Abstract
Typically, eukaryotic nuclei contain 10–30 prominent domains (referred to here as SC-35 domains) that are concentrated in mRNA metabolic factors. Here, we show that multiple specific genes cluster around a common SC-35 domain, which contains multiple mRNAs. Nonsyntenic genes are capable of associating with a common domain, but domain “choice” appears random, even for two coordinately expressed genes. Active genes widely separated on different chromosome arms associate with the same domain frequently, assorting randomly into the 3–4 subregions of the chromosome periphery that contact a domain. Most importantly, visualization of six individual chromosome bands showed that large genomic segments (∼5 Mb) have striking differences in organization relative to domains. Certain bands showed extensive contact, often aligning with or encircling an SC-35 domain, whereas others did not. All three gene-rich reverse bands showed this more than the gene-poor Giemsa dark bands, and morphometric analyses demonstrated statistically significant differences. Similarly, late-replicating DNA generally avoids SC-35 domains. These findings suggest a functional rationale for gene clustering in chromosomal bands, which relates to nuclear clustering of genes with SC-35 domains. Rather than random reservoirs of splicing factors, or factors accumulated on an individual highly active gene, we propose a model of SC-35 domains as functional centers for a multitude of clustered genes, forming local euchromatic “neighborhoods.”
Keywords: cell nucleus; chromosome banding; genome; splicing factor SC-35; chromosome structure
Introduction
Here, we investigate whether there is higher level organization of the genome such that specific protein-encoding genes cluster together at common nuclear structures. Interphase nuclei contain 15–30 prominent “speckles” (0.5–3 μm in diameter), the SC-35 domains, which are enriched in numerous mRNA metabolic factors (for review see Moen et al., 1995). Select active genes have been shown to position at the edge of an SC-35 domain with high consistency, and their transcripts are often within that domain (Xing et al., 1995; Smith et al., 1999; Shopland et al., 2002). This arrangement could be interpreted simply as factors accumulated on nascent transcripts of a single highly active gene (Huang and Spector, 1996). However, an important and unresolved alternative is that SC-35 domains are structures around which multiple, specific genes cluster in a cell type–specific arrangement (Johnson et al., 2000; Moen et al., 2003). This would indicate a higher level organization of protein-coding genes, in some ways similar to the clustering of rDNA loci at the nucleolus. The clustering of mRNA-encoding genes into specific euchromatic “neighborhoods” has not been demonstrated to date, and would have significant implications for the organization of genes and chromosomes throughout the nucleus, as well as for sequences within the chromosome territory.
Previously, we have determined the distribution of 21 specific active genes relative to SC-35 domains in a large cell population, and of these, 10 loci show a highly nonrandom positioning at the edge of an SC-35 domain, whereas the remaining active (and all inactive) genes do not (Smith et al., 1999; Shopland et al., 2002). These observations imply that a substantial fraction of all active mRNA-encoding genes might be similarly positioned. However, the association of multiple specific genes with a common domain has not been directly demonstrated, nor has the organization of individual chromosome segments or bands been investigated in this way. There is evidence that SC-35 domains occupy a more interior, largely euchromatic region of the nucleus (Carter et al., 1993; Ferreira et al., 1997; Sadoni et al., 1999), and that proteins associated with active chromatin may be around SC-35 domains (Hendzel et al., 1998), although their interiors likely do not contain DNA (Carter et al., 1993; Thiry, 1993; Belmont and Bruce, 1994; Hendzel et al., 1998). Labeling of transcription sites en masse by short pulses of Br-UTP reveals thousands of small foci throughout the nucleus, with a subset at the edges of SC-35 domains (Wei et al., 1999); however, these observations do not address whether multiple specific genes or specific chromosome segments cluster or align with individual SC-35 domains.
We begin this work by addressing whether individual SC-35 domains associate with multiple specific genes and multiple RNAs, and then expand the work to examine some genomic features that might relate to a clustered organization of genes with these domains. Localized clustering of genes in nuclei may be largely dependent on chromosomal context. Most importantly, genes have long been known to cluster more in chromosomal reverse bands (R-bands) rather than in Giemsa dark bands (G-bands; Goldman et al., 1984). However the reason for this fundamental aspect of chromosome organization remains obscure. The distribution of genes also must be affected in some way by the restriction of a chromosome's DNA into a relatively discrete chromosome territory (Cremer and Cremer, 2001). Our findings suggest that individual SC-35 domains are situated in local regions of euchromatin, and that these specific associations relate to features of chromosome sequence organization.
Results
Two different genes can associate with a common SC-35 domain containing multiple mRNAs
First, we directly tested the hypothesis that SC-35 domains are structures around which multiple, specific genes cluster by examining whether two nonsyntenic genes, collagen type I, α1 (COL1A1; chromosome 17) and collagen type I, α2 (COL1A2; chromosome 7), can simultaneously associate with the same SC-35 domain. Individually, each of these loci is known to associate with an SC-35 domain at high frequencies (98 and 87% of loci in diploid fibroblasts, respectively; Table I), and these are significantly greater than predicted or empirically observed random interactions (Xing et al., 1995; Shopland et al., 2002). We performed three-color analyses of these two genes and SC-35 in diploid fibroblasts. Results show that COL1A1 and COL1A2 position at the edge of the same SC-35 domain in a significant subset of cells (∼10%; Fig. 1 A, Table I, and see following section).
Table I. Frequencies at which two different genes/RNAs associate with a common SC-35 domain.
| RNA 1 (location, % domain-associated loci) |
RNA 2 (location, % domain-associated loci) |
Predicted random co-association (% cells)a |
Number of cells scored | Percentage of cells with co-associated signalsb |
|---|---|---|---|---|
| COL1A1 (17q21.3, 99%) |
COL1A2 (7q21.3, 87%) |
12 ± 3.6 | 666 | 7.9 ± 4.0 |
| Col1A1 (17q21.3, 99%) |
ACTB (7p12–p15, 89%) |
12 ± 3.5 | 439 | 11.3 ± 4.7 |
| COL1A1 (17q21.3, 99%) |
LMNA (1q21.3, 70%) |
9.6 ± 2.8 | 270 | 7.7 ± 2.6 |
| COL1A2 (7q21.3, 87%) |
ACTB (7p12–p15, 89%) |
11 ± 3.5 | 214 | 28.7 ± 8.4 |
Derivation for each pair of genes/RNAs is described in Materials and methods.
Cells with >1 pair of different, associated RNAs. Means ± SD are reported.
LMNA, lamin A/C.
