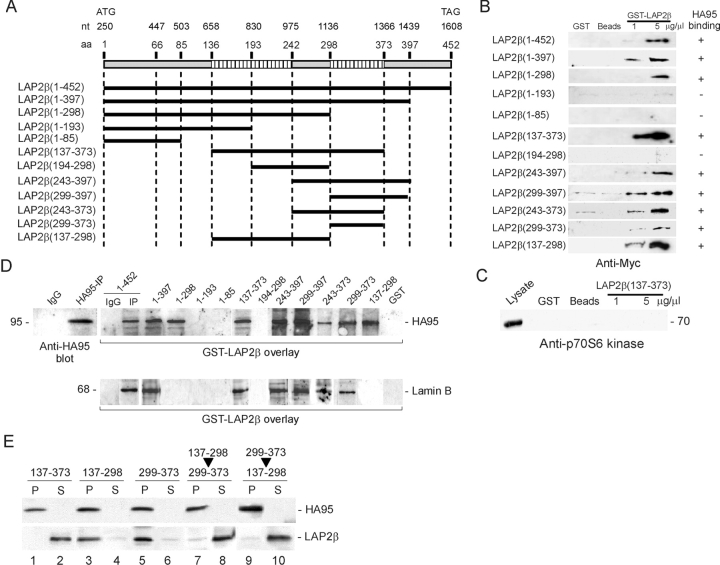
To map the domains of LAP2β involved in the interaction with HA95, GST–LAP2β fusion polypeptides were produced (Fig. 2 A) (Furukawa et al., 1995, 1997, 1998). Binding of each peptide to HA95–Myc was determined in GST precipitations after incubation of the peptides in a nuclear extract from Bjab cells expressing HA95–Myc. Control extracts were incubated with GST or glutathione beads alone. GST precipitates were immunoblotted using anti-Myc antibodies. Fig. 2 B shows that LAP2β(1–452), (1–397), (1–298), (137–373), (137–298), (243–397), (243–373), (299–397), and (299–373) precipitated HA95–Myc. In contrast, LAP2β(1–193), (1–85), or (194–298) did not precipitate HA95–Myc. Thus, a first HA95-binding domain localizes to amino acids 137–242 of LAP2β and a second domain coincides with the lamin B–binding domain at residues 299–373. We designated these domains HA95-NBD (for HA95 NH2-terminal binding domain) and HA95-CBD (HA95 COOH-terminal binding domain), respectively.
Figure 2.
LAP2β interacts with HA95 via two distinct domains. (A) GST–LAP2β deletion peptides. (B) Indicated peptides were incubated in nuclear extracts of Bjab cells expressing HA95–Myc and sedimented by GST precipitation. Binding of HA95–Myc to peptides was analyzed by immunoblotting using anti-Myc antibodies. Control incubations contained GST or glutathione beads alone. (C) GST–LAP2β(137–373) was incubated in a nuclear extract of Myc-tagged p70S6 kinase-expressing cells, and binding (or lack thereof) of p70S6 kinase to the peptide was detected by GST precipitation and immunoblotting. (D) GST–LAP2β overlays of immunoprecipitated HA95 or B-type lamins. Peptide binding was detected using anti-GST antibodies. (E) Peptide-mediated dissociation of LAP2β from HA95. HA95-IPs were incubated with 100 μM GST–LAP2β peptides, HA95-IPs were sedimented, and pellets and supernatants were immunoblotted using the indicated antibodies. Arrows indicate incubation with LAP2β(137–298) followed by addition of LAP2β(299–373) (lanes 7 and 8) or vice versa (lanes 9 and 10).
The Myc tag itself did not bind GST or LAP2β. LAP2β(137–373), which binds HA95–Myc, did not coprecipitate a stably expressed Myc-tagged p70S6 kinase (Fig. 2 C). Furthermore, anti-Myc antibodies coprecipitated LAP2β (1–397) but not the nonbinding fragment LAP2β(1–193) from a nuclear extract of Bjab cells expressing HA95–Myc (unpublished data). Altogether, the results indicate that HA95 interacts with recombinant LAP2β.
Direct association of LA2β with HA95 was demonstrated in an overlay assay. HA95-IP proteins were resolved by SDS-PAGE, blotted, and overlaid with 50 μM of each GST–LAP2β fragment. The same GST–LAP2β peptides found to precipitate HA95 also bound HA95 in the overlay, as detected with anti-GST antibodies (Fig. 2 D, top). Moreover, all LAP2β fragments containing the lamin B–binding domain (residues 299–373) bound to immunoprecipitated and immobilized lamin B (Fig. 2 D, bottom). However, two of the HA95-binding peptides, LAP2β(1–298) and LAP2β(137–298), did not bind lamin B, confirming the existence of an HA95-binding domain (HA95-NBD) distinct from the lamin B–binding region.
To determine whether HA95-binding LAP2β peptides would disrupt endogenous HA95–LAP2β association, dissociation experiments were performed using GST–LAP2β peptides harboring both HA95-binding domains (LAP2β [137–373]) or either HA95-NBD (LAP2β[137–298]) or HA95-CBD (LAP2β[299–373]). HA95-IPs were incubated for 1 h with 100 μM GST–LAP2β peptides, HA95-IPs were sedimented, and dissociation of LAP2β from HA95 was monitored by immunoblotting. LAP2β(137–373) completely dissociated LAP2β from HA95-IP (Fig. 2 E, lanes 1 and 2), however peptides containing HA95-NBD or HA95-CBD were ineffective (lanes 3–6). Nevertheless, two sequential 30-min incubations of HA95-IPs with LAP2β(137–298) followed by LAP2β(299–373) led to dissociation of the complex (lanes 7 and 8), and reversing the order of peptide addition produced similar results (lanes 9 and 10). We concluded that disruption of both HA95-binding domains was required for dissociation of LAP2β from HA95 in vitro. The data also argue that LAP2β(137–298) and LAP2β(299–373) peptides are capable of dissociating HA95 from HA95-NBD and HA95-CBD, respectively.
Inhibition of LAP2β binding to HA95 does not affect nuclear reassembly in vitro
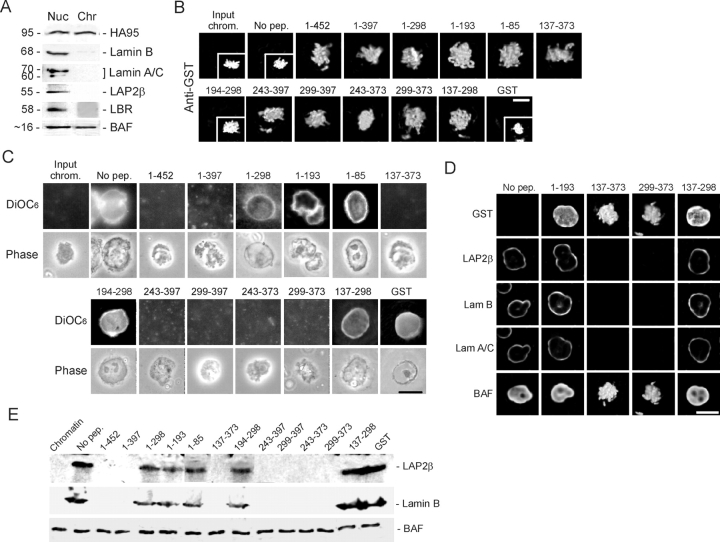
To determine whether interaction between HA95 and LAP2β was involved in NE assembly, we assessed the ability of HA95-binding GST–LAP2β peptides to compete with LAP2β for membrane targeting to chromosomes. Purified HeLa nuclei were disassembled in mitotic extract. The resulting condensed chromosomes contained HA95 and BAF, but no A- or B-type lamins, LAP2β, or lamin B receptor (LBR) (Fig. 3 A). After preincubation with 10 μM of each GST–LAP2β peptide, chromosomes were sedimented and peptide binding was examined by immunofluorescence using anti-GST antibodies. All peptides except GST–LAP2β(194–298) bound chromatin (Fig. 3 B), as anticipated from their ability to bind HA95 (Fig. 2), DNA, or chromatin (see Introduction). Chromosomes were resuspended in interphase extract (Fig. 3 C, Input chrom.) under conditions promoting nuclear assembly. Nuclear morphology was examined by phase contrast microscopy and membrane labeling with DiOC6 after 2 h (Fig. 3 C). Without peptide or with GST alone, >80% of chromatin masses supported nuclear reformation. In contrast, LAP2β fragments (1–452), (1–397), (137–373), (243–397), (299–397), (243–373), and (299–373) inhibited nuclear assembly. Each of these peptides contained the lamin B–binding domain/HA95-CBD. Peptides that do not bind HA95 (nor lamin B) (LAP2β[1–193], [1–85], and [194–298]) did not block membrane assembly, neither did LAP2β(1–298) or (137–298), which both contain HA95-NBD.
