Abstract
Localization of signaling complexes to specific microdomains coordinates signal transduction at the plasma membrane. Using immunogold electron microscopy of plasma membrane sheets coupled with spatial point pattern analysis, we have visualized morphologically featureless microdomains, including lipid rafts, in situ and at high resolution. We find that an inner-plasma membrane lipid raft marker displays cholesterol-dependent clustering in microdomains with a mean diameter of 44 nm that occupy 35% of the cell surface. Cross-linking an outer-leaflet raft protein results in the redistribution of inner leaflet rafts, but they retain their modular structure. Analysis of Ras microlocalization shows that inactive H-ras is distributed between lipid rafts and a cholesterol-independent microdomain. Conversely, activated H-ras and K-ras reside predominantly in nonoverlapping, cholesterol-independent microdomains. Galectin-1 stabilizes the association of activated H-ras with these nonraft microdomains, whereas K-ras clustering is supported by farnesylation, but not geranylgeranylation. These results illustrate that the inner plasma membrane comprises a complex mosaic of discrete microdomains. Differential spatial localization within this framework can likely account for the distinct signal outputs from the highly homologous Ras proteins.
Keywords: cholesterol; lipid rafts; immunogold; electron microscopy; statistical analysis
Introduction
Current models of the plasma membrane predict the existence of a patchwork of specialized microdomains coordinating a variety of cellular functions including signaling, cell adhesion, and membrane trafficking. A well-characterized plasma membrane microdomain is the lipid raft comprised of sphingolipids packed together with cholesterol, although the existence of other lipid-based microdomains has also been proposed (Simons and Ikonen, 1997; Brown and London, 1998; Kurzchalia and Parton, 1999; Murray et al., 1999). Sophisticated fluorescence techniques strongly support the existence of outer-leaflet plasma membrane lipid rafts (Varma and Mayor, 1998; Pralle et al., 2000), but rafts need to be cross-linked in order to visualize clustering of putative inner leaflet raft-associated proteins (Harder et al., 1998). A major difficulty with accurately characterizing lipid rafts and other cell surface microdomains is imaging resolution resulting in relatively large discrepancies in estimates of microdomain parameters and composition, dependent on the techniques used to probe them (Kenworthy et al., 2000; Anderson and Jacobson, 2002). In addition, the interactions of signaling proteins with rafts are dynamic and regulated. The tools available to accurately assign proteins to specific domains in intact membranes are limited (Simons and Toomre, 2000). The microlocalization of some signaling proteins has been visually mapped at high resolution with electron microscopy. However, the assignments have been limited to morphologically identifiable plasma membrane structures such as caveolae (Mineo et al., 1999; Prior et al., 2001) or osmiophilic patches (Wilson et al., 2000, 2001), all of which account for a relatively minor fraction of the total cell surface. Therefore, in this study, we investigated whether electron microscopic techniques can also be used to assign proteins to morphologically featureless surface domains.
Ras GTPases play critical roles in transducing extracellular signals (Campbell et al., 1998). The Ras isoforms (H-ras, N-ras, and K-ras) are ubiquitously expressed in mammalian cells, and although highly homologous, these proteins generate different signal outputs (Bos, 1989; Umanoff et al., 1995; Koera et al., 1997; Yan et al., 1998). This biochemical diversity may result from differential lateral segregation of Ras proteins across plasma membrane microdomains that is regulated by their different COOH-terminal membrane anchors (Roy et al., 1999; Prior et al., 2001; Niv et al., 2002). We have directly investigated this hypothesis by mapping the cell surface distributions of Ras proteins at high resolution.
Results and discussion
Ras microlocalization was examined on intact 2-D sheets of apical plasma membrane, ripped off from adherent cells directly onto EM grids (Sanan and Anderson, 1991; Parton and Hancock, 2001). First, we investigated the distributions of GFP fused to the minimal plasma membrane targeting motifs of H-ras, GFP-tH (Fig. 1). GFP-tH, targeted by a combination of palmitoylation and a farnesylated CAAX motif, is an excellent lipid raft marker because even on high expression, it is localized exclusively to low density fractions on sucrose gradients (Prior et al., 2001). We visualized GFP-tH in plasma membrane sheets with anti-GFP–5 nm gold (Fig. 1 a). GFP-tH is extensively distributed in small, morphologically featureless patches. To analyze the complete gold patterns as rigorously and objectively as possible, we used statistical methods for point pattern analysis that are well established, but rarely used in cell biology. Ripley's K-function (Ripley, 1977, 1979; Philimonenko et al., 2000) evaluates an exhaustive map of all interparticle distances over the study area and compares the observed distribution with that expected from complete spatial randomness (CSR).* The analysis, which focuses on the second-order properties of the gold pattern, has the advantage that it analyzes spatial structure at multiple ranges simultaneously (Ripley, 1977, 1979; see Materials and methods and supplementary data).