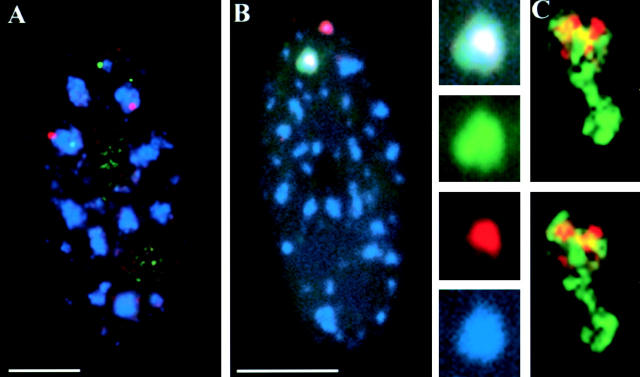
Figure 1.
COL1A1 and COL1A2 genes associate with a single, common domain. (A) WI-38 diploid fibroblasts were hybridized with differently labeled genomic probes of COL1A1 (red) and COL1A2 gene (green) and stained for SC-35 (blue). One homologue of each gene is simultaneously associated with the same SC-35 domain in the cell shown. (B) Transcripts from the COL1A1 (green) and COL1A2 (red) genes, detected with differentially labeled cDNA probes, intermingle within an SC-35 domain (blue). Overlap between the three colors appears white. (C) Three-dimensional deconvolution shows intermingling COL1A1 (green) and COL1A2 (red) transcripts in two focal planes. Regions of colocalization appear yellow. To view a three-dimensional reconstruction of this stack, see supplemental material (available at http://www.jcb.org/cgi/content/full/jcb.200303131/DC1). Bars, 5 μm.
Are the COL1A1 and COL1A2 genes associated with one common or two closely abutting structures? These genes, which are restricted to the SC-35 domain edge, both produce transcripts that accumulate within the domain interior (Xing et al., 1995; Shopland et al., 2002). We found that their transcripts can intermingle within the same SC-35 domain (Fig. 1 B), demonstrating that they occupy a common structure. This was also observed for COL1A2 and β-actin (ACTB) transcripts (Fig. 2 B, see following section). Three-dimensional deconvolution to remove out of focus light further confirmed these observations (Fig. 1 C; see also Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200303131/DC1). These data provide direct evidence that SC-35 domains are structures around which multiple mRNA-encoding genes cluster, and which contain multiple mRNAs. The COL1A1 and COL1A2 genes provide the most rigorous test of our hypothesis because they produce very highly expressed and heavily spliced nuclear RNA accumulations (Smith et al., 1999), which could be thought more likely to generate the appearance of a “domain” from an individual gene (Huang and Spector, 1996). However, these findings directly demonstrate that even in these cases, there are multiple genes clustered with each individual domain.
Figure 2.
Transcripts from multiple genes can associate with the same SC-35 domain. (A) One focus of lamin A/C RNA (red) associates with the edge of an accumulation of COL1A1 transcripts (green), which serves as a marker for an SC-35 domain. (B) Triple labeling also shows that transcripts from ACTB (green) and COL1A2 (red) accumulate within the same SC-35 domain (blue). (C) Chromosome 7 territories detected with a whole chromosome paint (red) contact 3–4 SC-35 domains (green) per nucleus. Bars, 5 μm.
Domain “choice” is random: coordinately expressed type 1 collagen genes do not preferentially associate with the same SC-35 domain
Might the association of COL1A1 and COL1A2 with the same SC-35 domain be related to their tightly coordinated expression (Karsenty and Park, 1995), or does their co-association frequency fit the expectation for two domain-associating genes that randomly “choose” one of the 15–30 domains? To answer this, we further examined the frequency of co-association of these and other unrelated genes scored in a large sample of cells. To facilitate scoring, COL1A1 and COL1A2 transcripts were detected in two different colors. Because COL1A1 RNA is virtually 100% coincident with its associated SC-35 domain, it can substitute as a marker for the SC-35 domain associated with the COL1A1 gene. Scoring indicated that the COL1A1 and COL1A2 RNA co-association frequency (7.9% of cells) falls within the range of the frequency calculated for random domain choice (Materials and methods; Table I). Next, we determined the frequency of co-association between COL1A1 and transcripts from two other active genes unrelated to collagens, ACTB and lamin A/C (Fig. 2 A). These comparisons again indicate similar frequencies of co-association and random domain choice (Table I). Although domain choice is apparently random because both of these genes are localized to one of the 15–30 SC-35 domains, in the context of the entire nucleus, they are not necessarily randomly distributed with respect to each other, and have a closer structural association than expressed genes that localize to distinct compartments. Smith et al. (1999) previously showed that certain transcripts essentially never localize to the same domain or compartment.
Effects of the chromosome territory: genes distantly located on different arms of a chromosome frequently co-associate with the same domain
To further explore parameters that might influence the association of two genes with a common SC-35 domain, we tested whether two genes on the same chromosome associate with the same domain more often than genes on different chromosomes. Alternatively, syntenic genes might be restricted to different neighborhoods within the chromosome territory, and thus rarely or never co-associate with a domain. To discriminate between these two possibilities, we determined the co-association frequency of ACTB and COL1A2 on chromosome 7p and 7q, respectively, and separated by >70 Mb. Because genes as little as 1 Mb apart are separated by an average of 1 μm in nuclei, genes on separate chromosome arms can be quite distant (Lawrence et al., 1990; Trask et al., 1991). As shown in Table I and Fig. 2 B, ACTB and COL1A2 co-associate with the same SC-35 domain in ∼28% of cells, three times more frequently than do pairs of genes from different chromosomes (e.g., ACTB and COL1A1; Table I, Fig. 2 B). In addition, when the entire chromosome 7 territory (∼30% of which is gene sequence) is detected with a whole chromosome paint, only 3–4 subregions of the territory contact SC-35 domains, whereas the vast majority of the chromosome is not in direct contact with these compartments (Fig. 2 C). This suggests that SC-35 domain co-association is specific for a small subset of chromosome 7 loci.
Recently, there has been much interest in the possible enrichment of genes at the chromosome territory surface. Our data indicate that the chromosome territory surface consists of functionally distinct subregions that contact the high concentration of metabolic factors in an SC-35 domain (Fig. 2 C) and contain specific genes. Moreover, the finding that ACTB and COL1A2 co-associate in 28% of cells (Table I) suggests that these distantly linked genes distribute randomly to one of the 3–4 domain-associated regions of the chromosome 7 territory surface. This indicates that their wide separation on different arms of the chromosome does not constrain them to any one of these particular subregions.