Figure 3.
Disruption of HA95 interaction with LAP2β does not interfere with nuclear assembly in vitro. (A) Purified HeLa nuclei (Nuc) were disassembled in mitotic extract, and chromosomes (Chr) were immunoblotted using the indicated antibodies. (B) Indicated GST–LAP2β peptides were allowed to bind to chromosomes, and binding was analyzed by immunofluorescence using anti-GST antibodies. Insets, DNA labeled with Hoescht 33342. (C) Chromosomes harboring the indicated peptides were incubated in interphase extract under conditions promoting nuclear formation. Membrane assembly was examined by DiOC6 labeling and phase contrast microscopy. (D) At the end of incubation, the nuclear distribution of GST–LAP2β peptides, LAP2β, A- and B-type lamins, and BAF was examined by immunofluorescence. (E) Nuclei or chromatin masses were also sedimented through sucrose, and proteins were immunoblotted using the indicated antibodies. Bars, 10 μm.
To address whether disruption of the of HA95–LAP2β interaction altered global nuclear organization, localization of LAP2β, A- and B-type lamins, and BAF was analyzed by immunofluorescence in nuclei reconstituted in the presence of either no peptide, a peptide not binding to HA95 (LAP2β[1–193]), or peptides containing both HA95-NBD and HA95-CBD (LAP2β[137–373]), HA95-NBD only (LAP2β[137–298]), or HA95-CBD only (LAP2β[299–373]). Anti-GST immunolabeling shows peptide distribution in the resulting nuclei or chromatin masses (Fig. 3 D). Distribution of LAP2β, lamins A/C and B, or BAF was not distinctively altered in nuclei reconstituted with HA95-binding or nonbinding LAP2β peptides (Fig. 3 D). LAP2β targeting to chromatin and nuclear assembly of A- and B-type lamins, or lack thereof, were verified by immunoblotting nuclei or chromatin purified from the extract (Fig. 3 E). The blots also showed that levels of chromatin-associated BAF were not significantly altered by GST–LAP2β fragments (Fig. 3 E). We concluded that LAP2β fragments containing the lamin B–binding domain and HA95-CBD prohibit nuclear reformation. However, peptides harboring HA95-NBD only are not inhibitory and do not notably affect the distribution of NE proteins or BAF in the reassembled nuclei. Thus, we could not attribute an effect of HA95 binding per se on NE assembly in vitro.
LAP2β(137–298) inhibits initiation of DNA replication in intact nuclei in vitro
To determine whether interphase nuclear functions were affected by the HA95–LAP2β association, we monitored the effect of disrupting the LAP2β–HA95 interaction on DNA replication in purified G1-phase nuclei. To this end, we adapted an in vitro replication assay from that of Krude et al. (1997).
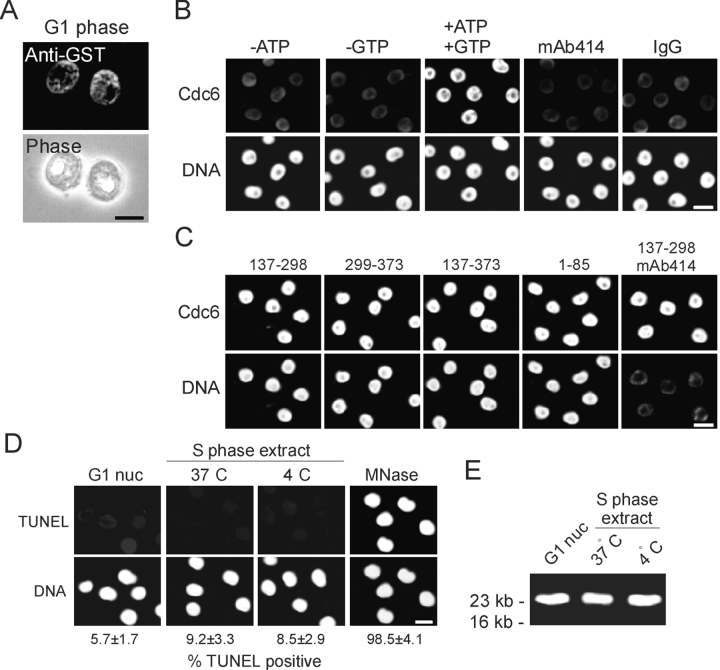
Nuclei were isolated from G1-phase HeLa cells. GST–LAP2β peptides were introduced into the nuclei after mild treatment with lysolecithin. Lysolecithin was previously shown not to affect dynamic properties of isolated nuclei in in vitro nuclear disassembly assays (Collas et al., 1999). Peptides were taken up by ∼90% of the nuclei, as shown by immunofluorescence using anti-GST antibodies (Fig. 4 A; GST–LAP2β[1–452] is shown). Control and peptide-loaded nuclei were incubated for 3 h in a concentrated (25–30 mg/ml) nuclear and cytosolic extract from S-phase HeLa cells containing [α32P]dCTP, dNTPs, GTP, and an ATP-regenerating system to promote replication. Under these conditions, G1 nuclei loaded with GST–LAP2β(1–452) were capable of importing an exogenous BSA–nuclear localization signal conjugate (unpublished data) or the replication factor Cdc6 (Fig. 4 B). Import was ATP and GTP dependent and blocked by preincubation of the nuclei with antibodies against nucleoporins (Fig. 5 B, mAb414). These results indicate that import took place through nuclear pores rather than passively through a damaged NE, and confirmed a previous report of physiological import of transcription factors by nuclei purified as previously described (Landsverk et al., 2002). We also tested whether peptides containing HA95-NBD or HA95-CBD introduced into G1 nuclei would inhibit nuclear import under the conditions described above, as this would be expected to affect DNA replication. Fig. 4 C shows that none of the peptides distinctly impaired import of Cdc6. Notably, import was permitted by the HA95-NBD–containing peptide LAP2β(137–298) and blocked by mAb414. Lastly, the nuclear DNA did not undergo any detectable degradation upon incubation of the G1 nuclei in the extract at 4°C or 37°C, as judged by TUNEL analysis (Fig. 4 C) and DNA agarose gel electrophoresis (Fig. 4 D). These results indicate that isolated G1 nuclei are functional in import, can be manipulated to introduce peptides, and do not undergo detectable DNA degradation in S-phase extract, and that LAP2β fragments containing either HA95-binding domain do not block nuclear import of Cdc6 in vitro.
Figure 4.
