Abstract
A gene density–related difference in the radial arrangement of chromosome territories (CTs) was previously described for human lymphocyte nuclei with gene-poor CT #18 located toward the nuclear periphery and gene-dense CT #19 in the nuclear interior (Croft, J.A., J.M. Bridger, S. Boyle, P. Perry, P. Teague, and W.A. Bickmore. 1999. J. Cell Biol. 145:1119–1131). Here, we analyzed the radial distribution of chromosome 18 and 19 chromatin in six normal cell types and in eight tumor cell lines, some of them with imbalances and rearrangements of the two chromosomes. Our findings demonstrate that a significant difference in the radial distribution of #18 and #19 chromatin is a common feature of higher order chromatin architecture in both normal and malignant cell types. However, in seven of eight tumor cell lines, the difference was less pronounced compared with normal cell nuclei due to a higher fraction of nuclei showing an inverted CT position, i.e., a CT #18 located more internally than a CT #19. This observation emphasizes a partial loss of radial chromatin order in tumor cell nuclei.
Keywords: nuclear architecture; tumor cells; nuclear shape; chromosome territories; radial chromatin arrangement
Introduction
Nuclei of a defined cell type are normally characterized by a distinct morphology and chromatin texture, which can change during malignant transformation. Changes of the chromatin density as well as the peripheral accumulation of heterochromatin have been observed since the early days of light microscopy, and have been used for the grading of malignancy (Keenan et al., 2000). However, these observations do not provide a detailed insight into the differences between the three-dimensional (3D) higher order chromatin architecture in tumor cells compared with their normal precursor cells.
For lymphocyte nuclei, it was shown that the positioning of chromatin relative to the nuclear periphery is correlated with gene density (Boyle et al., 2001). A striking example is the different nuclear location of chromosome territories (CTs) #19 and #18, which contain similar DNA amounts (77 and 60 Mbp, respectively), but differ profoundly in their gene content. CTs #19 consist mostly of gene-dense (28 genes/Mbp; http://www.ncbi.nlm.nih.gov/genome/guide/human) and early replicating chromatin, whereas CTs #18 consist mainly of gene-poor (8 genes/Mbp) and mid-replicating chromatin. 3D-FISH experiments demonstrated the positioning of CTs #19 toward the nuclear center, whereas CTs #18 were found close to the nuclear border (Croft et al., 1999; Cremer et al., 2001). A gene density–related radial dependence of the position of chromatin has been found to be conserved during genome evolution spanning a period of at least 300 million years (Tanabe et al., 2002a,b), irrespective of major chromosomal rearrangements that occurred in the phylogenetic lineages, which led to the present day mammals and birds. This conservation suggests a still-unknown functional relevance of a nonrandom higher order chromatin arrangement.
At present, it is unclear whether a gene density–related radial dependence of the position of distinct CTs is restricted to only a few cell types, or whether it represents a common feature of numerous cell types. There is also little knowledge about the extent to which the nonrandom arrangement of CTs in relation to gene density found in normal diploid cells is maintained or lost in nuclei of derivative tumor cells.
To answer these questions, we investigated the spatial distribution of CTs #18 and #19 in cell lines established from eight different tumors, and wherever available, in their normal diploid precursor cells by 3D-FISH, confocal optical serial sections, 3D reconstruction, and quantitative 3D evaluation. In addition, we analyzed the nuclear shape of all cell types to test whether this parameter has an influence on the distribution pattern of these CTs. Tumor cell lines included tumors with apparently normal karyotypes as well as tumors with highly rearranged karyotypes involving chromosome 18 and/or 19, with or without gains or losses of either of these chromosomes.
In all normal cell types, we found a significant difference in the radial distribution pattern for CTs #18 and #19. Chromosome 18 territories were located more peripherally compared with the territories of chromosome 19. This difference was maintained in all tumor cell lines, irrespective of the nuclear shape or the occurrence, for both structurally normal chromosomes #18 and #19 as well as rearranged #18 and #19 material, suggesting a basic stability of gene density–related radial chromatin arrangement in the malignant cell types studied. However, compared with nuclei of normal cells, in most tumor cell lines we found a higher fraction of nuclei with an inverted position of CT #18- and CT #19-specific material, i.e., a CT #18 located more interior than a CT #19, resulting in smaller average radial distances between these territories.
Results
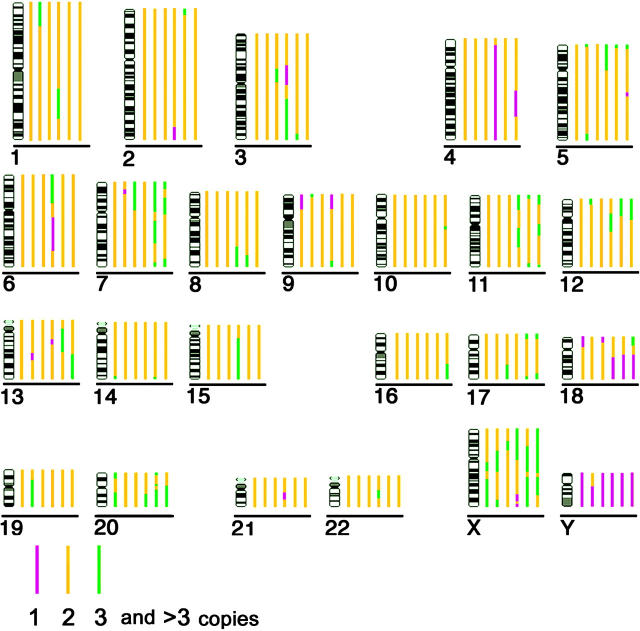
A short description of all normal cell types and tumor cell lines analyzed in this report, frequent structural aberrations, as well as copy number changes concerning chromosomes 18 and 19 are summarized in Table I. In cases where karyotype information on rearranged cell lines with heterogenic karyotype was based on data reported by other groups (Jurkat, HDLM-2, MelJuso, SW480, and SW620) we took care to perform our 3D-FISH analyses on nuclei from a cell passage that our providers had stored close to that used for karyotype analysis. Data on the copy number of chromosome segments were obtained by comparative genomic hybridization (CGH) analysis for each cell line with major chromosomal rearrangements, considering ploidy information from the karyotype (Fig. 1). The DNA, used for CGH analysis, was also obtained from a cell passage close to that used for 3D-FISH. Data for the assessment of the nuclear shape are listed in Table II.
Table I: Karyotype (K) and comparative genomic hybridization (CGH) analysis of chromosomes #18 and #19 in tumor cell nuclei.
| Description of cell lines and tissue origin | Structural aberrations andcopy number imbalances of chromosomes
|
References | |
|---|---|---|---|
| #18 | #19 | ||
| Jurkat | |||
| T cell–derived leukemic cell line; n = 46 |
K: 18/der(18)t(X;18) | K: 19/19 | Müller, S., personal communication |
| Stable karyotype | CGHa: monosomy 18p | balanced | |
| HDLM-2 | |||
| Hodgkin's disease–derived cell line; n < 46 |
K: 18/18 | K: 19/der(2)t(2;13;19;12)/ der(19)t(19;9;19;17) | Joos et al., 2003 |
| Heterogenic karyotype | CGH: balanced | CGH: partial trisomy 19 | |
| MelJuso | |||
| Melanoma-derived cell line; n = 40–44 |
K: 18/der(9;18)t(9;18) | K: 19/der(1;19)t(1;19)/der(4)t(4;19)b/der(19)t(19;22)b | Müller, S., personal communication |
| Heterogenic karyotype | CGH: monosomy 18p | CGH: balanced | |
| HeLa | |||
| Cervix carcinoma–derived cell line; n = 68–70 |
K: 18/del(18q)/der(14)t(14;18) | 19/der(19)t(13;19)/der(19)t(19;20) | Our own M-FISH data |
| Heterogenic karyotype | CGH: 18q under-represented | CGH: balanced | |
| RKO | |||
| Colon adenocarcinoma–derived cell line; n = 46 | K: 18/18 | K: 19/19 | Cahill et al., 1998 |
| Stable karyotype | CGH: not done | ||
| DLD1 | |||
| Colon adenocarcinoma–derived cell line; n = 46 | K: 18/18 | K: 19/19 | Abdel-Rahman et al., 2001 |
| Stable karyotype | CGH: not done | ||
| SW480 | |||
| Colon adenocarcinoma–derived cell line; n = 50–58 | K: 18/del(18q) | K: der(8;19)t(8;19)/der(19)t(19;8;19;5) | Müller, S., personal communication |
| Heterogenic karyotype | CGH: monosomy 18q | CGH: balanced | |
| SW620 | |||
| Colon adenocarcinoma–derived cell line (metastasis); n = 45–51 | K: 18/del(18q)/der(18)t(17;18) | K: 19/19 | Müller, S., personal communication |
| Heterogenic karyotype | CGH: monosomy 18q/partial trisomy 18p |
CGH: balanced | |
The CGH copy numbers were deduced from the karyotype data.