Nuclear organization of individual chromosome bands: R-bands are more intimately associated with SC-35 domains than are G-bands
Our data indicating that synteny can favor the association of specific genes with a common SC-35 domain led us to speculate that even closer chromosomal “linkage” might further correlate with domain co-association. Interestingly, genes are clustered along the chromosome and are enriched in certain cytogenetic bands (R-bands). Therefore, we tested whether individual chromosome bands show differences in their relationship to SC-35 domains, and whether gene-rich R-bands in particular tend to show more association with these structures than gene-poor G-bands. We developed an approach to detect specific chromosome bands in interphase nuclei relative to SC-35 domains (see Materials and methods). Probes generated from specific cytogenetic bands microdissected from metaphase chromosomes (Guan et al., 1993) were selected based on gene density (Table II). The chosen R-band probes were from 6p21.3, 17q21, and 19q13.3; G-band probes were from 3p14 and 7p21; and one probe, from 17q22–24, contained ∼1/3 R-band and 2/3 G-band material (Fig. 3). Two of these bands, 17q21 and 17q22–24, were also chosen because they are adjacent to one another and flank the COL1A1 gene, which maps to the telomeric end of 17q21 (Fig. 3 D), near its junction with 17q22–24.
Table II. Frequency of contacts between SC-35 domains and R- and G-bands.
| Band (type) | Estimated gene density (# of genes/Mb)a | Number of bands scored | Percentage of bands contacting 0, 1, or ≥2 SC-35 domains
|
Percentage of bands surrounding one half or more of a domain | Percentage of bands contacting nuclear peripheryb | ||
|---|---|---|---|---|---|---|---|
| 0 | 1 | ≥2 | |||||
| 3p14 (G) | 1.1 | 209 | 39 | 49 | 12 | 3 | 34 |
| 7p21 (G) | 1.5 | 184 | 37 | 52 | 11 | 2 | 44 |
| 17q22-24 (G/R/G) | 3.9 | 208 | 17 | 48 | 34 | 13 | 26 |
| 17q21 (R) | 10.1 | 193 | 1 | 34 | 63 | 24 | 11 |
| 6p21 (R) | 15.4 | 172 | 10 | 68 | 21 | 15 | 41 |
| 19q13.3 (R) | 12.5 | 228 | 8 | 62 | 30 | 12 | 36 |
Based on human genome map of known genes (http://genome.ucsc.edu).
Nuclear periphery defined by DAPI staining.
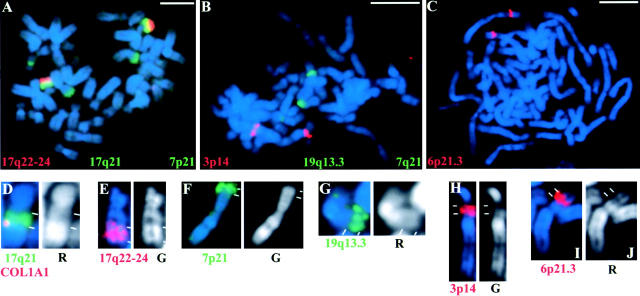
Figure 3.
Specificity of chromosome band probes. Probes from indicated bands were hybridized to spreads of human metaphase chromosomes (stained with DAPI, blue) of peripheral blood lymphocytes to assess their specificity. (A) 7p21, 17q21 (both green), and 17q22–24 (red); (B) 3p14 (red), 7q21 (green), and 19q13.3 (green); (C) 6p21.3 (red). Probes for 7p21 and 7q21 weakly detected additional bands, but 7p21 DNA was identified in interphase based on signal size, intensity, and cohybridization of the neighboring ACTB locus. Signals from 7q21 probe were not evaluated in interphase nuclei. (D–J) Enlarged views of single chromosomes, shown to scale, hybridized with different band probes as indicated and compared with DAPI bands (blue or white) indicates that they are largely either R- or G- band DNA. In D, the COL1A1 gene (red) is cohybridized with 17q21 (green), showing that this gene maps to the telomeric end of this band, proximal to 17q22–24 (E). Bars, 5 μm.
Visualization of these chromosome bands showed striking differences in their morphological organization relative to the SC-35 domains, demonstrating that different genomic segments have clearly distinct structural relationships to these compartments. Although we observed differences between all bands examined, in general, the organization of the R-bands relative to SC-35 domains was significantly different from that of the G-bands (Fig. 4). The vast majority of R-bands examined contact domains either at a single point (Fig. 4 E, right inset) or by extending for a considerable distance along the domain edge (Fig. 4 A, left; Fig. 4 E, left inset). In contrast, the G-bands most often contact domains at a single point, if at all (Fig. 4, F–I).
Figure 4.
Differential distribution of R- and G-band DNA with respect to SC-35 domains in interphase nuclei. (A and B) Probe of R-band 17q21 DNA (red in A, white in B) hybridized to WI-38 fibroblasts shows two highly extended homologues, each with different morphologies. The left homologue contacts four SC-35 domains (green). The right band surrounds half of one SC-35 domain and also contacts another. (C and D) Two 6p21.3 R-bands (red, white) each have a thin string of DNA extending from the more compact, main body of the band. In one case (top band), the extended region almost completely surrounds an SC-35 domain (green). (E) A tetraploid nucleus hybridized to detect 17q21 DNA (green) shows all four bands contacting multiple SC-35 domains (blue). The COL1A1 gene (red, right inset) is located at the tip of a DNA extension that contacts an SC-35 domain at a single point, in contrast to another homologue (left inset). (F and G) The G-band probe for 3p14 (red in F, white in G) shows two bands minimally contacting SC-35 domains (green). (H and I) Similar to other G-bands, probe for 7p21 (red, white) was detected in focal planes other than those containing SC-35 domains (green), which thus appear out of focus. (J) Hybridization to detect 17q22–24 DNA (red) and the adjacent COL1A1 gene (green) shows that although some of these bands can extensively contact the SC-35 domain (blue) associated with COL1A1 (top inset), others are clearly separated (bottom inset). (K) Three-dimensional deconvolution and reconstruction of 17q21 R-band signal (red) and surrounding SC-35 domains (green) shows that this band extends significantly in the X-Y plane (left), but when viewed along the Z-axis (right), appears largely restricted to the planes containing SC-35 domains. (L) A three-dimensional reconstruction as in K shows an example of a G-band, 3p14 (red), located above and separated from the nearest SC-35 domain (green). For rotational movies of K and L, see supplemental material (available at http://www.jcb.org/cgi/content/full/jcb.200303131/DC1). Bars, 5 μm.
The observed morphological differences between the two band types are difficult to fully capture by measurement of a single parameter; however, several morphometric criteria were scored in order to provide some quantitative assessment (Table II and Table III). For example, the differences in the intimacy of contact with a domain are demonstrated by the observations that substantially more of the R-bands were found surrounding more than half of an SC-35 domain (Fig. 4 A, right; Fig. 4 C, top; Table II). In addition, the numbers of domains contacted by each band are much greater for the R-bands than the G-bands. Chi square analyses indicated that this difference is statistically significant (P = 2 × 10−4 for 6p21.3 vs. 3p14, P = 2 × 10−7 for mean of two G-bands versus mean of three R-bands; Table II; unpublished data). In contrast, a correlation between band type and contact with the nuclear periphery was not observed (Table II). Also consistent with band-type differences in SC-35 domain contact, the probe for 17q22–24, which contains 2/3 G-band and 1/3 R-band DNA, displayed intermediate levels of association. In addition, morphometric analyses indicated that the length of SC-35 domain edges contacted by the R-bands is significantly greater than by the G-bands (P < 0.001 by two-tailed t test; Table III). The total amount of DNA in each band probe, determined by measuring the relative lengths of each band probe signal on multiple metaphase chromosomes, indicated that all but one band (17q22–24) contain similar amounts of DNA (Table III). Thus, DNA content does not account for the differences observed between the two band types.
