Abstract
Epithelial protein lost in neoplasm (EPLIN) is a cytoskeleton-associated protein encoded by a gene that is down-regulated in transformed cells. EPLIN increases the number and size of actin stress fibers and inhibits membrane ruffling induced by Rac. EPLIN has at least two actin binding sites. Purified recombinant EPLIN inhibits actin filament depolymerization and cross-links filaments in bundles. EPLIN does not affect the kinetics of spontaneous actin polymerization or elongation at the barbed end, but inhibits branching nucleation of actin filaments by Arp2/3 complex. Side binding activity may stabilize filaments and account for the inhibition of nucleation mediated by Arp2/3 complex. We propose that EPLIN promotes the formation of stable actin filament structures such as stress fibers at the expense of more dynamic actin filament structures such as membrane ruffles. Reduced expression of EPLIN may contribute to the motility of invasive tumor cells.
Keywords: actin stress fibers; lamellipodia; Rac1; actin nucleation; Arp2/3
Introduction
Motile cancer cells depend on assembly and disassembly of actin filaments to extend their leading edges as they move, so alterations in the expression or properties of actin binding proteins may influence their invasiveness (Ben-Ze'ev, 1997; Pawlak and Helfman, 2001). Tumor cell movements might be enhanced by overexpression or activating mutations of proteins that stimulate the actin system or lower expression or inactivation of proteins that act as negative regulators of the actin system. Some transformed cells express reduced levels of tropomyosin or α-actinin, proteins that stabilize actin filaments and actin filament bundles, and restoring their expression can reverse some transformed phenotypes (Gluck et al., 1993; Braverman et al., 1996). On the other hand, the increased expression of thymosin β15 and gelsolin, proteins implicated in the disassembly of the actin filaments, correlates with poor clinical outcome of cancer patients (Bao et al., 1996; Chakravatri et al., 2000; Thor et al., 2001).
The assembly of cellular actin filaments requires the creation of new barbed ends, either de novo or by removing the capping proteins or severing existing filaments (Pollard et al., 2000). Activation of Arp2/3 complex is one mechanism to create new barbed ends de novo. Arp2/3 complex has little actin nucleation activity (Welch et al., 1997; Mullins et al., 1998) unless activated by bacterial proteins (Welch et al., 1997) or WASp family proteins (Machesky et al., 1999; Rohatgi et al., 1999). Activated Arp2/3 complex forms new filaments on the side of preexisting filaments, which then elongate at their free barbed ends. WASp family proteins are key downstream targets for Rho family GTPases (Higgs and Pollard, 2000; Rohatgi et al., 2000) that link signals from chemotaxis or cell adhesion receptors to activation of Arp2/3 complex.
Epithelial protein lost in neoplasm (EPLIN)* was originally identified as the product of a gene that is down-regulated in prostate and breast cancer cell lines (Chang et al., 1998; Maul and Chang, 1999). In oral keratinocytes and mammary epithelial cells, it distributes to the cortical cytoskeleton, whereas in Saos-2 cells, it localizes to actin stress fibers and focal adhesions (Maul and Chang, 1999; Song et al., 2002; unpublished data). Two EPLIN isoforms (α and β) are generated by alternative promoter usage from a single gene on chromosome 12. EPLIN-β (85 kD) has an additional 160 residues at its NH2 terminus, but is otherwise identical to EPLIN-α (67 kD), the isoform down-regulated in transformed cells. The only domain recognizable in the amino acid sequence is a centrally located, 54-residue LIM domain, a putative protein–protein interaction module (Schmeichel and Beckerle, 1997) that may allow EPLIN to dimerize with itself or associate with other proteins.
We investigated the possibility that EPLIN might control cellular actin dynamics. Expression of EPLIN increases the number and size of cellular stress fibers and inhibits membrane ruffling induced by active Rac1. EPLIN has at least two actin binding domains. EPLIN stabilizes preformed filaments, a property attributed to its ability to cross-link and bundle actin filaments. Furthermore, EPLIN inhibits nucleation by Arp2/3 complex. These properties suggest that loss of EPLIN may contribute to transformation owing to a loss of stability of mature actin filament structures and more rapid turnover of filaments associated with motility.
Results
EPLIN expression increases stress fibers
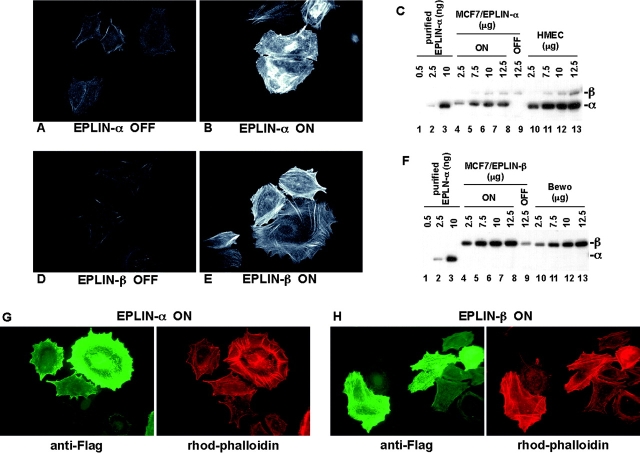
To test if EPLIN modifies actin organization in the cell, we expressed EPLIN in MCF-7 breast carcinoma cells using a tetracycline-regulated system. MCF-7 cells do not express EPLIN-α, but express EPLIN-β at a low level (Fig. 1 C, lane 9). Induction of EPLIN expression increased the number of actin stress fibers and the intensity with which they stained with rhodamine-phalloidin (Fig. 1 A, B, D, and E). Expression of EPLIN-α in MCF-7 cells increased the fluorescence intensity of rhodamine-phalloidin staining 2.1-fold, whereas expression of EPLIN-β increased actin staining 3.7-fold (see Materials and methods). The levels of EPLIN-α and -β overexpression in MCF-7 cells were equivalent to the levels of endogenous protein in HMEC and BeWo choriocarcinoma cells, respectively (Fig. 1, C and F). Staining for the induced EPLIN using anti-Flag antibodies revealed a positive correlation between the levels of EPLIN expression and the amount of stress fibers (Fig. 1, G and H).