Characterization of nuclei used in the in vitro DNA replication assay. (A) Nuclei purified from HeLa cells in G1 phase were loaded with GST–LAP2β(1–452). Peptide uptake was analyzed by immunofluorescence using anti-GST antibodies. (B) GST–LAP2β(1–452)-loaded G1 nuclei were exposed to an S-phase extract, and import of Cdc6 was visualized by immunofluorescence. Note that the G1 nuclei contain low detectable levels of Cdc6. (C) Cdc6 import was also examined in nuclei containing the indicated peptides. G1 nuclei were also analyzed by (D) TUNEL and (E) electrophoresis to monitor DNA degradation after exposure to replication extract at 4°C or 37°C. Bars, 10 μm.
Figure 5.
LAP2β(137–298) inhibits initiation of DNA replication in G1 nuclei. (A) Peptide-loaded G1 nuclei were incubated in S-phase extract containing [α32P]dCTP, dNTPs, GTP, and an ATP-regenerating system. [α32P]dCTP incorporation was analyzed by phosphorImager (cpm/105 nuclei). (B) Nuclei isolated from HeLa cells at the indicated cell cycle stages were incubated in extracts from S or G0 cells under conditions promoting replication. S-phase extracts also contained 1 mM olomoucine (+Olo) or H2O (−Olo) (right panel). Synthesized DNA was analyzed by autoradiography. (C) Relative BAF content of G1 nuclei loaded with the indicated peptides and exposed to S-phase extract was examined by immunoblotting. HA95 was also detected as a loading control in the gel. (D) Summary of GST–LAP2β peptides harboring HA95-NBD supporting (+) or inhibiting (−) replication in G1 nuclei. (E) BrdU density substitution experiment. G1 nuclei containing either no peptide (circles), GST alone (triangles), or GST–LAP2β(137–298) (squares) were allowed to replicate in S-phase extract containing BrdU and [α32P]dCTP. Substituted DNA was separated by centrifugation in CsCl gradients. cpm of each fraction was plotted against fractions of equal densities. HL indicates density of heavy-light replicated DNA. (F) Nuclei from S-phase HeLa cells were loaded with GST–LAP2β peptides and peptide uptake was analyzed as in Fig. 4 A (GST–LAP2β[1–452] is shown). Peptide-loaded nuclei were incubated in S-phase extract and incorporation of [α32P]dCTP was measured by phosphorImager (cpm/105 nuclei). Bar, 10 μM.
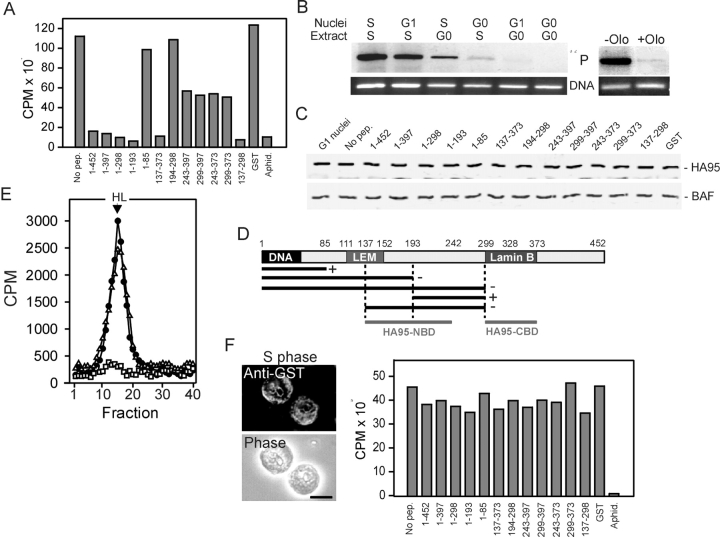
Having characterized the nuclei, the effect of GST–LAP2β peptides on replication was examined. Replication was assayed by incorporation of [α32P]dCTP, DNA electrophoresis, and phosphorImaging (Fig. 5 A). Absence of GST–LAP2β peptide or loading GST alone into G1 nuclei did not hinder replication. As expected, DNA synthesis was blocked with 50 μM aphidicolin in the extract. However, LAP2β(1–452) and (1–397) abolished replication and peptides containing HA95-CBD (LAP2β[243–397], [299–397], [243–373], and [299–373]) impaired replication efficiency by ∼50%. Furthermore, LAP2β(1–298), (1–193), (137–373), and (137–298) inhibited replication, whereas LAP2β(194–298) did not. We also ruled out the possibility that the DNA replication signal detected in G1 nuclei represented an elongation phase in already replicating nuclei, because G1 nuclei incubated in extract from G0-arrested cells did not replicate (Fig. 5 B).
Initiation of replication requires the cyclin A–Cdk2 complex (Stoeber et al., 1998). Thus, to provide evidence that DNA synthesis in G1 nuclei was due to true initiation of replication, we showed that DNA synthesis was inhibited with 10 μM of the Cdk2 inhibitor, olomoucine, in the assay (Stoeber et al., 1998) (Fig. 5 B). Similar results were obtained with 500 μg/ml dimethylaminopurine, a nonspecific protein kinase inhibitor (unpublished data). Altogether, the results indicate that peptides containing HA95-CBD partially inhibit replication in G1 nuclei. However, those containing HA95-NDB completely abolish replication. We ruled out an involvement of the GCL-binding domain of LAP2β (residues 219–298) in inhibition of replication by fragment 137–298 because LAP2β(194–298) was not inhibitory.
As described earlier in the nuclear reconstitution experiment, none of the peptides altered the immunofluorescence labeling pattern of LAP2β, A- and B-type lamins, or BAF in the G1 nuclei examined (unpublished data; see also below). In addition, Western blot analysis of the G1 nuclei loaded with the peptides shows that neither peptide affected BAF levels (or HA95) in the nuclei (Fig. 5 C). As LAP2β(1–193) and (137–298) block replication whereas LAP2β(194–298) and (1–85) allow replication (Fig. 5 D), the data suggest that region 137–193 of LAP2β is involved in replication initiation without affecting the distribution of NE proteins or the amount of BAF in the nuclei.
To establish that the LAP2β(137–298) peptide inhibited semiconservative DNA replication in G1 nuclei, BrdU substitution and density gradient centrifugation of 32P-labeled DNA was performed. G1 nuclei loaded with no peptide, GST alone, or LAP2β(137–298) were incubated for 3 h in S-phase extract containing BrdU and [α32P]dCTP to quantitate replication. DNA was analyzed by CsCl gradient centrifugation. In control nuclei, the cytosol promoted DNA synthesis, producing primarily hemisubstituted (heavy-light [HL]) DNA, indicative of semiconservative replication (Fig. 5 E). In contrast, no peak of hemisubstituted DNA was detected in nuclei loaded with LAP2β(137–298), consistent with replication inhibition detected by 32P incorporation. We concluded that LAP2β(137–298) interfered with semiconservative replication in G1 nuclei.
Disrupting LAP2β–HA95 interaction does not affect the elongation phase of DNA replication in vitro
We next determined whether the elongation phase of replication was also affected by LAP2β peptides. Nuclei isolated from S-phase HeLa cells were loaded with each GST–LAP2β peptide (Fig. 5 F, left; GST–LAP2β[1–452] is shown) and incubated in S-phase extract under conditions promoting replication. Remarkably, all peptides supported DNA synthesis to the same extent as a control without peptide (Fig. 5 F). In addition, to validate our DNA replication assay, we showed that DNA synthesis occurred in S-phase nuclei incubated in extract from G0 cells, reflecting the replicating state of these nuclei (Fig. 5 B). In contrast, G0 nuclei did not synthesize DNA in S-phase extract. Note, however, a faint 32P label in G0 nuclei in the S-phase extract, due to a minor proportion of slowly replicating nuclei in the G0 cell population (unpublished data).