These two translocations, each containing 19p and each observed in ∼50% of the cells, are mutually exclusive.
Figure 1.
Copy numbers of chromosome segments estimated by CGH analysis, considering ploidy information of the karyotypes from each cell line. Each bar represents the chromosome copy number in a particular cell line; different copy numbers are represented by different colors according to the key shown. Cell lines from left (nearest to chromosome ideogram) to right are: (1) Jurkat; (2) HDLM-2; (3) MelJuso; (4) HeLa; (5) SW480; (6) SW620 (furthest from chromosome ideogram). Note that the HeLa cell line is near triploid. Accordingly, the loss of material (i.e., chromosomes 4 and 18q) indicates a disomic status (compare with Table I).
Table II. Measurement parameters for the evaluation of the nuclear shape in different cell types.
| Cell type
|
Average length (min/max)
|
Average width (min/max)
|
Ratio (length/width) |
Average height (z-diameter) (min/max)
|
Ratio (height/length) |
|---|---|---|---|---|---|
| μm | μm | μm | |||
| Stimulated lymphocytes | 11,3 (9,6/13,3) | 11,0 (9,6/12,5) | 1,0 | 13,3 (12,0/15,2) | 1,2 |
| Hemopoietic progenitor cells | 9,8 (7,8/13,1) | 5,8 (3,0/8,0) | 1,8 | 12,3 (7,6/14,0) | 1,3 |
| Keratinocytes | 19,0 (14,3/25,0) | 14,2 (10,7/18,9) | 1,3 | 9,4 (6,8/12,4) | 0,5 |
| Cervix epithelium | 15,4 (11,2/20,1) | 6,9 (4,4/8,5) | 2,3 | 10,7 (9,0/12,2) | 0,7 |
| Colon epithelium (mid/upper part) | 13,8 (11,3/16,8) | 5,6 (2,9/8,0) | 2,6 | 9,86 (8,2/12,2) | 0,7 |
| Colon epithelium (bottom part) | 10,6 (8,8/13,4) | 8,1 (6,6/10,5) | 1,3 | 9,3 (7,8/12,6) | 0,9 |
| Jurkat | 12,2 (10,4/14,3) | 10,5 (9,5/13,0) | 1,2 | 10,0 (7,8/12,2) | 0,8 |
| HDLM-2 | 11,4 (8.9/14.4) | 9,5 (6,4/12,0) | 1,2 | 9,7 (7,6/11,6) | 1,0 |
| MelJuso | 16,9 (14,1/22,8) | 13,5 (10,0/16,7) | 1,3 | 14,7 (11,2/17,6) | 0,9 |
| HeLa | 17,8 (15,1/21,6) | 14,3 (9,4/17,6) | 1,3 | 9,8 (8,4/12,4) | 0,6 |
| RKO | 15,1 (12,4/18,8) | 12,5 (9,1/17,0) | 1,2 | 11,6 (9,2/14,2) | 0,8 |
| DLD-1 | 19,6 (13,7/30,3) | 14,0 (10,0/17,3) | 1,4 | 9,9 (8,0/13,2) | 0,5 |
| SW480 | 10,8 (8,3/14,7) | 8,8 (6,6/10,6) | 1,2 | 9,8 (8,1/12,9) | 0,9 |
| SW620 | 15,2 (12,1/18,5) | 11,4 (8,7/14,2) | 1,4 | 9,6 (7,0/14,8) | 0,6 |
Fig. 2 shows the curves for the distribution of chromosome 18–specific (red) and chromosome 19–specific (green) chromatin and for the nuclear counterstain (blue). For each cell type, 11–38 nuclei were evaluated. In each graph, the normalized DNA content, represented by its voxel intensity–weighted fluorochromes, is plotted against the relative radius within a nucleus (for details see Materials and methods). Thus, these curves explore the positioning of these CTs with regard to their distance from the geometrical center of the nucleus. We refer to this correlation as “radial chromatin arrangement.” 3D reconstructions of selected nuclei are displayed in Fig. 3.
Figure 2.
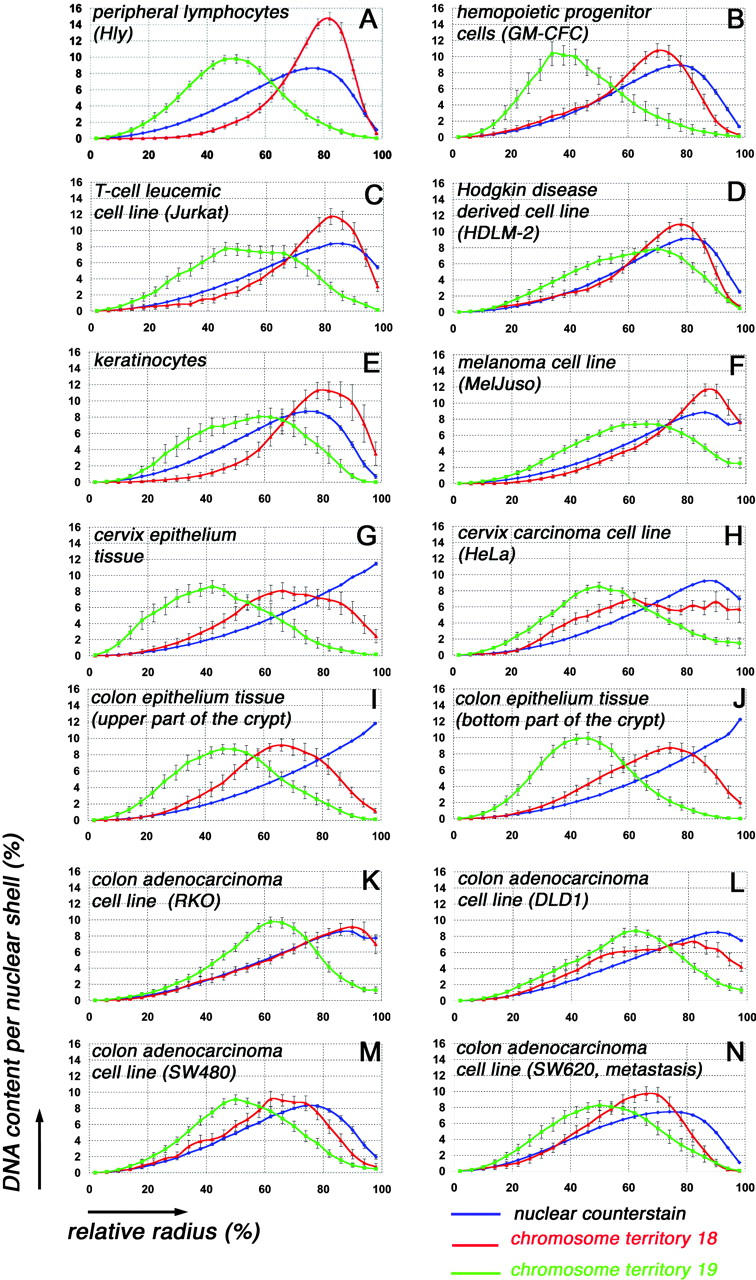
Quantitative 3D evaluation in the different cell types of radial CT #18 and CT #19 distribution in 25 concentric nuclear shells after painting with DNA probes specific for chromosome 18 (red) and chromosome 19 (green). Blue curves represent counterstained DNA. The abscissa denotes the relative radius r of the nuclear shells, the ordinate the normalized sum of intensities in the voxels for a respective fluorochrome assigned to a given shell. For normalization, the area underlying the curve for each color (total relative DNA content) was set to 100. Note the different curve for the DNA counterstain obtained in nuclei of cultivated cells (A–F, H, and K–N) and in nuclei obtained from tissue sections (G, I, and J). For explanation, see Materials and methods. All graphs show the different radial positioning of CT # 18 and CT #19 material. Bars indicate SEM.
Figure 3.