Table III. Length of SC-35 domain edges contacted by R- and G-bands.
| Band (type) | Number of cells | Percentage of SC-35 domain perimeters contacteda | Relative DNA content | Total band perimeter/cell (μm) |
|---|---|---|---|---|
| 3p14 (G) | 23 | 2.5 ± 0.4 | 0.8 ± 0.1 | 14 ± 1 |
| 7p21 (G) | 29 | 1.5 ± 0.2 | 1.0 ± 0.1 | 23 ± 1 |
| 17q22-24 (G/R/G) | 20 | 4.1 ± 0.5 | 1.4 ± 0.1 | 22 ± 1 |
| 17q21 (R) | 27 | 7.8 ± 0.7 | 1.0 ± 0.1 | 40 ± 3 |
| 6p21 (R) | 39 | 4.7 ± 0.4 | 0.9 ± 0.1 | 18 ± 1 |
| 19q13.3 (R) | 28 | 5.7 ± 0.5 | 0.8 ± 0.1 | 20 ± 1 |
Perimeter measurements for all domains in a given nucleus were summed and a mean of all nuclei was determined. Measurements ± SE are reported.
The clear statistically significant differences between the R- and G-bands examined suggest that SC-35 domains are more frequently, and probably more specifically, contacted by R-band rather than G-band DNA. At the very least, even if G-band contacts primarily represent random associations between domains and fairly large portions of the genome, then R-bands of similar genomic sizes must contact more specifically (see following two sections). Irrespective of other properties of these R-bands that may accompany this more intimate contact with SC-35 domains, these findings show that specific segments of the genome, corresponding to specific gene-rich R bands, have preferential association with nuclear domains enriched in RNA metabolic factors.
The relationship between band morphology and SC-35 domain associations
Because gene-dense R-bands are likely more active than G-bands, they may also be more decondensed (Li et al., 1998). This, in turn, may be related to the extent of SC-35 domain contact. To examine this possibility, we quantitatively estimated the relative compaction of each band by measuring the perimeters of each band signal, which approximates surface area (Table III). Results indicate that R- and G-bands with similar DNA content can have similar perimeters (e.g., 6p21 [R] and 7p21 [G], P = 0.03 by two-tailed t test), but very different degrees of contact with SC-35 domains (P = 2 × 10−9; Table III). Thus, the extent of SC-35 domain contact is not simply a function of the nuclear size of a given band. However, this may be a contributing factor. For example, band 17q21 is much less compact than the other R-bands examined (P < 10−7 by two-tailed t test) and has greater SC-35 domain contact. The difference in compaction even among R-bands may be related to the identities and activities of the genes in each band, as previously suggested (Volpi et al., 2000). Finally, we note that although we did not see a strong correlation between band type and condensation state, we do not rule out that there may be differences in the average condensation of R- and G-bands more generally (Croft et al., 1999).
Even for an R-band that shows a high degree of decondensation within the nucleus (e.g., 17q21), SC-35 domain association may well be primarily specific rather than a random interaction. If the increased associations of the R-bands with SC-35 domains in general are simply due to their greater distension in the nucleus, it would be expected that their distribution would be random relative to the 3-D distribution of SC-35 domains. However, three-dimensional analysis of specific bands indicated just the opposite. Previous findings showed that SC-35 domains essentially lie in a single focal plane, restricted to the bottom third of the nuclear volume in both cultured fibroblasts and myoblasts (Carter et al., 1993). Three-dimensional reconstructions showed that the R-bands probed here are largely restricted to the same focal plane containing SC-35 domains, in contrast to the G-bands studied (Fig. 4, H, K, and L; Video 2 and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200303131/DC1). This preferential positioning within the three-dimensional nuclear volume of the specific R-bands versus the G-bands strongly suggests specificity for associations with the SC-35 domains.
Individual genes may drive specific associations between chromosome bands and SC-35 domains
The data presented suggest that the R-band contacts with SC-35 domains are not simply nonspecific consequences of large portions of the genome distributed randomly throughout the nucleus. Providing still more direct evidence of this, certain genes have been shown to nonrandomly associate with SC-35 domains, such that at least a portion of the contact between a given band and a domain is likely specified by particular genes within that band. These loci may either have an affinity for SC-35 domains or initially nucleate the domain by drawing a critical mass of metabolic factors (Huang and Spector, 1996; Xing, et al., 1995). Indeed, the R-band 17q21 analyzed here contains COL1A1, which is virtually 100% associated with SC-35 domains, and therefore, this point of band contact must be highly specific.
We examined whether the flanking regions of COL1A1 are consistently domain associated, or whether COL1A1 can contact SC-35 domains independently, indicating that this locus is sufficient to drive the association. By direct comparisons of 17q21, COL1A1, and SC-35 in triple-label experiments, we found that the region of this band containing COL1A1 can contact an SC-35 domain at what appears to be a single point (20% of bands). Hence, nearby flanking sequences are not required for contact. Interestingly, half of these discrete contacts extend considerably (>0.5 μm) from the main body of the band (Fig. 4 E, right inset). The frequency of COL1A1 found in these extended regions indicates further specificity and organization. The COL1A1 gene signal, ∼0.3 μm in diameter, represents ∼0.8% of the 17q21 band perimeter (∼40 μm; Table III), and yet its frequency in extended band structures is at least 10-fold higher (10%). In fact, this is an underestimate of specificity because it does not include the potential localization of a gene to the band interior. Thus, 17q21 DNA may be nonrandomly organized to present COL1A1 on the extreme reaches of its surface.
We hypothesize that a highly expressed and spliced gene such as COL1A1 might have a dominant influence in determining the close association of its chromosomal neighborhood with the domain of mRNA metabolic factors. In support of this, our analysis of these triple-labeled cells further showed that 80% of the 17q21 bands extensively contact the SC-35 domain associated with COL1A1, rather than contacting at a single point (Fig. 4 E, left inset). Hence, close linkage to certain genes may favor SC-35 domain associations. These linked sites may be enriched for genes that have been shown to nonrandomly associate with SC-35 domains, but at frequencies (>70%) lower than that of COL1A1 (Smith, et al., 1999). In support of a linkage effect, the adjacent band probe, 17q22–24, was also found extensively contacting the SC-35 domain associated with COL1A1 (Fig. 4 J, top band), though not as frequently (40% of bands) as the region surrounding COL1A1 in 17q21.
