Abstract
Fragmentation of the mammalian Golgi apparatus during mitosis requires the phosphorylation of a specific subset of Golgi-associated proteins. We have used a biochemical approach to characterize these proteins and report here the identification of golgin-84 as a novel mitotic target. Using cryoelectron microscopy we could localize golgin-84 to the cis-Golgi network and found that it is enriched on tubules emanating from the lateral edges of, and often connecting, Golgi stacks. Golgin-84 binds to active rab1 but not cis-Golgi matrix proteins. Overexpression or depletion of golgin-84 results in fragmentation of the Golgi ribbon. Strikingly, the Golgi ribbon is converted into mini-stacks constituting only ∼25% of the volume of a normal Golgi apparatus upon golgin-84 depletion. These mini-stacks are able to carry out protein transport, though with reduced efficiency compared with a normal Golgi apparatus. Our results suggest that golgin-84 plays a key role in the assembly and maintenance of the Golgi ribbon in mammalian cells.
Keywords: Golgi apparatus; golgin; mitosis; Golgi structure; protein phosphorylation
Introduction
The Golgi apparatus of mammalian cells consists of stacked cisternae that are connected laterally by tubules to form a continuous ribbon (Rambourg and Clermont, 1997). Each face of the Golgi stack is flanked by tubulo-reticular networks that act as entry (cis-Golgi) or exit (trans-Golgi) sorting stations. Proteins that enter the cis-Golgi network are either recycled back to the ER or transported onwards through the Golgi stack where they can be modified by the sequential action of enzymes present in each of the individual cisternae. Upon arrival at the trans-Golgi network, proteins are packaged into carriers for delivery to the plasma membrane or endosomes. How the Golgi apparatus retains its identity and compartmental organization in spite of the extensive membrane and protein flux through this organelle is not clear, but at least two classes of protein complexes appear to be involved: the Golgi spectrin/ankyrin skeleton (De Matteis and Morrow, 2000) and the Golgi matrix proteins (Seemann et al., 2000a; Pfeffer, 2001).
The Golgi matrix was originally identified as a detergent-insoluble structure with the ability to bind Golgi enzymes (Slusarewicz et al., 1994). Several components of the Golgi matrix have now been identified. The best characterized are p115, the GM130–GRASP65 complex, and the integral membrane protein giantin (Linstedt and Hauri, 1993; Nakamura et al., 1995; Sapperstein et al., 1995; Barr et al., 1997). GRASP65 is a cisternal stacking protein that also acts as a receptor for the coiled-coil protein GM130, targeting it to the cis-Golgi (Barr et al., 1997, 1998). GM130, in turn, is a receptor for p115, a coiled-coil protein that is required for the tethering of transport vesicles to Golgi cisternae (Nakamura et al., 1997; Nelson et al., 1998). Giantin also binds to p115 and participates in vesicle tethering, and, interestingly, both p115 and giantin together with GM130 are required for stacking of Golgi cisternae in vitro (Sönnichsen et al., 1998; Shorter and Warren, 1999). The interactions between these proteins appear to be regulated by the rab GTPases because both p115 and GM130 can bind directly to active rab1, and rab1 is required for the recruitment of p115 to transport intermediates destined to fuse with the cis-Golgi (Allan et al., 2000; Moyer et al., 2001; Weide et al., 2001). A second GRASP complex is present on the medial Golgi, comprising GRASP55 and the coiled-coil protein golgin-45 (Short et al., 2001). This complex also appears to be regulated by rabs because golgin-45 binds directly to active rab2.
Several lines of evidence suggest that matrix proteins play a key role in maintaining Golgi structure. Blocking the interaction between p115 and GM130 in cells causes Golgi vesicles to accumulate (Seemann et al., 2000b) and complete removal of cellular p115 by antibody-induced degradation leads to a highly vesiculated Golgi apparatus (Puthenveedu and Linstedt, 2001). Depletion of golgin-45 has even more dramatic effects, resulting in the complete loss of Golgi structure and a redistribution of Golgi enzymes to the ER (Short et al., 2001). Upon treatment of cells with brefeldin A (BFA),* matrix proteins, unlike Golgi enzymes, remain distinct from the ER and accumulate in cytoplasmic structures, often referred to as BFA remnants (Nakamura et al., 1995; Seemann et al., 2000a). Upon removal of the drug, these remnants can assemble into a structure reminiscent of the Golgi ribbon even in the absence of other (nonmatrix) Golgi proteins (Seemann et al., 2000a). These remnants can be successfully partitioned into daughter cells during mitotic division and nucleate post-mitotic Golgi assembly (Seemann et al., 2002). Together, these results suggest that the matrix proteins form a structural scaffold that can exist and divide independently from Golgi enzyme–containing membranes. However, this view has recently been challenged with the finding that matrix proteins appear to constitutively cycle between the Golgi apparatus and the ER or cytosol, suggesting that the Golgi apparatus is a dynamic, steady-state system that may be capable of self-assembly (Miles et al., 2001; Ward et al., 2001). Recent work suggests that, at least in the yeast Pichia pastoris, de novo assembly of the Golgi apparatus can occur (Bevis et al., 2002). Thus it is still unclear whether a Golgi matrix exists, and if it does, whether such a structure is a stable or highly dynamic one. Interestingly, it has recently been shown that cis-Golgi matrix proteins can also cycle into the late intermediate compartment, suggesting that they might function in the incorporation of ER-derived membranes into the Golgi apparatus (Marra et al., 2001). Consistent with this, expression of a GM130 construct defective in p115 binding reduced delivery of membrane into the cis-Golgi.
During mitosis, membrane trafficking through the Golgi apparatus is arrested and the Golgi undergoes extensive fragmentation (Warren et al., 1995; Lowe et al., 1998a). This process is driven by protein phosphorylation, and, although it is poorly understood at present, some progress has been made in identifying the relevant kinases and their substrates. CDK1-cyclin B is required for Golgi fragmentation in vitro and one of its substrates has been identified as the cis-Golgi matrix protein GM130 (Lowe et al., 1998b). CDK1-mediated phosphorylation of GM130 on serine-25 abrogates binding to p115, providing a molecular explanation for the inhibition of vesicle docking seen in mitosis (Nakamura et al., 1997; Lowe et al., 1998b). Mitogen-activated protein kinase kinase I (MEK1) is also involved in mitotic fragmentation, but its effectors are not known (Acharya et al., 1998). One possibility is that a monophosphorylated form of ERK (pY-ERK), which has been localized to mitotic Golgi membranes (Cha and Shapiro, 2001), or perhaps diphospho (active) ERK, which can phosphorylate GRASP55 in vitro and in vivo (Jesch et al., 2001), is involved. A third kinase that appears to be required for mitotic fragmentation is Plk, which can phosphorylate GRASP65 (Lin et al., 2000; Sutterlin et al., 2001).
We have used a proteomics-based strategy to identify novel mitotic Golgi phosphoproteins with the reasoning that these will include important structural proteins and/or proteins involved in Golgi trafficking. Using this approach, we identified the coiled-coil membrane protein golgin-84 as a novel mitotic target and could show that this protein, although not a component of the putative Golgi matrix, plays an important role in the formation and maintenance of the Golgi apparatus.
Results
Identification of golgin-84 as a mitotic phosphoprotein
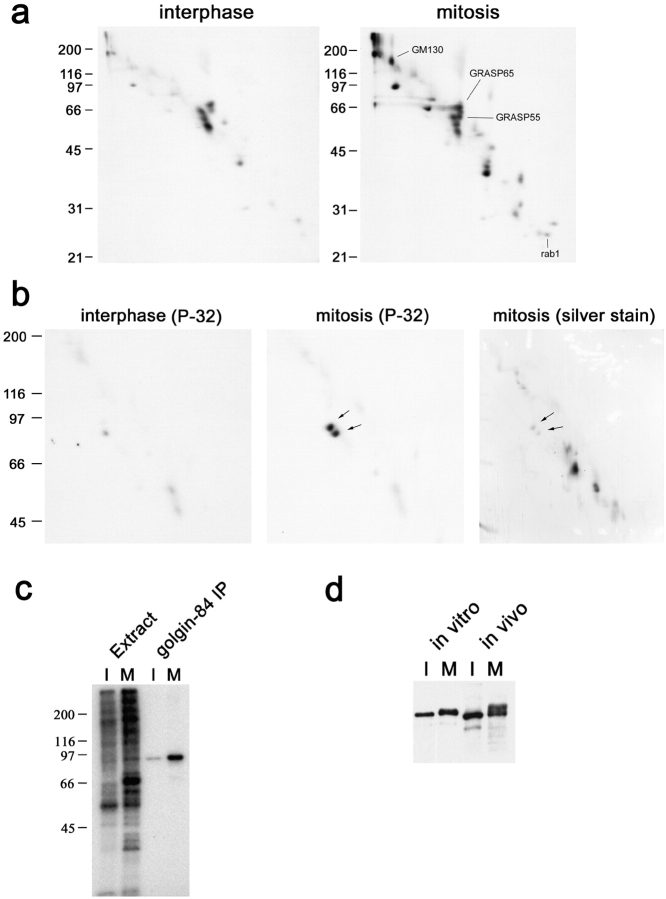
To identify mitotic Golgi phosphoproteins, Golgi membranes were incubated with interphase or mitotic cytosol in the presence of [γ-32P]ATP and analyzed by two-dimensional (2D) 16-BAC/SDS-PAGE followed by autoradiography. 20 spots were present exclusively in the mitotic sample (Fig. 1 a). The previously known mitotic phosphoproteins GM130 (Nakamura et al., 1997), GRASP65 (Barr et al., 1997), GRASP55 (Jesch et al., 2001), and rab1 (Bailly et al., 1991) were identified using a combination of mass spectrometry and Western blotting (Fig. 1 a; unpublished data). To identify the other mitotic phosphoproteins, we decided to first fractionate the membranes using sodium carbonate extraction and Triton X-114 phase partitioning in order to simplify the gel pattern before cutting out the spots and analyzing them by mass spectrometry. Analysis of the carbonate pellet/Triton X-114 aqueous phase revealed that only one mitotic phosphoprotein was present in this fraction (Fig. 1 b). This was identified using mass spectrometry as golgin-84, a previously identified coiled-coil protein of unknown function, localized to the Golgi apparatus (Bascom et al., 1999).
