Abstract
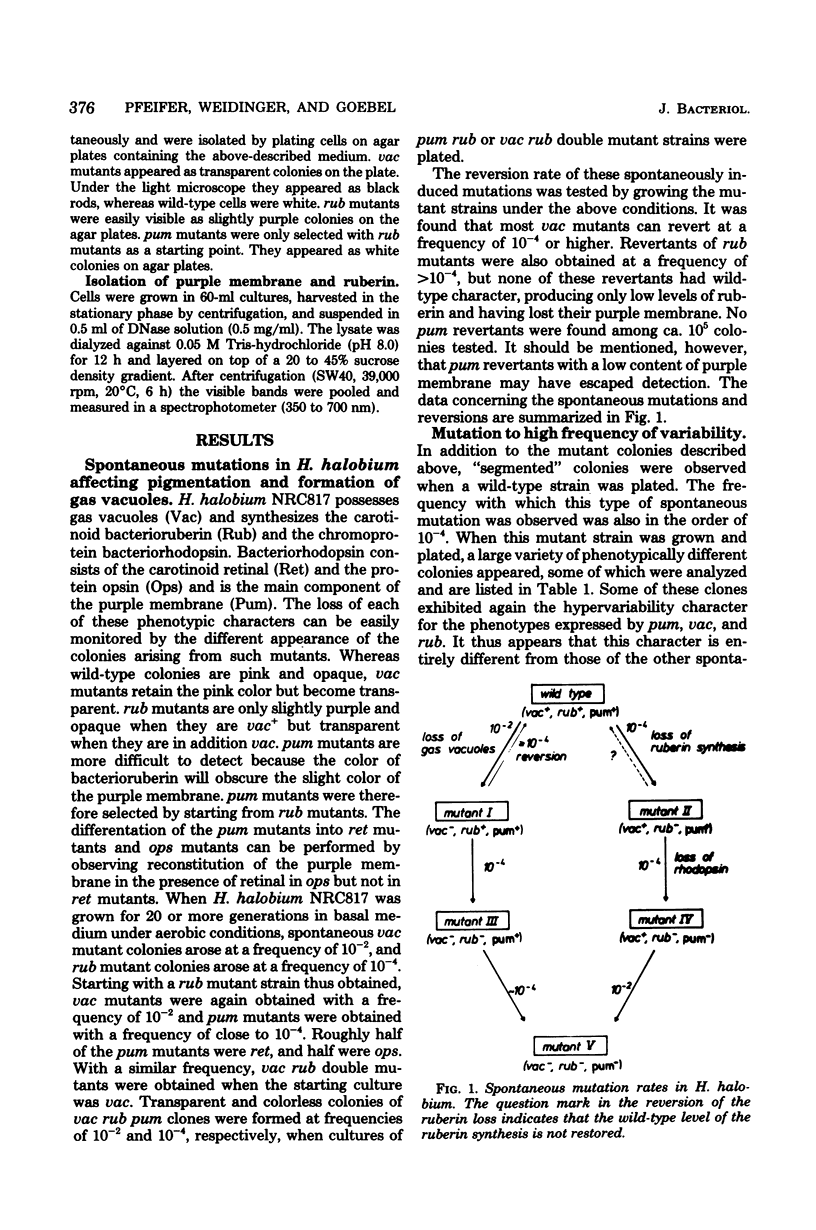
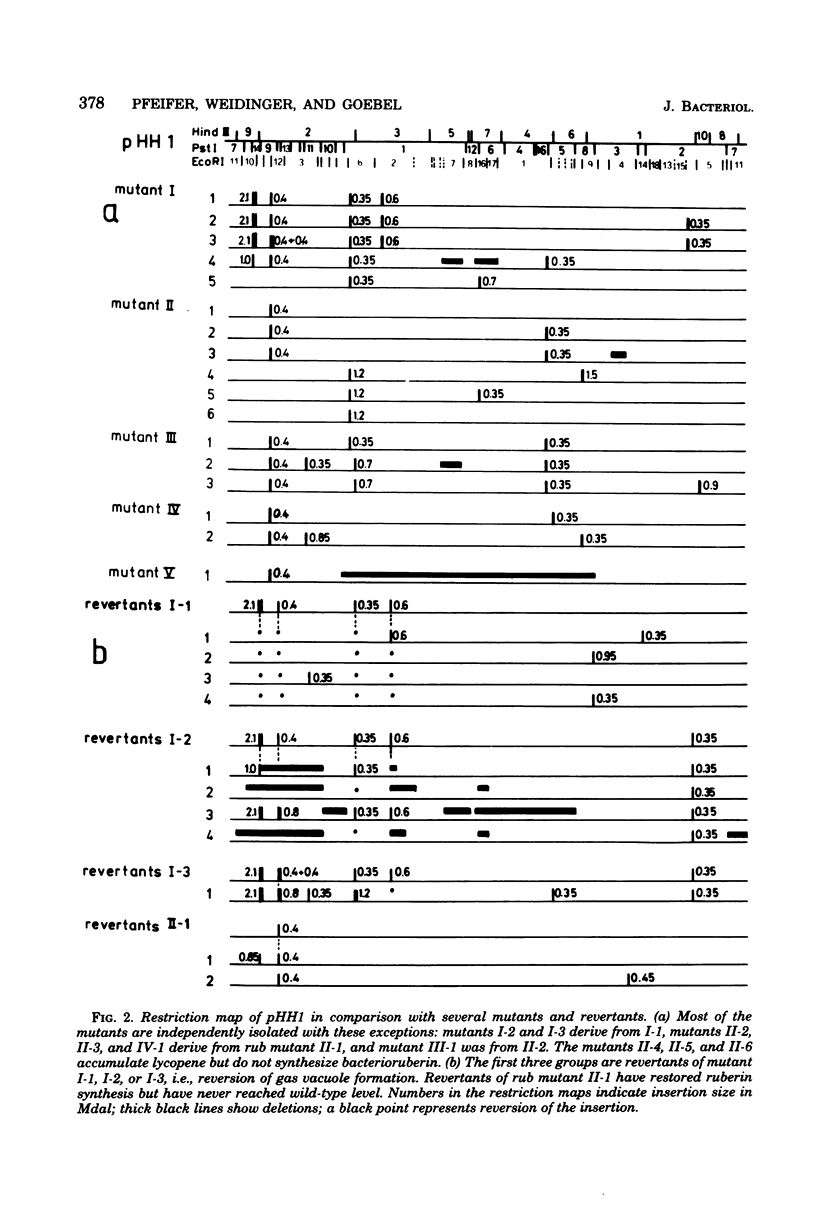
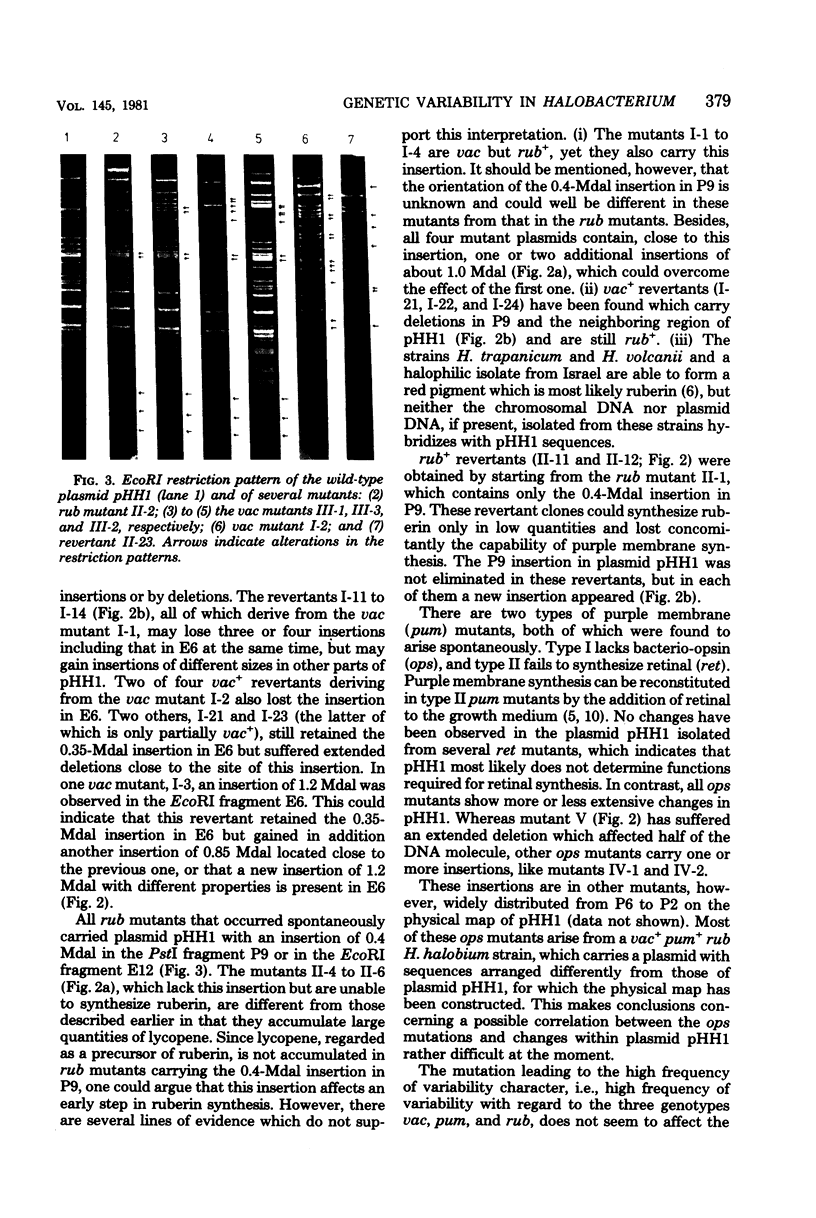
Halobacterium halobium exhibits an extraordinary degree of spontaneous variability. Mutants which are defective in the formation of gas vacuoles (vac) arise at a frequency of 10(-2). Other easily detectable phenotypes, like the synthesis of bacterioruberin (Rub) or the synthesis of retinal (Ret) and bacterio-opsin (Ops), the two components which form the purple membrane (Pum) of H. halobium, are lost at a frequency of about 10(-4). With the same frequency a mutant type appears which exhibits an extremely high variability in these phenotypes. With the exception of the ret mutants, all spontaneously arising mutants show alterations, i.e., insertions, rearrangements, or deletions, in the plasmid pHH1. It appears that the introduction of one insertion into pHH1 triggers further insertions, which makes the identification of relationships between phenotypic and genotypic alterations rather difficult. From the analysis of a large number of spontaneous vac mutants and their vac+ revertants it can be concluded that the formation of the gas vacuoles is determined or controlled by plasmid genes. No