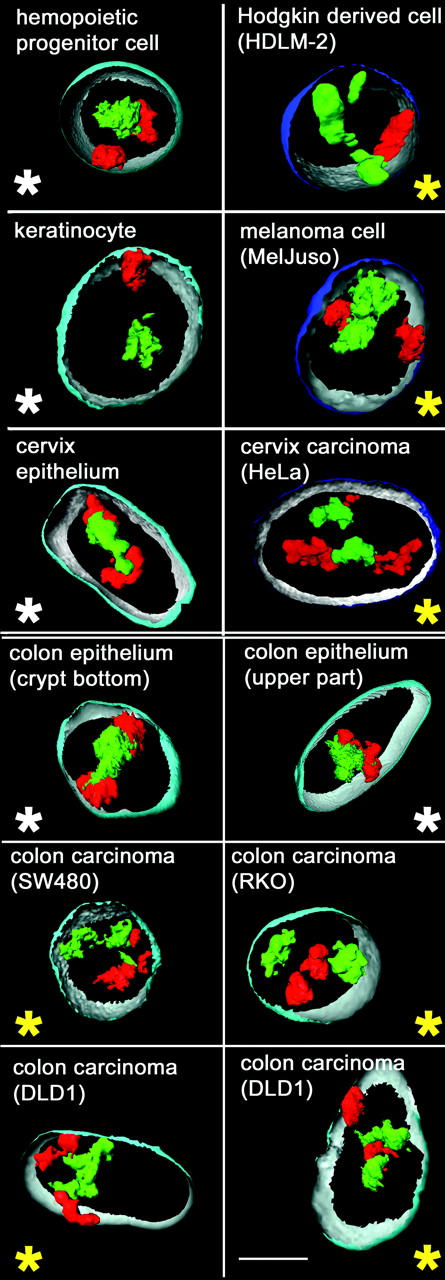
Visualization of CTs #18 (red) and CTs #19 (green) in 3D reconstructions of selected cell types. Part of the nuclear border is indicated by reconstruction of the counterstain periphery (outside, blue; inside, silver-gray). Note that in nuclei of normal cells (panels marked by a white asterisk), the CTs #19 are closely attached in the nuclear interior, whereas CTs #18 are found at the nuclear periphery, either side-by-side or at remote sites. In tumor cell nuclei (panels marked by a yellow asterisk), this radial distribution difference is often less apparent; note, for example, a peripheral CT #19 in a Hodgkin-derived cell nucleus (top right) or the internal position of a CT #18, which is located between two CTs #19 in one of the two DLD1 nuclei (bottom right panel) displayed. Bar, 5 μm.
Fig. 4 illustrates the shift of CT positions caused by a translocation event. Average relative radii (see Materials and methods) for all normal and malignant cell types are summarized in Fig. 5 and were used for a comparison of CT distribution between different cell types.
Figure 4.
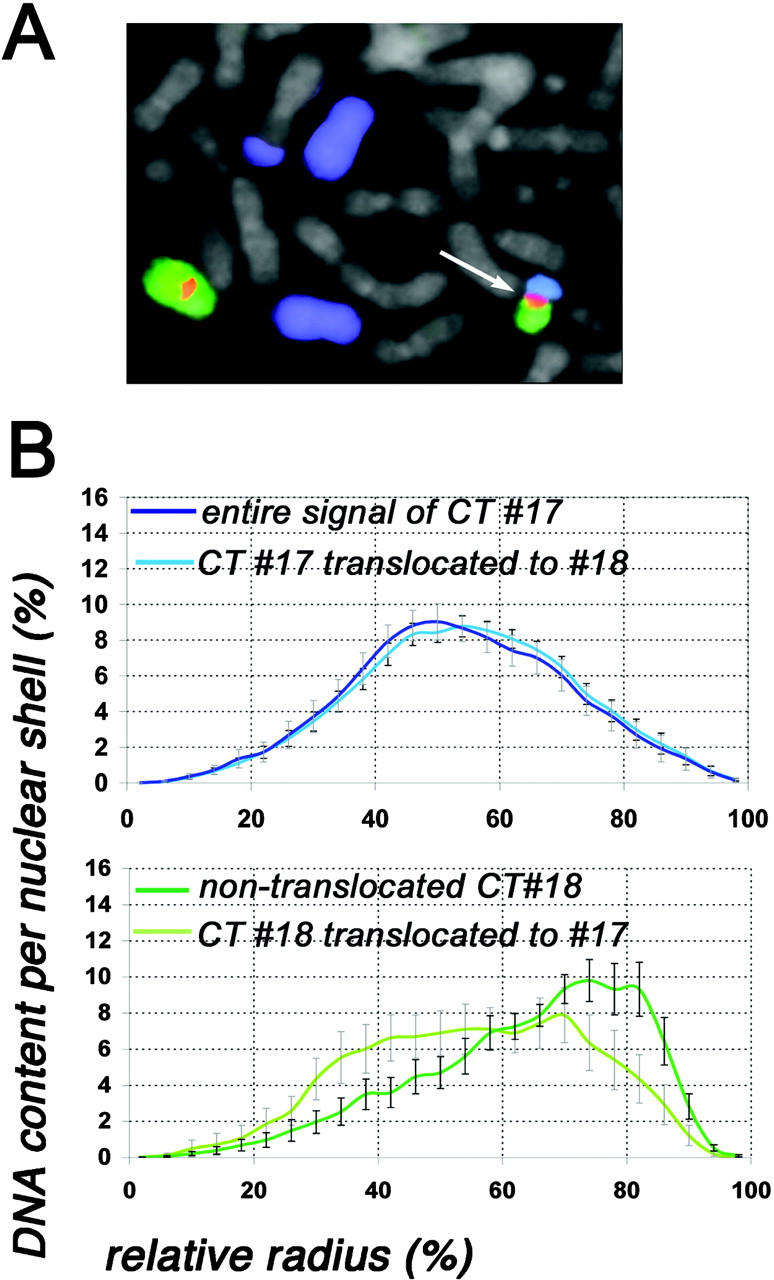
Evaluation of CT #17 and CT #18 distribution of the SW620 cell line carrying a translocation t(17;18). (A) Part of a metaphase spread after painting of chromosome 17 (visualized in blue) and chromosome 18 (visualized in green). The chromosome 18 centromere is visualized in red. In the metaphase shown, chromosome 17 is present as two free, normal copies and as two different translocation chromosomes, one of them forming the der(18)t(17;18) (arrow). The centromere of this rearranged chromosome is chromosome 18 specific. Chromosome 18 is also present as one normal copy. (B) Quantitative 3D evaluation of the radial CT #17 and CT #18 distribution in 22 nuclei. The abscissa denotes the relative radius r of the nuclear shells, the ordinate the normalized sum of intensities in the voxels for a respective fluorochrome assigned to a given shell. In the top panel, the dark blue curve denotes the radial distribution of the entire chromosome 17 material after painting with a DNA probe specific for chromosome 17, and the light blue curve denotes only chromosome 17 material translocated to chromosome 18. Note the almost identical curves for both fractions of CT #17 material. In the bottom panel, the dark green curve denotes the radial distribution of free CTs #18, and the light green curve denotes chromosome 18 material translocated to chromosome 17. Note the distinctly more peripheral positioning of the nontranslocated fraction of CTs #18 in comparison to the translocated fraction. Bars indicate SEM.
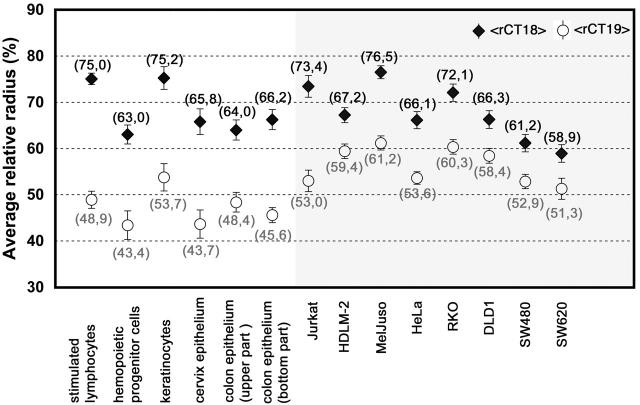
Figure 5.
Comparison of the average relative radii of the intensity-weighted fluorescence, represented by CT #18 (black rhombi) and CT #19 (white circles) material of all cell types analyzed. The left panel (white background) displays the data of normal cells, the right panel (gray background) the data of tumor cells. In all cell types, the average relative radius for CT #18 (<rCT18>) is larger compared with CT #19 (<rCT19>). The distances between <rCT18> and <rCT19> are smaller in the majority of tumor cell lines in comparison to normal cell nuclei (compare with Table III). Numbers in parentheses indicate the exact value of the average relative radii.
Hemopoietic cells and cell lines
Previous investigations have shown a peripheral localization of CTs #18 and an internal localization of CTs #19 in PHA-stimulated cycling lymphoblasts (Croft et al., 1999) and G0 lymphocytes from peripheral blood (Cremer et al., 2001), as well as in EBV-transformed lymphoblastoid B-cells (Croft et al., 1999; Boyle et al., 2001; Tanabe et al., 2002b). Here, we confirm the significant difference in the location of these territories (P = 0.0001 using the median quartile (MQ) test, and P = 0.001 using the two-sided Kolmogorov-Smirnov (KS) test; for details see Materials and methods) in 3D preserved nuclei from S-phase T-lymphocytes (Fig. 2 A), and also demonstrate the presence of a distinct distribution pattern in nuclei of hemopoietic progenitor cells (P = 0.01 [MQ] and P = 0.05 [KS]), as shown for the granulocyte-macrophage colony–forming cells (GM-CFC; Fig. 2 B). A 3D reconstruction of a typical nucleus from a hemopoietic progenitor cell is shown in Fig. 3. Both lymphocytes and GM-CFCs are derived from pluripotent hemopoietic stem cells. However, the GM-CFCs differentiate further into monocytes and a subpopulation of granulocytes (Gordon, 1993; Krause et al., 1996) and are thus not direct progenitor cells of lymphocytes. It would be interesting to test whether a difference in the distribution of CTs #18 and #19 is already present in the earliest pluripotent hemopoietic stem cells.