Figure 1.
Identification of golgin-84 as a mitotic phosphoprotein. (a–c) Rat liver Golgi membranes were incubated with interphase or mitotic HeLa cytosol in the presence of [γ-32P]ATP for 30 min at 30°C and reisolated by centrifugation. (a) Samples were subjected to 16-BAC/SDS-PAGE 2D electrophoresis and radiolabeled proteins were detected by autoradiography. Positions of the known mitotic phosphoproteins GM130, GRASP65, GRASP55, and rab1 are indicated. (b) The radiolabeled membranes were washed with sodium carbonate and the carbonate pellet was extracted with Triton X-114. Proteins partitioning into the aqueous phase were analyzed by 2D 16-BAC/SDS-PAGE and silver staining, followed by autoradiography. Arrows indicate the doublet of spots corresponding to golgin-84. The lower spot is a proteolytic cleavage product. (c) The radiolabeled membranes were solubilized in SDS and subjected to immunoprecipitation with antibodies to golgin-84. The membrane extracts and immunoprecipitates were analyzed by 1D SDS-PAGE followed by autoradiography. (d) Golgi membranes incubated with interphase or mitotic cytosol (in vitro) or total membrane fractions prepared from interphase and mitotic HeLa cells (in vivo) were analyzed by 1D SDS-PAGE and immunoblotting with antibodies to golgin-84.
To confirm that golgin-84 is phosphorylated specifically in mitosis, polyclonal antibodies were used to immunoprecipitate golgin-84 from interphase or mitotically-treated Golgi membranes. As shown in Fig. 1 c, golgin-84 was highly phosphorylated by mitotic cytosol but only poorly by interphase cytosol. Golgin-84 phosphorylation was stoichiometric because the mitotic form of the protein was shifted in apparent molecular weight compared with the interphase form upon SDS-PAGE (Fig. 1 d). Golgin-84 isolated from mitotic HeLa cells was also shifted in apparent molecular weight compared with that from interphase cells, confirming that phosphorylation occurs in vivo (Fig. 1 d).
Localization of golgin-84 to the cis-Golgi network
Previous work has shown that golgin-84 is present on the Golgi apparatus (Bascom et al., 1999), but the localization of this protein at the ultrastructural level has not been addressed. To localize golgin-84 within the Golgi apparatus, cryosections of A431 cells were labeled with polyclonal antibodies to golgin-84 and examined under the electron microscope. Golgin-84 was found predominantly on membranes at the cis side of the Golgi stack (Fig. 2). Quantitation revealed that 34% of golgin-84 labeling was on cisternae whereas 66% of labeling was on tubulo-vesicular profiles (often referred to as the cis-Golgi network [CGN]) (Fig. 2 f). Of the tubulo-vesicular profile labeling, the vast majority was on membranes at the lateral edges of the stack rather then on membranes underlying the stacked cisternae (Fig. 2 f). Interestingly, golgin-84 labeling could frequently be detected on tubulo-reticular elements apparently connecting adjacent Golgi stacks (Fig. 2 d). A similar golgin-84 distribution to that observed in A431 cells was also detected in HeLa cells, suggesting that the localization of this protein is not cell type dependent (Fig. 2 g).
Figure 2.
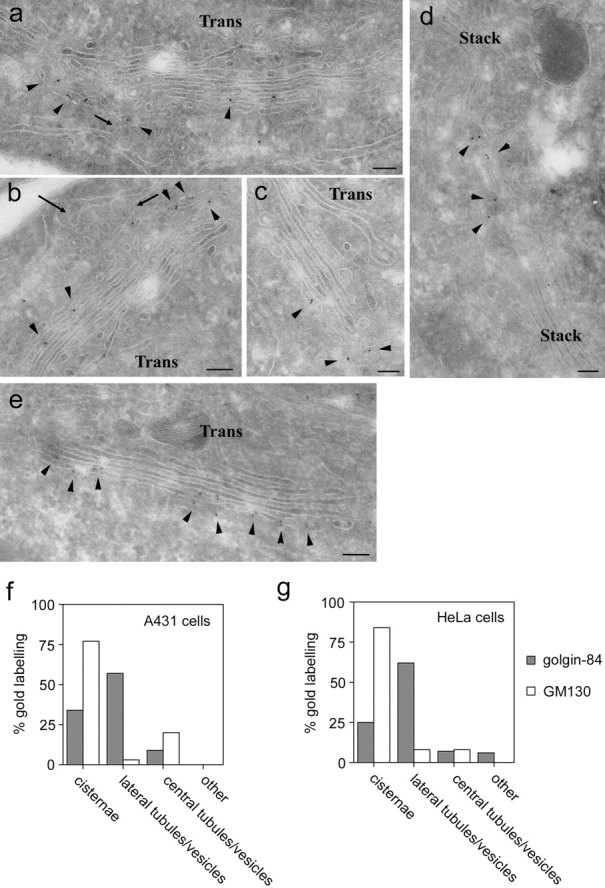
Golgin-84 is localized to the cis-Golgi network. (a–e) A431 cells were processed for cryoelectron microscopy and labeled with polyclonal antibodies to golgin-84 (a–d) or GM130 (e) followed by secondary antibodies coupled to 10-nm gold. Arrows indicate ER budding profiles, and arrowheads indicate golgin-84 (a–d) or GM130 (e) labeling. (f–g) Quantitation of golgin-84 and GM130 labeling of cryosections. The relative number of gold particles over Golgi cisternae, tubulo-vesicular profiles lateral to the Golgi stack, tubulo-vesicular profiles adjacent to the cis face of the stack (central), or over other structures was quantitated in gluteraldehyde-fixed A431 cells (f) and paraformaldehyde-fixed HeLa cells (g) as described in the Materials and methods section. Note that golgin-84 labeling is present on the cis-most cisternae and more predominantly on tubulo-vesicular profiles lateral to the stack, whereas GM130 labeling is mainly on the cis-most cisternae of the stack (a–c). Bars, 200 nm.
The localization of golgin-84 to the cis side of the Golgi apparatus is similar to that reported for the cis-Golgi matrix protein GM130 (Nakamura et al., 1995; Marra et al., 2001). We therefore analyzed the distribution of GM130 in cryosections of A431 cells and compared it to that of golgin-84. As shown in Fig. 2 e, strong labeling for GM130 was detected along the face of the cis-most Golgi cisterna. Quantitation revealed that ∼75% of GM130 labeling was present on cisternae with only ∼25% on tubulo-vesicular profiles in the vicinity of the Golgi stack (Fig. 2 f). Labeling of tubulo-vesicular profiles was predominantly on membranes underlying the cisternae, with little GM130 detected on membranes at the lateral edges of the stack. A similar GM130 distribution was also observed in HeLa cells (Fig. 2 g). Golgin-84 and GM130 therefore localize to distinct regions of the cis-Golgi, with golgin-84 more abundant at the lateral edges of the Golgi stack while GM130 is present on more central membranes.
Golgin-84 does not associate with cis-Golgi matrix proteins
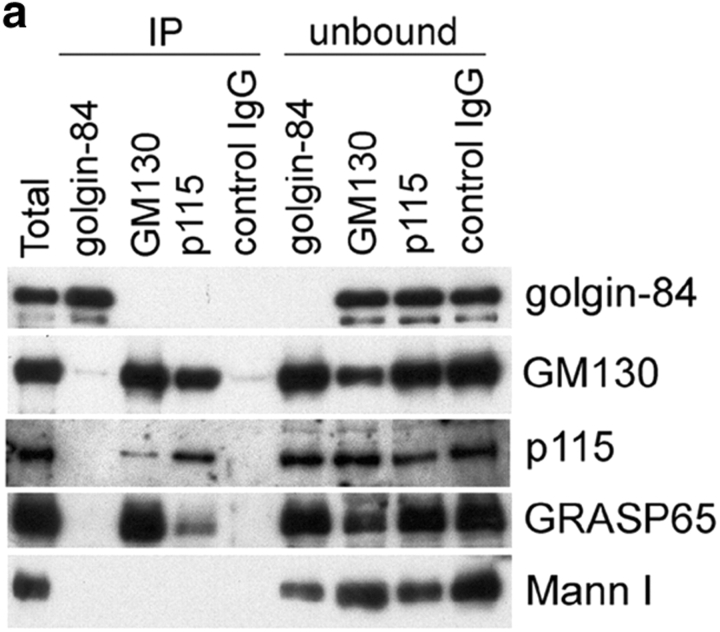
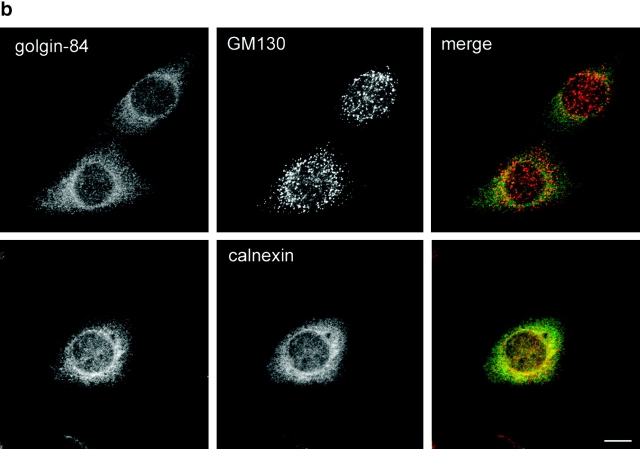
The different distributions of golgin-84 and GM130 in the cis-Golgi suggested that golgin-84 may not be part of the putative Golgi matrix described by Seemann et al. (2000a)(2002). We first tested whether golgin-84 can interact physically with cis-Golgi matrix proteins. Golgin-84 and the cis-Golgi matrix proteins GM130 and p115 were immunoprecipitated from Golgi extracts under mild conditions and the immunoprecipitates tested for the presence of golgin-84, the matrix proteins GM130, p115, and GRASP65, and the Golgi enzyme mannosidase I by immunoblotting. Even though golgin-84 was efficiently precipitated by its antibody (it was depleted from the unbound fraction), no matrix proteins could be detected in the immunoprecipitate. Similarly, no golgin-84 could be detected in the GM130 or p115 immunoprecipitates, which contained significant levels of GM130, p115, and GRASP65 (Fig. 3 a). Thus, golgin-84 does not appear to physically interact with cis-Golgi matrix proteins. To test more directly whether golgin-84 might exist as part of the putative Golgi matrix, we studied its behavior upon treatment of cells with BFA. As shown in Fig. 3 b, golgin-84 redistributed to the ER in BFA-treated cells, as suggested by previous work (Bascom et al., 1999). No golgin-84 was detected in the cytoplasmic GM130-containing punctate structures, indicating that golgin-84 is not part of the putative matrix described by Seemann et al. (2000a)(2002).