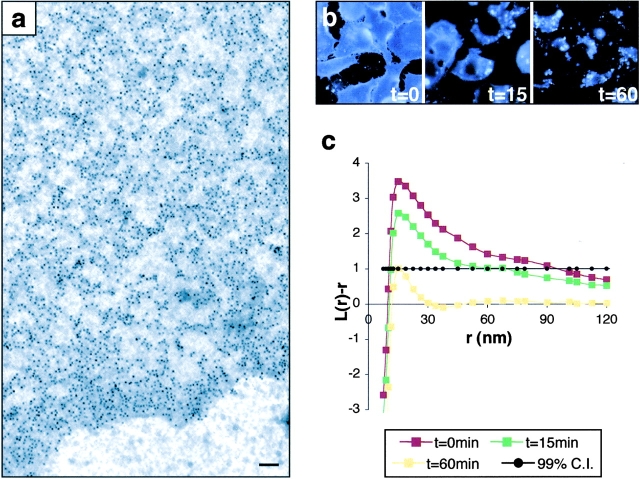
Figure 1.
Visualizing lipid rafts using electron microscopy and spatial point pattern analysis. (a) Anti-GFP labeling is specific, ending at the edge of a typical GFP-tH sheet. (b) 60 min of 1% cyclodextrin treatment depletes cell surface cholesterol, detected by filipin labeling (blue). (c) Pooled K-function analyses of the spatial distributions of GFP-tH; L(r) − r values above the 99% confidence interval for CSR (99% CI; closed circles) indicate clustering at that value of r. Untreated GFP-tH (t = 0, red line) shows maximal deflection from CSR at r = 22 nm. Cyclodextrin-treated cells show a time-dependent loss of GFP-tH clustering such that at t = 60 min, GFP-tH is not clustered. K-functions are means (n ≥ 9 for each condition) standardized on the 99% CI. Bar, 100 nm.
K-function analysis of GFP-tH sheets shows that the gold pattern is clustered (Fig. 1 c, red line), i.e., the curve shows significant positive deviation from the L(r) − r = 0 value expected for a random point pattern. The maximum deviation of the GFP-tH L(r) − r curve from CSR occurs at a radius of 22 nm. From these data, we can model the size and distribution of GFP-tH microdomains (Figs. S1 b and S2, available at http://www.jcb.org/cgi/content/full/jcb.200209091/DC1); we estimate that they are domains with mean radius of 22 ± 4 nm that occupy 35% of the plasma membrane. The mean radius of 22 nm is within the range of lipid raft size derived by other methods, which estimated diameters of <70 nm (Friedrichson and Kurzchalia, 1998; Varma and Mayor, 1998; Pralle et al., 2000). Detection of clustering is extremely sensitive to fixation and labeling procedures. Glutaraldehyde fixation must be used to eliminate short-range antibody-induced aggregation into larger clusters (Fig. S3 a), and clustering is only evident when gold particles ≤6 nm in diameter are directly conjugated to primary antibody (Fig. S3 b).
We tested if GFP-tH microdomains are cholesterol-dependent by treating cells with methyl-β-cyclodextrin. Depletion of cell surface cholesterol, visualized by filipin staining (Fig. 1 b), did not cause any loss of GFP-tH from the plasma membrane, assessed qualitatively by fluorescence or quantitatively by immunogold labeling (not depicted). However, K-function analysis of the gold patterns reveals a time-dependent loss of GFP-tH clustering in cyclodextrin-treated cells (Fig. 1 c). After 60 min of cyclodextrin treatment, L(r) − r tracks at zero over most of the r range analyzed, indicating a random distribution. These results confirm that GFP-tH is localized to cholesterol-rich lipid rafts and reveals that their disruption disperses GFP-tH over the plasma membrane, rather than driving association with other microdomains. The presence of rafts in the extracellular leaflet of the plasma membrane was supported by studies showing cholesterol-dependent clustering of glycophosphatidylinositol (GPI)-anchored proteins (Friedrichson and Kurzchalia, 1998; Harder et al., 1998; Varma and Mayor, 1998), but similar data for rafts in the intracellular leaflet have been lacking until now.
Next, we examined the relationship between inner- and outer-leaflet lipid rafts using a variation of the K-function analysis. When plasma membrane sheets are labeled for two different antigens with 2 nm and 4–5 nm gold, colocalization can be assessed using bivariate K-functions that determine whether one gold population is clustered with respect to the other (Diggle, 1986; see Materials and methods and supplementary data). We compared the distribution of GFP-tH with the outer-leaflet raft marker GFP-GPI; Fig. 2). Both proteins are GFP-tagged, but because only one membrane surface is exposed at any point in the labeling and rip-off procedure, no leakage of gold probes occurs (unpublished data). We used two protocols to induce different degrees of GFP-GPI aggregation, as revealed by univariate K-function analysis of the 2-nm gold patterns (Fig. 2 c). The semi-patched technique induces relatively little GFP-GPI aggregation (univariate K-function shows a mean cluster radius of 50 nm), whereas the patched protocol, routinely used to visualize lipid rafts by immunofluorescence, induces very large GFP-GPI aggregates (univariate K-function shows a mean radius of 180 nm). It is not possible to completely evaluate unpatched GFP-GPI because this necessitates ripping off apical membranes from prefixed cells, a technique that has to date proven unsuccessful. The bivariate K-function shows that there is significant colocalization of GFP-tH with GFP-GPI only when GFP-GPI is aggregated into very large patches (Fig. 2 d); this is indicated by significant positive deflections of the L biv(r) − r curve from zero. Interestingly, despite the major reorganization of GFP-GPI, GFP-tH remains in small clusters (Fig. 2 b; univariate K-function for GFP-tH). This result indicates that inner leaflet rafts, when aggregated by cross-linking GPI-anchored proteins, still retain their modular structure. Incomplete colocalization of inner- and outer-leaflet raft markers has been observed previously (Harder et al., 1998; Prior et al., 2001), but our new data show that inner and outer leaflet rafts are only loosely associated at steady state. This offers a potential new mechanism for regulating signaling by modulating the extent of coupling between inner and outer leaflet rafts.