Figure 1.
Expression of EPLIN increases actin filaments in MCF-7 cells. (A, B, D, and E) Expression of Flag-EPLIN-α or -β was induced by removing doxycycline from culture media. 72 h after protein induction, cells were fixed and stained with rhodamine-phalloidin to visualize actin filaments. A and D, − induction; B and E, + induction. Digital images were captured at the same exposure time (248 ms for A and B; 308 ms for D and E). (C and F) The levels of EPLIN overexpression in MCF-7 cells were estimated. Varying amounts (2.5–12.5 μg) of protein lysates from MCF-7 cells before and after protein induction, human mammary epithelial cells (HMEC; EPLIN-α positive), and BeWo choriocarcinoma cells (EPLIN-β positive) were fractionated by SDS-PAGE and then immunoblotted with anti-EPLIN antisera. To estimate the absolute level of expression, known amounts (0.5–10 ng) of purified his-EPLIN-α protein were also included in the immunoblot. (G and H) Flag-EPLIN-α or -β expressing MCF-7 cells were stained with rhodamine-phalloidin and the M2 anti-Flag mAb to stain the induced proteins. The increase in actin filaments correlated with the amount of EPLIN expressed in these cells.
EPLIN inhibits membrane ruffling
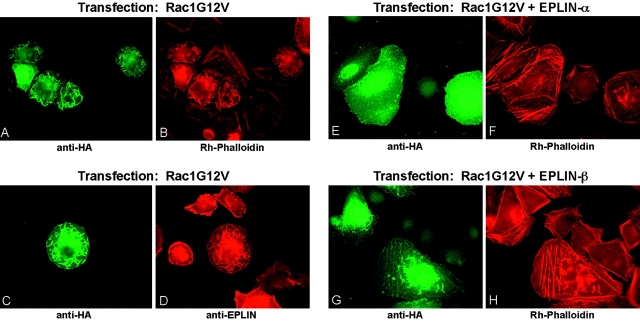
Activated Rac1 recruits WAVE/Scar (a member of WASp family of proteins) from the cytoplasm to the plasma membrane to induce actin filament assembly (Miki et al., 1998). To test if EPLIN influences the formation of actin filaments in vivo, we examined the effect of EPLIN expression on Rac1-induced membrane ruffling. Expression of constitutively active Rac1 (HA-tagged Rac1G12V) in HeLa cells produced extensive membrane ruffles containing Rac1G12V, actin filaments, and endogenous EPLIN (Fig. 2, A–D). Co-transfection of either EPLIN-α or EPLIN-β with Rac1G12V strongly inhibited membrane ruffling, producing thick actin stress fibers instead (Fig. 2, E–H). When EPLIN was coexpressed, transfected Rac1G12V was diffusely distributed in the cells, rather than localizing to membrane ruffles.
Figure 2.
EPLIN inhibits membrane ruffling induced by Rac1. Transfected HeLa cells were stained with anti-HA (A, C, E, and G), rhodamine-phalloidin (B, F, and H), or anti-EPLIN (D) to visualize the transfected HA-tagged Rac1G12V, actin filaments, or endogenous EPLIN, respectively. In A–D, HeLa cells were transfected with HA-tagged Rac1G12V alone. In E–H, HeLa cells were co-transfected with HA-tagged Rac1G12V and EPLIN-α (E and F) or EPLIN-β (G and H).
EPLIN binds to actin
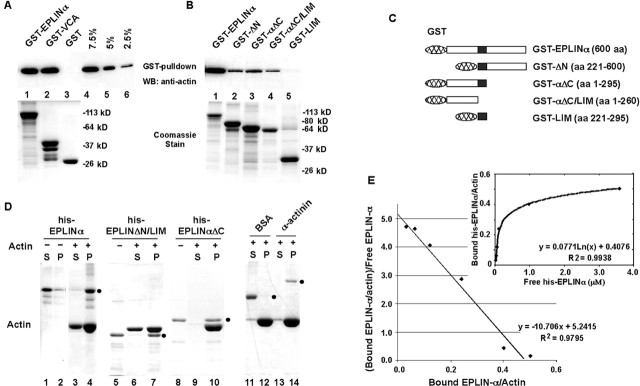
A recombinant full-length EPLIN-α GST-fusion protein purified from bacteria bound actin monomers in a pull-down assay (Fig. 3 A). Actin monomer binding was efficient and comparable to the control GST-VCA protein, pelleting ∼10% of the actin monomers. Truncated EPLIN containing either the NH2- or COOH-terminal region (GST-αΔC and GST-ΔN) also bound monomeric actin, but less efficiently than the full-length EPLIN-α (Fig. 3 B). GST-αΔC/LIM, lacking the central LIM domain, also bound to monomeric actin, but the LIM domain (GST-LIM) failed to bind monomeric actin. These findings indicate that EPLIN-α contains at least two actin-binding sites, one on each side of the centrally located LIM domain. Equivalent loading for the GST-fusion proteins in these experiments was confirmed by Coomassie staining.
Figure 3.