Figure 3.
Golgin-84 is not part of the Golgi matrix. (a) Rat liver Golgi extracts were immunoprecipitated under native conditions with antibodies to either golgin-84, GM130, or p115, or with a control IgG. The immunoprecipitated (IP) and unbound fractions were analyzed by Western blotting with antibodies to golgin-84, GM130, p115, GRASP65, and mannosidase I as indicated. (b) NRK cells were incubated with 5 μg/ml BFA for 1 h and then fixed and double labeled with antibodies to golgin-84 and GM130 or the ER marker calnexin. Bar, 10 μm.
Golgi-84 is a binding partner for active rab1
Several Golgi-associated coiled-coil proteins interact with the active or GTP-bound forms of the rab family of small GTPases involved in membrane traffic. Specifically, p115, a cis-Golgi vesicle tethering protein, binds to active rab1, and GM130 binds to active rab1 and less efficiently to active rab2, whereas golgin-45, a medial Golgi protein, binds only to active rab2 (Allan et al., 2000; Moyer et al., 2001; Short et al., 2001; Weide et al., 2001). Because golgin-84 shares similarities with these coiled-coil rab-binding proteins, we analyzed whether this protein may itself be a novel rab effector. Golgi extract was incubated with immobilized rab proteins in the inactive (GDP) or active (GTPγS) conformation and bound proteins were analyzed by immunoblotting. As expected, GM130 bound specifically to active rab1, but we could not detect any binding to rab2 (Fig. 4 a). Rab2 did however bind to golgin-45, as previously reported (Short et al., 2001), demonstrating the functionality of this protein in the binding assay (Fig. 4 a). Interestingly, golgin-84 bound specifically to active rab1, with no detectable binding to either rab2 or rab6. To confirm the interaction with rab1, the yeast two-hybrid system was employed. As previously shown, GM130 interacted directly with activated mutants of rab1, rab2, and rab33b (Short et al., 2001; Valsdottir et al., 2001; Weide et al., 2001) (Fig. 4 b). In contrast, golgin-84 interacted only with activated rab1, and not with any of the other rab proteins tested. Therefore, golgin-84 is a specific binding partner for active rab1. The rab1 binding site was mapped using the two-hybrid system to the coiled-coil region of golgin-84 (Fig. 4 c).
Figure 4.
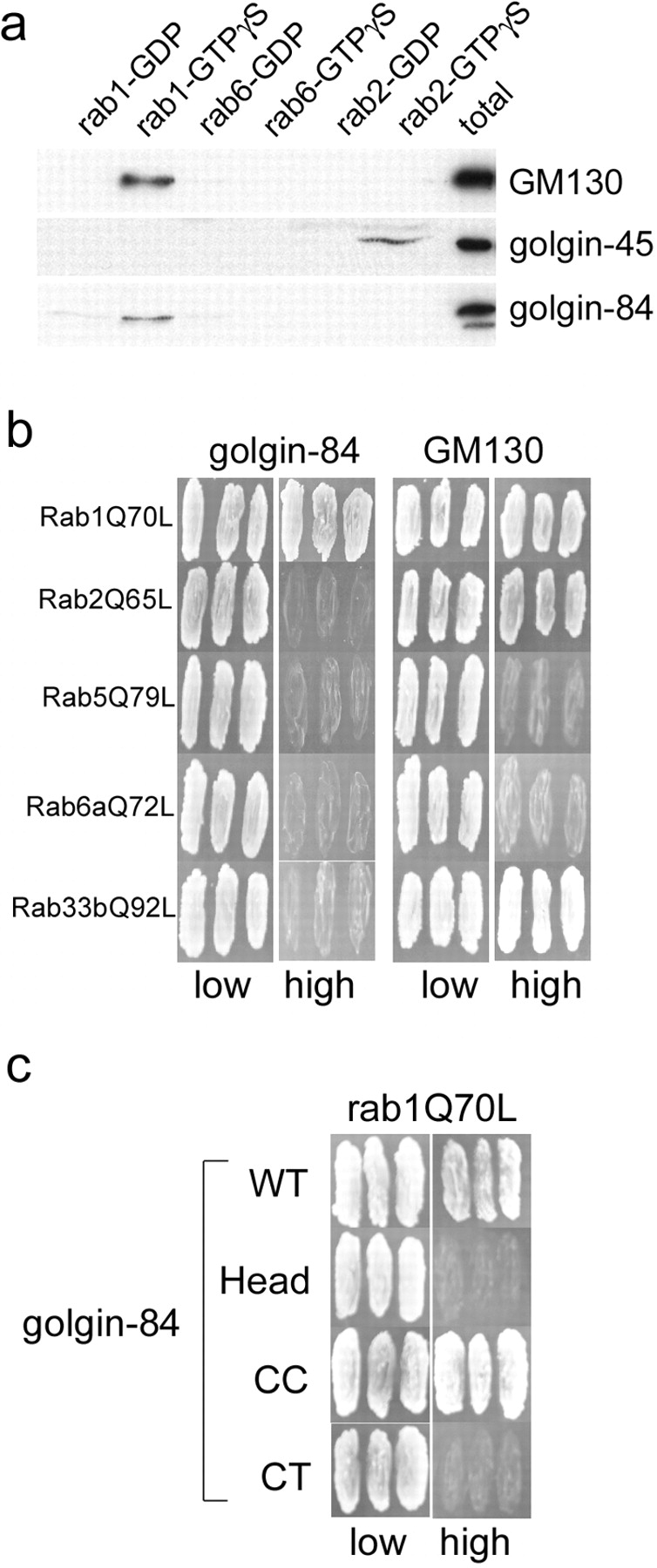
Golgin-84 is a specific binding partner for rab1. (a) GST-tagged rab1, rab2, and rab6 were loaded with GDP or GTPγS and incubated with Golgi extract, and specifically eluted proteins were analyzed by Western blotting with antibodies to GM130, golgin-45, and golgin-84. (b) Full-length GM130 and golgin-84 lacking the transmembrane domain were tested for interaction in the yeast two-hybrid system with the following rab proteins carrying activating point mutations: rab1Q70L, rab2Q65L, rab5Q79L, rab6Q72L, and rab33bQ92L. Interactions between the indicated proteins results in growth on high selection medium. (c) Full-length and truncation mutants of golgin-84 were tested for interaction with rab1Q70L in the yeast two-hybrid system. The CT mutant corresponds to the ΔHead + CC construct described in Fig. 5 without the membrane anchor.
Overexpression or depletion of golgin-84 fragments the Golgi ribbon
The stiochiometric phosphorylation of golgin-84 in mitosis together with its binding to active rab1 suggested that it may play a role in Golgi structure and/or membrane trafficking through the Golgi apparatus. To address a possible structural role for golgin-84, GFP-tagged full-length and truncated versions of the protein were expressed in HeLa cells, and effects upon Golgi structure were analyzed by immunofluorescence microscopy (Fig. 5 a). At moderate expression levels, none of the golgin-84 constructs elicited any significant change in Golgi structure (unpublished data). Furthermore, constructs that failed to target to the Golgi apparatus had no effects upon Golgi structure even at very high levels of expression (Fig. 5 a; unpublished data). In contrast, expression at high levels of both full-length and golgin-84 lacking the head region had dramatic effects upon Golgi structure, converting the ribbon into punctate structures dispersed throughout the cytoplasm (Fig. 5 b). EM analysis of the Golgi fragments in cells overexpressing full-length golgin-84 revealed that they are similar in overall organization to the Golgi apparatus in control cells, comprising three to four stacked cisternae and vesiculo-tubular profiles that label heavily for the overexpressed protein (Fig. 5 c). Thus, overexpression of golgin-84 does not lead to vesiculation of Golgi stacks, just break-up of the ribbon. Fragmentation was not due to the presence of GFP at the NH2 terminus because identical results were obtained with golgin-84 constructs containing an NH2-terminal myc tag instead of GFP (unpublished data). Fragmentation appeared more extensive with the construct lacking the head region, suggesting that the head may play some role in regulating Golgi structure. Interestingly, golgin-84 lacking most of the coiled-coil region in addition to the head domain had only a minor effect upon Golgi structure, even at extremely high expression levels. Together, the results suggest that the coiled-coil domain is important for the fragmentation observed, and that it is only able to induce fragmentation when properly targeted to the Golgi membranes.