Figure 2.
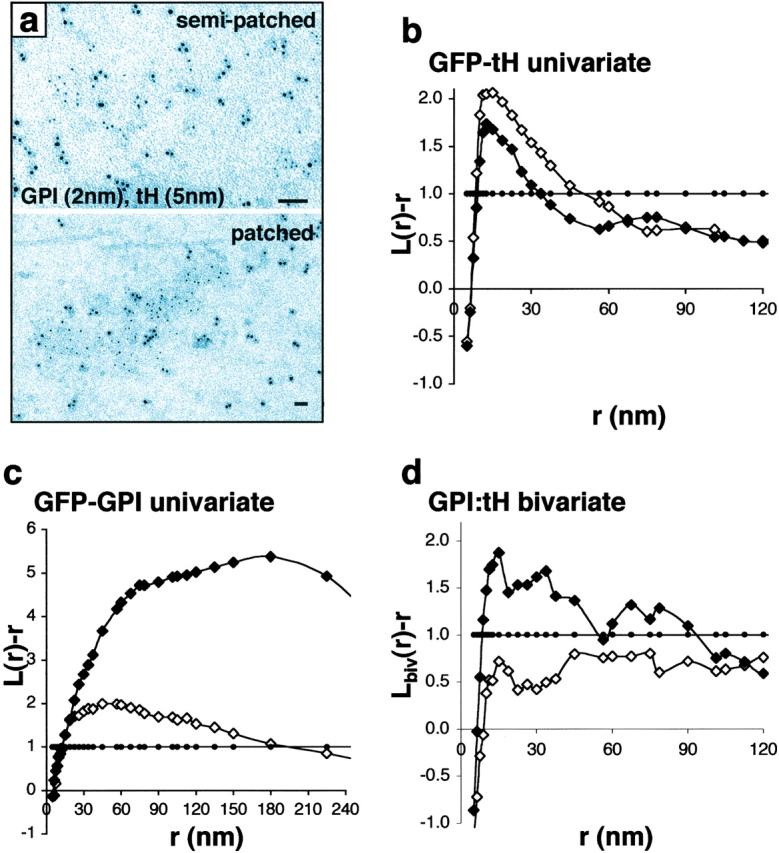
Analysis of inner- and outer-leaflet lipid raft markers. (a) Inner-leaflet GFP-tH (5 nm gold) and outer-leaflet GFP-GPI (2 nm gold) were specifically labeled to visualize individual microdomains. Univariate K-function analysis of GFP-tH (b) and GFP-GPI (c) show that extensive GFP-GPI aggregation is induced by the patched protocol (closed diamonds), but that GFP-tH remains clustered in small microdomains. Bivariate K-function analysis shows that GFP-tH and GFP-GPI co-cluster when GFP-GPI is aggregated into large patches (d, closed diamonds). There is a tendency for GFP-tH and GFP-GPI to colocalize with the semi-patched technique (open diamonds), but this is not statistically significant. K-functions are means (n ≥ 9 for each condition) standardized on the 99% CI (closed circles). Bars, 50 nm.
Full-length GFP–H-ras and GFP–H-rasG12V also have clustered distributions when expressed in BHK cells (Fig. 3). However, in contrast to GFP-tH, cyclodextrin treatment has relatively small effects on the clustering of GDP-bound H-ras (Fig. 3 a), GTP-bound H-rasG12V (Fig. 3 b) or GFP-targeted by the COOH-terminal 25 amino acids of H-ras (GFP-CTH; unpublished data). These data are consistent with a model where full-length H-ras has affinity for, and is resident in at least two plasma membrane microdomains; lipid rafts, and a noncholesterol-dependent nonraft microdomain (Jaumot et al., 2001; Prior et al., 2001). To formally test this hypothesis, we used bivariate K-function analysis to study the interaction of H-ras with lipid rafts. GFP-tH was coexpressed with untagged H-ras and plasma membrane sheets labeled with anti-GFP–2 nm and anti-Ras–4 nm gold. Bivariate analysis of the gold patterns (Fig. 3 d) reveals extensive colocalization of GDP-bound H-ras with GFP-tH in serum-starved cells, and shows that this decreases with serum stimulation. Constitutively activated H-rasG12V gives an even clearer picture of this phenomenon in that L biv(r) − r trends around zero, indicating no detectable colocalization of GFP-tH with H-rasG12V (Fig. 3 d). Together with the recent FRAP analysis of GFP-H-Ras in living cells, our results strongly suggest that wild-type H-ras is normally in a dynamic equilibrium between lipid rafts and other noncholesterol-dependent microdomains. Interaction with the nonraft site is favored experimentally by disrupting lipid rafts, i.e., removing one of the H-ras interaction sites, or physiologically by GTP loading. Thus, constitutively active H-ras is predominantly resident in the nonraft microdomain. The interaction of H-ras with the nonraft domain requires protein sequences in the hypervariable region because GFP-tH clustering is abolished by cyclodextrin treatment, whereas GFP-CTH is not.