HA95 coimmunoprecipitates with the Cdc6 protein in G1 phase
Initiation of DNA replication requires the assembly of preRCs at origins of replication in G1. The chromatin-bound ORC complex recruits Cdc6, which in turn promotes targeting of MCM proteins. HA95 and Cdc6 were found to coimmunoprecipitate from G1-phase HeLa cells; nevertheless, whereas anti-HA95 antibodies precipitated all detectable Cdc6, a substantial fraction of HA95 did not associate with the Cdc6 immune precipitate (Fig. 6 A). In S phase however, Cdc6 and HA95 did not coprecipitate (Fig. 6 A), despite the reported persistence of a fraction of Cdc6 on chromatin beyond G1 (Mendez and Stillman, 2000).
Figure 6.
Dissociation of HA95 from HA95-NBD in G1 elicits degradation of Cdc6. (A) HA95 (top three panels) or Cdc6 (bottom three panels) was immunoprecipitated from G1- or S-phase HeLa cells, and immune precipitates (P) and supernatants (S) were immunoblotted using either precipitating antibody. (B) Nuclei from G1-phase cells (lane 1) or G1 nuclei loaded with the indicated GST–LAP2β peptides (lanes 2–5) and incubated in nuclear isolation buffer for 1 h were immunoblotted using the indicated antibodies. In lanes 6–8, G1 nuclei loaded with LAP2β(137–298) were incubated in buffer for 1 h together with 25 μM β-lactone, LLnL, or LLM. In lane 9, whole samples (WS; nuclei + buffer) were immunoblotted. (C) G1 nuclei were loaded with GST–LAP2β(137–298) or GST–LAP2β(299–373) in the presence of no inhibitor (−), β-lactone, LLnL, or LLM. Nuclei were allowed to replicate in S-phase extract containing [α32P]dCTP, and synthesized DNA was analyzed by autoradiography.
Disruption of the interaction between HA95 and LAP2β via HA95-NBD in G1 induces proteolysis of Cdc6
The role of Cdc6 on preRC assembly led us to investigate the fate of Cdc6 in G1 nuclei after disruption of the HA95–LAP2β association via HA95-NBD or HA95-CBD. GST–LAP2β peptides were introduced into purified G1 HeLa nuclei and nuclei were incubated in nuclear isolation buffer for 1 h. Immunoblotting analyses of the nuclei show that nuclei loaded with GST alone or with LAP2β(299–373) contained Cdc6 (Fig. 6 B, lanes 1–3). However, nuclei loaded with LAP2β(137–298) or LAP2β(1–452) harbored no detectable Cdc6 (lanes 4 and 5). Additionally, blots of the whole incubation mix (nuclei and buffer) revealed no Cdc6 either, indicating that Cdc6 was degraded under these conditions (lane 9; see below). Note that p53 was also degraded in nuclei containing peptides harboring HA95-NBD, however, Orc2, a component of the preRC, was not affected (lanes 4 and 5).
The ORC large subunit has recently been shown to be degraded by ubiquitin-mediated proteolysis in human cells (Mendez et al., 2002). Thus degradation of Cdc6 by the 25S proteasome was examined. Inhibition of the proteasome by incubation of nuclei containing LAP2β(137–298) with 25 μM of the proteasome inhibitors LLnL or β-lactone during peptide loading and incubation in buffer prevented degradation of Cdc6 (Fig. 6 B, lanes 6 and 7). However, the calpain inhibitor LLM, used as a control, did not block Cdc6 proteolysis (lane 8). Inhibition of degradation of p53, a known substrate of ubiquitin-mediated proteolysis, with LLnL or β-lactone confirmed the efficacy of the inhibitors (Fig. 6 B). Introduction of LAP2β(137–298) in S-phase nuclei did not elicit degradation of Cdc6 (or p53 or Orc2; unpublished data). These results suggest that dissociation of HA95 from HA95-NBD in G1- but not S-phase nuclei triggers degradation of Cdc6 by the proteasome. Because p53 is also proteolyzed by LAP2β(137–298), the data also suggest that the peptide promotes degradation of a subset of nuclear proteins.
As G1 nuclei containing LAP2β(137–298) do not replicate DNA in S-phase extract, we determined whether proteasome inhibitors would rescue S-phase entry of these nuclei. G1 nuclei were loaded with LAP2β(137–298) in the presence of LLnL or β-lactone and incubated for 3 h in S-phase extract containing [α32P]dCTP under conditions promoting replication. Fig. 6 C shows that LLnL and β-lactone relieved the inhibition of DNA replication imposed by LAP2β(137–298). The calpain inhibitor LLM, however, had no effect and, as expected, LLnL or β-lactone did not affect replication of LAP2β(299–373)-loaded G1 nuclei (Fig. 6 C). Therefore, inhibition of the proteasome enables entry of the G1 nuclei into a replication phase.
Injection of LAP2β(137–298) into G1 nuclei inhibits entry into S phase in vivo
The significance of HA95 interaction with HA95-NBD or HA95-CBD was further investigated in vivo by injections of ∼5 nM GST–LAP2β peptides into the nuclei of HeLa cells in early G1 (2 h after release from mitotic arrest). Injections were verified by nuclear retention of a 150-kD FITC–dextran (see below and Fig. 8 A).
Figure 8.

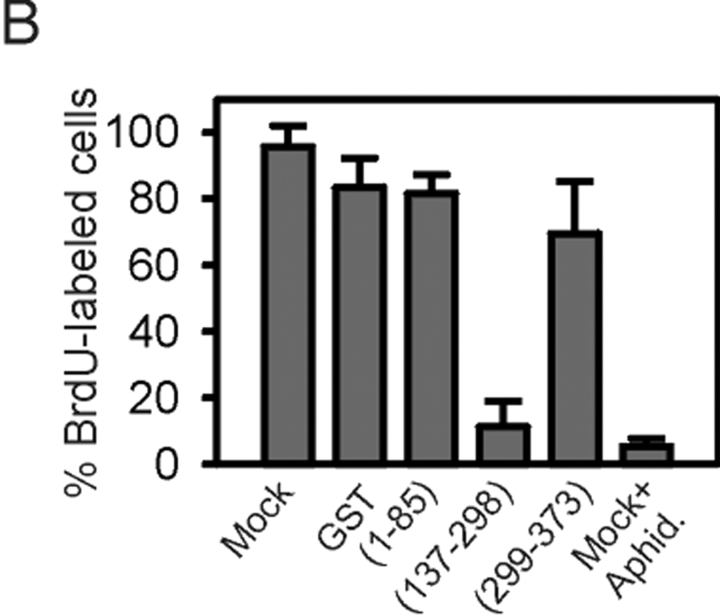
Nuclear injection of GST–LAP2β(137–298) in G1 inhibits replication. Nuclei of G1 HeLa cells were injected with 5 ng of the indicated GST–LAP2β peptide or GST alone together with a 150-kD FITC–dextran to visualize injections, and cells were cultured for 9–10 h with BrdU. Mock (buffer)-injected cells were also cultured with BrdU and 50 μM aphidicolin. BrdU incorporation was detected using TRITC-conjugated anti-BrdU antibodies. DNA was labeled with Hoechst 33342. (B) Percentages of cells labeled with BrdU 9–10 h after peptide injection (n = 45–50/treatment in two replicates). (C) Immunofluorescence analysis of Cdc6 and Orc2p in G1 cells injected as in A. Cells were fixed 2 h after peptide injection. Arrows point to noninjected cells. Bars, 10 μm.