Our findings suggest that SC-35 domains are contacted more by specific gene-rich R-bands because they are enriched with certain domain-associating sequences, and that these loci may further affect the organization of closely linked sequences. Differences between R-bands may also reflect differences in the propensity of specific genes within a given R-band to associate with SC-35 domains.
SC-35 domains are rarely contacted by later-replicating (G-band) DNA, and are typically surrounded by a subset of early-replicating (R-band) DNA
Because we were practically limited in the number of specific R- and G-bands we could examine directly, we expanded our analyses to obtain a general overview of the genome's gene-rich and gene-poor DNA. When cells are pulse labeled with halogenated dNTPs during S-phase, gene-rich early-replicating DNA, typically in R-bands, is detected in hundreds of small foci throughout the nuclear interior (Fig. 5 A, green), whereas the gene-poor middle- and late-replicating DNA (most G-bands) correspond respectively to similar small foci at the nucleolar and nuclear periphery (Fig. 5 B, green) and to ∼30 larger patches at the periphery and in the interior (Fig. 5 C, green; Ferreira et al., 1997). We found that SC-35 domains are rarely contacted by the two later-replicating classes of DNA (Fig. 5, B and C), even those signals within the nuclear interior. In contrast, SC-35 domains are typically surrounded by early-replicating DNA foci (Fig. 5 A, insets), though these foci do not exclusively localize around SC-35 domain peripheries. This is consistent with our findings that only a portion of any given R-band directly abuts SC-35 domains, and the fact that even early-replicating R-band DNA is substantially comprised of intergenic sequences and nonexpressed genes. In addition, some active genes are consistently expressed in the nucleoplasm between the SC-35 domains (Smith et al., 1999).
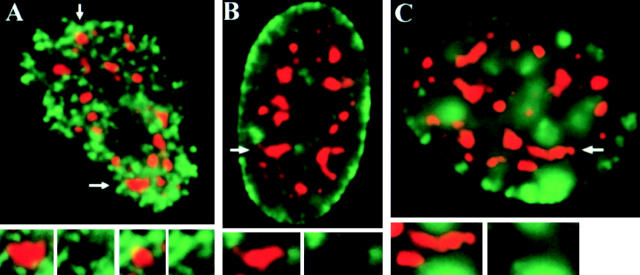
Figure 5.
Differential organization of early- and later-replicating DNA relative to SC-35 domains. (A) An early S-phase nucleus pulse labeled with BrdU (green) shows the typical pattern of hundreds of small early-replication foci throughout the nuclear interior, excluding the nucleolus. All SC-35 domains (red) are contacted by multiple early-replicating foci, which correspond to gene-rich DNA. Nuclei in mid (B) and late (C) S-phase are identified by replication sites of gene-poor DNA at the nuclear periphery or in large clumps that are more interior but on different focal planes than SC-35 domains. Arrows indicate SC-35 domains enlarged in insets.
Using morphometric analysis, we compared the lengths of contact between SC-35 domains and differentially labeled early- versus middle- and late-replicating DNA (see Materials and methods). Measurements indicate that approximately eightfold more of SC-35 domain perimeters per nucleus are contacted by early- rather than later-replicating DNA (the latter included patterns depicted in Fig. 5, B and C; see Materials and methods and Table IV). Because G- and R-bands make up approximately equal amounts of the genome, the observed differences in early- and later-replicating DNA contacts are not due to differences in their abundance, but rather to their specific partitioning within the nucleus and in relation to SC-35 domains.
Table IV. Amount of SC-35 domain edges contacted by early and later replicating DNA.
| Replication timing of DNA | Number of cells measured | Mean percentage of SC-35 domain perimeters contacteda |
|---|---|---|
| Early S-phase | 41 | 24 ± 7 |
| Later S-phaseb | 65 | 3 ± 3 |
Sum of all SC-35 domain perimeters per cell ± SD.
Includes replication patterns depicted in Fig. 5, B and C.
Discussion
This paper provides evidence for a fundamental and new concept relating nuclear and chromosome organization: genes cluster around individual SC-35 domains, and this organization is related to the clustering of genes into chromosomal bands. Our collective findings support a hierarchy of nuclear compartments in which early-replicating DNA and SC-35 domains are circumscribed to the broad nuclear interior, specific chromosome R-bands preferentially align with individual SC-35 domains, and each aligned band contains specific active genes that cluster at the immediate edge of the factor-rich domain. Thus, the region immediately surrounding the SC-35 domains may be considered a euchromatic neighborhood of multiple genes, many of which are linked on the same chromosome.
The number of genes associated with a given SC-35 domain may be quite large, indicating a complex organization. The four pairs of specific genes examined here each associate with a common SC-35 domain, with their transcripts intermixing in the domain interior. Although only a limited number of genes in the genome (21/∼30,000) have been surveyed for localization with respect to SC-35, half of these loci are nonrandomly domain associated. The clustering of multiple specific genes and extensions of R-band DNA around the periphery of SC-35 domains is clearly reflective of organization around a structure, rather than of factors simply collected on the transcripts of a highly expressed gene (summarized in Fig. 6). This is consistent with our earlier suggestion that each SC-35 domain corresponds at the ultrastructural level to a cluster of interchromatin granules surrounded by perichromatin fibrils, which represent nascent transcripts of individual genes (Xing et al., 1995). Indeed, the extensive intermingling of two different RNAs within a common SC-35 domain strongly suggests that these are not simply two very close, large perichromatin fibrils.
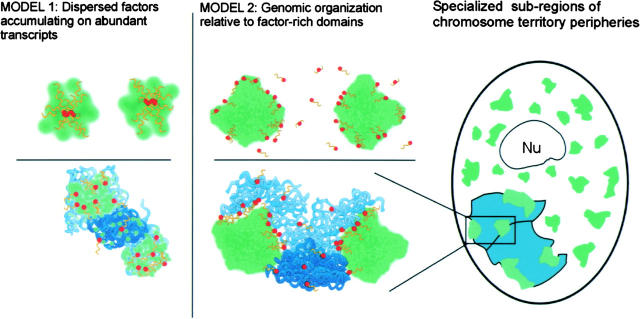
Figure 6.