Figure 3.
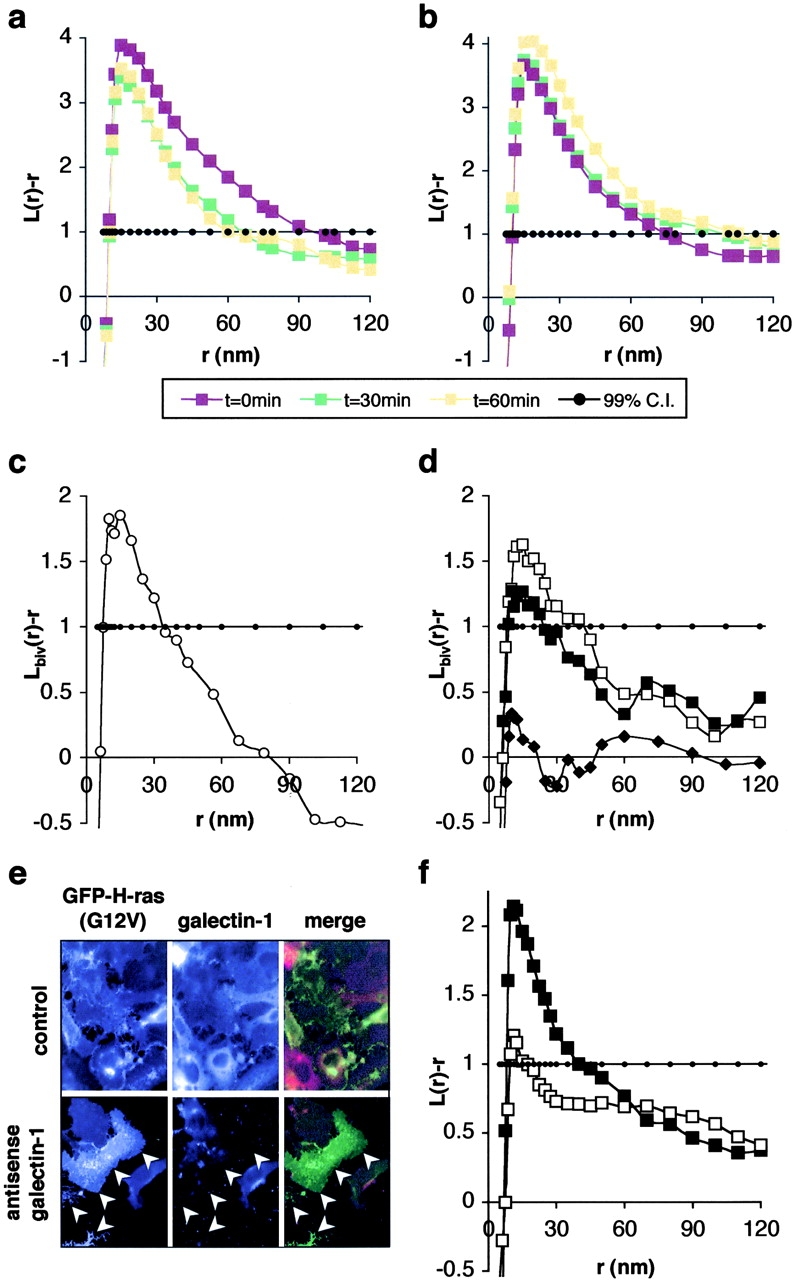
H-ras also occupies nonraft microdomains. Clustering of GDP-bound H-ras (a; GFP-HG12) and activated H-ras (b; GFP-HG12V) changes little with cyclodextrin treatment. (c) Plasma membrane sheets expressing GFP-H-ras were labeled with anti-GFP–2 nm and anti-Ras–4 nm gold to derive expected values for L biv(r) − r when there is complete colocalization of antigens under these assay conditions. Plasma membrane sheets from cells coexpressing GFP-tH and H-ras were then labeled with anti-GFP–2 nm and anti-Ras–4 nm gold. Bivariate analysis shows extensive colocalization of wild-type, GDP-bound H-ras with GFP-tH (d, open squares); serum stimulated GTP-loading of H-ras decreases coclustering (d, closed squares). Constitutively active H-rasG12V shows no colocalization with GFP-tH, i.e., L biv(r) − r trends around zero (closed diamonds). (e) Transfection with antisense galectin-1 DNA results in loss of endogenous galectin-1 expression. Note the loss of galectin-1 labeling in the transfected cells (arrowheads) compared with control. (f) Activated H-rasG12V clustering is significantly reduced in the absence of galectin-1 expression (open squares) compared with control (closed squares). K-functions are means (n ≥ 8 for each condition) standardized on the 99% CI for univariates and 95% CI for bivariates (closed circles in all panels).
