Abstract
Plakophilin 3 (PKP3) is a recently described armadillo protein of the desmosomal plaque, which is synthesized in simple and stratified epithelia. We investigated the localization pattern of endogenous and exogenous PKP3 and fragments thereof. The desmosomal binding properties of PKP3 were determined using yeast two-hybrid, coimmunoprecipitation and colocalization experiments. To this end, novel mouse anti-PKP3 mAbs were generated. We found that PKP3 binds all three desmogleins, desmocollin (Dsc) 3a and -3b, and possibly also Dsc1a and -2a. As such, this is the first protein interaction ever observed with a Dsc-b isoform. Moreover, we determined that PKP3 interacts with plakoglobin, desmoplakin (DP) and the epithelial keratin 18. Evidence was found for the presence of at least two DP–PKP3 interaction sites. This finding might explain how lateral DP–PKP interactions are established in the upper layers of stratified epithelia, increasing the size of the desmosome and the number of anchoring points available for keratins. Together, these results show that PKP3, whose epithelial and epidermal desmosomal expression pattern and protein interaction repertoire are broader than those of PKP1 and -2, is a unique multiprotein binding element in the basic architecture of a vast majority of epithelial desmosomes.
Keywords: armadillo; cell adhesion; desmosomes; protein interaction; two-hybrid system
Introduction
Desmosomes are adhering junctions that are involved in cell–cell adhesion, differentiation, and signal transduction (Hatzfeld, 1997; Green and Gaudry, 2000). They are responsible for strong intercellular adhesion and are as such indispensable in tissues that undergo mechanical stress, for example in the skin (McGrath et al., 1997). Recently, evidence was found for an active role of desmogleins (Dsgs)*in delineating the differentiation-specific properties of the epidermis (Elias et al., 2001; Ishii and Green, 2001). Desmosomal adhesion was also shown to be involved in epithelial morphogenesis to an extent comparable to the role of adherens junctions (Runswick et al., 2001). These results indicate a wider role for desmosomes than the mere establishment of rigid cell–cell adhesion.
Desmosomes are assembled on a scaffold of transmembrane desmosomal cadherins, the Dsgs and desmocollins (Dscs), both existing in three isoforms (Koch et al., 1992; King et al., 1997; Green and Gaudry, 2000). Dscs occur in two splice variants, a longer “a” and a shorter “b” type, as a result of alternative splicing. The corresponding genes display a tightly regulated expression pattern, dependent on cell type and cell differentiation (King et al., 1997; Green and Gaudry, 2000; Messent et al., 2000).
Desmosomal cadherins interact with a complex of plaque molecules, comprising plakoglobin (Pg), plakophilins (PKPs), and desmoplakin (DP). This complex binds intermediate filaments through DP (Green and Gaudry, 2000). The importance of DP and Pg in desmosome function has been shown in the last few years (Bierkamp et al., 1996; Ruiz et al., 1996; Gallicano et al., 1998; Armstrong et al., 1999; McKoy et al., 2000). Using mice lacking DP expression in skin epithelium, it was recently shown that functional desmosomes are required for epithelial sheet assembly (Vasioukhin et al., 2001). PKP1 is an essential desmosomal plaque molecule of the uppermost differentiated layers of stratified epithelium (Hatzfeld et al., 1994; Heid et al., 1994). Its importance is shown in patients lacking PKP1 expression, resulting in a skin fragility syndrome (McGrath et al., 1997; McGrath, 1999). PKP2 is found in basal cell layers of stratified epithelia and in single-layered epithelium, as well as in nonepithelial desmosomes (Mertens et al., 1996). Recently, PKP2-interacting desmosomal proteins have been identified, and their involvement in β-catenin–mediated signaling pathways was shown (Chen et al., 2002). Both PKP1 and PKP2 occur as widespread nuclear proteins whose functions are still unknown (Mertens et al., 1996; Schmidt et al., 1997). Finally, the latest PKP identified (PKP3) was shown to be generally expressed in epithelia, with the exception of hepatocytes (Bonné et al., 1999; Schmidt et al., 1999).
Here, we report that PKP3 binds directly to the desmosomal cadherins Dsg1, Dsg2, Dsg3, Dsc3a, and Dsc3b, and possibly also to Dsc1a and Dsc2a. This identifies PKP3 as the first interaction partner reported for a Dsc-b isoform. Moreover, a direct interaction was observed between PKP3 on the one hand and Pg, DP, and the epithelial cytokeratin 18 (CK18) on the other hand. We also found evidence for the presence of at least two DP-binding sites in the PKP3head domain. Binding of more than one DP molecule by PKPs might explain how desmosomes are laterally enlarged in upper layers of stratified epithelia, as previously hypothesized (Kowalczyk et al., 1999a). The broad spectrum of PKP3 interaction partners in desmosomes, combined with its ubiquitous expression in epithelia, suggests that PKP3 is an important building block in the continuously changing composition of epithelial and epidermal desmosomes.
Results
Detection of desmosomal PKP3 in simple and complex epithelia by use of novel mAbs
In immunofluorescence detection experiments using the newly generated anti-PKP3 mAbs 23E3/4 and 12B11F8 raised against different PKP3-specific peptides, we detected PKP3 in a desmosomal punctate pattern along cell–cell contacts, besides weak immunopositivity in the cytoplasm. This observation was made in both methanol- and formaldehyde-fixed cells (exemplified in Fig. 1 , a–c). Control immunodetections of DP (unpublished data) or PKP2 (Fig. 1 d) revealed a similar pattern along cell–cell contacts. The punctate localization of PKP3 and PKP2 along cell–cell contacts is also obvious from the larger magnifications (Fig. 1, a′–d′). Western blot experiments showed that these novel mouse anti-PKP3 mAbs do not cross react with either PKP1 or PKP2 (Fig. 1 e). However, using these mAbs we could not confirm nuclear localization of PKP3 observed previously with a rabbit pAb (Bonné et al., 1999). Intriguingly, mAb 12B11F8 was raised against the same antigenic peptide as this rabbit pAb. Therefore, the reported nuclear PKP3 localization using the rabbit pAb may be caused by additional immunoreactivity, unrelated to PKP3 despite the fact that preincubation of the polyclonal antiserum with the antigenic peptide could block its activity (Bonné et al., 1999).
Figure 1.
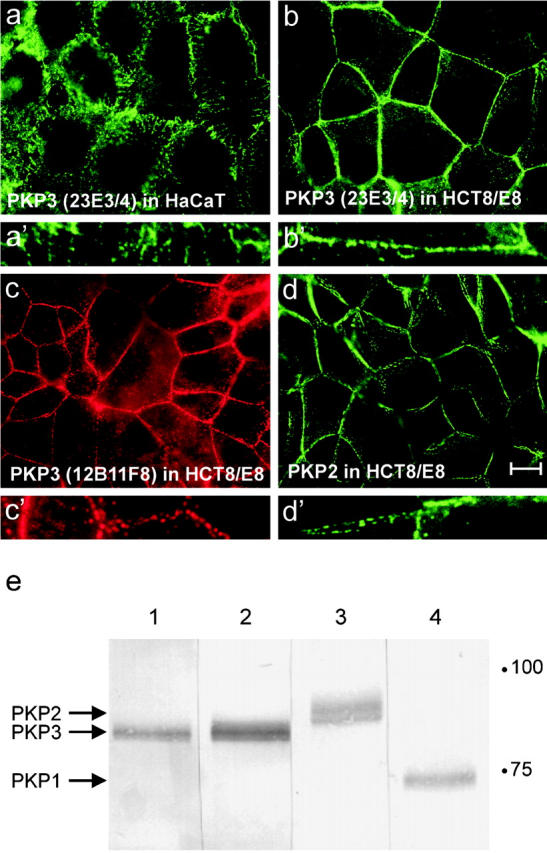
Immunodetection of PKPs using different mouse mAbs. PKP3, detected with mAb 23E3/4 (a and b), 12B11F8 (c), or PKP2 (d) show a similar expression pattern along cell–cell contacts in methanol-fixed HaCaT (a) and HCT8/E8 (b–d) cells. Larger magnifications display the punctate localization of these proteins along cell–cell contacts (a′–d′). Bar, 10 μm. (e) Western blot detection of PKP3 (lane 1, mAb 12B11F8; lane 2, 23E3/4), PKP2 (lane 3), and PKP1 (lane 4) in HaCaT protein lysates. Equal amounts of protein were loaded in each lane.
mAb 23E3/4 was also used on paraffin sections of formalin-fixed human skin and colon. PKP3 is detected in all living layers of the human epidermis, but not in the stratum corneum or dermis (Fig. 2 a). At higher magnifications, the punctate localization of PKP3 along cell–cell contacts, typical of desmosomal components, is clearly visible (Fig. 2 b). Epidermal cells of the hair follicle also synthesize PKP3 (Fig. 2 c), and PKP3 protein was observed in both the inner and outer root sheats of hair follicles (unpublished data). No signal could be detected in the negative control sections, in which the primary antibody was omitted (Fig. 2 d). PKP3 was also detected in vivo in simple epithelia such as colon (Fig. 2 e), whereas no signal was observed in the corresponding negative control sections (Fig. 2 f). In line with previously reported expression data on PKP3 (Bonné et al., 1999; Schmidt et al., 1999), the present results show that PKP3 is cosynthesized with the differentiation- and cell type–specific desmosomal cadherins, and with both PKP1 and PKP2, in single- and multilayered epithelia.