For an investigation of leukemic tumor cells, we chose two cell lines. First, we analyzed nuclei of the leukemic T cell–derived Jurkat cell line. The Jurkat cell line shows a stable karyotype with only few aberrations. One chromosome #18 is involved in a translocation t(X;18) (see Table I and Fig. 1). The radial positioning of both CTs #18 and #19 was again significantly different (P = 0.0001 [MQ] and P = 0.05 [KS]; Fig. 2 C), and the DNA maxima contents were observed at relative radii similar to those found in nuclei of normal diploid T-lymphocytes (compare Fig. 2 A with Fig. 2 C). However, the values of the DNA peaks of CT #18 and CT #19 are lower in these tumor cell nuclei compared with normal T lymphocytes, suggesting some decline of radial order.
Second, we analyzed the cell line HDLM-2, established from a patient with Hodgkin's disease. HDLM-2 cells have been characterized as Hodgkin and Reed-Sternberg analogous cells, which represent, in most instances, clonal populations of transformed germinal center B cells; in rare cases they are derived from T cells (Kuppers et al., 2002). In the HDLM-2 cell line, chromosome 19 is present in one normal copy, but is also involved in complex rearrangements with different chromosomes leading to the formation of huge marker chromosomes and resulting in a trisomic state of #19 material (Joos et al., 2003; compare Table I and Fig. 1). Rearrangements of #19 material involve gene-poor chromosomes such as #2, #13, and #9, which are known to have a distinctly peripheral localization in lymphocytes (Boyle et al., 2001; unpublished data). Compared with normal lymphocytes, the DNA maximum content of CT #19 material is shifted toward the nuclear periphery (compare Fig. 2 A with Fig. 2 D), but in comparison to CT #18, its more internal position is still maintained (Fig. 2 D). The peripheral localization of CTs #18, although present as two normal copies, is less pronounced compared with normal lymphocytes (compare Fig. 2 A with Fig. 2 D). This different distribution was significant for the MQ test (P = 0.01), but not significant for the more stringent KS test (P = 0.2). A 3D reconstruction of a typical nucleus is depicted in Fig. 3.
Epidermal cells: normal diploid keratinocytes and melanoma cells
CT #18 and #19 distribution was analyzed in two epidermal cell types: in normal diploid differentiated keratinocytes and in a melanoma cell line, MelJuso (Johnson et al., 1981). Keratinocytes and melanocytes form the stratum germinativum of the epidermis and both cell types are of ectodermal origin. However, the melanocytes emigrate during early embryogenesis from the neural crest into the epidermis. Although normal diploid melanocyte and premelanocyte cultures have been described previously (Bennett et al., 1985; Herlyn et al., 1988), they could not be obtained for the present work.
The quantitative 3D evaluation in differentiated keratinocytes revealed again a clearly distinct distribution pattern (Fig. 2 E) with an internal positioning of CTs #19 and a peripheral positioning of CTs #18, which were significantly different (P = 0.0001 [MQ] and P = 0.01 [KS]). A 3D reconstruction of a typical nucleus is shown in Fig. 3.
The MelJuso cell line carries numerous rearrangements involving both chromosome 18 and 19. The latter is involved in numerous translocations with different chromosomes (Table I). However, a balanced state for chromosome 19 material is maintained as shown by CGH analysis (Fig. 1). Irrespective of these complex translocation events, the distinct peripheral distribution of #18 material and the internal distribution of #19 material was maintained (Fig. 2 F) and shown to be significant, both for the MQ (P = 0.0001) and the KS test (P = 0.05). A 3D reconstruction of a typical nucleus is shown in Fig. 3.
Cervix epithelial cells
The distribution of CTs #18 and #19 in cervix epithelium was analyzed in the columnar epithelial cells from a tissue section of normal endocervix and in the (hypo) triploid HeLa cell line derived from a cervix carcinoma. As illustrated in Fig. 2 G, in nuclei of normal cervix tissue, CTs #19 showed an internal positioning and CTs #18 a peripheral positioning with a significant distribution difference (P = 0.01) using the MQ test and a nonsignificant difference for the KS test (P = 0.1). In HeLa cells, CT #19 material is involved in two translocations (Table I); however, its distribution pattern is largely maintained (Fig. 2 H). In comparison to CT #19, the rearranged CT #18 material was found to be more peripheral, although without a marked DNA maximum peak. The distribution difference between CT #18 and CT #19 material was again significant (P = 0.001) for the MQ test but not for the KS test (P = 0.1). Examples of 3D reconstructed nuclei from a normal cervix epithelium cell and from a HeLa cell are shown in Fig. 3.
Colon epithelial cells and colon adenocarcinoma cell lines
CT #18 and #19 distribution in colon epithelial cells was evaluated separately in cells from the bottom part of the crypt containing mitotically active stem cells, and in cells from the mid to the upper lumenal part containing terminally differentiated, mitotically inactive cells, demonstrated by their negative reaction with the cell cycle–associated nuclear protein Ki67 (unpublished data). Stem cells undergo 4–6 rounds of cell division generating a cell progeny, which migrates toward the mucosal surface where the mitotic activity stops at around half way toward the gut lumen (Bach et al., 2000).
The radial distribution pattern for CTs #18 and CTs #19 is displayed in Fig. 2 I for nuclei of the mid/upper part and in Fig. 2 J for nuclei from the bottom part of the crypt. Both graphs show a distinctly peripheral positioning of CTs #18 and an internal localization of CTs #19, which differs significantly for both tests in the near-spherical nuclei of the bottom part (P = 0.0001 for MQ and P = 0.05 for the KS test). In the strongly elongated nuclei (Table II) originating from the mitotically inactive upper part of the colon crypt, the difference was less pronounced (P = 0.01 for the MQ test and P = 0.1 for the KS test, respectively). A 3D reconstruction for both cell types is presented in Fig. 3.
Our analysis of colon carcinoma cell nuclei included four cell lines. RKO and DLD1 cell lines have stable, near diploid karyotypes with two normal copies for both chromosomes 18 and 19 (Cahill et al., 1998; Abdel-Rahman et al., 2001). In both cell lines, CT #19 holds an interior position compared with CTs #18 (Fig. 2, K and L), with distribution differences that are significant for the MQ test (P = 0.001). In the RKO cell line, CT #18 fits a random distribution as shown by the fairly identical curve with the nuclear counterstain (Fig. 2 K), suggesting that, compared with nuclei from normal colon epithelium, the arrangement of the gene-dense CT #19 material in the nuclear interior of RKO cells was maintained more stringently than that of CT #18 material. The SW480 and its metastasis-derived cell line SW620 show complex (but not identical) chromosomal rearrangements and a karyotype heterogeneity (Abdel-Rahman et al., 2001). Yet both cell lines show similar graphs for their CT #18 and #19 distribution, with an internal position for CT #19 and a more peripheral position for CT #18 material (Fig. 2, M and N). Again, the distribution differences were significant only for the MQ test (P = 0.01). 3D reconstructions for an SW480, an RKO, and two DLD1 nuclei are shown in Fig. 3.
In the passages used for our investigation, chromosome 19 is involved in complex translocations in SW480, but not in SW620, where it retains two nontranslocated copies. Irrespective of these differences, the radial distribution patterns for CT #19 material yielded almost identical graphs in both cell lines. Chromosome 18 is present as two nontranslocated copies (18/del(18q); Table I) in SW480. However, in SW620, a der(18)t(17;18) was observed in ∼20% of metaphase spreads. Three-color FISH on metaphase chromosomes of this cell line, using painting probes specific for chromosomes 17 and 18 and an alphoid DNA probe specific for the pericentromeric region of chromosome 18, confirmed that this translocation chromosome has a chromosome 18–specific centromere (Fig. 4 A). To further test whether this translocation influences the position of CT #18 material in this cell line, we separately analyzed the radial distribution pattern of the nontranslocated CTs #18 and the translocated CT #18 material in a set of 22 nuclei, where the translocation chromosome was identified by a close association and broad attachment of a small painted CT #17 and #18 (for details of evaluation, see Materials and methods). The gene-dense CT #17 material (21 genes/Mb), represented in addition to the der(18)t(17;18) by two normal copies and another translocation chromosome (Fig. 4 A), holds an internal position similar to that of CTs #19 with an average relative radius of 54.7% (Fig. 4 B, top graph, dark blue curve). A comparison of the distribution patterns of the entire CT #17 material and the CT #17 segment forming the der(18)t(17;18) chromosome showed that the translocated CT #17 material maintains a position almost identical to that found for the entire CT #17 material. The separate analysis of the radial distribution of the nontranslocated and the translocated portion of CT #18 material, on the other hand, revealed a distinct difference. The free CTs #18 are more peripherally located (Fig. 4 B, bottom graph, dark green curve), than the translocated CT #18 material (Fig. 4 B, bottom graph, light green curve).