Recent studies have shown that galectin-1 interacts with the processed COOH terminus of activated H-ras and may stabilize plasma membrane binding (Paz et al., 2001). Therefore, we examined whether galectin-1 might also be involved in regulating the microlocalization of activated H-ras. Transfection with antisense galectin-1 reduces endogenous galectin-1 expression (Fig. 3 e; Paz et al., 2001) and significantly reduces the clustering of H-rasG12V (Fig. 3 f). These data suggest that galectin-1 may have a critical role in stabilizing H-ras interactions with nonraft microdomains that in turn may also be required for stable plasma membrane association of activated H-ras.
Little is known about the microlocalization of K-ras targeted by a polylysine domain and a farnesylated CAAX motif. Therefore, we investigated the distribution of GFP fused to the minimal plasma membrane targeting motifs of K-ras, GFP-tK. Analysis of the GFP-tK–labeled gold patterns reveals that they are clustered, but with different characteristics from GFP-tH (Fig. 4 a and Fig. S1 a). Modeling establishes that the GFP-tK domains have a mean radius of 16 ± 3 nm and occupy 20% of the plasma membrane (Fig. S1 c). In contrast to GFP-tH, cholesterol depletion causes a small rise in GFP-tK clustering after 60 min of cyclodextrin treatment. The subtle effect of cyclodextrin on GFP-tK microdomains may reflect a general role of cholesterol in maintaining overall plasma membrane integrity. Wild-type and constitutively active K-ras show identical clustering to GFP-tK, both in the presence or absence of cyclodextrin (unpublished data), and bivariate analysis of plasma membranes coexpressing GFP-tH and activated K-rasG12V showed no significant colocalization of the lipid raft marker with K-ras (Fig. 4 b). Clustering of GFP-tK and K-ras was unexpected, although biophysical studies have shown that myristoylated polybasic peptides can sequester negatively charged lipids to generate novel membrane domains (Murray et al., 1999). Intriguingly, clustering was strikingly reduced when the wild-type K-ras CVIM motif was replaced with CCIL, to direct geranylgeranylation rather than farnesylation (Fig. 4 c, GFP-tKCCIL). Therefore, our data show that nature of the prenoid group profoundly affects the ability of polybasic K-ras to organize into specific microdomains, an observation that may have functional implications. Overall, our results clearly demonstrate for the first time the existence of K-ras microdomains within disordered plasma membranes that are distinct from classical lipid rafts.
Figure 4.
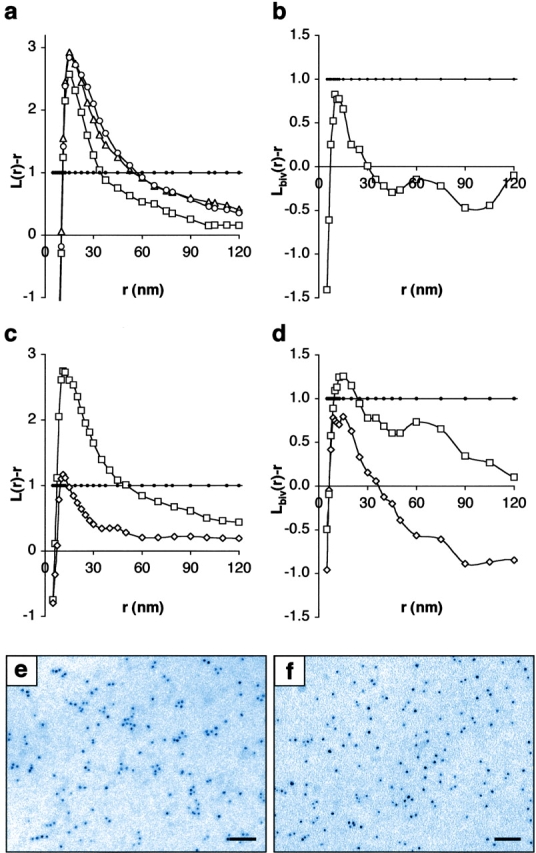
K-ras clusters in nonraft microdomains distinct from H-ras microdomains. Peak clustering of GFP-tK occurs at 16 nm; clustering is increased slightly with cyclodextrin treatment (a; t = 0 min, open squares; t = 15 min, open triangles; t = 60 min, open circles). Bivariate K-function analysis indicates no significant colocalization of activated K-ras with the lipid raft marker GFP-tH (b). (c) Replacement of the farnesyl group of GFP-tK (open squares) with a geranylgeranyl group results in a significant reduction in clustering (GFP-tKCCIL, open diamonds). Bivariate analysis of the association of activated Ras with microdomains marked by GFP-tK (d) shows significant colocalization of K-rasG12V with GFP-tK (open squares), but no significant colocalization of H-rasG12V with GFP-tK (open diamonds). Representative examples of electron microscopic images of GFP-tK (e) and GFP-tKCCIL (f) are shown. K-functions are means (n ≥ 9 for each condition) standardized on the 99% CI for univariates and 95% CI for bivariates (closed circles). Bars, 50 nm.