Figure 2.
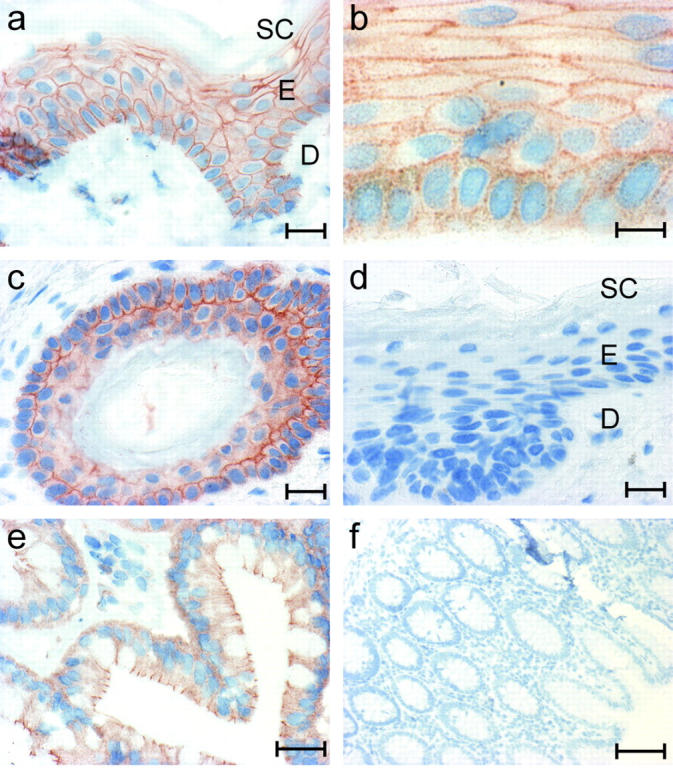
Immunohistochemical detection of PKP3 in paraffin sections of formalin-fixed human skin and colon using mAb 23E3/4. PKP3 is expressed in the living cell layers of the epidermis, but not in the stratum corneum and the dermis (a). At higher magnification, the interrupted PKP3 localization along cell–cell contacts is visible (b). The epidermal cells of the hair root also express PKP3 (c); no signal can be detected in the negative control sections, in which the primary antibody was omitted (d). PKP3 is expressed in simple colon epithelium (e); no signal is observed in the negative control sections (f). SC, stratum corneum; E, epidermis; D, dermis. Bars: 20 μm (a and c–e), 10 μm (b), and 100 μm (f).
The head and arm domains of PKP3 localize preferentially in the cytoplasm and nucleus, respectively
To study the intracellular distribution of full-length PKP3 and fragments thereof, transfection of PKP3 expression constructs was performed in HCT8/E8, MCF7/AZ, COS1, and PtK2 cells, yielding consistent data. A schematic overview of the protein fragments encoded by the different constructs is shown in Fig. 3 . In HCT8/E8 cells, full-length human PKP3 tagged with GFP displays an intracellular distribution similar to endogenous PKP3 detected by mAbs, i.e., a clear-cut localization in desmosomes, weaker in the cytoplasm, and absent from the cell nucleus (Fig. 4 a). Exogenous PKP3 colocalizes with endogenous DP in desmosomes (Fig. 4, b and c). Larger magnifications emphasize the punctate (co)localization of these proteins along cell–cell contacts of transfected cells (Fig. 4, a′–c′). GFP-tagged PKP3ΔHR2 protein displays a localization very similar to full-length PKP3 (Fig. 4 d). The HR2 domain is the only conserved sequence stretch in the head domain of PKPs (Bonné et al., 1999). The intense accumulation in desmosomes of this mutated protein overlaps with endogenous DP (Fig. 4, e and f). Larger magnifications clearly display the punctate (co) localization along cell–cell contacts of transfected cells (Fig. 4, d′–f'′ and d′′–f'′′). In contrast, the GFP-tagged PKP3arm fragment localizes in the cell nucleus, whereas only a faint localization is observed at cell–cell contacts (Fig. 4 g); DP detection in the same cell field is shown in Fig. 4 h. The GFP-tagged PKP3head domain is often observed as discrete aggregates in the cytoplasm and localizes only weakly in cell–cell contacts (Fig. 4 i); control detection of DP in the same field is shown in Fig. 4 j. In MCF7/AZ cells, a similar distribution of the GFP-PKP3head domain is observed (Fig. 4 k); detection of endogenous PKP3 in the same field using mAb 23E3/4 is shown in Fig. 4 l. In MCF7/AZ cells, cell–cell contacts are often less pronounced compared with HCT8/E8 cells, and endogenous PKP3 was sometimes found to colocalize with GFP-PKP3head aggregates (Fig. 4, k and l). Finally, exogenous PKP3headΔHR2 displayed an intracellular localization pattern like PKP3head (unpublished data).
Figure 3.
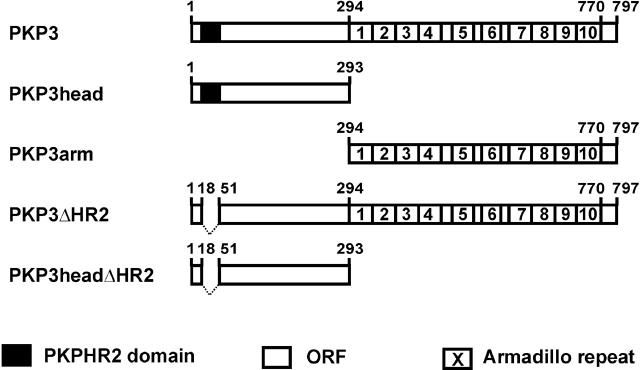
PKP3 fragments encoded by the eukaryotic expression and two-hybrid constructs used. All PKP3 constructs contain human PKP3 cDNA or fragments thereof. GFP and myc tags are located at the carboxy-terminal ends of the proteins. The PKP HR2 domain (PKPHR2) comprises the only conserved region in the head domains of PKPs-1 to -3. ORF, open reading frame.
Figure 4.
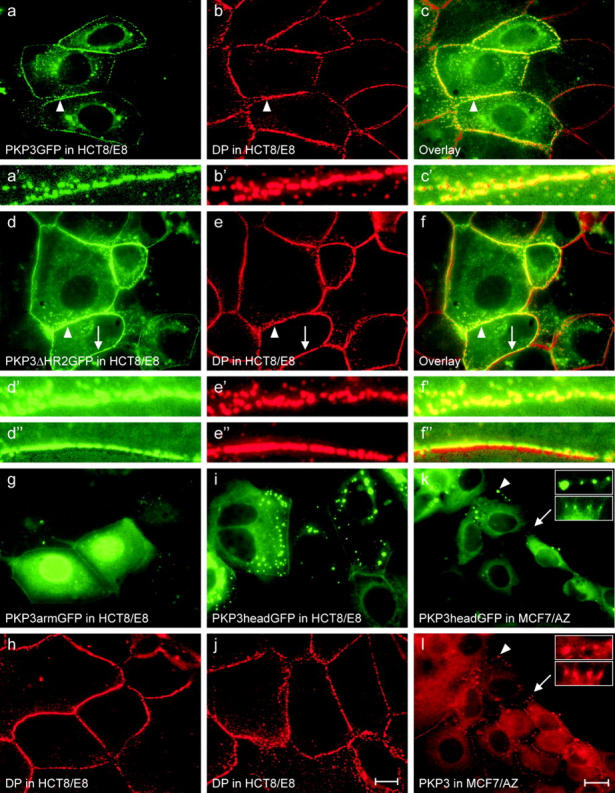
Intracellular localization of exogenous GFP-tagged PKP3 and fragments thereof in HCT8/E8 and MCF7/AZ cells. In HCT8/E8 cells, full-length GFP-tagged PKP3 localizes predominantly in desmosomes and to a lesser extent in the cytoplasm (a). PKP3GFP and DP (b) overlap in desmosomes of transfected cells (c). The punctate localization of these proteins along cell–cell contacts is clear from the larger magnifications (a′–c′) of the fields indicated by arrowheads in pictures a–c. GFP-tagged PKP3ΔHR2 displays a similar intracellular localization as the full-length PKP3 (d), and colocalizes with DP (e) in desmosomes of transfected cells (f). The punctate localization of these proteins along cell–cell contacts is obvious from the larger magnifications (d′–f′) of the fields indicated by arrowheads in pictures d–f. As is clear from images d′′–f′′ (fields indicated by arrows in pictures d–f), GFP-tagged PKP3 overlaps only half of the DP signal if transfected cells contact untransfected cells. PKP3arm GFP accumulates to high levels in the cell nucleus, but almost not at all at cell–cell contacts (g); control immunodetection of DP in the same cell field (h). GFP-tagged PKP3head protein accumulates as discrete cytoplasmic aggregates, in addition to a more diffuse cytoplasmic localization and less intense accumulation at cell–cell contacts (i); control detection of DP in the same field (j). GFP-tagged PKP3head is observed in a similar pattern in MCF7/AZ cells (k); endogenous PKP3 is detected using antibody 23E3/4 (l). Arrowheads, occasional endogenous PKP3 localization in PKP3headGFP aggregates; arrows, colocalization of exogenous fusion protein and endogenous PKP3 at sites of cell–cell contact. The marked regions are enlarged in the insets. Bars: 10 μm (a–j) and 20 μm (k and l).