Does nuclear shape predict the relative positions of CTs #18 and #19?
To test whether shifts in the relative radial distribution pattern of CTs #18 and #19 are correlated with differences of the nuclear shape, we measured the length, width, and height in all nuclei evaluated for each cell type. The average size and shape of nuclei were defined by the ratios of length/width and of height/length (Table II). The comparison between cell types with different nuclear shapes did not suggest such a correlation. We found very similar distribution patterns of CTs #18 and #19 in nuclei with a widely diverging morphology like in the relatively small spherical nuclei from lymphocytes with a height/length ratio of ∼1 and in the much larger ellipsoid nuclei of keratinocytes with a height/length ratio of ∼0.5. On the other hand, nuclei from keratinocytes and from DLD1 cells that have an almost identical shape showed considerable deviations in their relative arrangement of CTs #18 and #19 (compare Table II with Fig. 2, E and M, and Fig. 4).
Comparison of CT18 and CT19 distribution between different cell types
For a comprehensive comparison of the radial CT #18 and CT #19 arrangements in different cell types, we calculated from all nuclei within each cell type the average relative radii of CT #18 (<rCT18>) and CT #19 (<rCT19>) material. The data are shown in Fig. 5, and confirm the internal position for CT #19 material in comparison to CT #18 in all normal and malignant cell types included in this work. The difference between <rCT18> and <rCT19> was greater than 15% of the normalized relative nuclear radius in all normal cell types, ranging from 15.6 to 26.1% (mean value = 20.9%). For tumor cell lines, these values ranged from 7.6 to 20.4% of the normalized relative nuclear radius (mean value = 11.4%). The difference between nuclei from normal and malignant cell types was significant (P = 0.02). In addition, we determined in each cell type the fraction of individual nuclei where the average relative radius of CT #18 material (rCT18) was smaller than that of CT #19 material (rCT19). Values of rCT18 < rCT19 would indicate that at least one CT #18 within an individual nucleus was localized more internally compared with CT #19 material. As shown in Table III (right column), from the six normal cell types analyzed, such an inverse constellation was found in 9.1% of nuclei from the cervix epithelium and in 11.8% of nuclei from the mid/upper part of the colonic crypt, but not in the other cell types. In contrast, in all tumor cell lines investigated, nuclei with an “inverted” position of CTs #18 and #19 were found, with fractions ranging from 8.7% in Jurkat cell nuclei to 31% in nuclei of the HDLM-2 cell line (Table III). This difference between normal and malignant cell types indicates a higher variation and a decline of the nonrandom radial order of gene-dense and gene-poor CT arrangement in tumor cell nuclei compared to normal cell nuclei.
Table III.
Distances between the average relative radii of CT #18 and CT #19 (<rCT18> − <rCT19>) and fraction of individual nuclei with rCT18< rCT19 in normal and tumor cell types
| Cell typea | Distance <rCT18> − <rCT19>b |
Fraction of nuclei with rCT18 > rCT19 c |
|---|---|---|
| Stimulated peripheral lymphocytes | 26,1 | 0/19 (0%) |
| Cervix epithelium tissue | 22,1 | 1/11 (9,1%) |
| Normal diploid keratinocytes | 21,5 | 0/22 (0%) |
| Colon epithelium tissue (bottom part of crypt) | 20,6 | 0/19 (0%) |
| T cell leukemic cell line (Jurkat) | 20,4 | 2/23 (8,7%) |
| Hemopoietic progenitor cells | 19,6 | 0/14 (0%) |
| Colon epithelium tissue (mid/upper part of crypt) | 15,6 | 2/17 (11,8%) |
| Melanoma-derived cell line (MelJuso) | 15,3 | 3/32 (9,4%) |
| Cervix carcinoma (HeLa) | 12,5 | 7/38 (18,4%) |
| Colon carcinoma (RKO) | 11,8 | 7/29 (24,1%) |
| Colon carcinoma (SW480) | 8,3 | 7/31 (22,6%) |
| Colon carcinoma (DLD1) | 7,9 | 7/26 (26,9%) |
| Hodgkin-derived cell line (HDLM-2) | 7,8 | 9/29 (31,0%) |
| Colon carcinoma metastasis (SW620) | 7,6 | 7/26 (26,9%) |
Cell types are listed according to the decreasing distances of <rCT18> and <rCT19>, tumor cells are listed in italics.
Values indicate the distance between <rCT18> and <rCT19> within a given cell type.
Values indicate the number and percentage of nuclei within a cell type where rCT18 is smaller compared to rCT19.
Discussion
A nonrandom, internal nuclear position of the gene-rich CTs #19 and a peripheral position of the gene-poor CTs #18 was previously reported for human lymphocytes (Croft et al., 1999; Cremer et al., 2001). Here, we demonstrate that this difference in the radial arrangement of chromosome #18 and #19 material is a common feature, both in nuclei of normal cell types, originating from different embryonic discs and in nuclei of malignant cell lines.
In the normal cell types analyzed in this report, the average relative radii of CTs #18 exceeded those of CTs #19 at least by 15%; in most cell types by >20%. The distinct radial differences observed in hemopoietic progenitor cells indicate that a nonrandom radial arrangement is established early and is maintained during the differentiation of pluripotent blood stem cells. This observation is in agreement with Skalnikova et al. (2000), who found similar radial arrangements of different genetic loci at various stages of differentiation.
In the tumor cell lines analyzed here, the different distribution pattern for CTs #18 and #19 was maintained, irrespective of major karyotype changes, including gains and losses of chromosome 18 and 19 material and of chromosomal rearrangements involving these chromosomes. The maintenance of a radial arrangement of different genetic loci in normal and leukemic blood cells was also reported by Kozubek et al. (2002). However, with the exception of the Jurkat cell line, we found a decline in the gene density–related nuclear order of CTs in tumor cell lines compared with normal cells. This decline is reflected by the smaller differences between the average radial positioning of chromosome #18 and #19 material in tumor cell lines, and correlates with a considerably higher fraction of nuclei in most tumor cell lines with an inverted pattern of CT #18 and #19 arrangements, i.e., a CT #18 or translocated segments thereof was located more internally than #19 chromatin. In nuclei of normal cells, such an inverted pattern was observed much less frequently
Among potential factors that may contribute to the radial shifts of the CTs #18 and #19 arrangement observed in malignant cell nuclei in comparison to their normal progenitor cells, translocation events could play an essential role. Our result of a fairly similar location of CTs#18 in normal lymphocyte nuclei and in nuclei of the Jurkat cell line, showing an X/18 translocation, is consistent with the observation that CTs #18 and X, which belongs to the gene-poor chromosomes, have a similar peripheral distribution in lymphocyte nuclei (Weierich et al., 2003). The joining of a chromosome segment, holding an internal position within a normal diploid cell nucleus with a peripherally located segment of another chromosome, could result in a significant shift of the translocation chromosome either toward the nuclear interior or to the periphery. In the Hodgkin-derived cell line HDLM-2, the shift of CT #19 material toward the nuclear periphery compared with normal lymphocyte and hemopoietic stem cell nuclei may reflect multiple translocation events of chromosome 19 segments with other chromosomes, such as chromosomes 2 and 9, known to occupy a peripheral position in lymphocytes (Boyle et al., 2001). Gerlich et al. (2003) have recently put forward the hypothesis that global inheritance of CT position over different cell cycles depends on a chromosome-specific timing of centromere splitting and sister chromatid separation. As a consequence, the origin of the centromere should be a decisive parameter predicting the radial nuclear position of a translocation chromosome. We have tested this prediction in nuclei of the SW620 cell line carrying a der(18)t(17;18) marker chromosome with a centromere of proven #18 origin. Contrary to the prediction, the translocated CT #18 segment showed a distinct shift toward the nuclear interior in comparison to the free CTs #18, whereas the radial position of the gene-dense translocated CT #17 segment remained unchanged in comparison to the free CTs #17. It has previously been shown by Croft et al. (1999) in a two-dimensional analysis of lymphocyte nuclei carrying a reciprocal translocation t(18;19), that the translocated segment of chromosome 19 was more central than the translocated chromosome 18 segment. The latter tended to be slightly less peripheral than the normal chromosome 18. Our previous observations that chromosomal segments homologous to the human #19 are distributed in several large chromosomes of the Gibbon species Hylobates lar and positioned to the interior of Gibbon lymphoblastoid cell nuclei (Jauch et al., 1992; Tanabe et al., 2002b) support the hypothesis that the chromatin composition of a given chromosome segment, by itself, is important for its nuclear location.