Our analysis shows that neither activated H-ras nor K-ras localize to lipid rafts (Fig. 3 d and Fig. 4 b). A direct test of colocalization in cells coexpressing activated H- and K-ras is not possible at present because of a lack of suitable antibodies, and the fact that short NH2-terminal epitopes on Ras proteins attached to plasma membrane sheets are not visible to gold-conjugated antibodies (unpublished data). To indirectly address whether activated H-ras and K-ras occupy the same nonraft signaling domain, we used bivariate K-functions to examine colocalization to microdomains defined by the presence of GFP-tK. In this analysis, the L biv(r) − r curve for K-rasG12V coexpressed with GFP-tK shows significant deviation from zero, whereas the curve for H-rasG12V coexpressed with GFP-tK does not leave the 95% confidence interval (Fig. 4 d). This result indicates that a significant fraction of activated K-ras, but not H-ras, resides in microdomains labeled by GFP-tK. We can conclude from these data that the majority of activated H-ras does not colocalize with activated K-ras, indicating distinct (if not exclusive) locations for Ras microlocalization. The localization of activated H- and K-ras to different microdomains is important because if the distribution of effectors and/or concentration of cofactors for effector activation also varies across these distinct microenvironments, then differential signal output from H- and K-ras can be readily accounted for. For example, K-ras is a more potent activator of Raf than H-ras (Yan et al., 1998). Therefore, we might expect cofactors in Raf activation (such as phosphatidyl serine) to be more enriched in K-ras compared with H-ras microdomains. The challenge now is to better define the protein and lipid content of these microdomains.
A recent study has shown GFP dimerization compromises FRET analysis of protein–protein interactions in intact cells and results in lipid raft–targeted CFP and YFP continuing to display FRET when cells are treated with cyclodextrin (Zacharias et al., 2002). In contrast, we show here that GFP-tH clustering detected by immunogold labeling is completely abolished with cyclodextrin treatment; thus, if GFP is dimerized, then the gold-labeled antibody must detect only one GFP epitope in the dimer. Therefore, this loss of clustering is an important validation of the methodology, and clearly demonstrates that potential antibody or GFP dimerization–induced artifacts are not being observed. A similar argument applies to the loss of GFP-tK and GFP–H-rasG12V clustering induced by changing the prenoid group or reducing galectin expression, respectively, and an analysis of non-GFP tagged Ras clustering with anti-Ras–5 nm gold that yields very similar results to those observed with GFP-tagged Ras (unpublished data). Finally, we can also detect marked differences in the clustering of different transmembrane-anchored plasma membrane proteins, e.g., GFP-tagged EGF receptor is not clustered when expressed at relatively low levels (Fig. S3 c), whereas GFP-tagged Angiotensin II, Type 1 receptor, a G protein–coupled receptor, is highly clustered when expressed at comparable levels (unpublished data).
In summary, we have integrated powerful statistical analysis tools with high resolution imaging of morphologically featureless plasma membrane microdomains. This has allowed us to characterize both the size and distribution of lipid rafts and Ras signaling domains at steady state. Ras proteins are clustered irrespective of activation state, and occupy multiple raft and nonraft microdomains that are extensively distributed over the cell surface. K-ras requires the farnesyl moiety for correct microlocalization, whereas galectin-1 stabilizes H-ras interactions with nonraft microdomains. Our analysis shows that activated H-ras and activated K-ras largely occupy spatially distinct plasma membrane signaling domains; an observation that could account for differential signal outputs from these highly homologous proteins. Finally, the technique we have described has the potential to spatially map any epitope-tagged or endogenous signaling protein with respect to validated microdomain signposts. This will greatly facilitate studies dissecting the spatial organization and regulation of plasma membrane–based signaling pathways.
Materials and methods
Reagents
Affinity-purified polyclonal anti-GFP and monoclonal anti-Ras Y13–238 antibodies, and GFP-tH, GFP-tK, and GFP-ras plasmids have been described previously (Prior et al., 2001). GFP-GPI was a gift from Chiara Zurzolo (Università degli Studi di Napoli Federico II, Naples, Italy) and GFP-EGF receptor was a gift from Mark Philips (New York University, NY). Galectin-1 antisense expression plasmids and pAb was a gift from Yoel Kloog (Tel-Aviv University, Tel-Aviv, Israel). Gold-conjugated antibodies were prepared by the tannic acid/citrate method (Slot and Geuze, 1985). 2-nm and 4–5-nm gold antibody conjugates were further purified on a 10–40% glycerol gradient.
Electron microscopy and image analysis
LipofectAMINE™-transfected cells were incubated overnight in serum-free medium and, if indicated, treated with 1% methyl-β-cyclodextrin. Plasma membrane sheets were prepared, fixed with 4% PFA, 0.1% glutaraldehyde, labeled as described previously (Parton and Hancock, 2001), and photographed in an electron microscope (model 1010; JEOL USA, Inc.). 1.45-μm2 areas (for univariate analysis) or 0.725-μm2 areas (for bivariate analysis) of digitized negatives were processed using Adobe Photoshop® 5.0 Curves, Brightness/Contrast, and Airbrush tools to remove background. To ensure that all gold particles were counted, a 1-pixel line was inserted between conjoined particles. The coordinates of the gold particles were determined using NIH Image v1.82. For double-labeled areas, small gold was discerned from large gold by setting the limits for counting in NIH Image to values determined by precalibrating the gold fractions (see supplementary data). Gold densities were comparable within experiments, typically 500–800 gold/μm2 for single-labeled and 100–200 gold/μm2 for each gold probe in double-labeled areas.