Because COS cells were used in previous studies on PKP1 and PKP2 protein properties (Kowalczyk et al., 1999a; Chen et al., 2002), transfection experiments were repeated in this cell type. As shown in Fig. 5 , the intracellular localizations of full-length PKP3 (a and b), PKP3arm (c and d) and PKP3head (e and f) correspond with our observations made in MCF7/AZ, HCT8/E8 and PtK2 cells (unpublished observations in the latter cell type).
Figure 5.
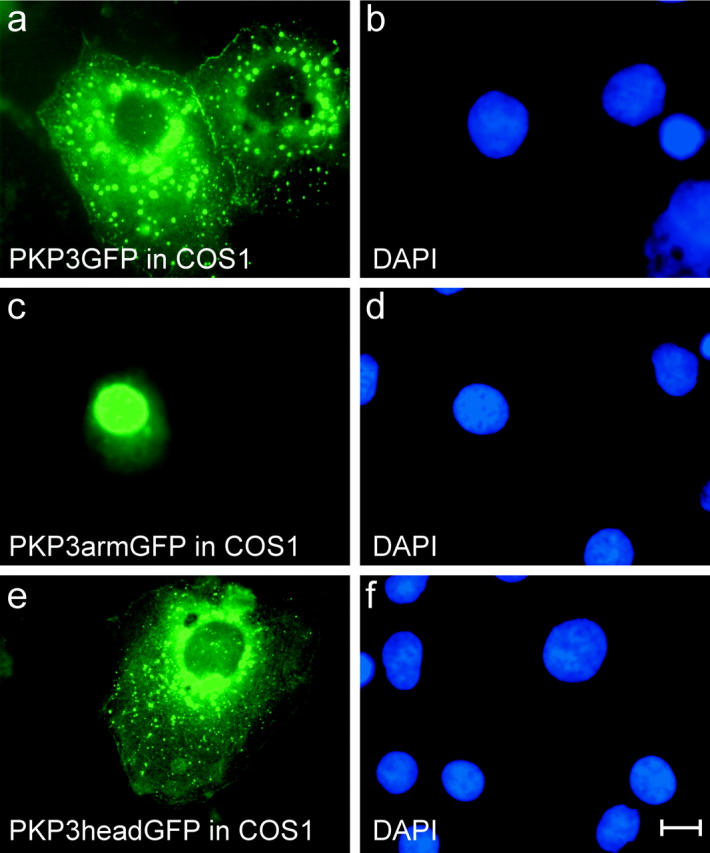
Intracellular localization of exogenous GFP-tagged PKP3 fragments in COS1 cells. Full-length PKP3 localizes at sites of cell–cell contact, and cytoplasmic aggregates are also observed (a). PKP3arm and head fragments localize predominantly in nucleus (c) and cytoplasm (e), respectively. Control DAPI stainings are shown (b, d, and f). Bar, 10 μm.
Exogenous PKP3head fragment binds DP and CK18 proteins; DP is recruited to cell borders by PKP3
MCF7/AZ, HCT8/E8, and COS cells were transfected with constructs encoding GFP-tagged PKP3 or fragments thereof and then subjected to immunofluorescence detection of desmosome-related proteins. PKP3head localized in MCF7/AZ cells as described in the previous paragraph (illustrated in Fig. 6 a). Immunodetection revealed DP not only to be present at sites of cell–cell contacts in MCF7/AZ cells, but also in cytoplasmic aggregates (Fig. 6 b), where it colocalizes with PKP3head (Fig. 6 c). This observation was more clear in MCF7/AZ cells than in HCT8/E8 cells (compare with Fig. 4, i and j). DP did not colocalize with GFP-tagged PKP3headΔHR2 in aggregates, as it was only detected in desmosomal cell–cell contacts (Fig. 6, d–f). This observation suggests that the HR2 domain is involved in PKP3 binding to DP. In contrast, CK18 is observed in aggregates containing either GFP-tagged PKP3head (unpublished data) or GFP-tagged PKP3headΔHR2 protein (Fig. 6, g–i), suggesting that the head domain of PKP3 (but not the HR2 domain) is involved in CK18 interaction. Apparently, oversynthesis of these head fragments results in disturbance of the keratin filament network. Both DP and CK18 bind the PKP3head domain in yeast two-hybrid experiments (see following paragraph). Other desmosomal proteins, like Dsg2 and PKP2, were not found to colocalize in PKP3head aggregates (unpublished data). Similar results were obtained using COS1 cells in transfection experiments, as exemplified by the association of CK18 with PKP3headGFP in COS1 cells (Fig. 6, j–l).
Figure 6.
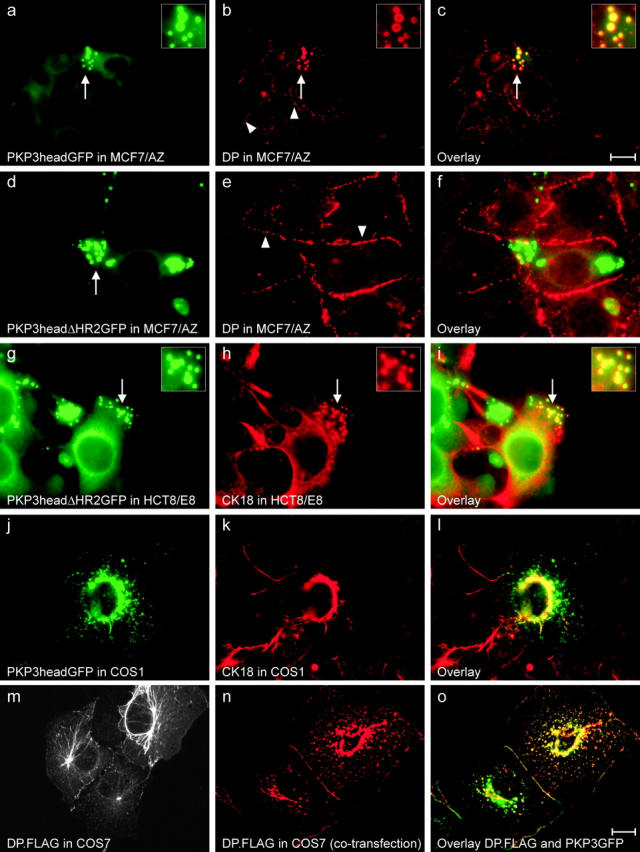
Localization of desmosome-related proteins in various transfected cells. GFP- and FLAG-tagged proteins wre expressed in MCF7/AZ (a–f), HCT8/E8 (g–i), COS1 (j–l), and COS7 (m–o) cells. PKP3head (a and j) and PKP3headΔHR2 (d and g) easily accumulate as discrete cytoplasmic aggregates (arrows in a and g). Endogenous DP (b) localizes to cell–cell contacts (arrowheads) and aggregates (arrows). There is clear colocalization with GFP-tagged PKP3head in these aggregates (c). Insets represent larger magnifications of the areas indicated with arrows. In contrast, GFP-tagged PKP3headΔHR2 (d) and DP (e) do not colocalize (f), whereas GFP-tagged PKP3ΔHR2 (g) and CK18 (h) still localize together in aggregates (i), as indicated by arrows (also see insets, representing larger magnifications). The same observation is made for PKP3head in COS1 cells (j–l). Oversynthesis of PKP3headΔHR2 or PKP3head results in disturbance of the keratin intermediate filament network as revealed by the anti-CK18 antibody (h and k). Single transfection of a DP.FLAG construct in COS7 results in decoration of the keratin network and a punctate localization along cell–cell contacts of the exogenous DP.FLAG protein (m). In DP.FLAG and PKP3GFP cotransfected cells, the DP.FLAG localization along cell contacts is more continuous (n), and DP.FLAG and PKP3GFP colocalize in these structures (o). Bars: 20 μm (a–c, m) and 10 μm (d–l, n–o).