Figure 5.
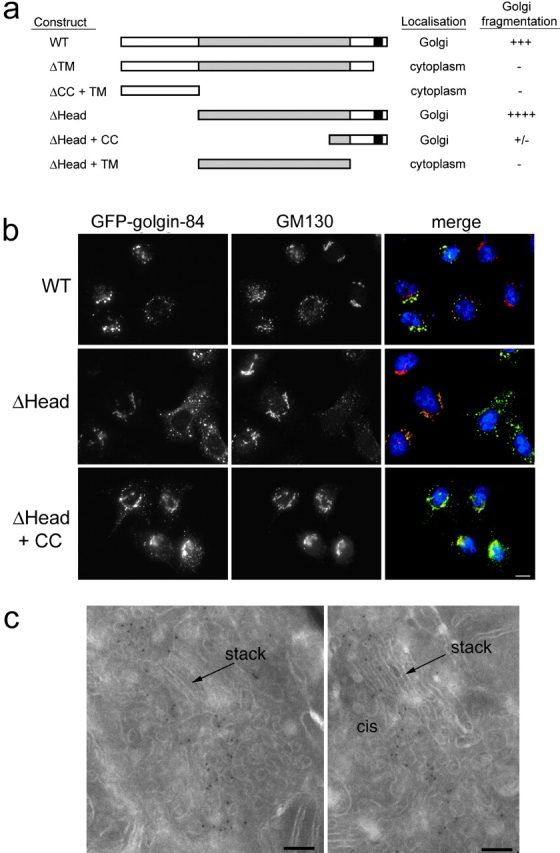
Overexpression of golgin-84 fragments the Golgi ribbon. (a) Schematic representation of the structure of golgin-84 showing the constructs that were expressed in HeLa cells. Each construct was tagged at the NH2 terminus with GFP. The predicted coiled-coil region is shaded gray and the predicted transmembrane region is black. A summary of the localization and effects upon Golgi structure of each construct is shown on the right. (b) HeLa cells transfected with GFP-tagged WT (top), ΔHead (middle), and ΔHead + CC (bottom) golgin-84 constructs were fixed and stained with antibodies to GM130. In the merged images on the right, DNA is blue, GFP–golgin-84 is indicated in green, GM130 is red, and yellow indicates regions of overlap between GFP–golgin-84 and GM130. Bar, 10 μm. (c) HeLa cells expressing GFP-tagged WT golgin-84 were processed for cryoelectron microscopy and labeled with polyclonal antibodies to GFP followed by rabbit anti–sheep antibodies and protein A coupled to 8-nm gold. Bars, 100 nm.
To further examine the role of golgin-84 in maintaining normal Golgi structure, we used small interfering RNA (siRNA) to deplete cellular golgin-84 (Elbashir et al., 2001). Transfection of HeLa cells with an siRNA duplex matching the nucleotide sequence of golgin-84 resulted in a reduction in golgin-84 levels of ∼90% after 3 d in culture (Fig. 6 a). This reduction was specific because levels of GM130 were unaffected, and transfection of cells with an siRNA duplex targeting lamin A had no effect upon golgin-84 (or GM130) levels. Immunofluorescence microscopy with antibodies to GM130 showed that there was little effect upon Golgi morphology in mock transfected cells or cells transfected with an siRNA that effectively depleted cellular lamin A (Fig. 6 b). Depletion of golgin-84, however, had a drastic effect upon Golgi structure, breaking the ribbon into large fragments dispersed in the cytoplasm (Fig. 6 b). The extent of Golgi fragmentation correlated well with loss of golgin-84 over time (Fig. 6 c). This, combined with the lack of fragmentation induced by prolonged exposure to the control lamin A oligo, suggests that fragmentation is a consequence of golgin-84 depletion and not cellular toxicity resulting from the RNA interference (RNAi) treatment itself. Although predominantly punctate in appearance, the Golgi fragments occasionally resembled short tubules. Fragments resulting from golgin-84 depletion contained markers of the cis- (GM130), medial (GalNacT2; Fig. 6 b), and trans-Golgi (TGN46; unpublished data), suggesting that some degree of Golgi organization may be retained in these cells. The Golgi fragments did not extensively colocalize with the ER exit site marker mSec23p, suggesting that they arise directly from the break-up of the Golgi ribbon rather than the cycling through the ER (Fig. 6 b).
Figure 6.
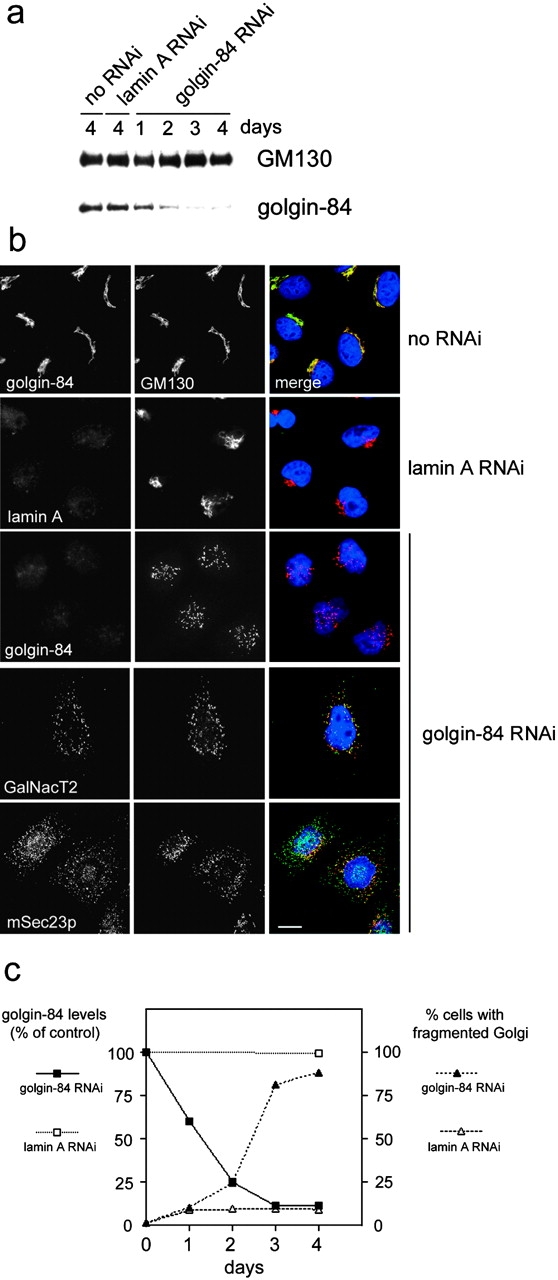
Depletion of golgin-84 using RNAi fragments the Golgi ribbon. (a) HeLa cells were either mock transfected (no RNAi) or transfected with duplex RNA to target lamin A or golgin-84 and, after 1–4 d, were subjected to Western blotting with antibodies to GM130 or golgin-84. (b) Mock transfected or RNAi-treated HeLa cells were fixed and double labeled with antibodies to golgin-84, lamin A, GalNacT2, mSec23p, and GM130. In the merged images on the right, DNA is blue, golgin-84, lamin A, GalNacT2, mSec23p are in green, and GM130 is red, with regions of overlap between these proteins indicated in yellow. Bar, 10 μm. (c) Quantitation of golgin-84 levels and Golgi fragmentation in RNAi-treated cells. Golgin-84 levels were measured by quantitating Western blots of RNAi-treated HeLa cells as described in the Materials and methods section. Golgi fragmentation was measured by immunofluorescence microscopy using antibodies to GM130 to assess Golgi morphology. For each time point, 200 cells were counted. The data are an average of two independent experiments.
To characterize the effects of golgin-84 depletion upon Golgi morphology in more detail, siRNA-treated cells were analyzed by electron microscopy. Interestingly, the Golgi apparatus appeared significantly smaller in golgin-84–depleted cells compared with control cells (Fig. 7, a and b). Quantitation revealed that ∼75% of Golgi cisternae and ∼70% of Golgi tubulo-vesicular membranes were lost upon golgin-84 depletion (Fig. 7 d). The overall morphology was not, however, dramatically affected. The number of stacked cisternae per stack and the ratio of membrane in stacked cisternae versus tubulo-vesicular profiles were similar in golgin-84–depleted cells and control cells (unpublished data). Depletion of golgin-84 also had a marked effect upon the ER, which was grossly exaggerated compared with control cells, forming an elaborated network extending throughout the cytoplasm (Fig. 7 c). The lumen of the ER often appeared swollen with electron-dense material, suggesting that an accumulation of proteins had occurred there. Quantitation revealed that the ER was ∼1.7 times larger in golgin-84–depleted cells compared with control cells, while the nuclear envelope was only marginally affected. Thus, depletion of golgin-84 results in a dramatic loss of Golgi membranes and a corresponding increase in the size of the ER.
Figure 7.