EPLIN binds directly to both G- and F-actin. (A) Actin monomer pull-down assay with GST-EPLIN. Glutathione-Sepharose beads carrying 2 μg of GST-EPLIN-α (lane 1), GST-VCA (lane 2), and GST (lane 3) were incubated with 50 nM G-actin in 0.5 ml. G-actin bound to the beads was assayed by immunoblotting with anti-actin antibodies (top). In lanes 4–6, varying amounts of G-actin (2.5–7.5% of the input) were included to estimate the amount of bound G-actin. A duplicate gel was stained with Coomassie blue to estimate the amounts of GST proteins present in each lane (bottom). GST-VCA contains a G-actin–binding verprolin (V) homology domain, a cofilin (C) homology domain, and an acidic segment (A) of bovine N-WASp. (B) Actin monomer pull-down assays were performed with GST-EPLIN-α (lane 1), GST-ΔN (lane 2), GST-αΔC (lane 3), GST-αΔC/LIM (lane 4), and GST-LIM (lane 5). (C) Schematic diagrams of EPLIN truncations used in the actin monomer pull-down assay are shown. (D) Actin filament pelleting assay. Samples containing 16 μM polymerized actin and 2 μM EPLIN-α were centrifuged at high speed. Control proteins (BSA, negative controls; α-actinin, positive control) were also used at 2 μM. The presence (+) or absence (−) of actin in the pelleting assay is denoted. The supernatant (S) and pellet (P) were analyzed by SDS-PAGE and stained with Coomassie blue. Positions of control proteins and EPLIN-α, or its truncated derivatives, his-EPLINΔN/LIM and his-EPLINαΔC, are indicated by filled circles. (E) Binding of his-EPLIN-α to actin filament. His-EPLIN-α (0–8 μM) was mixed with 10 μM polymerized actin, followed by ultracentrifugation. Amounts of the free and bound his-EPLIN-α in the supernatant and pellet fractions were determined from a digitized Coomassie-stained gel.
EPLIN-α also pelleted with purified actin filaments (Fig. 3 D, lanes 3 and 4). EPLIN-α alone did not pellet (Fig. 3 D, lanes 1 and 2), indicating that the pelleting is due to binding to F-actin. Two nonoverlapping truncations of EPLIN, ΔN/LIM (Fig. 3 D, lanes 5–7) and αΔC (Fig. 3 D, lanes 8–10), both pelleted with F-actin, indicating the presence of two independent F-actin binding sites on EPLIN-α. BSA, used as negative controls, remained in the supernatant, whereas α-actinin pelleted with F-actin (Fig. 3 D, lanes 11–14). The stoichiometry of binding was calculated to be two actin molecules per one his-EPLIN-α (Fig. 3 E), consistent with the presence of at least two actin-binding sites per EPLIN-α polypeptide. Quantitation of free and pelleted his-EPLIN-α as a function of actin filament concentration gave a dissociation equilibrium constant of ∼0.1 μM.
EPLIN cross-links and/or bundles F-actin
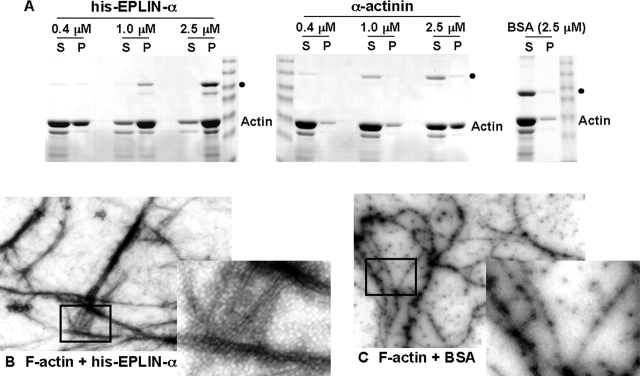
The increase in actin stress fibers observed in EPLIN-overexpressing cells, together with its localization to the actin cytoskeleton, raised the possibility that it may bundle actin filaments. Bundled or cross-linked actin filaments can be detected by sedimentation at low speed where free actin filaments remain in the supernatant. Because GST can form a homodimer (Ji et al., 1992), bundling assays were performed with his-EPLIN-α. His-EPLIN-α caused the majority of actin filaments to pellet during low speed centrifugation (Fig. 4 A). At the same molar concentration, EPLIN-α pelleted actin filaments more efficiently than α-actinin, a dimeric cross-linking protein with two actin-binding sites. A control protein, BSA, remained in the supernatant with the actin filaments. We used electron microscopy of negatively stained specimens to confirm that bundling was responsible for actin filaments pelleting with EPLIN during low speed centrifugation. Actin filaments formed thick, tightly packed bundles in the presence of EPLIN-α (Fig. 4 B). In the presence of BSA (used as a negative control), the F-actin remained as thin wavy filaments (Fig. 4 C).
Figure 4.
EPLIN cross-links and bundles F-actin. (A) Low speed pelleting assay. Samples containing 10 μM F-actin and 0.4–2.5 μM his-EPLIN-α or 0.4– 2.5 μM α-actinin were centrifuged at 8,000 g for 20 min. The negative control, BSA, was used at 2.5 μM. The supernatant (S) and pellet (P) were analyzed by SDS-PAGE and stained with Coomassie blue. Positions of his-EPLIN-α, α-actinin, and BSA are indicated by filled circles. (B and C) Electron microscopy. 2 μM actin was polymerized with 1 μM his-EPLIN-α (B) or 1 μM BSA (C), negatively stained with uranyl acetate and visualized by electron microscopy. The boxed areas were enlarged (insets) to better illustrate actin filaments.
EPLIN inhibits actin depolymerization in vitro
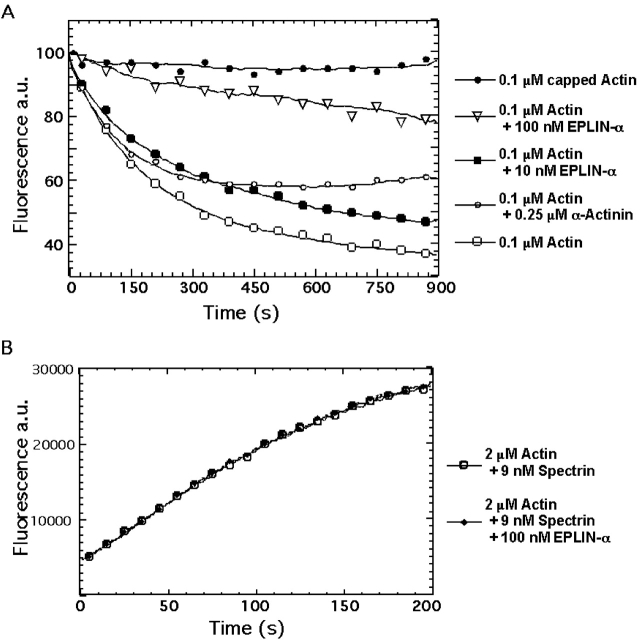
We used the fluorescence of pyrene-labeled actin to monitor polymerization and to test if EPLIN affects actin assembly or disassembly. The time course of spontaneous polymerization was the same in the presence or absence of EPLIN-α (see Fig. 6). On the other hand, EPLIN significantly delayed actin filament depolymerization (Fig. 5 A). When diluted below the critical concentration, actin filaments depolymerized with a half-time of 7 min. EPLIN-α slowed depolymerization in a concentration-dependent fashion. Addition and loss of actin monomers occurs exclusively at the filament ends, with the barbed ends displaying higher rates of turnover (Pollard et al., 2000). We tested the possibility that EPLIN-α stabilizes filaments by capping barbed ends (Fig. 5 B). The rate of barbed end elongation from spectrin-actin seeds was not affected by EPLIN-α, indicating that the EPLIN-α does not cap barbed ends.