Previously, it has been shown that both PKP1 and PKP2 are able to recruit DP to cell–cell contacts in experiments with COS cells (Kowalczyk et al., 1999a; Chen et al., 2002). In similar experiments using DP.FLAG- and PKP3GFP-encoding plasmids, we could also observe such colocalization at cell–cell contacts. In single transfection experiments, DP.FLAG is found mainly along intermediate filaments, but is also detectable in a punctate pattern along cell–cell contacts (Fig. 6 m). In cotransfection experiments, DP.FLAG localized more continuously along cell–cell contacts (Fig. 6 n) where it colocalized with PKP3GFP (Fig. 6 o). All three PKPs can apparently recruit DP to cell–cell contacts, although this is probably not the main function of PKP3 (see Discussion).
PKP3 interacts with a majority of desmosomal cadherins in the yeast two-hybrid system
Direct interactions between PKP3 and desmosomal cadherins were investigated by yeast two-hybrid analysis. The PKP3 protein fragments used are depicted in Fig. 3. Results are shown in Fig. 7 and summarized in Table I. Full-length PKP3 interacts with all three Dsgs (Fig. 7 a). Using deletion constructs, the Dsg1-binding site was confined to the head domain of PKP3, whereas the binding site of Dsg2 and Dsg3 apparently encompass (parts of) both the head and arm domain of PKP3 (Table I). Deletion of the HR2 domain had no effect on PKP3 binding with Dsgs (Table I). Also, binding of PKP3 with Dsc1a, Dsc2a, and Dsc3a was observed (Fig. 7 a). Interaction of full-length PKP3 with Dsc1b and Dsc2b remained unclear (colonies grew slower and turned only light blue), whereas the PKP3 interaction with Dsc3b was seemingly weaker than its interactions with Dsc-a forms. Nonetheless, the PKP3–Dsc3a interaction observed was not completely convincing because all PKP3 deletion constructs cotransformed with Dsc3a interacted with this desmosomal cadherin, for which we have no explanation (Table I). Both arm and head domains of PKP3 were necessary for interaction with Dscs, but deletion of the HR2 domain did not influence the interaction with Dscs (Table I). Using deletion constructs of the Dsg1 cytoplasmic domain in the bait plasmid pGADT7, we found two separate PKP3 interaction sites. Although the intracellular anchoring domain (IA) and catenin-binding segment (CBS) domains alone did not interact with PKP3, combination of both did (Fig. 7 b). On the other hand, interaction was also observed with the Dsg domain of Dsg1, i.e., the desmoglein-specific domain composed of a proline-rich linker segment, a repeat unit domain, and a terminal domain (Hatzfeld et al., 2000; see Fig. 11), implying the presence of two physically separable PKP3 interaction sites in the Dsg1 cytoplasmic domain (Fig. 7 b). The presence of two independent interaction sites was also observed with constructs of Dsg2 in pGADT7, as we found that both aa domains 583–721 and 882–1069 interact with full-length PKP3 (Fig. 7 b). As controls, all PKP3 constructs were cotransformed with several constructs including one encoding the mouse E-cadherin cytoplasmic tail, whereas desmosomal cadherin constructs were cotransformed with human p120ctn isoform 3AC. None of these cotransformed yeast clones grew on interaction-selective medium, whereas E-cadherin interacted with p120ctn (Fig. 7 a; Table I). Absence of growth of yeast clones was also observed when PKP3 bait plasmids were cotransformed with empty prey vectors, and when empty bait plasmids were cotransformed with prey constructs encoding desmosomal cadherins (Table I).
Figure 7.
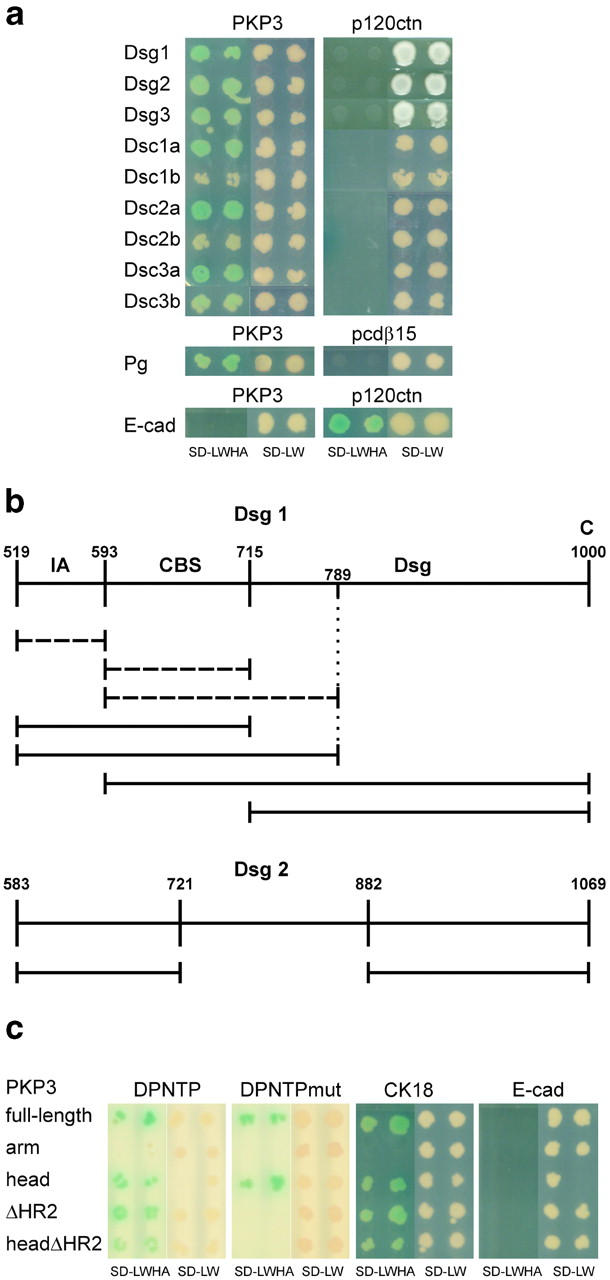
Yeast two-hybrid results using PKP3 and fragments thereof as baits. Two colonies are shown for each cotransformation, including negative controls. SD-LWHA is the medium indicative of protein interactions, as it is supplemented with Xα-gal, whereas SD-LW is indicative of successful cotransformation of bait and prey plasmids. (a) Full-length PKP3 interacts with Dsg1, Dsg2, Dsg3, Dsc1a, Dsc2a, and Dsc3a. Interaction with Dsc1b and Dsc2b is unclear, whereas PKP3 interaction with Dsc3b seems to be less strong than its interaction with Dsc-a proteins. No interaction is observed between desmosomal cadherins and p120ctn. Pg also interacts with PKP3 but not with protocadherin-β15 (pcdβ15). No interaction is observed between PKP3 and E-cadherin (E-cad), whereas E-cadherin and p120ctn clearly interact. (b) Defining the PKP3 interaction sites within the Dsg1 (aa 519–1000) and Dsg2 (aa 583–1069) cytoplasmic domains. Non-interacting Dsg1 protein fragments are indicated by broken lines. Separate IA and CBS domains do not interact with PKP3, and neither does a construct containing aa 593–789 of Dsg1 (i.e., the CBS domain plus part of the Dsg domain). Together, the IA and CBS domains interact with PKP3. The Dsg domain alone is also sufficient for interaction with PKP3. As such, two independent PKP3-interacting domains can be identified in the Dsg1 cytoplasmic domain. In the case of Dsg2, again two separate PKP3 interaction sites can be detected, i.e., aa 583–721 and aa 882–1069. IA, intracellular anchor; CBS, catenin-binding segment; Dsg, desmoglein-specific domain; C, carboxy terminus. (c) Full-length PKP3 and PKP3head, but not PKP3arm, interact with both DPNTP and DPNTPmut. PKP3ΔHR2 and PKP3headΔHR2 bind to DPNTP, but not to DPNTPmut. Only PKP3arm does not interact with CK18. None of the PKP3 constructs interacted with E-cadherin in the yeast two-hybrid system.
Table I. Interactions between human PKP3 and fragments thereof with other desmosomal proteins as observed in the yeast two-hybrid system.
| Bait→Prey↓ | PKP3 | PKP3arm | PKP3head | PKP3ΔHR2 | PKP3headΔHR2 | p120ctn 3AC | Empty bait |
|---|---|---|---|---|---|---|---|
| Dsg1 | + | − | + | + | + | − | − |
| Dsg2 | + | − | − | + | − | − | − |
| Dsg3 | + | − | − | + | − | − | − |
| Dsc1a | + | − | − | + | − | − | − |
| Dsc1b | ± | − | − | ± | − | − | − |
| Dsc2a | + | − | − | + | − | − | − |
| Dsc2b | ± | − | − | ± | − | − | − |
| Dsc3a | + | + | + | + | + | − | − |
| Dsc3b | + | − | − | ± | − | − | − |
| Pg | + | − | − | + | − | ND | − |
| DPNTP | + | − | + | + | + | ND | − |
| DPNTPmut | + | − | + | − | − | ND | − |
| CK18 | + | − | + | + | + | ND | − |
| mEcad | − | − | − | − | − | + | − |
| Empty prey | − | − | − | − | − | − | − |
+, growth and blue staining of cotransformed yeast colonies on medium selective for interaction; −, no growth of cotransformed yeast colonies on medium selective for interaction; ±, severe reduction in both growth and blue staining of cotransformed yeast colonies on medium selective for interaction.