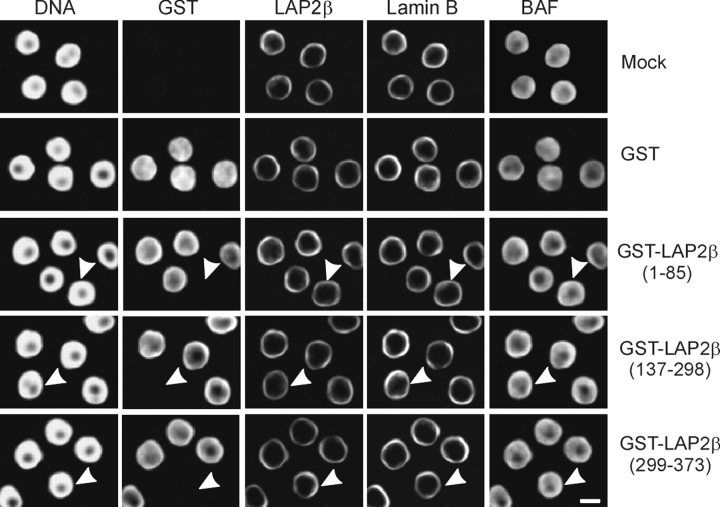
We first assessed whether the distribution of INM and lamina proteins was altered in the injected nuclei. As seen earlier in in vitro–reconstituted nuclei, GST–LAP2β peptides were detected throughout the nucleus for the most part, with a propensity of the anti-GST antibody to decorate the nuclear periphery more strongly (Fig. 7, GST). This, however, was not specific for the peptide injected (Fig. 7, bottom three rows). Immunofluorescence analysis of peptide- and mock (buffer)-injected cells indicated that LAP2β and B-type lamins remained localized at the NE 2–3 h after injection with either peptide (Fig. 7). Similar results were obtained for LBR, emerin, and A-type lamins (unpublished data). Additionally, no alteration in the localization of BAF in peptide-injected and control cells was detected (Fig. 7). BAF remained distributed throughout the nucleoplasm with an enrichment around the periphery. Thus, we could not attribute a noticeable effect of intranuclear peptide injection in G1 on overall nuclear architecture.
Figure 7.
Nuclear injection of GST–LAP2β(137–298) in G1 does not alter distribution of NE proteins or BAF. Nuclei of G1 HeLa cells were injected with 5 ng of the indicated GST–LAP2β peptide or GST alone, cultured for 2–3 h, and analyzed by immunofluorescence using anti-GST, LAP2β, lamin B (goat polyclonal), or BAF antibodies. Arrows point to noninjected cells. Bar, 10 μm.
To examine the effect of peptide injection in G1 on DNA replication, injected G1-phase cells were cultured with 10 μM BrdU for 10 h and DNA synthesis was monitored using anti-BrdU antibodies. Fig. 8 (A and B) shows that 95% of mock-injected cells underwent DNA synthesis, which was inhibited by 50 μM aphidicolin. Likewise, cells injected with LAP2β(1–85) or GST alone replicated DNA. However, LAP2β(137–298) abolished DNA synthesis in 90% of the cells, whereas LAP2β(299–373) had no effect. Injection of LAP2β(137–298) in S-phase nuclei was not inhibitory (unpublished data), indicating that once DNA synthesis is initiated, disruption of the LAP2β–HA95 interaction via HA95-NBD has no effect.
Additional immunofluorescence analysis of injected G1 HeLa cells indicated that Cdc6 was undetectable in LAP2β (137–298)-injected cells, whereas intranuclear labeling was evident in noninjected cells (Fig. 8 C, arrow) or in cells injected with GST alone or LAP2β(299–373) (Fig. 8 C). Note that Orc2 was not degraded in LAP2β(137–298)-injected cells, as shown by intranuclear immunolabeling (Fig. 8 C). The results indicate that, as shown biochemically in vitro, LAP2β(137–298) inhibits S-phase entry in vivo. Inhibition correlates with the degradation of Cdc6, but is not due to a displacement of NE proteins or a change in BAF distribution during G1.
Discussion
Anchoring of the INM to chromatin
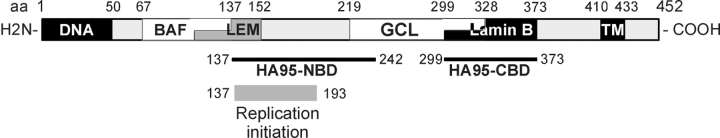
This study provides evidence for a novel direct interaction of the NE with the genome, via the INM protein LAP2β and the chromatin- and nuclear matrix–associated protein HA95. The nucleoplasmic 410 amino acids of LAP2β are prompt to interactions with multiple intranuclear ligands (Fig. 9). The first 50 residues are common to all LAP2 proteins and bind DNA (Cai et al., 2001), in consistency with the chromatin-binding property of this region, which causes post-mitotic association of LAP2α with chromosomes (Vlcek et al., 1999). LAP2 proteins also bind the DNA-bridging protein BAF through residues 67–137 (Furukawa, 1999; Shumaker et al., 2001), which include most of the LEM domain (Fig. 9). The significance of this interaction in interphase remains unclear, although a role in chromatin decondensation and nuclear expansion in vitro has been proposed (Segura-Totten et al., 2002). LAP2β also binds GCL, a transcription regulator that interacts with components of the E2F transcription machinery (Nili et al., 2001). LAP2β is capable of reducing the transcription activity of the E2F complex alone or with GCL (Nili et al., 2001), suggesting an involvement of the NE in transcription regulation. It will be interesting to determine whether mutations in NE proteins that cause disease (Vigouroux and Bonne, 2002) also lead to defects in transcription regulation in affected tissues.
Figure 9.
Functional domains of LAP2β. Identified functional regions of LAP2β (LEM and transmembrane [TM]) and domains binding to indicated ligands are shown. Numbers refer to amino acid positions. See text for references to binding domains.
We now show that LAP2β also directly interacts with the chromatin-associated protein HA95 via two HA95-binding domains that are distinct from the chromosome-binding region. The NH2-terminal domain (HA95-NBD) localizes within amino acids 137–242, while the COOH-terminal domain (HA95-CBD) is restricted to residues 299–373 (Fig. 9). HA95-NBD partially overlaps with the 187 NH2-terminal amino acids common to all LAP2 proteins, thus HA95-NBD may not be restricted to NE-associated LAP2β and may also exist in intranuclear LAP2 proteins. HA95-CBD coincides with the lamin B–binding domain of LAP2β and is unique to LAP2β, nevertheless LAP2β binds HA95 independently of lamin B in vitro. The functional implications of each HA95-binding domain of LAP2β are important, as disruption of only one or the other has distinct effects on NE assembly or DNA replication (see below). As the binding domains to each ligand are distinct, interaction of LAP2β with either domain may not be exclusive, and it is tempting to speculate that a single protein of the INM may regulate chromatin function at multiple levels in interphase.
Interaction between HA95 and LAP2β is dispensable for nuclear membrane assembly
We previously reported that anti-HA95 antibody-mediated blocking of HA95 function on condensed chromosomes had no inhibitory effect on nuclear membrane assembly in vitro (Martins et al., 2000). This suggested that HA95 may not be involved in membrane targeting to chromatin. Our results indicate that any LAP2β fragment that can bind lamin B inhibits nuclear assembly, however peptides that bind HA95, but not lamin B, fail to inhibit assembly. Thus, competition for LAP2β binding to HA95 has no detectable effect on nuclear formation. Although our results show that the lamin B–binding domain of LAP2β is involved in NE formation (Yang et al., 1997; Gant et al., 1999), they document a lack of effect of HA95 binding activity on nuclear assembly.