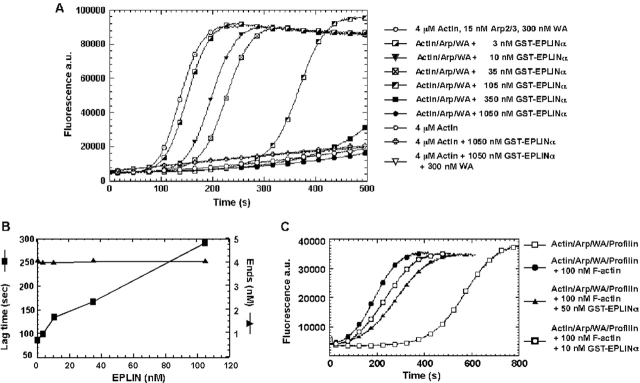
Figure 6.
EPLIN inhibits actin nucleation by Arp2/3 complex. (A) The time course of actin polymerization was monitored by pyrene fluorescence. Samples contained 4 μM G-actin (5% pyrene labeled), ± 15 nM Arp2/3 complex, or ± 300 nM Scar-WA domain, ± varying concentrations of GST-EPLIN-α in polymerization buffer. (B) Dependence of the lag time and concentration of ends produced by Arp2/3 complex on the concentration of EPLIN-α. The polymerization lag time was defined as the time required to achieve 10% of the maximum fluorescence. The concentration of barbed ends was calculated from elongation rate (measured by the rate of polymerization where 80% of monomers were polymerized) using the equation: [barbed ends] = elongation rate/(k + [actin monomer]), where k + = 10 μM−1 s−1. At GST-EPLIN-α concentrations >105 nM, polymerization was incomplete in the time course of the experiment, disallowing determination of the lag time or the concentration of barbed ends. (C) The effect of EPLIN on the ability of preformed actin filaments to stimulate nucleation by Arp2/3 complex. Samples in polymerization buffer contained 2 μM G-actin (5% pyrene labeled), 15 nM Arp2/3 complex, 300 nM Scar-WA domain, and 8 μM profilin-I, ± 100 nM actin filament, ± 10 or 50 nM GST-EPLIN-α. Profilin-I was added to accentuate the polymerization lag time.
Figure 5.
Effect of EPLIN on actin filament depolymerization and barbed-end elongation. (A) Time course of the depolymerization of actin filaments in the presence of EPLIN, capping protein, and α-actinin. Actin was polymerized with capping protein, EPLIN, or α-actinin, and was then diluted 20-fold into polymerization buffer to initiate depolymerization. The final protein concentration in the samples were 0.1 μM actin, ± 0.15 μM capping protein, ± 10 or 100 nM GST-EPLIN-α, and ± 0.25 μM α-actinin. The actin polymer concentration was monitored over time using pyrene fluorescence. (B) Time course of actin filament elongation from barbed ends provided by spectrin-actin seeds. Samples in polymerization buffer contained 2 μM actin (5% pyrene labeled), 9 nM spectrin-actin seeds, and ± 100 nM EPLIN-α. In the presence of capping protein, actin polymerization was inhibited by <90% (not depicted).
EPLIN inhibits nucleation of actin polymerization by Arp2/3 complex
The ability of EPLIN-α to bind the sides of actin filaments and bundle them suggested that EPLIN might influence the ability of filaments to serve as secondary activators of actin filament nucleation by Arp2/3 complex. Arp2/3 complex activated by Scar-WA stimulates actin assembly after a short lag time (Fig. 6 A). This lag is attributed to the time required to form enough actin filaments to serve as secondary activators of branching nucleation (Higgs et al., 1999; Machesky et al., 1999; Pantaloni et al., 2000).
EPLIN-α had no effect on the time course of spontaneous polymerization of actin alone or with Scar-WA. However, it increased the initial lag when actin was polymerized with Arp2/3 complex and Scar-WA (Fig. 6 A). The lag depended on the concentration of EPLIN-α. A concentration of 35 nM GST-EPLIN-α nearly doubled the lag time (Fig. 6 B, filled square). The concentration of barbed ends created by Arp2/3 complex remained unaffected at up to 105 nM GST-EPLIN-α (Fig. 6 B, filled triangle). His-EPLIN-α fusion protein similarly increased the lag period in a concentration-dependent manner at nanomolar concentrations (unpublished data).
These observations suggest that binding of EPLIN-α to the sides of actin filaments might prevent secondary activation of nucleation, such that nucleation mediated by Arp2/3 complex is delayed until polymerization saturates the filament-binding capacity of EPLIN. Consistent with this interpretation, addition of preformed actin filaments decreased the lag time, and this effect was partially inhibited by EPLIN-α in a concentration-dependent manner (Fig. 6 C). In this set of experiments, profilin was included to suppress spontaneous nucleation (Machesky et al., 1999; Rohatgi et al., 1999), thereby increasing the lag time to accentuate the effect of added filaments. Together, these experiments suggest that actin filaments bound by EPLIN-α are poor secondary activators of nucleation mediated by Arp2/3 complex.