Figure 11.
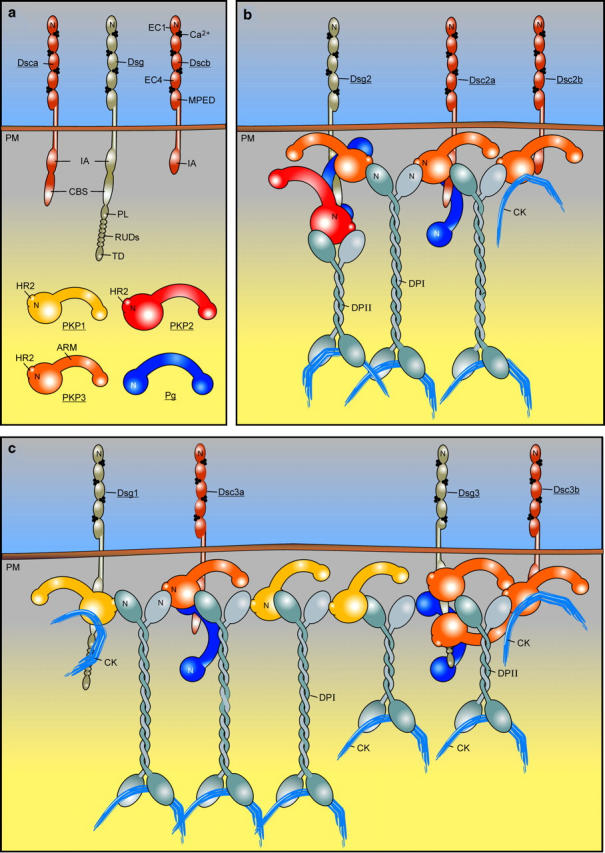
Desmosome model showing interactions between selected molecular components of simple and stratified epithelia (modified after Nollet et al., 2000). (a) Some representative desmosomal proteins are depicted. CBS, catenin-binding segment; CK, cytokeratin; EC, ectodomain module; IA, intracellular anchor domain; MPED, membrane-proximal extracellular domain; N, amino-terminal domain; PL, proline-rich linker; PM, plasma membrane; RUD, repeat unit domain; TD, terminal domain. In the present work, the combination (PL-RUDs-TD) was designated Dsg domain (Hatzfeld et al., 2000). (b) The localizations in simple epithelia of PKP1, PKP2, and PKP3 as compared with Pg are mainly based on observations made by others (Mertens et al., 1996; North et al., 1999; Schmidt et al., 1999). DP occurs as two splice variants, DPI and the shorter DPII that is expressed in epithelia, but not in heart (Kowalczyk et al., 1999b). The stoichiometry of the interactions between desmosomal plaque molecules is unclear. For the Pg–Dsg1 interaction, ratios >1:1 have been reported (Bannon et al., 2001). It is also unclear which and how many proteins can bind at the same time to a single PKP protein. Here, we have shown that at least the PKP3 head domain contains two DP interaction sites. (c) The location and multimolecular interactions of PKP1 in stratified epithelia are adapted from a model proposed by Kowalczyk et al. (1999a). According to immunoelectron localization studies, the carboxy-termini of DPI and DPII are localized at the same distance from the cell membrane that is not reflected here. In epidermis, PKP1, Dsc1, and Dsg1 are enriched in the superficial layers, whereas Dsg3, Dsc3, and PKP2 are concentrated in the basal layers. PKP3 is expressed throughout all living cell layers of the epidermis (Fig. 2).
PKP3 interacts with Pg, DP, and CK18 proteins in the yeast two-hybrid system
Interaction between the armadillo proteins PKP3 and Pg was clearly observed in the yeast two-hybrid system (Fig. 7 a). Like PKP1 and PKP2 (Kowalczyk et al., 1999a; Chen et al., 2002), PKP3 also interacts with a DP head domain of 584 aa (desmoplakin amino-terminal fragment, DPNTP; Fig. 7 c). This DPNTP fragment binds the head domain of PKP3, and deletion of this head domain (PKP3arm) abrogates interaction with DPNTP. Using a truncated version of DPNTP, encoding only 63 aa of the DP head domain (mutated desmoplakin amino-terminal fragment, DPNTPmut), we found that deletion of the HR2 domain of PKP3 completely abolished binding with this DPNTPmut, though this was not the case for interaction with DPNTP (Fig. 7 c). The same observation was made when the PKP3headΔHR2 construct was used. Double missense mutation of the HR2 domain (P23A, R45A) resulted in a weaker interaction with DPNTPmut, but not in complete inhibition (unpublished data). These results indicate that the PKP3head domain has at least two DP binding sites interacting with at least two different sites in the DP head domain. Finally, an interaction was observed with epithelial CK18 that binds the nonarmadillo head domain of PKP3 (Fig. 7 c).
PKP3 coimmunoprecipitates Dsg1 to -3, Dsc3a and -b, DP, and Pg
To confirm the interactions observed in the yeast two-hybrid system, coimmunoprecipitation (CoIP) experiments were performed upon cotransfection of the relevant expression plasmids in HEK293T fibroblasts. These cells have no functional desmosomes and their endogenous protein levels of desmosomal components, such as Dsg2, PKP3, DP, and Pg, are very low or nil (unpublished data). We noticed that the full-length PKP3 protein sticks nonspecifically to protein G Sepharose beads, and because more stringent washings did not significantly reduce this binding, CoIP experiments were performed in one direction only. The PKP3 protein was immunoprecipitated using mAb 23E3/4, whereas all other proteins were myc-tagged. Incompletely reduced primary mouse antibody was sometimes detected in CoIP samples (Fig. 8 a, star). Full-length PKP3 protein precipitates the desmosomal cadherins Dsg1, Dsg2, Dsg3, Dsc3b (Fig. 8 a), and Dsc3a (Fig. 8 c). In parallel, lysates were incubated with protein G Sepharose beads only, and no signal could be observed in these samples (Fig. 8, a and c). Using Dsc3b deletion constructs (depicted in Fig. 8 d), the PKP3 interaction site was confined to the membrane-proximal domain of Dsc3b (Fig. 8 c, left). Control detection of proteins in total lysates is shown in Fig. 8 (b and c). In a similar experiment, we tried to CoIP Dsc1a and Dsc2a with full-length PKP3. Unfortunately, no detectable levels of Dsc protein could be observed in cell lysates of appropriately cotransfected HEK293T cells (unpublished data). Finally, we found that full-length PKP3 immunoprecipitates the desmosomal plaque proteins DP and Pg (Fig. 8 a). Again, no signal is observed in the negative control lanes (Fig. 8 a). As such, these results confirm our data obtained using the yeast two-hybrid system.
Figure 8.