Translocations are obviously neither the only determining factor for shifts of CT positions, nor do they necessarily lead to changes in the radial arrangements of the chromosomes involved. For example, a comparison of CT #19 positions in the colon carcinoma lines SW480 and SW620 shows nearly identical internal arrangements of CTs #19, although chromosome 19 retained two free copies in SW620, but is involved in complex rearrangements with chromosomes 5 and 8 in SW480. The latter are known to occupy a peripheral position in lymphocytes (Boyle et al., 2001). Nuclei from the colon carcinoma cell lines RKO and DLD1 have nearly normal diploid karyotypes. Although their #18 CTs are more peripherally located than their #19 CTs, both CTs show changes in their radial nuclear location compared with normal colon epithelial cells (Fig. 2 and Fig. 5), suggesting that tumor cell nuclei may exhibit changes of their higher order chromatin arrangements even in the absence of karyotype changes.
The conservation of a nonrandom side-by-side CT positioning in normal cells and cancer cells was recently described by Parada et al. (2002), who found CTs #12, 14, and 15 clustered in nuclei of normal mouse splenocytes and also in a mouse lymphoma cell line where these three chromosomes are involved in two translocation chromosomes. Both translocation chromosomes were found in close spatial association in nuclei of this cell line, suggesting a conservation of CT arrangements between normal and derivative tumor cells.
Our findings of a different positioning of CTs #18 and #19 suggest a gene density–related radial arrangement of chromatin in nuclei with a widely diverging morphology, ranging from rather spherical nuclei, as in lymphocytes, the Hodgkin-derived tumor cells HDLM-2, colon carcinoma cells SW480, and melanoma cells MelJuso, to ellipsoidal nuclei as in keratinocytes and the colon carcinoma cells DLD1.
However, it should be kept in mind that the different positions of CTs #18 and #19 do not apply to all human cell types. In nuclei of human diploid fibroblasts and cultured amniotic fluid cells, the territories of the small chromosomes #17, #18, #19, #20, and Y are found in the nuclear center, irrespective of their different gene density, suggesting a chromosome size-correlated, rather than a gene density–correlated radial chromosome arrangement (Bridger et al., 2000; Cremer et al., 2001). Cultured amniotic fluid cells are of a different origin to fibroblasts because they are derived from the fetal urogenital epithelium (Ochs et al., 1983). Although the average z-diameter of nuclei from all cell types included in the present report exceeds 9 μm, the ellipsoidal nuclei of human fibroblasts and amniotic fluid cells are much flatter, with z-diameters of only 3–5 μm (unpublished data). Whether the marked differences of radial CT arrangements noted between human fibroblasts or amniotic fluid cells and nuclei of the other cell types studied so far is causally connected with the distinct differences in nuclear shape of these cell types is not clear. In attempting a possible explanation, one should keep in mind a common feature of nuclear architecture observed in all primary, immortal, as well as transformed mammalian cells: mid-replicating, gene-poor chromatin domains form a typical shell both at the nuclear periphery and around the nucleoli (Dimitrova and Berezney, 2002). This distinct distribution is possibly triggered by HP1, a protein that is recruited at specific methylated transcriptionally inactive (hetero)chromatic sites, and which interacts with the lamin B receptor, an integral component of the inner nuclear membrane (for review see Lachner and Jenuwein, 2002). During mitosis, the mass centers of small chromosomes such as #18 and #19 are located close to the central spindle axis, irrespective of their gene density (Habermann et al., 2001). When cell nuclei expand in late telophase/early G1, the bulk of the gene-poor transcriptionally inactive chromatin of chromosome 18 should be broadly fixed at the nuclear lamina, and therefore pulled toward the nuclear periphery. For CTs #19, we would expect that their predominant mass of early-replicating, gene-dense and transcriptionally active chromatin is left in the nuclear interior, and only some of its relatively small amount of gene-poor, mid-replicating chromatin may assemble around the nucleoli or is also moved to the nuclear periphery. The fact that the formation of fibroblast nuclei is accompanied only by a small nuclear extension along the z-axis may then explain that both #19 and #18 CTs retain a central nuclear location in fibroblast nuclei, still allowing for the possibility that the CTs #18 are more broadly attached to the nuclear envelope compared with CTs #19. This is in contrast to the formation of nuclei with much larger z-diameters observed in the cell types analyzed here. It will be interesting to analyze the shape of sarcoma cell nuclei and answer the question whether such nuclei show a size-related or gene density–related pattern of radial CT arrangements. Such analyses could provide further information on whether the transformation of fibroblasts into sarcoma cells shows a switch from a size-related to a gene density–related CT arrangement possibly correlated with a switch in nuclear shape.
In conclusion, this report presents evidence for a radial higher order chromatin arrangement in a variety of different normal and malignant cell types, but suggests a decline in the gene density–correlated radial nuclear order of CTs in several tumor cell lines compared with their normal progenitor cells. The mechanisms leading to such a decline and to changes in heterochromatin patterns, such as increased amounts of facultative heterochromatin in the nuclear periphery (Dufer et al., 2000), are not known. It is still an open question to which extent these changes are a consequence of or a cause for an altered gene expression pattern associated with changes of chromatin methylation and histone acetylation. Although possible functional implications of changes of the higher order chromatin architecture in malignant cells have not been proven so far, a reliable description of this architecture in normal and malignant cell types provides the necessary basis to explore the potential impact of higher order chromatin organization for epigenetic mechanisms that control gene expression and gene silencing.
Materials and methods
Cell materials
PHA-stimulated lymphocytes from fresh peripheral blood were obtained from a healthy donor and grown for 60–70 h in RPMI 1640 medium with 10% FCS.
Circulating hemopoietic stem and progenitor cells were obtained from cryopreserved low density mononuclear cells separated by Ficoll-Hypaque density gradient. Cryopreserved mononuclear cells were plated at a concentration of 1–4 × 105/ml for cultivation. Clonal cell cultures were obtained in 1 ml Iscove's modified Dulbecco's medium (Sigma-Aldrich) containing 1% methylcellulose (Fluka), 30% FCS (Sebak), 1% BSA (A2153; Sigma-Aldrich), 10 μg/ml iron-saturated human transferrin (Boehringer Mannheim), 10−4 M α-thioglycerol (Sigma-Aldrich), and recombinant human hemopoietic growth factors as follows: 30 ng/ml stem cell factor (IC chemicals), 20 ng/ml Interleukin-3 (Genzyme), and 20 ng/ml granulocyte/macrophage colony–stimulating factor (Behringwerke AG). Colonies containing >50 cells formed by GM-CFCs were identified microscopically.
The human T cell line Jurkat (American Type Culture Collection [ATCC] no. TIB-152), was derived from a patient with an acute lymphoblastic T cell leukemia (Schneider et al., 1977). The passage studied here was provided by S. Müller from our department. The human Hodgkin's lymphoma derived cell line HDLM-2 (DSMZ no. ACC 17) was provided by S. Joos (German Cancer Research Center, Heidelberg, Germany). A HeLa cell line expressing histone H2B-GFP (Kanda et al., 1998) originally derived from a cervix carcinoma, was provided by K. Sullivan (The Scripps Research Institute, La Jolla, CA). These cell lines were grown in RPMI 1640 medium with 10% FCS at 37° and 5% CO2. Cell lines derived from human primary colorectal adenocarcinomas included DLD1, RKO (ATCC no. 2577; kindly provided by C. Lengauer, John Hopkins Hospital, Baltimore, MD), SW480 (ATCC no. CCL 228) and SW620 (ATCC no. CCL 227). SW480 was established from the primary tumor and SW620 from a metastasis from the same patient (Leibovitz et al., 1976; provided by S. Müller). A cell line derived from a malignant melanoma, MelJuso (Johnson et al., 1981), was provided by S. Müller. The latter cell lines were grown in complete DME containing 10% FCS at 37° and 5% CO2.
For the identification of S phase, cells were pulse-labeled with BrdU (5 μM final concentration) 1 h before fixation. Only BrdU-positive nuclei indicating their S phase stage were selected for further evaluation. Fixed diploid epidermal keratinocytes with different grades of differentiation grown on a fibroblast feeder layer were provided by P. Boukamp (Deutsches Krebsforschungszentrum, Heidelberg, Germany).
Samples of paraffin-embedded sections from normal colon and cervix tissue with an average thickness of 15 μm were prepared from surgical resections. Normal histology was confirmed by HE-staining.
Slide preparation of cultured cells and tissue samples
For the preparation of 3D preserved cell nuclei, cells growing in suspension (lymphocytes, single GM-CFC colonies, Jurkat, and HDLM-2 cell lines) were seeded onto polylysine-coated slides and fixed in 4% PFA/0.3× PBS. Adherently growing cells (HeLa, MelJuso, keratinocytes, and all colon cancer cell lines) were fixed in 4% PFA/1× PBS. Permeabilization steps for all cell cultures included treatment in 0.5% Triton X-100 in PBS, 20% glycerol in PBS, repeated freezing/thawing in liquid nitrogen, incubation in 0.1 M HCl, and pepsinization (2 mg/ml pepsin in 0.01 N HCl at 37°C for 7–10 min). Slides were stored at 4°C in 50% formamide/2× SSC (pH = 7.0) until use. Care was taken to avoid any drying of cells. For a detailed description, see Solovei et al. (2002).