EPLIN reduces the formation of branched actin filaments
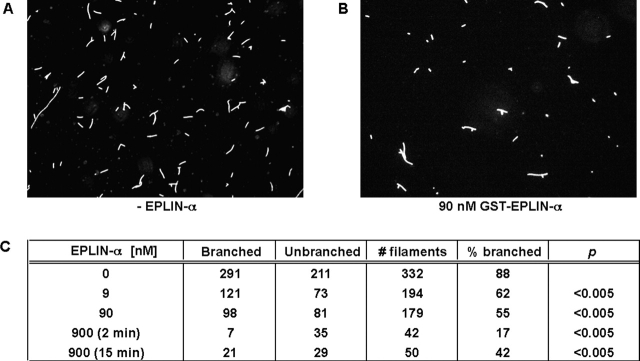
If EPLIN delays actin filament nucleation mediated by Arp2/3 complex by binding and sequestering actin filaments, the formation of branched filaments should be inhibited. By fluorescence microscopy, we measured branching of the products produced by Arp2/3 complex. After 2 min of polymerization in the presence of activated Arp2/3 complex, most (88%) of the products were short, branched filaments (Fig. 7). EPLIN-α reduced the fraction of branched filaments in a concentration-dependent fashion. Similar to the pyrene fluorescence assay, the inhibition occurred at nanomolar concentrations of EPLIN-α, reducing the fraction of branched filaments to 62% at 9 nM, 55% at 90 nM, and 17% at 900 nM. As seen in the kinetic studies, the inhibition of branch formation was transient; even with 900 nM EPLIN-α, the frequency of branches increased to 42% after 15 min of polymerization.
Figure 7.
Effect of EPLIN on the morphology of filaments formed by Arp2/3 complex. (A and B) Fluorescence micrographs of actin filaments nucleated by Arp2/3 complex in the absence (A) or presence (B) of EPLIN. Samples contained 4 μM G-actin (5% pyrene labeled), 30 nM Arp2/3 complex, 100 nM Scar-WA domain, and varying concentrations of GST-EPLIN-α (0–900 nM) in polymerization buffer. Polymerization was performed in 4 μM rhodamine-phalloidin, which stabilizes branched structure by inhibiting phosphate release from ADP-Pi-actin filaments and allows visualization of filaments by fluorescence microscopy. Polymerization was allowed to proceed for 2 min before being quenched by dilution. (C) Quantitation of branched and unbranched filaments. The chi-squared test was used to calculate the p-values.
Discussion
Our work provides evidence that EPLIN regulates actin-based structures in cells. Expression of EPLIN in MCF-7 cells increased the number and size of stress fibers. In addition, expression of EPLIN inhibited Rac1-induced membrane ruffling in HeLa cells. Biochemical studies revealed three important properties of EPLIN that provide clues about its functions. First, EPLIN-α contains more than one actin binding site and is capable of cross-linking and bundling actin filaments. Second, EPLIN-α stabilizes actin filaments in vitro. Third, EPLIN-α inhibits actin nucleation by Arp2/3 complex. These properties may favor bundles of actin filaments in cells relative to protrusion of the leading edge.
Both GST-pull-down and F-actin cosedimentation assays indicate that the NH2-terminal half and COOH-terminal half of EPLIN-α bind independently to actin monomers. The two halves share no recognizable repeats in the primary sequence, indicating that the two actin-binding domains are unrelated. Both actin-binding domains are also capable of binding filaments, although the filament binding is more efficient for the COOH-terminal half of EPLIN-α (unpublished data). Therefore, the two actin-binding domains of EPLIN-α may have different roles in the cell.
Low speed pelleting and electron microscopy showed that EPLIN-α bundles actin filaments. Cross-linking and bundling require a minimum of two actin-binding sites per cross-linking unit. His-EPLIN-α elutes in size-exclusion chromatography with Stokes radius of ∼61 Å (unpublished data). This unusually high Stokes radius corresponds to a globular particle with a maximum molecular mass of 781 kD; an asymmetric particle would have a lower molecular mass. Sedimentation equilibrium analytical ultracentrifugation of three concentrations of his-EPLIN-α gave an average molecular mass of 621 kD. This initial characterization indicates that his-EPLIN-α is a slightly asymmetric oligomer of ∼8 polypeptides. Additional work is required to determine how much of this aggregation is attributable to self-association of LIM domains or if the his-tag contributes. The presence of two independent F-actin binding sites per EPLIN molecule and the tendency of naive EPLIN to form an oligomer in solution could explain the observed cross-linking.
EPLIN-α also stabilizes actin filaments from depolymerization. We ruled out capping of barbed ends, because EPLIN-α does not affect the barbed end elongation or spontaneous polymerization. Filament-side binding proteins may stabilize filaments by altering their structure (Stokes and DeRosier, 1987) or inhibiting phosphate release from ADP-actin (Korn et al., 1987), or more speculatively, by forming bundles. We cannot rule out any of these possibilities, but favor the idea that EPLIN-α stabilizes filaments by binding to more than one actin subunit, similar to tropomyosin, which lowers the monomer dissociation rate constant at the pointed end (Broschat et al., 1989).
Also similar to tropomyosin (Blanchoin et al., 2001), EPLIN inhibits secondary activation of branching nucleation by Arp2/3 complex. This is revealed by the ability of EPLIN-α to increase the lag at the outset of polymerization in the presence of Arp2/3 complex and Scar-WA, without significantly affecting the rate of spontaneous polymerization or the number of barbed ends created by Arp2/3 complex. This inhibition occurs early in the reaction when the actin polymer concentration is below 200 nM and is evident at nanomolar concentrations of EPLIN. Under these conditions, EPLIN could inhibit secondary activation of Arp2/3 complex either by blocking binding sites on the sides of the initially formed filaments or by aggregating these filaments. At these early time points, EPLIN-α reduced the frequency of branches, but our fluorescence microscopy assay did not reveal large bundles. High concentrations of EPLIN-α also reduce the number of barbed ends, an indication that fewer Arp2/3 complexes have initiated polymerization.