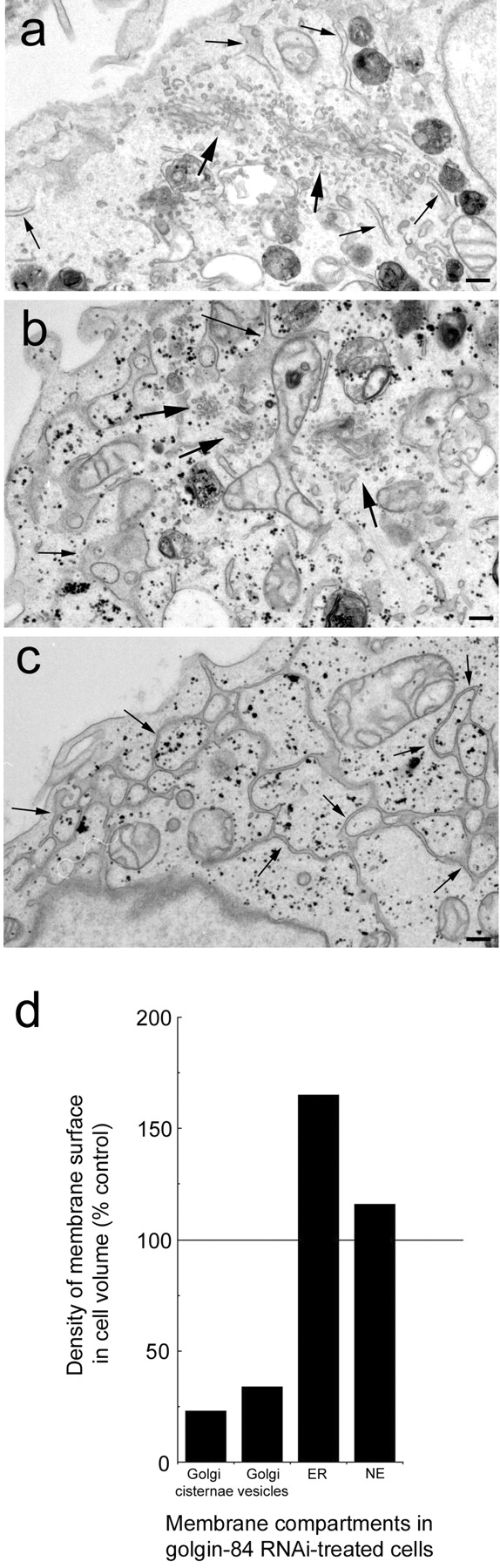
Golgin-84 depletion results in a significant loss of membrane from the Golgi apparatus and an exaggerated ER. Control (a) or golgin-84 RNAi–treated HeLa cells (b and c) were fixed and embedded in Epon for electron microscopy. Large arrows indicate Golgi membranes whereas small arrows indicate the ER. Note the decrease in size of the Golgi apparatus and the exaggerated and swollen ER in golgin-84–depleted cells. (d) Quantitation of the amount of membrane in Golgi cisternae, Golgi vesicles, ER, and nuclear envelope (NE) in golgin-84–depleted cells relative to control cells expressed as density of membrane surface in cell volume. For controls and RNAi samples, n = 23 and 22 micrographs, respectively. Bars, 200 nm.
Depletion of golgin-84 partially inhibits protein transport from the ER to the cell surface
To study whether golgin-84 plays any role in protein trafficking through the Golgi apparatus, we used a GFP-tagged temperature-sensitive allele of the vesicular stomatitis virus G protein (ts045 VSV-G). This well-characterized secretory cargo marker accumulates in the ER at the nonpermissive temperature of 39.5°C due to a reversible folding defect. When shifted to the permissive temperature (31°C), correctly folded ts045G VSV-G is rapidly transported from the ER, through the Golgi apparatus to the cell surface, where its appearance can be monitored using an antibody against the lumenal domain of the glycoprotein (Pepperkok et al., 1993; Seemann et al., 2000b). As expected, VSV-G was efficiently transported to the cell surface when control cells were shifted to 31°C (Fig. 8 a). In cells depleted of golgin-84, VSV-G could also be detected on the cell surface at 31°C (Fig. 8 a). Golgin-84 depletion therefore does not block VSV-G transport to the cell surface. To determine whether there is a more subtle effect, transport of VSV-G to the cell surface was quantitated in golgin-84–depleted cells relative to control cells. This revealed that VSV-G transport to the cell surface was inhibited by 15%, 39%, and 42% in golgin-84–depleted cells after 30, 60, and 90 min at 31°C, respectively (Fig. 8 b). These results suggest that golgin-84 is required for efficient protein trafficking through the Golgi apparatus.
Figure 8.
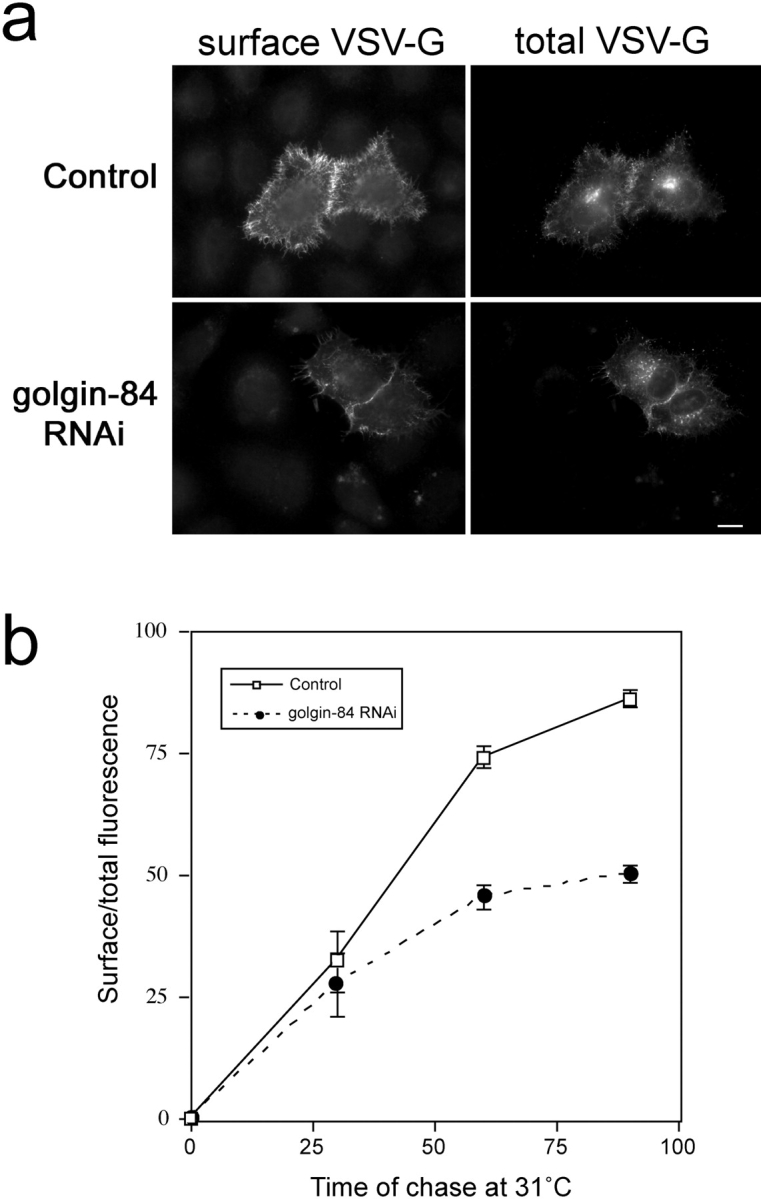
Depletion of golgin-84 partially inhibits transport of VSV-G from the ER to the cell surface. Control or golgin-84 RNAi–treated HeLa cells were transfected with a plasmid encoding GFP-tagged ts045G VSV-G protein. Cells were incubated at 39.5°C to arrest ts045G VSV-G in the ER, and then chased at 31°C for various times to allow transport before fixation and labeling for cell surface VSV-G. (a) An example of cells shifted to 31°C for 60 min and labeled for cell surface VSV-G. (b) The extent of VSV-G transport was measured as indicated in the Materials and methods and is expressed as the ratio of cell surface to total VSV-G fluorescence. The data shown are representative of two experiments with n = 15 for all data points in each experiment.
Discussion
In this report, we have identified golgin-84 as an important structural component of the Golgi apparatus. Overexpression of golgin-84 or depletion of the protein by RNAi results in extensive breakdown of the Golgi ribbon. The Golgi structures formed upon golgin-84 depletion retain their stacked organization and contain Golgi resident proteins, but their overall size is significantly smaller than that of a normal Golgi. This suggests that golgin-84 is required for the incorporation of membranes into the Golgi apparatus. This would be consistent with the cis-Golgi network localization of golgin-84. This highly pleiomorphic structure is where ER-derived transport intermediates (vesiculo-tubular clusters or intermediate compartment) fuse and incorporate into the cis side of the Golgi apparatus (Klumperman, 2000; Marra et al., 2001). This process is at least partially dependent upon the cis-Golgi matrix proteins GM130 and p115 because perturbation of the interaction between these proteins inhibits transport into the CGN (Marra et al., 2001). Our results suggest that golgin-84 also participates in the incorporation of membranes into the CGN.
How might golgin-84 function in building up the CGN? At the structural level, golgin-84 is similar to the coiled-coil proteins GM130, p115, giantin, and CASP, which have been implicated in vesicle tethering at the Golgi apparatus (Linstedt and Hauri, 1993; Nakamura et al., 1995; Sapperstein et al., 1995; Bascom et al., 1999; Gillingham et al., 2002). In addition, we found that golgin-84 specifically interacts with active rab1. Rab GTPases have an established role in regulating membrane tethering in both the endocytic and exocytic pathways (Waters and Hughson, 2000; Whyte and Munro, 2002). We therefore believe that golgin-84 is a tethering factor required for tethering incoming membranes to the CGN and thereby promoting their fusion with this compartment. Because golgin-84 is not part of the Golgi matrix and is located to a region of the CGN devoid of the matrix protein GM130, it is likely that golgin-84 participates in a tethering reaction different than that mediated by the cis-Golgi matrix proteins.