Tissue sections were deparaffinized by two successive 30-min incubations in Xylole and rehydrated in a descending series of ethanol. Sections were then incubated for 30 min in 1 M NaSCN (80°C) and subsequently treated with 8 mg/ml pepsin in 0.01 N HCl for 25 min at 37°C to permeabilize the tissue. To reduce autofluorescence, sections were incubated in 100 mM CuSO4 for 60 min at 37°C (Schnell et al., 1999) and in 0.2% NaBH4 for 10 min at RT (Clancy and Cauller, 1998). The sections were then dehydrated in ethanol (70 and 100%) and air dried.
DNA probe preparation and detection
Whole-chromosome paint probes from sorted chromosomes and the L1.84 probe, an alphoid DNA probe specific for the pericentromeric heterochromatin of chromosome 18 were amplified and hapten-labeled by DOP-PCR as described previously (Schermelleh et al., 1999). Two- or three-color FISH on metaphase chromosomes and on morphologically preserved nuclei (3D-FISH), detection of labeled probes and of incorporated BrdU by fluorochrome-conjugated antibodies or fluorochrome-conjugated avidin was performed according to protocols described in detail elsewhere (Cremer et al., 2001; Solovei et al., 2002). Nuclei were counterstained for 5 min with 1 μM ToPro-3 (Molecular Probes, Inc.).
Tissue sections were incubated before hybridization with the denatured hybridization mixture overnight at 37°C to allow complete tissue penetration of the probe, and were then denatured for 20 min at 80°C on a hot block. Hybridization of the tissue was performed for another 5 d at 37°C. Detection of DNA probes was done in the same way as for cell cultures; counterstain was performed with 10 μM Syto 16.
CGH analysis
CGH analysis was performed in all tumor cells with known aberrations concerning chromosomes 18 and 19 according to protocols described previously (du Manoir et al., 1993). In brief, 200 ng biotinylated tumor DNA was mixed with the same amount of digoxigenin labeled (male) reference DNA in the presence of 50 μg COT-1 DNA and 50 μg sonicated salmon sperm DNA in a 10-μl hybridization mix. Hybridization to normal metaphases was performed for 3 d at 37°C. Probes were detected as described previously (Kraus et al., 2003). Image acquisition was done using a microscope (DMRXA; Leica) equipped with a CCD camera (Sensys; Photometrics). CGH was evaluated with the QCGH-software package (Leica Microsystems Imaging Solutions).
Confocal microscopy
Nuclei were scanned with an axial distance of 200 nm between light optical sections using a three-channel laser scanning confocal microscope (LSM 410; Carl Zeiss MicroImaging, Inc.). For each optical section, images were collected sequentially for all three fluorochromes. Stacks of 8-bit gray-scale two-dimensional images were obtained with a pixel size of 66–80 nm. Confocal images were processed with ImageJ software (http://rsb.info.nih.gov/ij/). 3D reconstructions of CT image stacks were performed using Amira 2.3 TGS.
Quantitative evaluation of the nuclear positioning of painted territories by assessment of the 3D relative radial distribution
A detailed description of the quantitative radial 3D evaluation of light optical serial sections by a voxel (volume element) based algorithm was published elsewhere (Cremer et al., 2001). In brief, as a first step, the geometrical center and the border of the nucleus were determined using the 3D data set of the DNA-counterstain fluorescence. For segmentation, we defined all voxels not belonging to the nuclear interior as image background. For comparison of nuclei with different shape and size, the distance between the nuclear center and each point located on the segmented nuclear border was given as the relative radius (r0 = 100). A decline of the curve for the nuclear counterstain in the most peripheral shells observed by this approach results in part from the Gaussian filtering of the data and in part from irregularities of the nuclear border (Cremer et al., 2001). In tissue sections, an additional prior step for the determination of the nuclear border was necessary. In the 15-μm thick sections of cervix and colon epithelium, nuclei overlapped each other and are so closely attached that confocal images of the nuclear counterstain had to be segmented before further evaluation. The optical isolation of an individual nucleus from the surrounding tissue and a 3D reconstruction was achieved with the Amira 2.3 TGS program. With this program, a 3D reconstructed image stack can be displayed in all three space dimensions and further processed. After marking the region of interest in selected images of the stack, the different perpendicular planes were used as a scaffold to form a 3D body representing the nuclear counterstain. This reconstruction can be stored as an image stack and applied to the 3D relative radial distribution program. By this approach, the DNA counterstain curve results in a near linear increase from the nuclear center to the border. In the second step, segmentation of CTs was performed in each 3D stack representing the color channels for painted CTs. All voxel intensities below an automatically set threshold were set to zero. Using an iterative procedure, a threshold value was estimated for each 3D data set for CT thresholding. The segmented nuclear space was divided into 25 equidistant shells with a thickness of Δr = 1/25 r0. For each voxel located in the nuclear interior, the relative distance r from the nuclear center was calculated as a fraction of r0. For each shell, all voxels assigned to a given CT were identified and the fluorescence intensities derived from the respective emission spectrum were summed up. This procedure yielded the individual relative DNA content within each shell for painted CTs as well as the overall DNA content reflected by the DNA counterstain. The sum of the voxel intensities measured in each nucleus was set to 100% for each fluorochrome. Using this normalization, the relative DNA content within a nuclear shell as a function of the relative distance r from the 3D center in the entire set of evaluated nuclei was plotted as a graph. For a separate quantitative evaluation of the radial distribution from the translocated and the nontranslocated portions of CT #17 and #18 material in the SW620 cell line, nuclei were selected, where the translocation chromosome could be identified by a close association and broad attachment of a small CT #17 and a small CT#18. In 3D data stacks of painted territories, created by the ImageJ program, either the voxels representing the free portions of homologous CTs or the voxels representing the translocated part of a CT were erased before quantitative evaluation.
To test for significant differences (P ≤ 0.05) in the distribution curves for each fluorochrome, two different tests were applied: (1) the MQ test (Bauer, 1962), a test which measures the distribution differences of two independent samples in a nonparametric mode, was applied to record the distribution of the average radial values for each fluorochrome in each nucleus; and (2) the more stringent two-sample KS test was applied to test for significant differences in the cumulative frequencies of voxel intensities for each fluorochrome plotted against the relative radius. For a comparison between different normal and malignant cell types, the average relative radius of the voxel distribution of painted chromosome #18 material, referred to as <rCT18>, and for chromosome #19 material, referred to as <rCT19>, was calculated from all evaluated nuclei of each cell type. The fraction of individual nuclei within a cell type, where CT #18 material was located more centrally compared with CT #19 material, was determined by measuring the average radial value of voxels assigned to a CT within a single nucleus, referred to as rCT18 and rCT19, respectively. The frequency of nuclei with rCT18 < rCT19 (inversed pattern) was divided by the total number of nuclei in a series. From this fraction, we estimated a confidential interval (variance) under the assumption of a binomial distribution.
Measurement of nuclear size and shape
The maximum length and width of each counterstained nucleus was measured using the ImageJ program. The height of each nucleus (z-axis) was determined by the number of serial sections encompassing the nuclear counterstain. For an estimation of the nuclear morphology, the ratio of length/width and of height/length was determined.
Acknowledgments
The authors are grateful to Dr. S. Müller for providing unpublished data on the karyotype of the Jurkat, the MelJuso, and the SW480/620 cell lines and helpful technical support. The authors also thank I. Jentsch for performing M-FISH analysis of the HeLa cell line used in this report and L. Schermelleh for stimulating discussions.
This work was supported by a grant from the Wilhelm Sander Stiftung (2001.079.1) to M. Speicher, T. Cremer, and J. Diebold, and by a grant from the Deutsche Forschungsgemeinschaft (Cr59/20-1) to T. Cremer.
Abbreviations used in this paper: 3D, three-dimensional; CGH, comparative genomic hybridization; CT, chromosome territory; GM-CFC, granulocyte-macrophage colony–forming cells; KS, Kolmogorov-Smirnov; MQ, median quartile.