In cells, branching nucleation is tightly regulated spatially, occurring primarily in the forefront of lamellipodia (Svitkina et al., 1997; Svitkina and Borisy, 1999). Rac regulates the formation of lamellipodia in mammalian cells by activating WAVE/Scar (Miki et al., 1998, 2000), presumably by displacing a complex of inhibitory proteins from WAVE/Scar (Eden et al., 2002). Localization of EPLIN to the membrane ruffles places it at the site of active remodeling of actin filaments. Furthermore, the finding that EPLIN can inhibit membrane ruffling induced by Rac1G12V directly supports our hypothesis that EPLIN inhibits remodeling of the actin cytoskeleton. The mechanism for this inhibition may rest on the inhibition of branching nucleation by Arp2/3, as shown by our biochemical analysis. Alternatively, EPLIN might sequester Rac1 or dissociate Rac1 from the plasma membrane, but EPLIN is not known to interact with Rac1.
The net effect of EPLIN on actin filaments, based on its known properties, is to inhibit actin turnover. Although any process that affects either actin assembly or actin disassembly alters filament turnover, EPLIN is capable of influencing both assembly (actin nucleation) and disassembly (filament stability). The mechanism of EPLIN appears to be surprisingly simple, and centers on the presence of two (perhaps more) actin-binding domains, which allows EPLIN to bind more than one subunit along the side of an actin filament. This provides the basis for stabilizing filaments and interfering with branching by Arp2/3 complex, similar to tropomyosin (Broschat et al., 1989; Blanchoin et al., 2001). Other multivalent actin-binding proteins, such as dystrophin (Rybakova et al., 1996), may influence actin dynamics in a similar way.
Cellular events leading to the loss of cytoarchitecture and enhanced motility in malignant cells have not been fully elucidated. Although these events are frequently attributed to aberrant activation of receptor tyrosine kinases or Rho GTPases, our study raises the possibility that actin filament–binding proteins may function to limit dynamic remodeling of actin filaments to regulate cell motility. The loss of expression or function of EPLIN in cancer cells would then remove this regulatory mechanism to enhance cell motility and/or invasion.
Materials and methods
EPLIN expression in cells
The coding region of EPLIN-α and -β was excised from the pCMV-FlagEPLIN plasmids (Maul and Chang, 1999) and cloned into the pTRE vector (CLONTECH Laboratories, Inc.). The pTRE/EPLIN plasmids and a puromycin resistance marker were transfected into the MCF-7 cells (Tet-Off™; CLONTECH Laboratories, Inc.), and puromycin-resistant clones were selected in the presence of 2 μg/ml tetracycline. Clones that expressed EPLIN in a tightly regulated manner (expression off in the presence of doxycycline) were expanded for analysis. For transient expression, HeLa cells were plated in six-well dishes at a density of 2 × 105 cells per well and cultured for 18 h. Two micrograms of plasmid DNA were transfected using 6 μl of Dosper (Roche) according to the manufacturer's instructions. In co-transfection experiments, pCMV-FlagEPLIN and Rac1G12V plasmids (Kuroda et al., 1996) were used at a ratio of 5.6:1. 40 h later, the cells were replated on fibronectin-coated glass coverslips for 3 h before fixing them in 3.7% formaldehyde in PBS. HA-epitope–tagged Rac1G12V was detected with monoclonal anti-HA antibody (1:300; Santa Cruz Biotechnology, Inc.), whereas endogenous EPLIN was detected with polyclonal anti-EPLIN sera (1:300; Maul and Chang). Actin filaments were stained with rhodamine-conjugated phalloidin (Molecular Probes Inc.).
Measurement for actin stress fibers
The quantitative changes in actin stress fibers in MCF-7 cells after the induction of EPLIN were estimated from the fluorescence intensities of rhodamine-phalloidin staining. Images were digitally captured at a fixed exposure time (248 ms for EPLIN-α, and 308 ms for EPLIN-β) and analyzed by using IQMac v1.2 software (Amersham Biosciences). Arbitrary units of fluorescence intensities per cell were 2,314,281 ± 1,268,755 (MCF-7/EPLIN-α ON; n = 20); 1,107,451 ± 536,852 (MCF-7/EPLIN-α OFF; n = 28); 3,659,482 ± 1,649,809 (MCF-7/EPLIN-β ON; n = 13); and 988,320 ± 321,784 (MCF-7/EPLIN-β OFF; n = 16).
Protein purification
The coding region of EPLIN-α was cloned into the pGEX-4T1 (Amersham Biosciences) and pQE30 (QIAGEN) vectors to generate GST-EPLIN-α and his-EPLIN-α, respectively. Transformed bacterial cells (BL21) were cultured in LB-amp at 37°C to an early exponential phase of growth (OD600 ≈ 0.5) before the addition of 1 mM IPTG. After an additional 3 h, bacterial cells were harvested and lysed by sonication in a PBS buffer containing 1% Triton X-100, 500 mM NaCl, and 25 mM EDTA. The cleared lysates were incubated with glutathione-Sepharose beads (Sigma-Aldrich). Bound GST-fusion protein was eluted from the beads with 10 mM free glutathione in 25 mM Tris, pH 8.0, for ∼1 h at RT. His-EPLIN-α was prepared by incubating cleared lysates with Ni-NTA beads (QIAGEN). The Ni-NTA beads were extensively washed in 40 mM imidazole and 300 mM NaCl in 50 mM Tris, pH 8.0, and the fusion protein was eluted with 100 mM imidazole in 50 mM Tris, pH 8.0. His-EPLIN-α in 100 mM imidazole, 50 mM Tris, pH 8.0, and 0.5 mM DTT eluted from a Superose 6 gel filtration column as a single, symmetrical peak with a Stokes radius of 61Å between catalase (RS 52.2 Å) and ferritin (RS 67 Å; de Haen, 1987). Sedimentation equilibrium analytical ultracentrifugation of his-EPLIN-α at three initial concentrations (A280 = 0.1, 0.2, and 0.4) in 100 mM imidazole, 50 mM Tris, pH 8.0 for 24 h at 8°C gave an average mol wt of 621. Production and purification of Scar1-WA domain (Scar-WA), Arp2/3 complex, and profilin-I were described previously (Kaiser et al., 1986; Higgs et al., 1999). Spectrin-actin seeds were prepared from human erythrocytes using the method of Lin and Lin (Lin and Lin, 1980). α-actinin was purchased from Cytoskeleton Inc.