Role of association of HA95 with the NH2-terminal HA95-binding domain of LAP2β in DNA replication
Our results suggest that the HA95–LAP2β interaction via HA95-NBD may represent an additional form of control for replication initiation, at least in transformed cells. We ruled out the possibility that disruption of the HA95–LAP2β interaction upon intranuclear introduction of LAP2β fragments affected overall nuclear organization, which in itself might compromise DNA replication (Gant et al., 1999; Moir et al., 2000). Notably, the localization of endogenous LAP2β, A- and B-type lamins, and BAF does not seem to be perturbed by disruption of the interaction of HA95 with HA95-NBD upon nuclear reassembly in vitro, in isolated G1 nuclei, or in G1-phase nuclei in vivo. Additionally, BAF levels remain unaffected in G1 nuclei, arguing that defects in replication induced by LAP2β disruptor peptides do not result from changes in BAF protein levels (Segura-Totten et al., 2002). The HA95-NBD of LAP2(β) partially overlaps with the LEM domain, which interacts with BAF. BAF distribution and levels are not altered by LAP2β peptides containing HA95-NBD, and regions nonoverlapping with the HA95-binding domain were sufficient for BAF binding in yeast two-hybrid assays (Furukawa, 1999). Nonetheless, whether manipulating the association of HA95 with LAP2β interferes with binding of LAP2 proteins to BAF remains to be examined (Shumaker et al., 2001).
How does the HA95–LAP2β interaction affect DNA replication? First, we should emphasize that the domain of HA95-NBD involved in replication (residues 137–193; Fig. 9) is almost entirely included in the 187 amino acids (1–187) common to all LAP2 proteins. Thus, the HA95–LAP2 interaction is probably not restricted to the NE. We propose a hypothesis whereby interaction of HA95 with the NH2-terminal domain of LAP2 proteins and with components of the preRC brings components of the preRC to replication origins, maintains integrity of the preRC, and/or protects preRC components from degradation. It is conceivable that destabilizing the HA95–LAP2β interaction in G1 may displace preRC components, triggering their proteasome-mediated degradation and, as a result, blocking replication initiation. These alternatives are compatible with the distribution of preRCs throughout the genome and the intranuclear localization of LAP2α, a nonmembrane-bound LAP2 isoform (Vlcek et al., 1999). Disruption of the interaction specifically abolishes S-phase entry but has no effect on the elongation phase of replication. Furthermore, HA95 and Cdc6 coimmunoprecipitate in G1 but not S phase, suggesting that HA95 (in)directly interacts with the preRC. The lack of interaction of HA95 with Cdc6 in S phase is consistent with nuclear export or degradation of a fraction of Cdc6 in mammalian cells (Saha et al., 1998; Coverley et al., 2000) and with disassembly of the preRC.
Disruption of HA95 interaction with HA95-NBD in G1 triggers proteasome-mediated degradation of Cdc6 (and p53), whereas proteasome inhibitors block LAP2β(137–298)-induced proteolysis and rescue the replication inhibitory effect of the LAP2β peptide in G1 nuclei. Yet, the inhibitors obviously did not rescue the LAP2β–HA95 interaction, indicating that this association is dispensable for replication. It is likely that the LAP2β(137–298) peptide causes degradation of a class of nuclear proteins, because p53 reacts similarly to Cdc6. This could result from a pleiotropic effect of the peptide, perhaps disturbing other proteins that interact through a similar domain. In any event, inhibition of DNA synthesis by LAP2β(137–298) may be indirect, for example by activating a checkpoint pathway. This speculative hypothesis remains to be tested.
Our results extend the notion that the NE contributes to regulating replication. In Xenopus, disruption of lamina organization alters the distribution of replication factors and inhibits the elongation phase of DNA synthesis (Moir et al., 2000). Thus, replication may be regulated at several levels by nuclear lamins and nuclear membrane–chromatin interactions, in addition to interactions between chromatin-associated proteins.
Multiple functions of HA95 dictated by its interaction with multiple ligands
By interacting with several intranuclear ligands, HA95 emerges as a multifunctional molecule. In addition to its involvement in replication, HA95 binds RHA (Westberg et al., 2000), suggestive of a role in transcription regulation. RHA binds to a CTE involved in nuclear export of unspliced viral RNA, and association of HA95 with RHA enhances CTE-mediated gene expression and promotes nuclear export of unspliced mRNA (Yang et al., 2001). By binding to LAP2β, HA95 may favor viral RNA export by tethering it near the NE.
HA95 also interacts with the catalytic subunit of PKA within the nucleus and localizes the catalytic subunit to sites where it can modulate transcription from specific promoters (Han et al., 2002). Thus, HA95 may act as a targeting molecule for the PKA catalytic subunit in the nucleus. Along this line, it is tempting to speculate that HA95–LAP2β, or HA95–LAP2, interactions may promote targeting or stabilization of signaling molecules in the vicinity of transcription sites and at origins of replication at discrete stages of the cell cycle.
Materials and methods
Antibodies and peptides
Anti-Myc antibodies were from Invitrogen. Anti-GST, anti-Cdc6 (H-304 and clone 180.2), anti-Orc2, goat anti–lamin B antibodies, and anti-p53 antibodies were from Santa Cruz Biotechnology, Inc. Anti-BrdU antibodies were from Sigma-Aldrich. Affinity-purified polyclonal antibodies against LBR, LAP2β, A- and B-type lamins (gifts from J.-C. Courvalin, Institut J. Monod, Paris, France), BAF (a gift from K. Wilson, Johns Hopkins University School of Medicine, Baltimore, MD), and HA95 were described elsewhere (Buendia and Courvalin, 1997; Orstavik et al., 2000; Haraguchi et al., 2001). LLnL, LLM, and β-lactone were from Calbiochem. GST–LAP2β peptides were expressed as previously described (Furukawa et al., 1995, 1997, 1998).
Cells and nuclei
Bjab cells were grown in RPMI 1640/10% FCS (GIBCO BRL) (Orstavik et al., 2000). HeLa cells were cultured in EMEM/10% FCS (GIBCO BRL). HeLa cells were synchronized in M phase with 1 μM nocodazole for 18 h. To allow cell cycle reentry, cells were plated at 2.5 × 106 cells in 162-cm2 flasks. G1- and S-phase cells were harvested (or microinjected) 2 and 12 h, respectively, after release from mitotic arrest. Cells were arrested in G0 by a 5-d culture under confluent conditions without serum. Nuclei were isolated from Bjab cells and from G0-, G1-, or S-phase HeLa cells by Dounce homogenization (Martins et al., 2000). For nuclear reassembly assays, nuclei were isolated from confluent HeLa cells and used fresh of frozen/thawed (Steen et al., 2000). Freshly isolated nuclei were used in replication assays.
Microinjection and BrdU incorporation
HeLa cells released from M-phase arrest were seeded on coverslips. Within 2 h, nuclei were microinjected with 25 pl PBS containing 10 μg/ml 150-kD FITC–dextran to visualize injections, and ∼5 nM indicated GST–LAP2β peptide. Cells were cultured in EMEM/10% FCS for up to 10 h without or with 100 μM BrdU and processed for immunofluorescence or BrdU incorporation analysis. S-phase cells were injected 10–12 h after release from M-phase arrest. Approximately 50 cells were injected per treatment in two to three replicates. To detect BrdU incorporation, cells were fixed with methanol for 5 min, blocked, and overlaid with anti-BrdU antibodies (1:200 dilution) and TRITC-conjugated anti-BrdU antibodies.