Data presented here support model 2 (center) rather than model 1 (left), and are further summarized at far right. (Model 1) mRNA metabolic factors, including SC-35 (green), accumulate on transcripts (yellow) of a single highly active gene (red, top left) or genomic clusters of genes (bottom left). This model requires no structural organization of genes relative to the large concentrations of splicing factors, which merely reflect the distribution of transcripts on different genes. (Model 2) Multiple genes (center top) cluster at the periphery of a single large accumulation of mRNA metabolic factors. R-band DNA (light blue), which is gene rich, is more intimately associated with these SC-35 domains than gene-poor G-band DNA (dark blue, bottom center). (Right) Each chromosome territory (aqua) associates with three or four SC-35 domains, indicating specialized regions at the chromosome territory periphery. These contain domain-associated genes that can even come from different chromosome arms. An SC-35 domain can also associate with genes from different chromosomes. Because domain choice for individual genes is often random, the relative positions of their respective chromosomes also may be highly variable.
With the exception of syntenic genes, all pairs of genes examined here associate with a common SC-35 domain at frequencies equal to that expected for genes that preferentially localized to one of the ∼15–30 SC-35 domains, but randomly “choose” a specific domain. Random domain choice was even observed for two coordinately expressed, functionally related genes, though it remains possible that other sets of coregulated genes may preferentially associate with the same SC-35 domain. In addition, a more defined three-dimensional organization of genes might occur in cell nuclei of tissues (Marshall et al., 1996), which are clearly different structurally from cultured cells. The largely random co-association frequencies observed here also have implications for the relative organization of chromosome territories in the nucleus.
Our analysis of chromosome 7 sequences importantly indicates that there may be specialized subregions on the surface of a chromosome territory that interact with SC-35 domains (Fig. 6, right). These subregions contain specific genes that are known to associate with SC-35 domains with high specificity (e.g., COL1A1, COL1A2, ACTB). Our data suggest that two syntenic, domain-associating genes (COL1A2 and ACTB) assort randomly into these chromosome territory subregions. These genes are in different and distant cytogenetic bands (>70 Mb apart), further indicating that sequences from different bands can intermingle within each subchromosomal region that contacts an SC-35 domain, though previous reports have suggested that different bands may be mostly separated from each other within the chromosome territory (Dietzel et al., 1998; Zink et al., 1999).
The COL1A1 gene was occasionally found at the tip of an extended piece of band DNA, suggestive of purposeful targeting to a preexisting structure. In contrast, our group and others have previously hypothesized that genes like COL1A1 might nucleate the formation of SC-35 domains (Moen et al., 1995; Xing et al., 1995; Huang and Spector, 1996; Misteli et al., 1997). The nuclear organization we have uncovered here, in fact, may be both a cause and a consequence of gene activity (Melcak et al., 2000), though the precise contribution of each mechanism is unknown.
Our data support the model that SC-35 domain neighborhoods may be nuclear hubs of enhanced mRNA metabolic activity (Carter et al., 1991). Other evidence suggests that SC-35 domains are more than factor reservoirs or assembly sites, but also may be intimately involved in mRNA maturation and export (Johnson et al., 2000; Melcak et al., 2000; Reed and Magni, 2001; Shopland et al., 2002). Although the factors required for mRNA transcription, processing, and transport are available throughout much of the nucleus and are presumably accessible to all active genes, concentration of these factors within SC-35 domains may serve to enhance the expression of the nearest genes. Further, we hypothesize that the enrichment of genes in chromosomal R-bands may have evolved to facilitate gene clustering in these expression-promoting neighborhoods. The fact that chromosome bands are often evolutionarily conserved strongly suggests that this level of organization is important and functional, but few links to gene function have been identified or postulated. Based on changes in the conformation of a highly repetitive, very large exogenous DNA sequence, one report has proposed that the close linkage of genes in R-bands might facilitate chromatin decondensation, and hence transcription (Tumbar et al., 1999). Our findings suggest that endogenous bands are specifically organized around structures replete with metabolic factors, thereby enhancing the processing and transport of their products as well (Johnson et al., 2000; Shopland et al., 2002).
Materials and methods
Cell culture
For interphase cell preparations, WI-38 fetal human diploid fibroblasts (American Type Culture Collection) were grown on glass coverslips at 37°C in Eagle's basal medium (Life Technologies) supplemented with 10% heat-inactivated FCS, and were fixed according to two previously established protocols that produced similar results (Johnson et al., 1991; Kurz et al., 1996). For BrdU labeling, cells were grown in fresh, prewarmed media containing 30 μg/ml BrdU (Roche) for 15 min, and were washed twice with prewarmed media before fixation. For metaphase chromosome preparations, phytohemagglutinin-stimulated human peripheral blood lymphocytes were grown in BrdU (Sigma-Aldrich) to enhance banding, and in methotrexate to enhance yields of mitotic cells. Chromosome spreads were prepared from hypotonically swollen cells according to standard cytogenetic methods (Johnson et al., 1991).
FISH
Specific gene probes were generated by nick translating the following plasmids with either biotin- or digoxigenin-dUTP (Roche; Johnson et al., 1991). CG103 contains genomic DNA with the entire COL1A1 gene (Barsh et al., 1984). pHVC1 contains full-length COL1A1 cDNA (American Type Culture Collection). Clone 7404 has genomic DNA containing the entire COL1A2 sequence (D. Prockop, MCP-Hahnemann School of Medicine, Philadelphia, PA; Korkko et al., 1998). pSTL14 includes full-length COL1A2 cDNA (D. Rowe, University of Connecticut Health Center, Farmington, CT; Lee et al., 1988). pHFβ-A-1 includes a full-length ACTB cDNA (J. Stein, University of Massachusetts Medical School, Worcester, MA; Ponte et al., 1984; Ng et al., 1985). A full-length cDNA of lamin A was obtained from H. Worman (Columbia University, New York, NY; Fisher et al., 1986). Human band-specific probes were purchased from ResGen, PCR amplified according to manufacturer's directions but in the absence of labeled nucleotide, and then nick translated with label as above. Biotinylated chromosome 7 StarFISH paint was obtained from CamBio.
Hybridization to DNA was performed as previously described (Johnson et al., 1991) or according to manufacturer's directions (CamBio). To detect only RNA in fibroblasts, cell denaturation steps were omitted and cells were hybridized with cDNA probes (Johnson et al., 1991). Hybridized probes were detected with TRITC- or FITC-sheep anti-digoxigenin antibodies (Jackson ImmunoResearch Laboratories), FITC-avidin (Roche), or Alexa® 594–streptavidin (Molecular Probes, Inc.). Where indicated, cells and chromosomes were counter-stained with 1 μg/ml DAPI (Sigma-Aldrich).
Immunostaining
FISH was coupled with immunofluorescence with anti-SC-35 antibody (Sigma-Aldrich) as described (Xing et al., 1995). To detect BrdU incorporated into DNA, cells first were heat denatured as above and then incubated with either a 1/500 dilution of mouse anti-BrdU (Partec) or 1/20 dilution of rat anti-BrdU (Harlan Bioproducts for Science) mixed with either 1/1,000 rabbit anti-SRm300 (B. Blencowe, University of Toronto, Toronto, Ontario, Canada; Blencowe et al., 1994) or 1/500 mouse anti-SC-35 antibodies. Primary antibodies were detected with the following: FITC-donkey or -goat anti–mouse IgG, AMCA-donkey anti–mouse IgG, TRITC-donkey anti–rabbit IgG, or TRITC-donkey anti–rat IgG (all obtained from Jackson ImmunoResearch Laboratories), or Alexa® 488–goat anti–mouse IgG (Molecular Probes, Inc.). All samples were mounted in Vectashield® (Vector Laboratories).