What might this tethering reaction be? One possibility is that golgin-84 tethers retrograde Golgi vesicles to the CGN. In this case, depletion of golgin-84 would be expected to cause accumulation of these vesicles, but no such accumulation was observed in our experiments. Perhaps in the absence of tethering to the CGN, these vesicles would by default fuse with the ER, causing an expansion of this compartment as we have observed. However, we believe this unlikely as no redistribution of Golgi enzymes, which have been detected in Golgi-derived COPI vesicles (Lanoix et al., 1999; Martinez-Menarguez et al., 2001), was detected in our experiments. A more likely role for golgin-84 is in the incorporation of incoming VTCs into the cis-Golgi. It could either act in a parallel pathway to that mediated by the cis-Golgi matrix proteins, or it may act at a temporally distinct stage. Cis-Golgi matrix proteins cycle into the intermediate compartment and are present on tubular connections between this pre-CGN compartment and the CGN itself (Marra et al., 2001). Nearly all (∼85%) of these GM130-containing structures label for the cargo protein VSV-G, suggesting that cis-Golgi matrix proteins mediate the first step in the incorporation of VTCs into the CGN (Marra et al., 2001). We could not detect any golgin-84 in the intermediate compartment under steady-state conditions or upon incubation at 15°C (unpublished data), suggesting that golgin-84 is not involved at such an early step. We therefore think it likely that golgin-84 operates after the cis-Golgi matrix proteins. The current model we favor is that golgin-84 tethers newly-forming cis-Golgi matrix-positive CGN elements and promotes their lateral fusion, which may be a homotypic event. The presence of golgin-84 at the rims of CGN elements places it in the ideal position for connecting these together laterally and promoting their fusion to form a continuous cisternal/ribbon structure.
In addition to the cis-Golgi matrix proteins and golgin-84, the cis-Golgi also contains the multisubunit tethering complexes TRAPP1, TRAPPII, and COG (for review see Whyte and Munro, 2002). Why have so many tethering complexes on one compartment? One reason may be that the cis-Golgi participates in multiple transport pathways. It receives membrane from the ER and from retrograde Golgi vesicles, and exports membrane back to the ER as well as forward into the Golgi stack. Thus, different tethering factors may be required for different transport steps. For example, COG appears to be required for the tethering of retrograde Golgi vesicles (Suvorova et al., 2002), whereas TRAPP1 is required for the tethering of anterograde ER-derived vesicles (Sacher et al., 2001). Another reason may be that the CGN is where Golgi cisternae are formed. The transition from pleiomorphic tubulo-vesicular clusters into a regular array of flattened and stacked cisternae is likely to involve multiple tethering and fusion events, with different proteins responsible for different reactions in the pathway. Rab1 (Ypt1 in yeast) may be a master regulator of this pathway, as it has been shown to interact with all of the cis-Golgi–associated tethering complexes (Whyte and Munro, 2002).
It was recently reported that the Golgi matrix proteins alone are sufficient to form a perinuclear Golgi-like ribbon (Seemann et al., 2000a). However, at the EM level, this structure was comprised predominantly of vesicles rather than cisternae (Seemann et al., 2000a), suggesting that additional factors are required for cisternae formation and Golgi apparatus assembly. One of these factors may be golgin-84, which would be expected to be absent from the matrix-containing structures described by Seemann et al. (2000a). This would be consistent with the predicted role of golgin-84 in promoting lateral fusion of Golgi membranes.
We found that protein transport was inhibited by only ∼40% in cells depleted of golgin-84. There are several possible explanations for this. One possibility is that golgin-84 is essential for transport, but the residual protein remaining after depletion, corresponding to only a few percent of the normal amount, is sufficient for transport to occur. Alternatively, golgin-84 may have no direct role in transport, with the transport inhibition merely reflecting the reduced amount of Golgi membranes present in golgin-84–depleted cells. In this case, golgin-84 would function purely as a structural protein. Finally, golgin-84 may improve the efficiency of transport without actually being essential for the process per se. This would be analogous to the situation with GM130 and p115. Blocking the interaction between these tethering proteins only partially inhibits protein transport through the Golgi apparatus (Seemann et al., 2000b; Marra et al., 2001). Currently, we cannot distinguish between these possibilities, and further experiments will be required to fully understand the role of golgin-84 in protein transport.
Golgin-84 is the second nonmatrix protein, after rab1 (Bailly et al., 1991), to be identified as a target for mitotic kinases. Further work is required to elucidate the role of golgin-84 phosphorylation and to determine whether this plays a part in the mitotic fragmentation process. One possibility is that phosphorylation is required in the early stages of mitosis when the Golgi ribbon is broken down to mini-stacks arranged around the nuclear envelope (Misteli and Warren, 1995; Shima et al., 1998). This would be most consistent with the predicted role of golgin-84 in linking membranes into a ribbon. Interestingly, there are no evolutionarily conserved candidate MAPK or CDK1 phosphorylation sites in golgin-84, suggesting that it is either a substrate for Plk (Sutterlin et al., 2001) or another kinase not known to be involved in the fragmentation process.
In summary, we have identified golgin-84 as a novel Golgi structural protein. The challenge now is to identify the molecular interactions of golgin-84 during both interphase and mitosis. This should not only lead to a greater understanding of Golgi apparatus assembly and maintenance but also illuminate how these processes are regulated during the cell cycle.
Materials and methods
Antibodies
Polyclonal antibodies to golgin-84 were raised in sheep using GST-tagged golgin-84 head or coiled-coil domain as immunogens. Antibodies were adsorbed against GST and affinity-purified against the corresponding fusion proteins covalently coupled to glutathione-sepharose (Amersham Biosciences). Polyclonal antibodies were raised in sheep against GST-tagged GFP and affinity-purified against the same protein coupled to glutathione-sepharose. The following antibodies were also used in this study: 4H1 monoclonal anti-p115; MLO7 (anti-N73pep) polyclonal anti-GM130 (Nakamura et al., 1997); 4A3 monoclonal anti-GM130 (Seemann et al., 2002); polyclonal anti-GM130 for electron microscopy (from Maria Antonietta De Matteis, Mario-Negri Institute, Santa Maria Imbaro, Italy); rabbit polyclonal antibodies to mannosidase I, GRASP65, and golgin-45 (from Francis Barr, Max-Planck Institute for Biochemistry, Martinsried, Germany); monoclonal anti-GalNacT2 (from Dr. Henrik Clausen, University of Copenhagen, Denmark); polyclonal anti-calnexin (from Dr. Stephen High, University of Manchester, Manchester, UK); and monoclonal anti–VSV-G lumenal domain (from Dr. Rainer Pepperkok, European Molecular Biology Laboratory, Heidelberg, Germany). Goat polyclonal antibodies to lamin A/C (N-18) were purchased from Santa Cruz Biotechnology, Inc. Rabbit polyclonal anti-mSec23p antibodies were purchased from Affinity BioReagents, Inc. Fluorophore and HRP-conjugated secondary antibodies were purchased from Molecular Probes and Tago Immunologicals, respectively.
In vitro phosphorylation of Golgi membranes and 16-BAC gel electrophoresis
Rat liver Golgi membranes were purified as described previously (Hui et al., 1998). Interphase and mitotic cytosols were prepared from spinner HeLa cells according to Sönnichsen et al. (1996) and desalted into buffer A (20 mM β-glycerophosphate, 15 mM EGTA, 50 mM KOAc, 10 mM MgOAc, 2 mM ATP, 1 mM DTT, 0.2 M sucrose). Golgi membranes were incubated with desalted interphase and mitotic HeLa cytosol (9 mg/ml) in the presence of 0.2 μCi/μl [γ-32P]ATP for 30 min at 30°C. The incubated membranes were then adjusted to 1.6 M sucrose, overlaid with 1.2 M sucrose, 1.0 M sucrose, and finally 0.4 M sucrose (all sucrose solutions were made in TKN buffer [20 mM Tris-Cl, pH 7.4, 0.1 M KCl, 0.1 M NaF and 1 mM DTT]), and centrifuged for 4 h at 55,000 rpm in a SW55 rotor. The Golgi membranes (at the 0.4 M/1.0 M interface) were collected and pelleted by centrifugation at 55,000 rpm for 30 min in a TLA55 rotor. The membranes were either solubilized directly into sample buffer and subjected to 2D 16-BAC/SDS-PAGE (Hartinger et al., 1996) or washed with 0.2 M sodium carbonate and the carbonate pellet was further extracted with 1% Triton X-114 (Nakamura et al., 1995) before electrophoresis. 16-BAC/SDS-PAGE gels were analyzed by silver staining and autoradiography.
Mass spectrometry
Radiolabeled proteins were excised from dried 2D gels with a protein-free razor blade. Excised spots were subjected to in-gel digestion with trypsin and the resulting peptides were analyzed using a MALDI-TOF instrument (M@LDI; Micromass) and probability-based database searching (Pappin, 2003). To confirm the identity of a protein, the digest extracts were analyzed by nano electrospray on an ion trap instrument (Finnigan LCQ Deca; Thermoquest). MS/MS data were obtained for a number of peptides and the spectra were used to query the MS/MS Ion Search program on MASCOT.