GFP-GPI clustering
Cells were cotransfected with GFP-GPI and GFP-tH constructs. For semi-patching, cells on coverslips were incubated with anti-GFP–2 nm gold for 45 min at 10–12°C. To induce extensive aggregation (patched protocol), washed cells were then further incubated with anti–rabbit-CY3 for 45 min at 10–12°C. After further washing, plasma membrane sheets were prepared and labeled with anti-GFP–5 nm.
Statistics
The first order property of a point pattern is its intensity λ (=N/A), where N = number of points in the study area A. The second order property is characterized by Ripley's K-function K(r), where the expected number of neighbors N(r) within a distance r of any point in A is given by N(r) = λK(r). Thus, K(r) = N(r)/λ, and it normalizes N(r) for the density of the pattern. Our question is whether gold particles are clustered or exhibit CSR. In random patterns, at any distance r, the expected value of N(r) is λπr2, so K(r) = πr 2. If K(r) > πr 2, the gold particles have more neighbors than expected from CSR, i.e., are clustered. We use a linear transformation of K(r) where L(r) = √K(r)/π (Besag, 1977). L(r) − r is readily interpreted because under CSR, the expected value of L(r) − r = 0 for all values of r. We estimate N(r) as the mean value calculated over A, taking into account border effects (Ripley, 1977), and calculate L(r) − r from the above equations. When there are big gold (N b) and small gold (N s) in A, their distributions are described by three K-functions. The univariate functions K b(r) and K s(r) are used as above. The bivariate function K biv(r) maps distances from each big to each small gold particle, and vice versa. K biv(r) examines whether either gold particle population, at a distance r, is clustered around the other; a test of colocalization (Diggle, 1986). Significance tests of L(r) − r and L biv(r) − r are performed by Monte Carlo methods (Besag and Diggle, 1977; for review see supplementary data).
Online supplemental material
Online supplemental material includes additional information on statistical methods, modeling of gold point patterns, and calibration of gold sizes. Figs. S1 and S2 show modeling data to estimate the size and distribution of GFP-tH and GFP-tK microdomains. Fig. S3 shows that glutaraldehyde fixation abolishes antibody-induced clustering, that probe size is important for detecting clustering, and that GFP-tagged EGF receptor is unclustered. Fig. S4 illustrates an example of gold size calibration, necessary for double-labeling studies. Online supplemental material available at http://www.jcb.org/cgi/content/full/jcb.200209091/DC1.
Acknowledgments
We thank Yoel Kloog, Mark Philips, and Chiara Zurzolo for gifts of reagents.
This work was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia to J.F. Hancock and R.G. Parton, and a UQ Early Career Research Grant to I.A. Prior. R.G. Parton is a Principal Research Fellow of the NHMRC. The Institute for Molecular Bioscience is a Special Research Centre of the Australian Research Council.
The online version of this report includes supplemental material.
Footnotes
Abbreviations used in this paper: CSR, complete spatial randomness; GPI, glycophosphatidylinositol.
References
- Anderson, R., and K. Jacobson. 2002. A role for lipid shells in targeting proteins to caveolae, rafts and other lipid domains. Science. 296:1821–1825. [DOI] [PubMed] [Google Scholar]
- Besag, J.E. 1977. Contribution to the discussion of Dr. Ripley's paper. J. R. Statist. Soc. B39:193–195. [Google Scholar]
- Besag, J., and P.J. Diggle. 1977. Simple Monte Carlo tests for spatial pattern. Appl. Stat. 26:327–333. [Google Scholar]
- Bos, J.L. 1989. ras oncogenes in human cancer: a review. Cancer Res. 49:4682–4689. [PubMed] [Google Scholar]
- Brown, D., and E. London. 1998. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 14:111–136. [DOI] [PubMed] [Google Scholar]
- Campbell, S.L., R. Khosravi-Far, K.L. Rossman, G.J. Clark, and C.J. Der. 1998. Increasing complexity of Ras signaling. Oncogene. 17:1395–1413. [DOI] [PubMed] [Google Scholar]
- Diggle, P.J. 1986. Displaced amacrine cells in the retina of a rabbit: analysis of a bivariate spatial point pattern. J. Neurosci. Methods. 18:115–125. [DOI] [PubMed] [Google Scholar]
- Friedrichson, T., and T. Kurzchalia. 1998. Microdomains of GPI-anchored proteins in living cells revealed by crosslinking. Nature. 394:802–805. [DOI] [PubMed] [Google Scholar]
- Harder, T., P. Scheiffele, P. Verkade, and K. Simons. 1998. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 141:929–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaumot, M., J. Yan, J. Clyde-Smith, J. Sluimer, and J.F. Hancock. 2001. The linker domain of the Ha-Ras hypervariable region regulates interactions with exchange factors, Raf-1 and phosphoinositide 3-kinase. J. Biol. Chem. 277:272–278. [DOI] [PubMed] [Google Scholar]