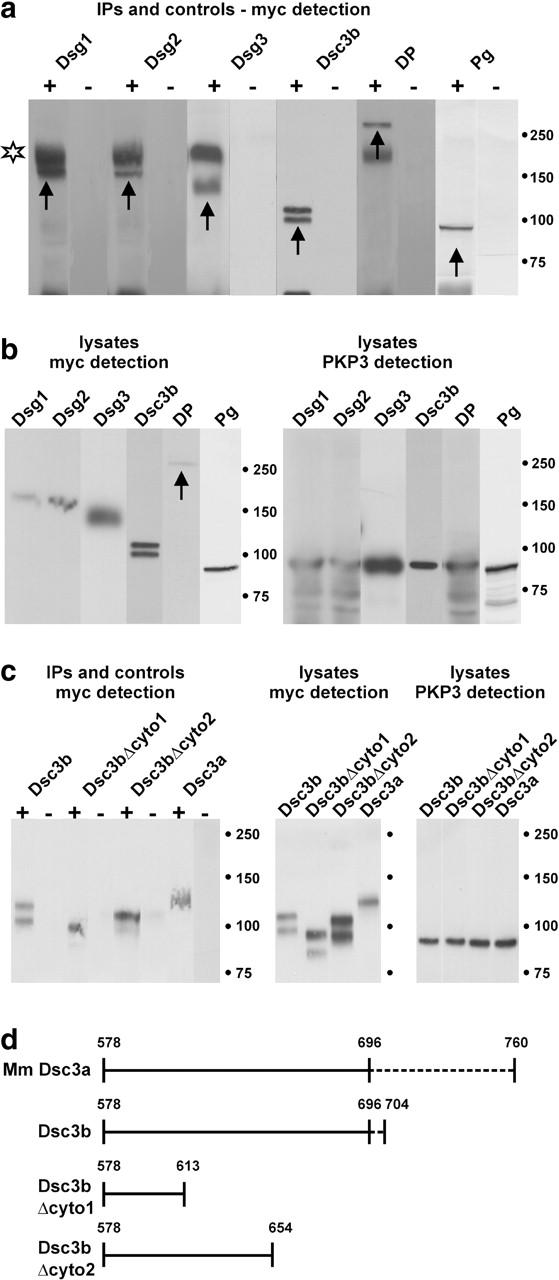
CoIP experiments using lysates of HEK293T cells cotransfected with plasmids of interest. A plasmid encoding full-length PKP3 (p1744) was cotransfected with plasmids encoding Dsg1-myc, Dsg2-myc, Dsg3-myc, Dsc3a-myc, Dsc3b-myc, Dsc3bΔcyto1-myc, Dsc3bΔcyto2-myc, DP-myc, or Pg-myc fusion proteins. (a) Using anti-PKP3 mAb 23E3/4, myc-tagged Dsg1, Dsg2, Dsg3, Dsc3b, DP, and Pg proteins (arrows) were coimmunoprecipitated with PKP3 (+ lanes). The nature of the Dsc3b doublet is unclear, but might represent incompletely processed protein in addition to mature protein. In the negative control lanes (lysates incubated with protein G Sepharose in the absence of mAb 23E3/4), no signal could be detected (− lanes). Incompletely reduced primary antibody was often detected (star). Identical exposure times were used for each set of +/− lanes. (b) Control Western blot detection of total cell lysates: Dsg1 (165 kD), Dsg2 (160 kD), Dsg3 (135 kD), Dsc3b (99 kD), DP (250 kD, arrow), and Pg (82 kD) in the left panel; PKP3 (87 kD) in the right panel. Mol wt markers (in kD) are indicated. (c) Using anti-PKP3 mAb 23E3/4, myc-tagged Dsc3b, two COOH-terminally truncated derivatives of Dsc3b and Dsc3a were coimmunoprecipitated with PKP3 (+ lanes in left panel). In the negative control lanes (lysates incubated with protein G Sepharose in the absence of mAb 23E3/4), no signal could be detected (− lanes in left panel). Expression of each of these fusion proteins was detected by Western blotting (right), using anti-Myc and anti-PKP3 antibodies. (d) Schematic representation of the cytoplasmic domains of mouse (Mm) Dsc3a, Dsc3b, and Dsc3b truncation mutants used in the CoIP experiments of (C). Dashed lines indicate isoform-specific domains. The fragment containing aa 578–696 is shared by Dsc3a and Dsc3b. Dsc3bΔcyto1 and Dsc3bΔcyto2 encompass, respectively, 36 and 77 membrane-proximal aa of both Dsc3a and -b.
PKP3 colocalizes in vitro with Dsc1a and Dsc2a
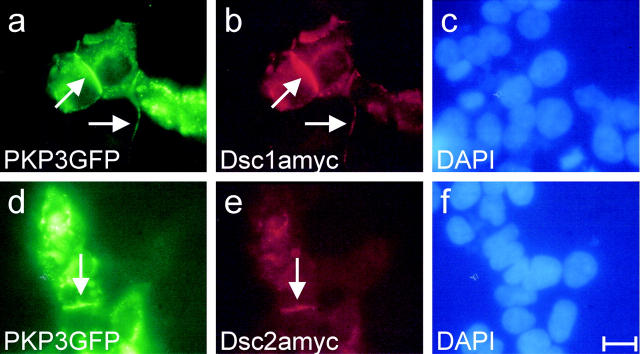
In vitro colocalization between PKP3 and Dscs was observed in cotransfection experiments. PKP3 was expressed as a fusion protein in-frame with a GFP tag, whereas Dsc proteins contained a myc tag. GFP-tagged PKP3 colocalized with exogenous Dsc1a (Fig. 9 , a–c) and Dsc2a (Fig. 9, d–f) at cell–cell contacts in cotransfected HEK293T cells. These results provide further evidence for direct in vivo interactions between PKP3 and Dscs.
Figure 9.
Colocalization of GFP-tagged PKP3 and myc-tagged Dsc1a and Dsc2a in cotransfected HEK293T cells. Exogenous GFP-tagged PKP3 colocalizes with Dsc1a and Dsc2a at cell–cell contacts (a vs. b and d vs. e, arrows). Control DAPI stainings are shown (c and f). Bar, 30 μm (a–f).
Discussion
Desmosomes are cell–cell adhesion structures that are especially important in tissues that are subject to mechanical stress, for example the skin (McGrath et al., 1997). Desmosomal plaque molecules include the PKPs, which display a differential expression pattern. Here, we report on desmosomal interactions of PKP3, of which the epithelial expression pattern overlaps that of both PKP1 and PKP2 (Bonné et al., 1999; Schmidt et al., 1999).
PKP3 is generally expressed in single- and multilayered epithelia and overlaps the more restricted expression patterns of desmosomal cadherins, PKP1 and PKP2
Using novel mouse mAbs, we investigated the protein synthesis pattern of PKP3 in vitro and in vivo. In immunofluorescence detection assays using epithelial cell lines, PKP3 was clearly detected in desmosomes. However, no nuclear PKP3 could be detected in these cell lines, in contrast to earlier observations recorded with a rabbit polyclonal anti-PKP3 antibody, and despite the fact that mAb 12B11F8 was generated against the same peptide as the pAb (Bonné et al., 1999). Neither was nuclear PKP3 detected in cells synthesizing exogenous full-length PKP3. Altogether, these data indicate that the previously reported nuclear PKP3 localization might be exceptional or even artificial.
Using mAb 23E3/4, PKP3 synthesis was observed in the desmosomes of all living cell layers of the human epidermis, whereas no PKP3 was found in the stratum corneum and dermis. In addition, PKP3 was also detected in simple colon epithelium. These results confirm and strengthen previous expression data on PKP3 in various epithelia and epithelial cell lines (Bonné et al., 1999; Schmidt et al., 1999). Although the expression patterns of desmosomal cadherins and of PKP1 and PKP2 reflect a tight regulation in function of cell type and differentiation state (King et al., 1993, 1997; Heid et al., 1994; Mertens et al., 1996; North et al., 1996), the PKP3 expression pattern, like the situation of Pg and DP, seems to indicate a basic role for PKP3 in epithelial and epidermal desmosomes with the exception of liver desmosomes (Schmidt et al., 1999). The functional consequence of the lacking PKP3 expression in hepatocytes is so far unclear, but considering the multiple interaction partners of PKP3, it is feasible that cell adhesive structures in the liver are quite different from the model we propose in the Discussion section below (see Fig. 11).
Intracellular localization of PKP3 protein fragments
To study the function and intracellular localization of PKP3 and fragments thereof, various epithelial cell lines were transfected with appropriate expression plasmids. Full-length PKP3 was clearly observed in desmosomal cell–cell adhesion complexes. In contrast, the GFP-tagged PKP3arm fragment accumulates to high levels in the cell nucleus. It is presently unclear which sequences from the PKP3arm fragment direct its nuclear localization, and how this is counteracted in the full-length protein. It has been reported earlier that the arm domain of β-catenin is sufficient to direct its nuclear localization and function (Funayama et al., 1995; Fagotto et al., 1998). On the other hand, an as yet unidentified nuclear export signal might be present in the PKP3head domain because this fragment remains mainly localized in (aggregates in) the cytoplasm, without any localization in the nucleus and also without prominent localization along cell–cell contacts. These results obtained with PKP3head and PKP3arm fragments are in obvious contrast with earlier reports on the intracellular localization of head and arm fragments from PKP1 and PKP2, of which the head domains preferentially localize in both the desmosomes and the nucleus, whereas the corresponding arm domains are dispersed throughout the cytoplasm (Klymkowsky, 1999; Kowalczyk et al., 1999a; Hatzfeld et al., 2000; Chen et al., 2002).
Aggregates of exogenous PKP3head fragments were shown to contain desmosome-associated molecules, such as DP and CK18, and sometimes endogenous PKP3, but not PKP2 or Dsg2. It is unlikely that the endogenous PKP3 found in these aggregates has been replaced in desmosomes by the DP-binding PKP3head domain. Indeed, only little PKP3head is detected in desmosomes, whereas PKP3 can still be readily detected in these junctions. Hypothetically, endogenous PKP3 might be forced in these aggregates by interaction with PKP3head-bound DP. Although a second DP interaction site in the PKP3headΔHR2 protein was observed in yeast two-hybrid experiments, only CK18 (but not DP) was found to localize in PKP3headΔHR2 aggregates. This might reflect the presence of more stringent DP interaction partners in desmosomes, and indicates also that the HR2 domain is involved in strong binding of PKP3 to DP. On the other hand, we could show that in DP and PKP3 cotransfections of COS cells, PKP3 is able to recruit DP to sites of cell–cell contact. COS cells contain only few, very small desmosomes. However, it appears that in cell types containing many and larger desmosomes this PKP3 function is less prominent.