Immunoprecipitation of golgin-84
For analysis of golgin-84 phosphorylation, 32P-labeled Golgi membranes were resuspended in TKN buffer containing 1% SDS and protease inhibitors, boiled for 3 min, mixed with an equal volume of ice-cold 4% Triton X-100, and clarified by centrifugation at 14,000 rpm for 10 min. 2 μg of polyclonal antibodies to golgin-84 and 10 μl protein G–sepharose beads were added and incubated at 4°C for 2–4 h at 4°C. After washing three times with IP buffer (TKN containing 0.5% TX-100), bound proteins were eluted by boiling in SDS sample buffer and analyzed by 1D SDS-PAGE followed by autoradiography. To test for coimmunoprecipitation of golgin-84 and matrix proteins, Golgi membranes were extracted in IP buffer lacking NaF for 30 min on ice, clarified, and incubated with 2 μg of anti–golgin-84, anti-GM130 (4A3), or anti-p115 (4H1) antibodies and 10 μl protein G–sepharose. Beads were washed with IP buffer lacking NaF, boiled in SDS sample buffer, and eluted proteins were analyzed by 1D SDS-PAGE and immunoblotting with the appropriate antibodies.
Rab effector binding assay
Binding of Golgi proteins to GST-tagged rab proteins was performed according to Short et al. (2001) except that 0.25 mg rab protein and 25 μl glutathione-sepharose beads were incubated with 100 μg Golgi extract in a final volume of 200 μl for 2 h at 4°C. After elution, bound proteins were precipitated with 10% TCA and analyzed by Western blotting.
Molecular biology and yeast two-hybrid analysis
Standard molecular biology techniques were used for all constructs; primer sequences are available upon request. The full-length and truncated versions of human golgin-84 cDNA were inserted into the BglII and EcoRI sites of the pEGFP-C1 vector (CLONTECH Laboratories, Inc.) or the BamHI and EcoRI sites of a modified pcDNA3.1 vector (Stratagene) containing an NH2-terminal myc tag. Full-length and truncated versions of human golgin-84 cDNA lacking the trans-membrane domain were inserted into the yeast two-hybrid activation domain vector pGADT7. All bait vector pGBT9/rab GTPase constructs were provided by Francis Barr. The pGADT7/golgin-84 and pGBT9/rab GTPase plasmids were cotransformed into the yeast reporter strain AH109 on synthetic medium lacking leucine and tryptophan (low selection) and then restreaked onto synthetic medium lacking leucine, tryptophan, histidine, and adenine with 2% glucose as the carbon source (high selection) according to the CLONTECH Laboratories, Inc. yeast protocol handbook.
Cell culture and drug treatments
HeLa, normal rat kidney (NRK), and A431 cells were cultured at 37°C and 5% CO2 in DME containing 10% FCS. NRK cells were incubated with 5 μg/ml BFA in tissue culture medium for 1 h at 37°C before fixation.
Transfections and RNAi
HeLa cells were transfected with DNA plasmids using Fugene 6 (Roche Biochemicals) according to the manufacturer's instructions. RNAi was performed on HeLa cells using oligofectamine (Life Technologies) with duplex RNA oligos (Dharmacon Research) for 1–4 d as described by Elbashir et al. (2001). Golgin-84 was targeted with the sequence AAGTAGGATCTCGGACACCAG and the lamin A control was described previously (Elbashir et al., 2001). Golgin-84 levels were quantitated from Western blots according to Sönnichsen et al. (1998).
Fluorescence microscopy
Cells were grown on coverslips and fixed in 100% methanol at −20°C for 4 min. Coverslips were incubated with primary antibodies diluted into PBS containing 0.5 mg/ml BSA for 20 min at RT, washed, and incubated with PBS/BSA containing fluorophore-conjugated secondary antibodies and 200 ng/ml Hoechst 33342 for an additional 20 min at RT. Coverslips were mounted in Mowiol and analyzed by conventional epifluorescence microscopy using an Olympus BX60 upright microscope with a MicroMax CCD camera (Roper Scientific) driven by Metamorph software. Confocal images were obtained using a Leica NT confocal microscope. All images are projections of optocal sections in the z axis at 0.5-μm intervals.
VSV-G transport assays
3 d after RNAi treatment, HeLa cells were transfected with a plasmid encoding GFP-tagged ts045G VSV-G (provided by Patrick Keller, Max-Planck Institute for Molecular Cell Biology and Genetics, Dresden, Germany) for 1 h at 37°C and then 12 h at 39.5°C. Cells were then incubated at 4°C for 30 min to promote VSV-G protein folding, the growth medium was replaced with prewarmed medium (31°C), and the cells were incubated for a further 0, 30, 60, or 90 min at 31°C. Cells were then fixed in 3.5% paraformaldehyde and cell surface VSV-G was detected with a monoclonal antibody to the VSV-G lumenal domain and a Texas red–conjugated anti–mouse secondary antibody and total VSV-G by GFP. The ratio of surface to total measured fluorescence was used to calculate the extent of VSV-G protein transport (Pepperkok et al., 1993; Seemann et al., 2000b).
Electron microscopy
Cells were fixed in 2% gluteraldehyde or 8% paraformaldehyde and processed for cryosectioning and immunogold labeling as described by Farmaki et al. (1999), except the sections were picked up using the modified pick up method (Liou et al., 1996); one part methyl cellulose and three parts 2.3 M sucrose in PBS. For quantitation of gold labeling, cell profiles contained in a randomly selected grid square were scanned systematically and Golgi areas were identified by the presence of cisternal stacks and/or vesicular profiles labeled for golgin-84 or GM130. Labeling was assigned to one of four categories: Golgi cisternae, tubulo-vesicular profiles lateral to the Golgi stack, tubulo-vesicular profiles on the cis face of the stack, and any other labeling detected. Golgi cisternae are defined as membrane-bounded profiles with a length/breadth ratio of 4 or more. Tubulo-vesicular profiles are noncisternal profiles with a diameter of <80 nm. The lateral aspect of the stack was delineated by drawing a line across the end of the stack orthogonal to the cisternae. For structural and quantitative analysis of RNAi-treated cells, samples were fixed in 2% gluteraldehyde, post-fixed with reduced osmium tetroxide, and embedded in epoxy resin according to Lucocq et al. (1989). The surface density of membranes in the cell were estimated using stereological methods from the formula 2I/L, in which I represents intersections of the lines on a square lattice grid with the membrane of interest and L is the total line length applied to the reference space (Lucocq, 1993).
Acknowledgments
We thank Drs. Francis Barr, Henrik Clausen, Maria Antonietta De Matteis, Viki Allan, Stephen High, Rainer Pepperkok, and Patrick Keller for generously providing antibodies and reagents as noted above. We are grateful to Joanna Woodburn for preparing the GFP antibodies and Drs. Philip Woodman and Viki Allan for critical reading of the manuscript.
This work was supported by a Medical Research Council Career Development Award to M. Lowe (G120/483).
Footnotes
Abbreviations used in this paper: 2D, two dimensional; BFA, brefeldin A; CGN, cis-Golgi network; NRK, normal rat kidney; RNAi, RNA interference; siRNA, small interfering RNA; VSV-G, vesicular stomatitis virus G protein.
References
- Acharya, U., A. Mallabiabarrena, J.K. Acharya, and V. Malhotra. 1998. Signaling via mitogen-activated protein-kinase kinase (MEK 1) is required for Golgi fragmentation during mitosis. Cell. 92:183–192. [DOI] [PubMed] [Google Scholar]
- Allan, B.B., B.D. Moyer, and W.E. Balch. 2000. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 289:444–448. [DOI] [PubMed] [Google Scholar]
- Bailly, E., M. McCaffrey, N. Touchot, A. Zahraoui, B. Goud, and M. Bornens. 1991. Phosphorylation of two small GTP-binding proteins of the Rab family by p34cdc2. Nature. 350:715–718. [DOI] [PubMed] [Google Scholar]
- Barr, F.A., M. Puype, J. Vandekerckhove, and G. Warren. 1997. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 91:253–262. [DOI] [PubMed] [Google Scholar]
- Barr, F.A., N. Nakamura, and G. Warren. 1998. Mapping the interaction between GRASP65 and GM130, components of a protein complex involved in the stacking of Golgi cisternae. EMBO J. 17:3258–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bascom, R.A., S. Srinivasan, and R.L. Nussbaum. 1999. Identification and characterization of golgin-84, a novel Golgi integral membrane protein with a cytoplasmic coiled-coil domain. J. Biol. Chem. 274:2953–2962. [DOI] [PubMed] [Google Scholar]
- Bevis, B.J., A.T. Hammond, C.A. Reinke, and B.S. Glick. 2002. De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat. Cell Biol. 4:750–756. [DOI] [PubMed] [Google Scholar]
- Cha, H., and P. Shapiro. 2001. Tyrosine-phosphorylated extracellular signal–regulated kinase associates with the Golgi complex during G2/M phase of the cell cycle: evidence for regulation of Golgi structure. J. Cell Biol. 153:1355–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis, M.A., and J.S. Morrow. 2000. Spectrin tethers and mesh in the biosynthetic pathway. J. Cell Sci. 113:2331–2343. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 411:494–498. [DOI] [PubMed] [Google Scholar]
- Farmaki, T., S. Ponnambalam, A.R. Prescott, H. Clausen, B.L. Tang, W. Hong, and J.M. Lucocq. 1999. Forward and retrograde trafficking in mitotic animal cells. ER-Golgi transport arrest restricts protein export from the ER into COPII-coated structures. J. Cell Sci. 112:589–600. [DOI] [PubMed] [Google Scholar]
- Gillingham, A.K., A.C. Pfeifer, and S. Munro. 2002. CASP, the alternatively spliced product of the gene encoding the CCAAT-displacement protein transcription factor, is a Golgi membrane protein related to giantin. Mol. Biol. Cell. 13:3761–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartinger, J., K. Stenius, D. Hogemann, and R. Jahn. 1996. 16-BAC/SDS-PAGE: a two-dimensional gel electrophoresis system suitable for the separation of integral membrane proteins. Anal. Biochem. 240:126–133. [DOI] [PubMed] [Google Scholar]
- Hui, N., N. Nakamura, P. Slusarewicz, and G. Warren. 1998. Purification of rat liver Golgi stacks. Cell Biology: A Laboratory Handbook. Vol. 2. J. Celis, editor. Academic Press Inc., Orlando, FL. 46–55.