Immunological procedures
Immunoblotting analysis was performed as previously described (Martins et al., 2000) using antibodies against HA95 (1:250 dilution), Myc (1:1,000), GST (1:1,000), B-type lamins (1:1,000), lamin A/C (1:500), LAP2β (1:500), LBR (1:500), BAF (1:500), p53 (1:500), Cdc6 (1:500), and Orc2 (1:500). For immunoprecipitations, cells or nuclei were sonicated in IP buffer (10 mM Hepes, pH 7.5, 10 mM KCl, 2 mM EDTA, 1% Triton X-100, 1 mM DTT, and protease inhibitors) and lysates were centrifuged at 15,000 g for 15 min. Immunoprecipitations were performed from the supernatants with relevant antibodies (1:50 dilutions) as described earlier (Martins et al., 2000).
GST precipitation
Nuclei isolated from Bjab cells (109 nuclei/ml) were sonicated in GST precipitation buffer (300 mM KCl, 20 mM Hepes, pH 7.6, 0.1% Triton X-100, 1 mM DTT, 5 mM benzamidine, and protease inhibitors) and the lysate was centrifuged at 10,000 g. The supernatant (nuclear extract) was incubated with specified GST–LAP2β peptides (1 or 5 μg/μl) overnight at 4°C with rotation. GST precipitations were performed using glutathione beads coated with 10 mg/ml BSA and pellets were washed in GST precipitation buffer. Proteins were eluted in SDS sample buffer.
Overlay assays
HA95-IPs proteins resolved by 10% SDS-PAGE were blotted onto nitrocellulose, blocked with 5% milk in Tris-buffered saline/0.01% Tween 20 (TBST) for 1 h, washed in TBST, and overlaid with 10 μM GST–LAP2β peptides for 2 h in TBST. Membranes were washed in TBST and peptide binding was detected using anti-GST antibodies and peroxidase-conjugated secondary antibodies.
Nuclear reconstitution assay
Condensed, membrane-free chromatin masses were prepared from HeLa nuclei disassembled in mitotic extract (Martins et al., 2000). After sedimentation through 1 M sucrose, chromosomes were resuspended in peptide-binding buffer (100 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 20 mM Hepes, pH 7.5, 1 mM DTT, and protease inhibitors) containing 5 μM of indicated GST–LAP2β peptides.
After 1 h at room temperature, chromosomes were sedimented, washed, and either immunologically analyzed or resuspended in nuclear reassembly extract. In this case, chromosomes (2 μl) were resuspended in 40 μl of an interphase HeLa cell 200,000 g cytosolic extract at 5,000 chromatin masses/μl, containing 4 μl mitotic membranes, an ATP-regenerating system (1.2 μl), and 100 μM GTP (0.4 μl) (Steen et al., 2000). After 2 h at 30°C, nuclear assembly was examined by phase contrast microscopy, membrane staining with 10 μg/ml DiOC6, or by immunofluorescence. Nuclei or chromatin masses were also sedimented through 1 M sucrose, washed, and solubilized in SDS sample buffer.
Loading of nuclei with GST–LAP2β peptides
Peptide loading into isolated HeLa nuclei was performed as described earlier (Collas et al., 1999). In brief, nuclei were mildly permeabilized with lysolecithin and incubated for 1 h with indicated GST–LAP2β peptides (100 μM) or GST alone. Nuclei were washed by sedimentation through 1 M sucrose and held on ice until use. Lysolecithin-treated nuclei were capable of active protein import in vitro (see Results).
In vitro replication and quantification of DNA synthesis
Replication was assayed by incorporation of [α32P]dCTP into newly synthesized DNA. Isolated G0-, G1-, or S-phase nuclei (preloaded with GST–LAP2β peptides, as indicated) were incubated for 3 h at 5,000 nuclei/μl in 40 μl of S-phase extract (unless indicated otherwise) containing the ATP-regenerating system, 100 μM GTP, a buffered mix of dNTPs (40 mM Hepes, pH 7.8, 7 mM MgCl2, 0.1 mM each of dATP, dGTP, dTTP, and dCTP; 2 μl) and 1 μl [α32P]dCTP (3,000 Ci/mmol; Amersham Biosciences). Concentrated S-phase whole cell extracts were prepared from S-phase HeLa cells collected 15 h after release from mitotic arrest. Cells were lysed by Dounce homogenization in cell lysis buffer (Martins et al., 2000), and then briefly sonicated on ice to lyse nuclei and release soluble nuclear components. The lysate was sedimented at 15,000 g for 15 min and then at 200,000 g for 2 h, both at 4°C. Protein concentration of the extract was 25–30 mg/ml.
At the end of incubation in extract, samples were mixed with 1 volume of 20 mM Tris (pH 7.5) and 1 mg/ml proteinase K and digested for 2 h at 37°C. Samples were mixed by pipetting and 5-μl aliquots were electrophoresed through 0.8% agarose. Gel loading was assessed by ethidium bromide staining. Samples contained equal numbers of nuclei and sedimentation steps were eliminated to avoid loss of nuclei (Gant et al., 1999). Signals were quantified by phosphorImaging or by autoradiography.
BrdU density substitution
Density substitutions were done as previously described (Gant et al., 1999) in reactions consisting of isolated G1 nuclei in S-phase extract containing [α32P]dCTP and 0.5 mM BrdU. Samples were digested with proteinase K and DNA was extracted with phenol–chloroform then chloroform, ethanol precipitated, and dissolved in 100 μl Tris-EDTA buffer. Samples were mixed with 12 ml of 1.75 g/ml CsCl and centrifuged for 45 h at 60,000 g. 40 fractions were collected, an aliquot was counted by liquid scintillation, and the refractive index of each fraction was measured (Gant et al., 1999).
Acknowledgments
We are grateful to Drs. K. Wilson and J.-C. Courvalin for antibodies.
This work was supported by the Portuguese Foundation for Science and Technology (S. Martins), the Research Council of Norway, the Norwegian Cancer Society, and the Human Frontiers Science Program (P. Collas).
Footnotes
Abbreviations used in this paper: BAF, barrier-to-autointegration factor; CBD, COOH-terminal binding domain; CTE, constitutive transport element; GCL, germ cell less; HA95-IP, anti-HA95 immune precipitate; INM, inner nuclear membrane; LBR, lamin B receptor; LEM, LAP2, emerin, MAN-1; MCM, minichromosome maintenance; NBD, NH2-terminal binding domain; NE, nuclear envelope; ORC, origin recognition complex; PKA, protein kinase A; preRC, prereplication complex; RHA, RNA helicase A.