The observations made in our transfection studies emphasize the different behavior of PKP3 compared with PKP1 and PKP2. Nuclear PKP3 could not be detected in contrast to PKP1 and PKP2 (Mertens et al., 1996; Schmidt et al., 1997). The subcellular distributions of PKP3head and arm fragments are very different from those of the corresponding PKP1 and PKP2 domains (Kowalczyk et al., 1999a; Chen et al., 2002). Unlike PKP1head and PKP2head, PKP3head does not colocalize with DP in desmosomes, although PKP3 recruits DP to cell–cell contacts in COS cells. As such, it appears that both head and arm domains of PKP3 are necessary for genuine desmosomal anchoring. Interaction with desmosomal cadherins and/or Pg, for which both head and arm domains of PKP3 are necessary on the basis of yeast two-hybrid data (Table I), is apparently imperative for desmosomal localization of PKP3 in epithelial cells.
PKP3 interacts with a majority of desmosomal proteins and, as such, may act as an essential scaffold
By use of the yeast two-hybrid system, we analyzed direct desmosomal PKP3 interactions and observed clear-cut PKP3 interactions with all desmosomal cadherins but Dsc1b and Dsc2b. The PKP3 protein domain that interacts with Dsg1 was confined to the nonarmadillo head domain of the protein, as has also been reported for the Dsg1–PKP1 and Dsg1–PKP2 interactions (Smith and Fuchs, 1998; Kowalczyk et al., 1999a; Hatzfeld et al., 2000; Chen et al., 2002). The exact sequence of the PKP3head domain responsible for this interaction is still unknown. For PKP1, it has been reported that aa 70–213, residing in the head domain of PKP1, are sufficient to interact with Dsg1 (Hatzfeld et al., 2000). Removal of the single conserved sequence stretch in the PKP head domains, i.e., the HR2 domain involved in DP binding, had no effect on Dsg1 binding in the yeast two-hybrid system. In contrast, several conserved sequence stretches can be observed in the head domains of the human, mouse, rat, and Xenopus laevis PKP3 orthologues, which might include PKP3-specific Dsg1 interaction sites (Fig. 10) . Using Dsg1 deletion constructs, we found two physically separable PKP3 interaction sites (namely the IA+CBS and the Dsg domain), whereas PKP1 was previously reported to bind the CBS+Dsg domain (Hatzfeld et al., 2000). Hence, it appears that PKP1 and PKP3 head domains bind different regions of the Dsg1 intracellular domain. Also in the Dsg2 tail, two separable PKP3 interaction sites were identified by us. The PKP3 interactions observed in the yeast two-hybrid system were confirmed, where possible, in CoIP and colocalization experiments. These experiments further strengthened the evidence for a direct interaction between PKP3, and Dsg1 to -3, Dsc1a, Dsc2a, Dsc3a, and Dsc3b. Although it was less clear from yeast two-hybrid experiments whether PKP3 and Dsc3a or Dsc3b interact, strong evidence for such interactions was provided by CoIP experiments. Moreover, the PKP3 binding site in the Dsc3a/Dsc3b cytoplasmic tails could be narrowed down to 36 membrane-proximal aa. As such, PKP3 is the first protein interaction partner of a Dsc-b isoform ever identified.
Figure 10.
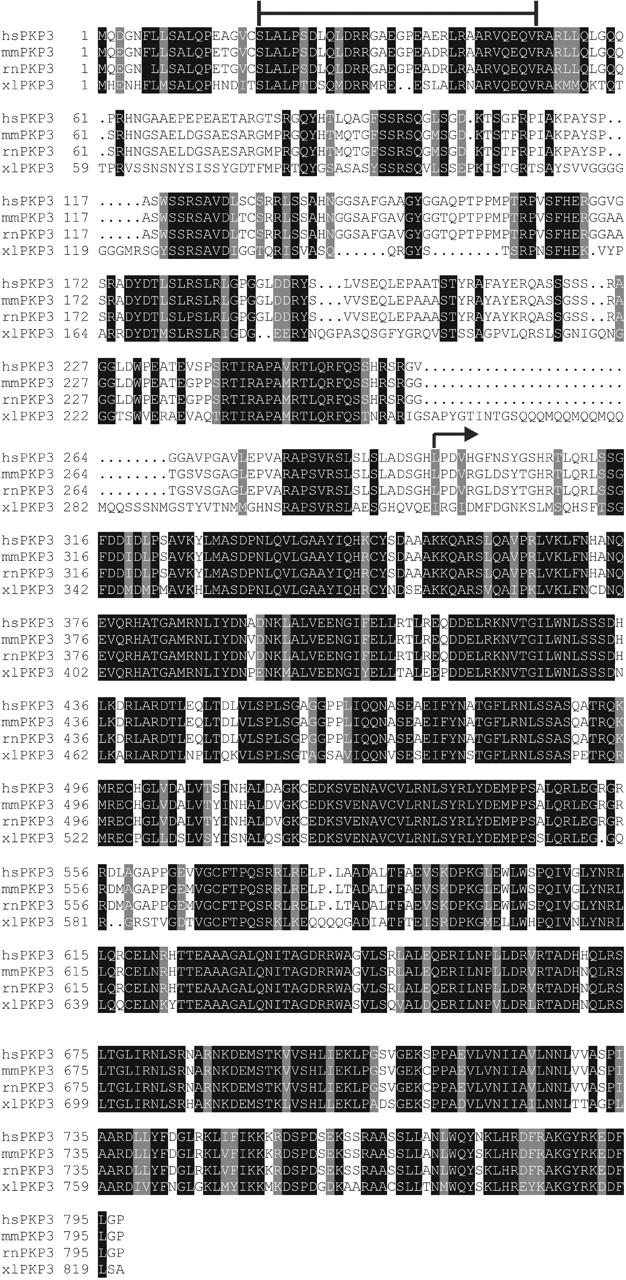
Clustal W alignment of the PKP3 protein orthologues from man (hsPKP3), mouse (mmPKP3), rat (rnPKP3), and X. laevis (xlPKP3) shaded using the BOXSHADE server (http://www.ch.embnet.org). Only those aa are shaded that are identical (black) or similar (gray) in each of the sequences. The HR2 domain is indicated by the top line, and the start of the arm domains is indicated by an arrow. The general sequence conservation in the arm domains is striking compared with the situation in the head domains, where only short sequence stretches are conserved. Database accession nos. are: AF053719 (hsPKP3), AF136719 (mmPKP3), and AX046097 (xlPKP3). The rat PKP3 protein sequence was predicted on the basis of the genomic sequences identified by BLAT search at http://genome.ucsc.edu/goldenPath (UCSC Rat Genome Project, November 2002 release).
In conclusion, the PKP3 interaction pattern with desmosomal cadherins is obviously different from those reported for PKP1 and PKP2 (Smith and Fuchs, 1998; Hatzfeld et al., 2000; Chen et al., 2002). A model displaying the different PKP3 interactions in the desmosomal plaque is depicted in Fig. 11 . The occurrence of PKP3 interactions with many desmosomal cadherins is consistent with the observed in vivo expression pattern of PKP3 that overlaps the differentiation-dependent expression of all desmosomal cadherins. Furthermore, like PKP1, PKP3 was reported to be present in the desmosomal plaque close to the cell membrane where it physically overlaps with all desmosomal cadherin tails (North et al., 1999; Schmidt et al., 1999). Together, these data further support the possibility of many PKP3–desmosomal cadherin interactions in vivo (Fig. 11).
In the yeast two-hybrid system, we also found an interaction between PKP3 and the plaque molecule Pg (depicted in Fig. 11), which was confirmed in CoIP experiments. As such, both PKP2 and PKP3 appear to interact with Pg, whereas PKP1 does not (Hofmann et al., 2000; Chen et al., 2002). In addition, we found the head domain of PKP3 interacting with both CK18 and DP, and this latter observation was also confirmed in CoIP experiments. Both PKP1 and PKP2 have been reported to establish interactions with CK18 and DP (Smith and Fuchs, 1998; Kowalczyk et al., 1999a; Hatzfeld et al., 2000; Hofmann et al., 2000; Chen et al., 2002). It has been unclear for quite some time whether PKPs can interact in vivo with keratin filaments, as they are localized in the outer dense plaque and seem to be inaccessible to keratin filaments. Recently, it has been reported that transfection of the DP head domain in DP-null keratinocytes restores not only cellular adhesion, but also partial keratin association with desmosomes (Vasioukhin et al., 2001). Because the DP head mutant lacks the intermediate filament interaction site, this effect is hypothesized to be mediated by PKPs, which bind the DP head protein and become optimally positioned to interact with intermediate filaments (Vasioukhin et al., 2001). Moreover, a recessive DP mutation was reported, resulting in dilated cardiomyopathy, woolly hair and keratoderma (Norgett et al., 2000). This mutant DP lacks the C subdomain of the carboxy-terminal domain responsible for interaction with intermediate filaments; in suprabasal keratinocytes of patients carrying this mutation, keratin filaments are indeed found to be perinuclear. However, such patients develop quite normally and in basal keratinocytes, desmosomal anchoring of intermediate filaments is observed. It is tempting to speculate that this cytokeratin anchoring is mediated by PKPs, but with lower efficiency because intermediate filament networks collapse when cells are stressed (Norgett et al., 2000).