- Jesch, S.A., T.S. Lewis, N.G. Ahn, and A.D. Linstedt. 2001. Mitotic phosphorylation of Golgi reassembly stacking protein 55 by mitogen-activated protein kinase ERK2. Mol. Biol. Cell. 12:1811–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman, J. 2000. Transport between ER and Golgi. Curr. Opin. Cell Biol. 12:445–449. [DOI] [PubMed] [Google Scholar]
- Lanoix, J., J. Ouwendijk, C.C. Lin, A. Stark, H.D. Love, J. Ostermann, and T. Nilsson. 1999. GTP hydrolysis by arf-1 mediates sorting and concentration of Golgi resident enzymes into functional COP I vesicles. EMBO J. 18:4935–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.Y., M.L. Madsen, F.R. Yarm, Y.J. Jang, X. Liu, and R.L. Erikson. 2000. Peripheral Golgi protein GRASP65 is a target of mitotic polo-like kinase (Plk) and Cdc2. Proc. Natl. Acad. Sci. USA. 97:12589–12594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt, A.D., and H.P. Hauri. 1993. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol. Biol. Cell. 4:679–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou, W., H.J. Geuze, and J.W. Slot. 1996. Improving structural integrity of cryosections for immunogold labeling. Histochem. Cell Biol. 106:41–58. [DOI] [PubMed] [Google Scholar]
- Lowe, M., N. Nakamura, and G. Warren. 1998. a. Golgi division and membrane traffic. Trends Cell Biol. 8:40–44. [DOI] [PubMed] [Google Scholar]
- Lowe, M., C. Rabouille, N. Nakamura, R. Watson, M. Jackman, E. Jamsa, D. Rahman, D.J. Pappin, and G. Warren. 1998. b. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 94:783–793. [DOI] [PubMed] [Google Scholar]
- Lucocq, J.M. 1993. Unbiased 3-D quantitation of ultrastructure in cell biology. Trends Cell Biol. 3:345–358. [DOI] [PubMed] [Google Scholar]
- Lucocq, J.M., E.G. Berger, and G. Warren. 1989. Mitotic Golgi fragments in HeLa cells and their role in the reassembly pathway. J. Cell Biol. 109:463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, P., T. Maffucci, T. Daniele, G.D. Tullio, Y. Ikehara, E.K. Chan, A. Luini, G. Beznoussenko, A. Mironov, and M.A. De Matteis. 2001. The GM130 and GRASP65 Golgi proteins cycle through and define a subdomain of the intermediate compartment. Nat. Cell Biol. 3:1101–1113. [DOI] [PubMed] [Google Scholar]
- Martinez-Menarguez, J.A., R. Prekeris, V.M. Oorschot, R. Scheller, J.W. Slot, H.J. Geuze, and J. Klumperman. 2001. Peri-Golgi vesicles contain retrograde but not anterograde proteins consistent with the cisternal progression model of intra-Golgi transport. J. Cell Biol. 155:1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles, S., H. McManus, K.E. Forsten, and B. Storrie. 2001. Evidence that the entire Golgi apparatus cycles in interphase HeLa cells: sensitivity of Golgi matrix proteins to an ER exit block. J. Cell Biol. 155:543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T., and G. Warren. 1995. Mitotic disassembly of the Golgi apparatus in vivo. J. Cell Sci. 108:2715–2727. [DOI] [PubMed] [Google Scholar]
- Moyer, B.D., B.B. Allan, and W.E. Balch. 2001. Rab1 interaction with a GM130 effector complex regulates COPII vesicle cis-Golgi tethering. Traffic. 2:268–276. [DOI] [PubMed] [Google Scholar]
- Nakamura, N., C. Rabouille, R. Watson, T. Nilsson, N. Hui, P. Slusarewicz, T.E. Kreis, and G. Warren. 1995. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 131:1715–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, N., M. Lowe, T.P. Levine, C. Rabouille, and G. Warren. 1997. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 89:445–455. [DOI] [PubMed] [Google Scholar]
- Nelson, D.S., C. Alvarez, Y.S. Gao, R. Garcia-Mata, E. Fialkowski, and E. Sztul. 1998. The membrane transport factor TAP/p115 cycles between the Golgi and earlier secretory compartments and contains distinct domains required for its localization and function. J. Cell Biol. 143:319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappin, D.J. 2003. Peptide mass fingerprinting using MALDI-TOF mass spectrometry. Methods Mol. Biol. 211:211–219. [DOI] [PubMed] [Google Scholar]
- Pepperkok, R., J. Scheel, H. Horstmann, H.P. Hauri, G. Griffiths, and T.E. Kreis. 1993. β-COP is essential for biosynthetic membrane transport from the endoplasmic reticulum to the Golgi complex in vivo. Cell. 74:71–82. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S.R. 2001. Constructing a Golgi complex. J. Cell Biol. 155:873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu, M.A., and A.D. Linstedt. 2001. Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. J. Cell Biol. 155:227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambourg, A., and Y. Clermont. 1997. Three-dimensional structure of the Golgi apparatus in mammalian cells. The Golgi Apparatus. E.G. Berger and J. Roth, editors. Birkhauser Verlag, Basel, Switzerland. 37–61.
- Sacher, M., J. Barrowman, W. Wang, J. Horecka, Y. Zhang, M. Pypaert, and S. Ferro-Novick. 2001. TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol. Cell. 7:433–442. [DOI] [PubMed] [Google Scholar]
- Sapperstein, S.K., D.M. Walter, A.R. Grosvenor, J.E. Heuser, and M.G. Waters. 1995. p115 is a general vesicular transport factor related to the yeast endoplasmic reticulum to Golgi transport factor Uso1p. Proc. Natl. Acad. Sci. USA. 92:522–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann, J., E. Jokitalo, M. Pypaert, and G. Warren. 2000. a. Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature. 407:1022–1026. [DOI] [PubMed] [Google Scholar]
- Seemann, J., E.J. Jokitalo, and G. Warren. 2000. b. The role of the tethering proteins p115 and GM130 in transport through the Golgi apparatus in vivo. Mol. Biol. Cell. 11:635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann, J., M. Pypaert, T. Taguchi, J. Malsam, and G. Warren. 2002. Partitioning of the matrix fraction of the Golgi apparatus during mitosis in animal cells. Science. 295:848–851. [DOI] [PubMed] [Google Scholar]
- Shima, D.T., N. Cabrera-Poch, R. Pepperkok, and G. Warren. 1998. An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J. Cell Biol. 141:955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short, B., C. Preisinger, R. Korner, R. Kopajtich, O. Byron, and F.A. Barr. 2001. A GRASP55–rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155:877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter, J., and G. Warren. 1999. A role for the vesicle tethering protein, p115, in the post-mitotic stacking of reassembling Golgi cisternae in a cell-free system. J. Cell Biol. 146:57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusarewicz, P., T. Nilsson, N. Hui, R. Watson, and G. Warren. 1994. Isolation of a matrix that binds medial Golgi enzymes. J. Cell Biol. 124:405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen, B., R. Watson, H. Clausen, T. Misteli, and G. Warren. 1996. Sorting by COP I–coated vesicles under interphase and mitotic conditions. J. Cell Biol. 134:1411–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen, B., M. Lowe, T. Levine, E. Jämsä, B. Dirac-Svejstrup, and G. Warren. 1998. A role for giantin in docking COPI vesicles to Golgi membranes. J. Cell Biol. 140:1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin, C., C.Y. Lin, Y. Feng, D.K. Ferris, R.L. Erikson, and V. Malhotra. 2001. Polo-like kinase is required for the fragmentation of pericentriolar Golgi stacks during mitosis. Proc. Natl. Acad. Sci. USA. 98:9128–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova, E.S., R. Duden, and V.V. Lupashin. 2002. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J. Cell Biol. 157:631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsdottir, R., H. Hashimoto, K. Ashman, T. Koda, B. Storrie, and T. Nilsson. 2001. Identification of rabaptin-5, rabex-5, and GM130 as putative effectors of rab33b, a regulator of retrograde traffic between the Golgi apparatus and ER. FEBS Lett. 508:201–209. [DOI] [PubMed] [Google Scholar]
- Ward, T.H., R.S. Polishchuk, S. Caplan, K. Hirschberg, and J. Lippincott-Schwartz. 2001. Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155:557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, G., T. Levine, and T. Misteli. 1995. Mitotic disassembly of the mammalian Golgi apparatus. Trends Cell Biol. 5:413–416. [DOI] [PubMed] [Google Scholar]
- Waters, M.G., and F.M. Hughson. 2000. Membrane tethering and fusion in the secretory and endocytic pathways. Traffic. 1:588–597. [DOI] [PubMed] [Google Scholar]
- Weide, T., M. Bayer, M. Koster, J.P. Siebrasse, R. Peters, and A. Barnekow. 2001. The Golgi matrix protein GM130: a specific interacting partner of the small GTPase rab1b. EMBO Rep. 2:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte, J.R.C., and S. Munro. 2002. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 115:2627–2637. [DOI] [PubMed] [Google Scholar]