References
- Bell, S.P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333–374. [DOI] [PubMed] [Google Scholar]
- Berger, R., L. Theodor, J. Shoham, E. Gokkel, F. Brok-Simoni, K.B. Avraham, N.G. Copeland, N.A. Jenkins, G. Rechavi, and A.J. Simon. 1996. The characterization and localization of the mouse thymopoietin/lamina-associated polypeptide 2 gene and its alternatively spliced products. Genome Res. 6:361–370. [DOI] [PubMed] [Google Scholar]
- Buendia, B., and J.-C. Courvalin. 1997. Domain-specific disassembly and reassembly of nuclear membranes during mitosis. Exp. Cell Res. 230:133–144. [DOI] [PubMed] [Google Scholar]
- Cai, M., Y. Huang, R. Ghirlando, K.L. Wilson, R. Craigie, and G.M. Clore. 2001. Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. EMBO J. 20:4399–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, T.R., P.B. Carpenter, and W.G. Dunphy. 1996. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 87:53–63. [DOI] [PubMed] [Google Scholar]
- Collas, P., K. Le Guellec, and K. Tasken. 1999. The A-kinase anchoring protein, AKAP95, is a multivalent protein with a key role in chromatin condensation at mitosis. J. Cell Biol. 147:1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley, D., C. Pelizon, S. Trewick, and R.A. Laskey. 2000. Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J. Cell Sci. 113:1929–1938. [DOI] [PubMed] [Google Scholar]
- Foisner, R., and L. Gerace. 1993. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 73:1267–1279. [DOI] [PubMed] [Google Scholar]
- Furukawa, K. 1999. LAP2 binding protein 1 (L2BP1/BAF) is a candidate mediator of LAP2-chromatin interaction. J. Cell Sci. 112:2485–2492. [DOI] [PubMed] [Google Scholar]
- Furukawa, K., and T. Kondo. 1998. Identification of the lamina-associated-polypeptide-2-binding domain of B-type lamin. Eur. J. Biochem. 251:729–733. [DOI] [PubMed] [Google Scholar]
- Furukawa, K., N. Panté, U. Aebi, and L. Gerace. 1995. Cloning of a cDNA for lamina-associated polypeptide 2 (LAP2) and identification of regions that specify targeting to the nuclear envelope. EMBO J. 14:1626–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, K., C. Glass, and T. Kondo. 1997. Characterization of the chromatin binding activity of lamina-associated polypeptide (LAP) 2. Biochem. Biophys. Res. Commun. 238:240–246. [DOI] [PubMed] [Google Scholar]
- Furukawa, K., C.E. Fritze, and L. Gerace. 1998. The major nuclear envelope targeting domain of LAP2 coincides with its lamin binding region but is distinct from its chromatin interaction domain. J. Biol. Chem. 273:4213–4219. [DOI] [PubMed] [Google Scholar]
- Gant, T.M., C.A. Harris, and K.L. Wilson. 1999. Roles of LAP2 proteins in nuclear assembly and DNA replication: truncated LAP2β proteins alter lamina assembly, envelope formation, nuclear size, and DNA replication efficiency in Xenopus laevis extracts. J. Cell Biol. 144:1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, I., Y. Xue, S. Harada, S. Orstavik, B. Skålhegg, and E. Kieff. 2002. Protein kinase A associates with HA95 and affects transcriptional coactivation by Epstein-Barr virus nuclear proteins. Mol. Cell. Biol. 22:2136–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi, T., T. Koujin, M. Segura-Totten, K.K. Lee, Y. Matsuoka, Y. Yoneda, K.L. Wilson, and Y. Hiraoka. 2001. BAF is required for emerin assembly into the reforming nuclear envelope. J. Cell Sci. 114:4575–4585. [DOI] [PubMed] [Google Scholar]
- Kelly, T.J., and G.W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829–880. [DOI] [PubMed] [Google Scholar]
- Krude, T., M. Jackman, J. Pines, and R.A. Laskey. 1997. Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell. 88:109–119. [DOI] [PubMed] [Google Scholar]
- Landsverk, H.B., A.M. Håkelien, T. Küntziger, J.M. Robl, B.S. Skålhegg, and P. Collas. 2002. Reprogrammed gene expression in a somatic cell-free extract. EMBO Rep. 3:384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F., D.L. Blake, I. Callebaut, I.S. Skerjanc, L. Holmer, M.W. McBurney, M. Paulin-Levasseur, and H.J. Worman. 2000. MAN1, an inner nuclear membrane protein that shares the LEM domain with lamina-associated polypeptide 2 and emerin. J. Biol. Chem. 275:4840–4847. [DOI] [PubMed] [Google Scholar]
- Martins, S.B., T. Eide, R.L. Steen, T. Jahnsen, B.S. Skålhegg, and P. Collas. 2000. HA95 is a protein of the chromatin and nuclear matrix regulating nuclear envelope dynamics. J. Cell Sci. 113:3703–3713. [DOI] [PubMed] [Google Scholar]
- Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez, J., X.H. Zou-Yang, S.Y. Kim, M. Hidaka, W.P. Tansey, and B. Stillman. 2002. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell. 9:481–491. [DOI] [PubMed] [Google Scholar]
- Moir, R.D., T.P. Spann, H. Herrmann, and R.D. Goldman. 2000. Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J. Cell Biol. 149:1179–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nili, E., G.S. Cojocaru, Y. Kalma, D. Ginsberg, N.G. Copeland, D.J. Gilbert, N.A. Jenkins, R. Berger, S. Shaklai, N. Amariglio, et al. 2001. Nuclear membrane protein LAP2β mediates transcriptional repression alone and together with its binding partner GCL (germ-cell-less). J. Cell Sci. 114:3297–3307. [DOI] [PubMed] [Google Scholar]
- Orstavik, S., T. Eide, P. Collas, I.O. Han, K. Tasken, E. Kieff, T. Jahnsen, and B.S. Skålhegg. 2000. Identification, cloning and characterization of a novel nuclear protein, HA95, homologous to A-kinase anchoring protein 95. Biol. Cell. 92:27–37. [DOI] [PubMed] [Google Scholar]
- Saha, P., J. Chen, K.C. Thome, S.J. Lawlis, Z.H. Hou, M. Hendricks, J.D. Parvin, and A. Dutta. 1998. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol. Cell. Biol. 18:2758–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura-Totten, M., A.K. Kowalski, R. Craigie, and K.L. Wilson. 2002. Barrier-to-autointegration factor: major roles in chromatin decondensation and nuclear assembly. J. Cell Biol. 158:475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, N., N. Ueki, K. Yano, T. Saito, Y. Masuho, and M. Muramatsu. 2000. cDNA cloning of a novel human gene NAKAP95, neighbor of A-kinase anchoring protein 95 (AKAP95) on chromosome 19p13.11-p13.12 region. J. Hum. Genet. 45:31–37. [DOI] [PubMed] [Google Scholar]
- Shumaker, D.K., K.K. Lee, Y.C. Tanhehco, R. Craigie, and K.L. Wilson. 2001. LAP2 binds to BAF-DNA complexes: requirement for the LEM domain and modulation by variable regions. EMBO J. 20:1754–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen, R.L., S.B. Martins, K. Tasken, and P. Collas. 2000. Recruitment of protein phosphatase 1 to the nuclear envelope by A-kinase anchoring protein AKAP149 is a prerequisite for nuclear lamina assembly. J. Cell Biol. 150:1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeber, K., A.D. Mills, Y. Kubota, T. Krude, P. Romanowski, K. Marheineke, R.A. Laskey, and G.H. Williams. 1998. Cdc6 protein causes premature entry into S phase in a mammalian cell-free system. EMBO J. 17:7219–7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux, C., and G. Bonne. 2002. One gene, two proteins, five diseases. Dynamics of the Nuclear Envelope in Embryos and Somatic Cells. P. Collas, editor. Kluwer Academic/Plenum Publishers, New York. 153–172.
- Vlcek, S., H. Just, T. Dechat, and R. Foisner. 1999. Functional diversity of LAP2α and LAP2β in postmitotic chromosome association is caused by an α-specific nuclear targeting domain. EMBO J. 18:6370–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westberg, C., J.-P. Yang, H. Tang, T.R. Reddy, and F. Wong-Staal. 2000. A novel shuttle protein binds to RNA helicase A and activates the retroviral constitutive transport element. J. Biol. Chem. 275:21396–21401. [DOI] [PubMed] [Google Scholar]
- Williams, R.S., R.V. Shohet, and B. Stillman. 1997. A human protein related to yeast Cdc6p. Proc. Natl. Acad. Sci. USA. 94:142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J.P., H. Tang, T.R. Reddy, and F. Wong-Staal. 2001. Mapping the functional domains of HAP95, a protein that binds RNA helicase A and activates the constitutive transport element of type D retroviruses. J. Biol. Chem. 276:30694–30700. [DOI] [PubMed] [Google Scholar]
- Yang, L., T. Guan, and L. Gerace. 1997. Lamin-binding fragment of LAP2 inhibits increase in nuclear volume during the cell cycle and progression into S phase. J. Cell Biol. 139:1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]