Binding assays
Binding of EPLIN-α to monomeric actin was tested in a GST-pull-down assay. Two μg of immobilized GST fusion proteins were incubated for 1 h at 4°C with 50 nM G-actin in 100 mM KCl, 1 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 0.1% (vol/vol) Tween 20, 0.2 mM ATP, and 0.5 mg/ml BSA in 10 mM Hepes, pH 7.6. This actin concentration is below the critical concentration. The GST-VCA construct was provided by M. Kirschner (Harvard Medical School, Boston, MA. Beads were washed three times, bound actin was eluted with SDS sample buffer, run on an SDS-PAGE, and immunoblotted with anti-actin antibody (Santa Cruz Biotechnology, Inc.).
Actin filament pelleting assay
G-actin and recombinant EPLIN proteins were first precleared by centrifugation at 150,000 g for 30 min at 4°C. Actin was polymerized in 50 mM KCl, 2 mM MgCl2, 0.2 mM ATP, and 0.2 mM DTT in 2 mM Tris, pH 7.5, at 24°C for 1 h and then incubated with EPLIN for 30 min at 24°C. After centrifuging at 150,000 g for 30 min at 24°C, the supernatant and pellet were separated and analyzed by SDS-PAGE and Coomassie blue staining. The quantity of protein in supernatant and pellet fractions was measured by digitizing stained gels.
Actin cross-linking/bundling assay
His-EPLIN-α or α-actinin was mixed with 10 μM freshly prepared F-actin for 30 min at RT, and was then centrifuged at 8,000 g for 20 min at 24°C. The supernatant and pellet were separated and analyzed by SDS-PAGE and Coomassie blue staining. For electron microscopy study, 2 μM F-actin was incubated in the presence of 1 μM his-EPLIN-α or 1 μM BSA for 30 min at 24°C. The mixture was absorbed onto carbon-coated electron microscope grids and negatively stained with 0.2% uranyl acetate. Electron micrographs were obtained using a transmission electron microscope (model 10C; Carl Zeiss MicroImaging, Inc.) at a magnification of 45,000.
Actin polymerization assays
Purified G-actin (5% pyrene-labeled) in Buffer G (0.1 mM CaCl2, 0.2 mM ATP, 0.5 mM DTT, and 2 mM Tris, pH 8.0) was converted to Mg-ATP-actin by incubation in 0.1 mM MgCl2 and 1 mM EGTA for 2 min at 22°C. Aliquots of various proteins (Arp2/3 complex, Scar-WA, and EPLIN fusion proteins) in Buffer G (as above, but with 2 mM imidazole, pH 7.0) were added, and polymerization was initiated with 0.1 vol of 10× polymerization buffer (500 mM KCl, 10 mM MgCl2, 10 mM EGTA, and 100 mM imidazole, pH 7.0). The polymer concentration was monitored over time with a Spectrofluorimeter (excitation at 365 nm and emission at 407 nm; Photon Technology Instruments). To measure depolymerization, polymerized filaments were diluted to the critical concentration with 20 vol of polymerization buffer. For barbed-end elongation, 9 nM spectrin-actin seeds were added.
Fluorescence microscopy of branched actin filaments
Mixtures of actin, Arp2/3 complex, Scar-WA, and GST-EPLIN-α were polymerized in the presence of 4 μM rhodamine-phalloidin (Amann and Pollard, 2001). After incubation for 2 min at 24°C, aliquots were diluted 500-fold in polymerization buffer with 0.5% methylcellulose, 20 μg/ml catalase, 100 μg/ml glucose oxidase, and 3 mg/ml glucose. 1.6-μl samples were applied to coverslips (22 × 22 mm) coated with 20 μg/ml poly-l-lysine and viewed with a microscope (model IX-70; Olympus). Branches were verified as not being merely artifacts of overlain filaments by visualizing their Brownian vibration. Random fields were selected and the number of branched filaments was enumerated.
Acknowledgments
We thank B. Sjostrand for assistance in the electron microscopy work and D. Kaiser for sedimentation equilibrium analytical ultracentrifugation analysis.
This work was supported by Public Health Service grants CA90498 (to D.D. Chang) and GM26338 (to T.D. Pollard). R.S. Maul was supported by a Susan G. Komen Foundation postdoctoral fellowship (PDF 2000 448) and K.J. Amann by a National Institutes of Health postdoctoral fellowship.
Footnotes
Abbreviation used in this paper: EPLIN, epithelial protein lost in neoplasm.