Microscopic analysis
Cells were examined with a microscope (Axioplan; Carl Zeiss MicroImaging, Inc.) equipped with a filter wheel, triple-bandpass epifluorescence filter set (Chroma Technology), and a 100×, 1.4 N.A. objective (Carl Zeiss MicroImaging, Inc.). Digital images were acquired by cameras (Series 200 or Quantix; Photometrics) and MetaMorph® imaging software (Universal Imaging Corp.). Where indicated, Z-series image stacks were acquired at 0.1-μm intervals using a Ludl stage controller (Hawthorne) and rendered in 3D with MetaMorph's® remove haze function, which uses a nearest neighbor algorithm, or by exhaustive photon reassignment (Fay et al., 1989; Carter et al., 1993). For presentation, image contrast was adjusted with Adobe Photoshop® v.6.0 to scale the most intense and weakest pixels between 255 and 0. Hence, they do not retain precise quantitative information for signal intensity, but the relative locations of signals are maintained.
Images were evaluated either by scoring through the microscope eyepiece or by measuring digital images. For scoring, cells were randomly selected and evaluated by at least two investigators. Signals were considered “associated” if they overlapped (appeared yellow) or contacted the edge of an SC-35 domain or each other, with no visible space between them (juxtaposed red and green). Given this definition and the resolution limits of the microscope, the distance between two associated objects is ≤ ∼300 nm. For scoring co-associations of RNA signals, those that abutted each other were also included because COL1A2 RNA was used to demark SC-35 domains in some experiments, and is restricted to the domain periphery in some cells (Shopland et al., 2002). Calculations for theoretical frequencies of random domain choice for a given gene relative to another were made for genes determined previously (Smith et al., 1999) to associate with any of the SC-35 domains at significantly greater than nonrandom frequencies. The expected frequency of random association for one copy of a gene (a) with a domain marked by another gene (b) is (1/nd)(fa)(fb), where nd is the number of domains per cell and fa and fb are the independent frequencies of domain association for each gene. In a cell with two copies of each gene, this increases fourfold, assuming that the number of cells in which more than two genes associate with the same domain is negligible, which is consistent with our observations.
Measurements of chromosome band contact lengths and perimeters were made in MetaMorph® from two-dimensional images of cells with two distinct band territories. The focal plane that showed the most contact between domains and bands was selected for imaging. Because SC-35 domains are largely situated within a single focal plane (Carter et al., 1993), two-dimensional analyses were sufficient to estimate the degree of SC-35 domain contact. A small fraction of cells (<20%) had DNA signals directly above or below domains and were not evaluated. For each cell, measurements of SC-35 domain contacts were summed together and normalized to the sum of all SC-35 domain perimeters in that cell. The average DNA content of each band probe was determined by length measurements of individual band signals on metaphase chromosomes relative to a cohybridized, standard band probe.
Online supplemental material
A three-dimensional rotational movie (Video 1) of the deconvolved and reconstructed image stack used to generate Fig. 1 C shows the relationship of COL1A1 (green) and COL1A2 (red) transcripts inside an SC-35 domain (not depicted). Rotational movies (Video 2 and Video 3) of three-dimensional reconstructions shown in Fig. 4 (K and L) illustrate the topological organization of R-band 17q21 (red, K) and G-band 3p14 (red, L) relative to the nearest SC-35 domains (green). Online supplemental material available at http://www.jcb.org/cgi/content/full/jcb.200303131/DC1.
Acknowledgments
We thank Drs. Kelly Smith and Lisa Hall for comments on this manuscript, Dr. Joel Graber for advice on statistics, and Drs. David Rowe, Darwin Prockop, Janet Stein, Howard Worman, and Ben Blencowe for reagents.
This work was supported by National Institutes of Health (NIH) grants to J.B. Lawrence (GM68138 and GM53234). The contents are solely the responsibility of the authors and do not necessarily reflect the official views of the NIH.
The online version of this article includes supplemental material.
L.S. Shopland's present address is The Jackson Laboratory, 600 Main Street, Bar Harbor, ME 04609.
Abbreviations used in this paper: ACTB, β-actin; COL1A1, collagen type 1, α1; COL1A2, collagen type 1, α2; G-band, Giemsa dark band; R-band, reverse band.
References
- Barsh, G.S., C.L. Roush, and R.E. Gelinas. 1984. DNA and chromatin structure of the human alpha-1 (I) collagen gene. J. Biol. Chem. 259:14906–14913. [PubMed] [Google Scholar]
- Belmont, A.S., and K. Bruce. 1994. Visualization of G1 chromosomes: a folded, twisted, supercoiled chromonema model of interphase chromatid structure. J. Cell Biol. 127:287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe, B.J., J.A. Nickerson, R. Issner, S. Penman, and P.A. Sharp. 1994. Association of nuclear matrix antigens with exon-containing splicing complexes. J. Cell Biol. 127:593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, K.C., K.L. Taneja, and J.B. Lawrence. 1991. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J. Cell Biol. 115:1191–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, K.C., D. Bowman, W. Carrington, K. Fogarty, J.A. McNeil, F.S. Fay, and J.B. Lawrence. 1993. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 259:1330–1335. [DOI] [PubMed] [Google Scholar]
- Cremer, T., and C. Cremer. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292–301. [DOI] [PubMed] [Google Scholar]
- Croft, J.A., J.M. Bridger, S. Boyle, P. Perry, P. Teague, and W.A. Bickmore. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145:1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzel, S., A. Jauch, D. Kienle, G. Qu, H. Holtgreve-Grez, R. Eils, C. Munkel, M. Bittner, P.S. Meltzer, J.M. Trent, and T. Cremer. 1998. Separate and variably shaped chromosome arm domains are disclosed by chromosome arm painting in human cell nuclei. Chromosome Res. 6:25–33. [DOI] [PubMed] [Google Scholar]
- Fay, F.S., W. Carrington, and K.E. Fogarty. 1989. Three-dimensional molecular distribution in single cells analysed using the digital imaging microscope. J. Microsc. 153:133–149. [PubMed] [Google Scholar]