such conclusion is yet possible for the rub mutants, although all mutants of this type so far analyzed exhibit a defined insertion. pum mutants which have lost the capability of forming bacterio-opsin carry insertions in the plasmid which are distributed over a rather large region of the plasmid. No strains of H. halobium could be obtained which had lost plasmid pHH1 completely.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Larsen H., Omang S., Steensland H. On the gas vacuoles of the halobacteria. Arch Mikrobiol. 1967;59(1):197–203. doi: 10.1007/BF00406332. [DOI] [PubMed] [Google Scholar]
- Moore R. L., McCarthy B. J. Base sequence homology and renaturation studies of the deoxyribonucleic acid of extremely halophilic bacteria. J Bacteriol. 1969 Jul;99(1):255–262. doi: 10.1128/jb.99.1.255-262.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore R. L., McCarthy B. J. Characterization of the deoxyribonucleic acid of various strains of halophilic bacteria. J Bacteriol. 1969 Jul;99(1):248–254. doi: 10.1128/jb.99.1.248-254.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D., Schuhmann L. Reconstitution of bacteriorhodopsin. FEBS Lett. 1974 Aug 30;44(3):262–265. doi: 10.1016/0014-5793(74)81153-1. [DOI] [PubMed] [Google Scholar]
- Pfeifer F., Weidinger G., Goebel W. Characterization of plasmids in halobacteria. J Bacteriol. 1981 Jan;145(1):369–374. doi: 10.1128/jb.145.1.369-374.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starlinger P., Saedler H. IS-elements in microorganisms. Curr Top Microbiol Immunol. 1976;75:111–152. doi: 10.1007/978-3-642-66530-1_4. [DOI] [PubMed] [Google Scholar]
- Sumper M., Herrmann G. Biogenesis of purple membrane: regulation of bacterio-opsin synthesis. FEBS Lett. 1976 Oct 15;69(1):149–152. doi: 10.1016/0014-5793(76)80673-4. [DOI] [PubMed] [Google Scholar]
- Toeckenius W., Kunau W. H. Further characterization of particulate fractions from lysed cell envelopes of Halobacterium halobium and isolation of gas vacuole membranes. J Cell Biol. 1968 Aug;38(2):337–357. doi: 10.1083/jcb.38.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger G., Klotz G., Goebel W. A large plasmid from Halobacterium halobium carrying genetic information for gas vacuole formation. Plasmid. 1979 Jul;2(3):377–386. doi: 10.1016/0147-619x(79)90021-0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Fox G. E. Archaebacteria. J Mol Evol. 1978 Aug 2;11(3):245–251. doi: 10.1007/BF01734485. [DOI] [PubMed] [Google Scholar]