References
- Amann, K.J., and T.D. Pollard. 2001. The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat. Cell Biol. 3:306–310. [DOI] [PubMed] [Google Scholar]
- Bao, L., M. Loda, P.A. Janmey, R. Stewart, B. Anand-Apte, and B.R. Zetter. 1996. Thymosin beta 15: a novel regulator of tumor cell motility upregulated in metastatic prostate cancer. Nat. Med. 2:1322–1328. [DOI] [PubMed] [Google Scholar]
- Ben-Ze'ev, A. 1997. Cytoskeletal and adhesion proteins as tumor suppressors. Curr. Opin. Cell Biol. 9:99–108. [DOI] [PubMed] [Google Scholar]
- Blanchoin, L., T.D. Pollard, and S.E. Hitchcock-DeGregori. 2001. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr. Biol. 11:1300–1304. [DOI] [PubMed] [Google Scholar]
- Braverman, R.H., H.L. Cooper, H.S. Lee, and G.L. Prasad. 1996. Anti-oncogenic effects of tropomyosin: isoform specificity and importance of protein coding sequences. Oncogene. 13:537–545. [PubMed] [Google Scholar]
- Broschat, K.O., A. Weber, and D.R. Burgess. 1989. Tropomyosin stabilizes the pointed end of actin filaments by slowing depolymerization. Biochemistry. 28:8501–8506. [DOI] [PubMed] [Google Scholar]
- Chakravatri, A., E.M. Zehr, A.L. Zietman, W.U. Shipley, W.B. Goggins, D.M. Finkelstein, R.H. Young, E.L. Chang, and C.L. Wu. 2000. Thymosin beta-15 predicts for distant failure in patients with clinically localized prostate cancer-results from a pilot study. Urology. 55:635–638. [DOI] [PubMed] [Google Scholar]
- Chang, D.D., N.H. Park, C.T. Denny, S.F. Nelson, and M. Pe. 1998. Characterization of transformation related genes in oral cancer cells. Oncogene. 16:1921–1930. [DOI] [PubMed] [Google Scholar]
- de Haen, C. 1987. Molecular weight standards for calibration of gel filtration and sodium dodecyl sulfate-polyacrylamide gel electrophoresis: ferritin and apoferritin. Anal. Biochem. 166:235–245. [DOI] [PubMed] [Google Scholar]
- Eden, S., R. Rohatgi, A.V. Podtelejnikov, M. Mann, and M.W. Kirschner. 2002. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 418:790–793. [DOI] [PubMed] [Google Scholar]
- Gluck, U., D.J. Kwiatkowski, and A. Ben-Ze'ev. 1993. Suppression of tumorigenicity in simian virus 40-transformed 3T3 cells transfected with alpha-actinin cDNA. Proc. Natl. Acad. Sci. USA. 90:383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs, H.N., and T.D. Pollard. 2000. Activation by Cdc42 and PIP(2) of Wiskott-Aldrich syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J. Cell Biol. 150:1311–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs, H.N., L. Blanchoin, and T.D. Pollard. 1999. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 38:15212–15222. [DOI] [PubMed] [Google Scholar]
- Ji, X., P. Zhang, R.N. Armstrong, and G.L. Gilliland. 1992. The three-dimensional structure of a glutathione S-transferase from the mu gene class. Structural analysis of the binary complex of isoenzyme 3-3 and glutathione at 2.2-A resolution. Biochemistry. 31:10169–10184. [DOI] [PubMed] [Google Scholar]
- Kaiser, D.A., M. Sato, R.F. Ebert, and T.D. Pollard. 1986. Purification and characterization of two isoforms of Acanthamoeba profilin. J. Cell Biol. 102:221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn, E.D., M.F. Carlier, and D. Pantaloni. 1987. Actin polymerization and ATP hydrolysis. Science. 238:638–644. [DOI] [PubMed] [Google Scholar]
- Kuroda, S., M. Fukata, K. Kobayashi, M. Nakafuku, N. Nomura, A. Iwamatsu, and K. Kaibuchi. 1996. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J. Biol. Chem. 271:23363–23367. [DOI] [PubMed] [Google Scholar]
- Lin, D.C., and S. Lin. 1980. A rapid assay for actin-associated high-affinity cytochalasin binding sites based on isoelectric precipitation of soluble protein. Anal. Biochem. 103:316–322. [DOI] [PubMed] [Google Scholar]
- Machesky, L.M., R.D. Mullins, H.N. Higgs, D.A. Kaiser, L. Blanchoin, R.C. May, M.E. Hall, and T.D. Pollard. 1999. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. USA. 96:3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul, R.S., and D.D. Chang. 1999. EPLIN, epithelial protein lost in neoplasm. Oncogene. 18:7838–7841. [DOI] [PubMed] [Google Scholar]
- Miki, H., S. Suetsugu, and T. Takenawa. 1998. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 17:6932–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, H., H. Yamaguchi, S. Suetsugu, and T. Takenawa. 2000. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature. 408:732–735. [DOI] [PubMed] [Google Scholar]
- Mullins, R.D., J.A. Heuser, and T.D. Pollard. 1998. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA. 95:6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni, D., R. Boujemaa, D. Didry, P. Gounon, and M.F. Carlier. 2000. The Arp2/3 complex branches filament barbed ends: functional antagonism with capping proteins. Nat. Cell Biol. 2:385–391. [DOI] [PubMed] [Google Scholar]
- Pawlak, G., and D.M. Helfman. 2001. Cytoskeletal changes in cell transformation and tumorigenesis. Curr. Opin. Genet. Dev. 11:41–47. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., L. Blanchoin, and R.D. Mullins. 2000. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545–576. [DOI] [PubMed] [Google Scholar]
- Rohatgi, R., L. Ma, H. Miki, M. Lopez, T. Kirchhausen, T. Takenawa, and M.W. Kirschner. 1999. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 97:221–231. [DOI] [PubMed] [Google Scholar]
- Rohatgi, R., H.Y. Ho, and M.W. Kirschner. 2000. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J. Cell Biol. 150:1299–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakova, I.N., K.J. Amann, and J.M. Ervasti. 1996. A new model for the interaction of dystrophin with F-actin. J. Cell Biol. 135:661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeichel, K.L., and M.C. Beckerle. 1997. Molecular dissection of a LIM domain. Mol. Biol. Cell. 8:219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, Y., R.S. Maul, C.S. Gerbin, and D.D. Chang. 2002. Inhibition of anchorage independent growth of transformed NIH3T3 cells by EPLIN is dependent on localization of EPLIN to the actin cytoskeleton. Mol. Biol Cell. 13:1408–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, D.L., and D.J. DeRosier. 1987. The variable twist of actin and its modulation by actin-binding proteins. J. Cell Biol. 104:1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina, T.M., and G.G. Borisy. 1999. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145:1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina, T.M., A.B. Verkhovsky, K.M. McQuade, and G.G. Borisy. 1997. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J. Cell Biol. 139:397–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor, A.D., S.M. Edgerton, S. Liu, D.H. Moore ii, and D.J. Kwiatkowski. 2001. Gelsolin as a negative prognostic factor and effector of motility in erbB-2-positive epidermal growth factor receptor-positive breast cancers. Clin. Cancer Res. 7:2415–2424. [PubMed] [Google Scholar]
- Welch, M.D., A. Iwamatsu, and T.J. Mitchison. 1997. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 385:265–269. [DOI] [PubMed] [Google Scholar]