- Ferreira, J., G. Paolella, C. Ramos, and A.I. Lamond. 1997. Spatial organization of large-scale chromatin domains in the nucleus: a magnified view of single chromosome territories. J. Cell Biol. 139:1597–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, D.Z., N. Chaudhary, and G. Blobel. 1986. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl. Acad. Sci. USA. 83:6450–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, M.A., G.P. Holmquist, M.C. Gray, L.A. Caston, and A. Nag. 1984. Replication timing of genes and middle repetitive sequences. Science. 224:686–692. [DOI] [PubMed] [Google Scholar]
- Guan, X.Y., J.M. Trent, and P.S. Meltzer. 1993. Generation of band-specific painting probes from a single microdissected chromosome. Hum. Mol. Genet. 2:1117–1121. [DOI] [PubMed] [Google Scholar]
- Hendzel, M.J., M.J. Kruhlak, and D.P. Bazett-Jones. 1998. Organization of highly acetylated chromatin around sites of heterogeneous nuclear RNA accumulation. Mol. Biol. Cell. 9:2491–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., and D.L. Spector. 1996. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J. Cell Biol. 133:719–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C., D. Primorac, M. McKinstry, J. McNeil, D. Rowe, and J.B. Lawrence. 2000. Tracking COL1A1 RNA in osteogenesis imperfecta. splice-defective transcripts initiate transport from the gene but are retained within the SC35 domain. J. Cell Biol. 150:417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C.V., R.H. Singer, and J.B. Lawrence. 1991. Fluorescent detection of nuclear RNA and DNA: Implication for genome organization. Methods Cell Biol. 35:73–99. [PubMed] [Google Scholar]
- Karsenty, G., and R.W. Park. 1995. Regulation of type I collagen genes expression. Int. Rev. Immunol. 12:177–185. [DOI] [PubMed] [Google Scholar]
- Korkko, J., L. Ala-Kokko, A. De Paepe, L. Nuytinck, J. Earley, and D.J. Prockop. 1998. Analysis of the COL1A1 and COL1A2 genes by PCR amplification and scanning by conformation-sensitive gel electrophoresis identifies only COL1A1 mutations in 15 patients with osteogenesis imperfecta type I: identification of common sequences of null-allele mutations. Am. J. Hum. Genet. 62:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz, A., S. Lampel, J.E. Nickolenko, J. Bradl, A. Benner, R.M. Zirbel, T. Cremer, and P. Lichter. 1996. Active and inactive genes localize preferentially in the periphery of chromosome territories. J. Cell Biol. 135:1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, J.B., R.H. Singer, and J.A. McNeil. 1990. Interphase and metaphase resolution of different distances within the human dystrophin gene. Science. 249:928–932. [DOI] [PubMed] [Google Scholar]
- Lee, S.T., B.D. Smith, and D.S. Greenspan. 1988. Construction of a full-length cDNA encoding human pro-alpha 2(I) collagen and its expression in pro-alpha 2(I)-deficient W8 rat cells. J. Biol. Chem. 263:13414–13418. [PubMed] [Google Scholar]
- Li, G., G. Sudlow, and A.S. Belmont. 1998. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning. J. Cell Biol. 140:975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W.F., A.F. Dernburg, B. Harmon, D.A. Agard, and J.W. Sedat. 1996. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol. Biol. Cell. 7:825–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcak, I., S. Cermanova, K. Jirsova, K. Koberna, J. Malinsky, and I. Raska. 2000. Nuclear pre-mRNA compartmentalization: trafficking of released transcripts to splicing factor reservoirs. Mol. Biol. Cell. 11:497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli, T., J.F. Caceres, and D.L. Spector. 1997. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 387:523–527. [DOI] [PubMed] [Google Scholar]
- Moen, P.T. Jr., C.V. Johnson, M. Byron, L.S. Shopland, I. de la Serna, A. Imbalzano, and J.B. Lawrence. 2003. Repositioning of muscle-specific genes to the periphery of SC-35 domains during skeletal myogenesis. Mol. Biol. Cell. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen, P.T. Jr., K.P. Smith, and J.B. Lawrence. 1995. Compartmentalization of specific pre-mRNA metabolism: an emerging view. Hum. Mol. Genet. 4:1779–1789. [DOI] [PubMed] [Google Scholar]
- Ng, S.Y., P. Gunning, R. Eddy, P. Ponte, J. Leavitt, T. Shows, and L. Kedes. 1985. Evolution of the functional human beta-actin gene and its multi-pseudogene family: conservation of noncoding regions and chromosomal dispersion of pseudogenes. Mol. Cell. Biol. 5:2720–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponte, P., S.Y. Ng, J. Engel, P. Gunning, and L. Kedes. 1984. Evolutionary conservation in the untranslated regions of actin mRNAs: DNA sequence of a human beta-actin cDNA. Nucleic Acids Res. 12:1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R., and K. Magni. 2001. A new view of mRNA export: separating the wheat from the chaff. Nat. Cell Biol. 3:E201–E204. [DOI] [PubMed] [Google Scholar]
- Sadoni, N., S. Langer, C. Fauth, G. Bernardi, T. Cremer, B.M. Turner, and D. Zink. 1999. Nuclear organization of mammalian genomes. Polar chromosome territories build up functionally distinct higher order compartments. J. Cell Biol. 146:1211–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland, L.S., C.V. Johnson, and J.B. Lawrence. 2002. Evidence that all SC-5 domains contain mRNAs and that transcripts can be structurally constrained within these domains. J. Struct. Biol. 140:131–139. [DOI] [PubMed] [Google Scholar]
- Smith, K.P., P.T. Moen, K.L. Wydner, J.R. Coleman, and J.B. Lawrence. 1999. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol. 144:617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry, M. 1993. Differential location of nucleic acids within interchromatin granule clusters. Eur. J. Cell Biol. 62:259–269. [PubMed] [Google Scholar]
- Trask, B.J., H. Massa, S. Kenwrick, and J. Gitschier. 1991. Mapping of human chromosome Xq28 by two-color fluorescence in situ hybridization of DNA sequences to interphase cell nuclei. Am. J. Hum. Genet. 48:1–15. [PMC free article] [PubMed] [Google Scholar]
- Tumbar, T., G. Sudlow, and A.S. Belmont. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi, E.V., E. Chevret, T. Jones, R. Vatcheva, J. Williamson, S. Beck, R.D. Campbell, M. Goldsworthy, S.H. Powis, J. Ragoussis, et al. 2000. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113:1565–1576. [DOI] [PubMed] [Google Scholar]
- Wei, X., S. Somanathan, J. Samarabandu, and R. Berezney. 1999. Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J. Cell Biol. 146:543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y., C.V. Johnson, P.T. Moen Jr., J.A. McNeil, and J. Lawrence. 1995. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J. Cell Biol. 131:1635–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink, D., H. Bornfleth, A. Visser, C. Cremer, and T. Cremer. 1999. Organization of early and late replicating DNA in human chromosome territories. Exp. Cell Res. 247:176–188. [DOI] [PubMed] [Google Scholar]