- Kenworthy, A., N. Petranova, and M. Edidin. 2000. High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes. Mol. Biol. Cell. 11:1645–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koera, K., K. Nakamura, K. Nakao, J. Miyoshi, K. Toyoshima, T. Hatta, H. Otani, A. Aiba, and M. Katsuki. 1997. K-ras is essential for the development of the mouse embryo. Oncogene. 15:1151–1159. [DOI] [PubMed] [Google Scholar]
- Kurzchalia, T.V., and R.G. Parton. 1999. Membrane microdomains and caveolae. Curr. Opin. Cell Biol. 11:424–431. [DOI] [PubMed] [Google Scholar]
- Mineo, C., G. Gill, and R. Anderson. 1999. Regulated migration of epidermal growth factor receptor from caveolae. J. Biol. Chem. 274:30636–30643. [DOI] [PubMed] [Google Scholar]
- Murray, D., A. Arbuzova, G. Hangyás-Mihályné, A. Gambhir, N. Ben-Tal, B. Honig, and S. McLaughlin. 1999. Electrostatic properties of membranes containing acidic lipids and adsorbed basic peptides: theory and experiment. Biophys. J. 77:3176–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niv, H., O. Gutman, Y. Kloog, and Y. Henis. 2002. Activated K-ras and H-ras display different interactions with saturable nonraft sites at the surface of live cells. J. Cell Biol. 157:865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton, R.G., and J.F. Hancock. 2001. Caveolin and Ras function. Methods. Enzymol. 333:172–183. [DOI] [PubMed] [Google Scholar]
- Paz, A., R. Haklai, G. Elad-Sfadia, E. Ballan, and Y. Kloog. 2001. Galectin-1 binds oncogenic H-ras to mediate Ras membrane anchorage and cell transformation. Oncogene. 20:7486–7493. [DOI] [PubMed] [Google Scholar]
- Philimonenko, A., J. Janacek, and P. Hozak. 2000. Statistical evaluation of colocalization patterns in immunogold labeling experiments. J. Struct. Biol. 132:201–210. [DOI] [PubMed] [Google Scholar]
- Pralle, A., P. Keller, E. Florin, K. Simons, and J. Horber. 2000. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 148:997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior, I.A., A. Harding, J. Yan, J. Sluimer, R.G. Parton, and J.F. Hancock. 2001. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nat. Cell Biol. 3:368–375. [DOI] [PubMed] [Google Scholar]
- Ripley, B.D. 1977. Modelling spatial patterns. J. R. Statist. Soc. B39:172–192. [Google Scholar]
- Ripley, B.D. 1979. Tests of randomness for spatial point patterns. J. R. Statist. Soc. B41:368–374. [Google Scholar]
- Roy, S., R. Luetterforst, A. Harding, A. Apolloni, M. Etheridge, E. Stang, B. Rolls, J.F. Hancock, and R.G. Parton. 1999. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nat. Cell Biol. 1:98–105. [DOI] [PubMed] [Google Scholar]
- Sanan, D., and R. Anderson. 1991. Simultaneous visualization of LDL receptor distribution and clathrin lattices on membranes torn from the upper surface of cultured cells. J. Histochem. Cytochem. 39:1017–1024. [DOI] [PubMed] [Google Scholar]
- Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature. 387:569–572. [DOI] [PubMed] [Google Scholar]
- Simons, K., and D. Toomre. 2000. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 1:31–39. [DOI] [PubMed] [Google Scholar]
- Slot, J.W., and H.J. Geuze. 1985. A new method of preparing gold probes for multiple-labelling cytochemistry. Eur. J. Cell Biol. 38:87–93. [PubMed] [Google Scholar]
- Umanoff, H., W. Edelmann, A. Pellicer, and R. Kucherlapati. 1995. The murine N-ras gene is not essential for growth and development. Proc. Natl. Acad. Sci. USA. 92:1709–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma, R., and S. Mayor. 1998. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 394:798–801. [DOI] [PubMed] [Google Scholar]
- Wilson, B., J. Pfeiffer, and J. Oliver. 2000. Observing FcepsilonR1 signaling from the inside of the mast cell membrane. J. Cell Biol. 149:1131–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, B., J. Pfeiffer, Z. Surviladze, E. Gaudet, and J. Oliver. 2001. High resolution mapping of mast cell membranes reveals primary and secondary domains of Fc(epsilon)R1 and LAT. J. Cell Biol. 154:645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, J., S. Roy, A. Apolloni, A. Lane, and J.F. Hancock. 1998. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. J. Biol. Chem. 273:24052–24056. [DOI] [PubMed] [Google Scholar]
- Zacharias, D., J. Violin, A. Newton, and R. Tsien. 2002. Partitioning of lipid-modified GFPs into membrane microdomains of live cells. Science. 296:913–916. [DOI] [PubMed] [Google Scholar]