References
- Abdel-Rahman, W.M., K. Katsura, W. Rens, P.A. Gorman, D. Sheer, D. Bicknell, W.F. Bodmer, M.J. Arends, A.H. Wyllie, and P.A. Edwards. 2001. Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proc. Natl. Acad. Sci. USA. 98:2538–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach, S.P., A.G. Renehan, and C.S. Potten. 2000. Stem cells: the intestinal stem cell as a paradigm. Carcinogenesis. 21:469–476. [DOI] [PubMed] [Google Scholar]
- Bauer, R.K. 1962. Der median-quartile-test. Metrika. 5:1–16. [Google Scholar]
- Bennett, D.C., K. Bridges, and I.A. McKay. 1985. Clonal separation of mature melanocytes from premelanocytes in a diploid human cell strain: spontaneous and induced pigmentation of premelanocytes. J. Cell Sci. 77:167–183. [DOI] [PubMed] [Google Scholar]
- Boyle, S., S. Gilchrist, J.M. Bridger, N.L. Mahy, J.A. Ellis, and W.A. Bickmore. 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211–219. [DOI] [PubMed] [Google Scholar]
- Bridger, J.M., S. Boyle, I.R. Kill, and W.A. Bickmore. 2000. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr. Biol. 10:149–152. [DOI] [PubMed] [Google Scholar]
- Cahill, D.P., C. Lengauer, J. Yu, G.J. Riggins, J.K. Willson, S.D. Markowitz, K.W. Kinzler, and B. Vogelstein. 1998. Mutations of mitotic checkpoint genes in human cancers. Nature. 392:300–303. [DOI] [PubMed] [Google Scholar]
- Clancy, B., and L.J. Cauller. 1998. Reduction of background autofluorescence in brain sections following immersion in sodium borohydride. J. Neurosci. Methods. 83:97–102. [DOI] [PubMed] [Google Scholar]
- Cremer, M., J. von Hase, T. Volm, A. Brero, G. Kreth, J. Walter, C. Fischer, I. Solovei, C. Cremer, and T. Cremer. 2001. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 9:541–567. [DOI] [PubMed] [Google Scholar]
- Croft, J.A., J.M. Bridger, S. Boyle, P. Perry, P. Teague, and W.A. Bickmore. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145:1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova, D.S., and R. Berezney. 2002. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 115:4037–4051. [DOI] [PubMed] [Google Scholar]
- du Manoir, S., M.R. Speicher, S. Joos, E. Schrock, S. Popp, H. Dohner, G. Kovacs, M. Robert-Nicoud, P. Lichter, and T. Cremer. 1993. Detection of complete and partial chromosome gains and losses by comparative genomic in situ hybridization. Hum. Genet. 90:590–610. [DOI] [PubMed] [Google Scholar]
- Dufer, J., M.F. Poupon, and S. Yatouji. 2000. Nuclear DNA content and chromatin pattern of rat rhabdomyosarcoma cell sublines with different metastatic potentials. Anal. Cell. Pathol. 20:41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich, D., J. Beaudouin, B. Kalbfuss, N. Daigle, R. Eils, and J. Ellenberg. 2003. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 112:751–764. [DOI] [PubMed] [Google Scholar]
- Gordon, M.Y. 1993. Human haemopoietic stem cell assays. Blood Rev. 7:190–197. [DOI] [PubMed] [Google Scholar]
- Habermann, F.A., M. Cremer, J. Walter, G. Kreth, J. von Hase, K. Bauer, J. Wienberg, C. Cremer, T. Cremer, and I. Solovei. 2001. Arrangements of macro- and microchromosomes in chicken cells. Chromosome Res. 9:569–584. [DOI] [PubMed] [Google Scholar]
- Herlyn, M., M.L. Mancianti, J. Jambrosic, J.B. Bolen, and H. Koprowski. 1988. Regulatory factors that determine growth and phenotype of normal human melanocytes. Exp. Cell Res. 179:322–331. [DOI] [PubMed] [Google Scholar]
- Jauch, A., J. Wienberg, R. Stanyon, N. Arnold, S. Tofanelli, T. Ishida, and T. Cremer. 1992. Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc. Natl. Acad. Sci. USA. 89:8611–8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J.P., M. Demmer-Dieckmann, T. Meo, M.R. Hadam, and G. Riethmuller. 1981. Surface antigens of human melanoma cells defined by monoclonal antibodies. I. Biochemical characterization of two antigens found on cell lines and fresh tumors of diverse tissue origin. Eur. J. Immunol. 11:825–831. [DOI] [PubMed] [Google Scholar]
- Joos, S., M. Granzow, H. Holtgreve-Grez, R. Siebert, L. Harder, J.I. Martin-Subero, J. Wolf, M. Adamowicz, T.F. Barth, P. Lichter, and A. Jauch. 2003. Hodgkin's lymphoma cell lines are characterized by frequent aberrations on chromosomes 2p and 9p including REL and JAK2. Int. J. Cancer. 103:489–495. [DOI] [PubMed] [Google Scholar]
- Kanda, T., K.F. Sullivan, and G.M. Wahl. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8:377–385. [DOI] [PubMed] [Google Scholar]
- Keenan, S.J., J. Diamond, W.G. McCluggage, H. Bharucha, D. Thompson, P.H. Bartels, and P.W. Hamilton. 2000. An automated machine vision system for the histological grading of cervical intraepithelial neoplasia (CIN). J. Pathol. 192:351–362. [DOI] [PubMed] [Google Scholar]
- Kozubek, S., E. Lukasova, P. Jirsova, I.I. Koutna, M. Kozubek, A. Ganova, E. Bartova, M. Falk, and R. Pasekova. 2002. 3D Structure of the human genome: order in randomness. Chromosoma. 111:321–331. [DOI] [PubMed] [Google Scholar]
- Kraus, J., K. Pantel, D. Pinkel, D.G. Albertson, and M.R. Speicher. 2003. High-resolution genomic profiling of occult micrometastatic tumor cells. Genes Chromosomes Cancer. 36:159–166. [DOI] [PubMed] [Google Scholar]
- Krause, D.S., M.J. Fackler, C.I. Civin, and W.S. May. 1996. CD34: structure, biology, and clinical utility. Blood. 87:1–13. [PubMed] [Google Scholar]
- Kuppers, R., I. Schwering, A. Brauninger, K. Rajewsky, and M.L. Hansmann. 2002. Biology of Hodgkin's lymphoma. Ann. Oncol. 13:11–18. [DOI] [PubMed] [Google Scholar]
- Lachner, M., and T. Jenuwein. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14:286–298. [DOI] [PubMed] [Google Scholar]
- Leibovitz, A., J.C. Stinson, W.B. McCombs, III, C.E. McCoy, K.C. Mazur, and N.D. Mabry. 1976. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 36:4562–4569. [PubMed] [Google Scholar]
- Ochs, B.A., W.W. Franke, R. Moll, C. Grund, M. Cremer, and T. Cremer. 1983. Epithelial character and morphologic diversity of cell cultures from human amniotic fluids examined by immunofluorescence microscopy and gel electrophoresis of cytoskeletal proteins. Differentiation. 24:153–173. [DOI] [PubMed] [Google Scholar]
- Parada, L.A., P.G. McQueen, P.J. Munson, and T. Misteli. 2002. Conservation of relative chromosome positioning in normal and cancer cells. Curr. Biol. 12:1692–1697. [DOI] [PubMed] [Google Scholar]
- Schermelleh, L., S. Thalhammer, W. Heckl, H. Posl, T. Cremer, K. Schutze, and M. Cremer. 1999. Laser microdissection and laser pressure catapulting for the generation of chromosome-specific paint probes. Biotechniques. 27:362–367. [DOI] [PubMed] [Google Scholar]
- Schneider, U., H.U. Schwenk, and G. Bornkamm. 1977. Characterization of EBV-genome negative “null” and “T” cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int. J. Cancer. 19:621–626. [DOI] [PubMed] [Google Scholar]
- Schnell, S.A., W.A. Staines, and M.W. Wessendorf. 1999. Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J. Histochem. Cytochem. 47:719–730. [DOI] [PubMed] [Google Scholar]
- Skalnikova, M., S. Kozubek, E. Lukasova, E. Bartova, P. Jirsova, A. Cafourkova, I. Koutna, and M. Kozubek. 2000. Spatial arrangement of genes, centromeres and chromosomes in human blood cell nuclei and its changes during the cell cycle, differentiation and after irradiation. Chromosome Res. 8:487–499. [DOI] [PubMed] [Google Scholar]
- Solovei, I., J. Walter, M. Cremer, F. Habermann, L. Schermelleh, and T. Cremer. 2002. FISH on three-dimensionally preserved nuclei. FISH: A Practical Approach. J. Squire, B. Beatty, and S. Mai, editors. Oxford University Press, Oxford. 119–157.
- Tanabe, H., F.A. Habermann, I. Solovei, M. Cremer, and T. Cremer. 2002. a. Non-random radial arrangements of interphase chromosome territories: evolutionary considerations and functional implications. Mutat. Res. 504:37–45. [DOI] [PubMed] [Google Scholar]
- Tanabe, H., S. Muller, M. Neusser, J. von Hase, E. Calcagno, M. Cremer, I. Solovei, C. Cremer, and T. Cremer. 2002. b. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl. Acad. Sci. USA. 99:4424–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weierich, C., A. Brero, S. Stein, J.v. Hase, C. Cremer, T. Cremer, and I. Solovei. 2003. Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res. 11:485–502. [DOI] [PubMed] [Google Scholar]