Upon use of DPNTPmut, encoding only the first 63 aa of the DP head domain, as a two-hybrid prey, we observed that binding of PKP3ΔHR2 protein to this DP fragment was completely abolished, although the same PKP3ΔHR2 protein could still bind to the larger DPNTP protein. Deletion of the HR2 domain of PKP3 had no effect on any other interaction observed in the yeast two-hybrid system. On the other hand, deletion of the HR2 domain abolished interaction with DP in cytoplasmic aggregates in transfected cells. Together, these data indicate that the HR2 domain of PKPs is involved in interaction with a site located in the first 63 aa of the DP head domain (Fig. 11). Previously, it was shown that the first 86 aa of DP are sufficient to direct desmosomal localization of this protein fragment, whereas for PKP1, aa 1–70 are essential to interact with DP (Smith and Fuchs, 1998; Hatzfeld et al., 2000). Interestingly, at least one additional DP-binding site is present more carboxy-terminally in the head domain of PKP3, interacting with an unidentified region between aa 64–584 of DP. Apparently, at least in the yeast two-hybrid system, the presence of only one DP–PKP3 interaction site is sufficient to establish an interaction. Nevertheless, the presence of more than one DP binding site in the head domain of PKPs would support a model for increasing desmosome size in the upper layers of complex epithelia through a mechanism of lateral DP–PKP interactions (Kowalczyk et al., 1999a; Green and Gaudry, 2000; Bornslaeger et al., 2001; Fig. 11 c). Consequently, more DP and PKP molecules would be available for intermediate filament anchoring, conferring robust cell–cell adhesion to keratinocytes. Because the HR2 domain is the only region conserved in the head domains of all three known PKPs (Bonné et al., 1999; Schmidt et al., 1999), it may serve as a general DP interaction site for PKPs. The molecular binding mechanism of the other DP interaction(s), on the other hand, is likely to differ between PKPs, mirroring distinct PKP roles in desmosomes and desmosomal interactions. Protein sequence alignments of the human, mouse, rat, and X. laevis orthologues of PKP3 reveal several strikingly conserved sequence stretches in the head domains besides the HR2 domain (Fig. 10), which might represent additional DP binding sites. Such a second PKP3 interaction site, as evidenced for PKP3 interactions with DP, Dsg1, or Dsg2, might not be sufficient on its own to achieve PKP3 binding, but rather act as an accessory interaction site to strengthen PKP3 binding under certain conditions or to modulate the interaction with other desmosomal proteins (Bornslaeger et al., 2001). This would explain why no DP is found in PKP3ΔHR2GFP aggregates.
Altogether, we have shown that the desmosomal plaque protein PKP3 is a generally expressed epithelial and epidermal PKP, unlike PKP1 or PKP2. PKP3 was shown to interact with most of the principal desmosomal components, including Dsc3b. This is the first interaction partner ever observed for a Dsc-b isoform. Thus, PKP3 provides for various physical links between the often selectively synthesized desmosomal proteins. Interaction with desmosomal cadherins and/or Pg (but not DP) appears necessary for proper desmosomal localization of PKP3. We propose a model (Fig. 11) in which PKP3 is involved in the basic assembly of a vast majority of epithelial and epidermal desmosomes by multiple interactions within the desmosomal plaque. The remarkably broad spectrum of PKP3 binding partners might indicate that cooperative PKP3 interactions with several desmosomal proteins are more effective in the generation of stable desmosomes than the strength of each of the discrete interactions.
Materials and methods
Antibodies
C57/BL6 mice were immunized with a human PKP3-specific synthetic peptide (aa sequence FTPQSRRLRELPLAADALTF), spanning mostly the spacer region between armadillo repeats 6 and 7 of the human PKP3 sequence (Bonné et al., 1999). The immunization protocol used was essentially as described previously (Schäfer et al., 1996). Alternatively, mice were immunized with a PKP3-specific synthetic peptide localized at the extreme carboxy terminus of the protein (aa sequence KLHRDFRAKGYRKED), yielding antibody 12B11F8.
Other antibodies used were mouse anti-myc tag (Oncogene Research Products), mouse anti-CK18 antibody RGE53 (Euro-Diagnostics), mouse anti-DP and anti-PKP2 (Progen), mouse anti-PKP1 (Zymed Laboratories), polyclonal OctA-Probe™ (Santa Cruz Biotechnology, Inc.). Secondary antibodies used in immunofluorescence microscopy were Alexa® 594- or Alexa® 488-conjugated anti–mouse Ig antibodies (Molecular Probes, Inc.).
Immunodetection assays
Proteins were detected in Western blot experiments by ECL detection (Amersham Biosciences) or NBT/BCIP detection (Sigma-Aldrich). Antibody dilution used for Western blots was 1:5000 (23E3/4) and 1:100 (12B11F8).
Immunofluorescence assays were performed essentially as described previously (Bonné et al., 1999). Formaldehyde-fixed cells were treated for 15 min with 0.2% Triton X-100 before incubation with primary antibodies. 23E3/4 antibody dilution was 1:300. Fluorescence image results were captured using a camera (MicroMAX; RS Photometrics) and processed using the MetaMorph® software (Universal Imaging Corp.). Alternatively, images were taken using a standard camera.
Immunohistochemical detection of PKP3 was performed essentially as described by Mertens et al. (1999). In these experiments, the DAKO LSAB2 system HRP (ready-to-use AEC) was used according to the manufacturer's instructions (DakoCytomation). 23E3/4 antibody dilution ranged from 1:100 to 1:500. Images were captured using a CoolSNAP camera and dedicated software (RS Photometrics).
Cell lines and transfections
The ileocecal adenocarcinoma cell line HCT8/E8 was obtained by subcloning of the cell line HCT8 (Vermeulen et al., 1995). MCF7/AZ is derived from the MCF-7 human mammary carcinoma cell line (Bracke et al., 1991) and HaCaT is a human keratinocyte cell line (Boukamp et al., 1988). MCF7/AZ cells were transiently transfected with FuGENE™ 6 (Roche); HCT8/E8 cells were transiently transfected with LipofectAMINE™ PLUS (Life Technologies). Transfected cells were fixed 26–30 h later. HEK293T cells (Simmons, 1990), COS1 and COS7 cells (Gluzman, 1981) and PtK2 cells (Basehoar and Berns, 1973) were transfected using the calcium phosphate method and fixed 48 h later.
Eukaryotic expression plasmids
All human PKP3 constructs were made by PCR using Pfu DNA polymerase (Stratagene) and primers containing appropriate additional restriction sites, which are underlined in the primer sequences. Silent mutations present in primers are in bold. All constructs (Fig. 3) were fully sequenced to ensure that mutations had not occurred during PCR. Full-length human PKP3 was amplified with forward primer 5′-atacgctagcccaggcccggtggacct-3′ (P1) and reverse primer 5′-atacgaattc aggacccaggaagtcctcct-3′ (P2), using the full-length human PKP3 cDNA cloned in pGEM11 as template DNA (Bonné et al., 1999). The resulting PCR product was NheI/EcoRI-cut and cloned in the SpeI/EcoRI sites of pEF6/Myc-His B (Invitrogen), out of frame with the Myc and His tags of the vector, resulting in plasmid p1744. The PCR product was also cloned in the NheI/EcoRI sites of the pEGFP-N2 vector (CLONTECH Laboratories, Inc.) resulting in p1910, encoding full-length human PKP3 in-frame with the GFP tag.
PKP3head and PKP3arm expression constructs were obtained as follows: PCR was performed using the full-length human PKP3 cDNA cloned in pGEM11 as template DNA. The PKP3head fragment was amplified using forward primer P1 and reverse primer 5′-ataagaattcgtgacccgagtcagccaggc-3′; the PKP3arm fragment with forward primer 5′-ataagctagcgccatgttgccggacgtgcatggg-3′ and reverse primer P2. PCR products were NheI/EcoRI-cut and cloned in the NheI/EcoRI sites of pEGFP-N2.
PKP3ΔHR2 was obtained as follows: two primer sets were developed that do not span the HR2-coding cDNA part of PKP3 and which introduce a silent BssHII site in the PKP3 cDNA. The PCR products, amplified with primer set 1 (forward primer P1 and reverse primer 5′-aatagcgcgcgacacacgccggcctcagg-3′) and primer set 2 (forward primer 5′-aatagcgcgcgactcttgcagctg-3′ and reverse primer P2), were respectively NheI/BssHII- and BssHII/EcoRI-cut, and simultaneously cloned in the SpeI/NheI sites of pEF6Myc-HisA (Invitrogen) to obtain p1912. After sequencing, this insert was again amplified with the full-length PKP3 primers for cloning in pEGFP-N2, resulting in p1911. The PKP3headΔHR2 fragment was amplified with the primer set used for amplification of the PKP3head domain and cloned accordingly.
Mouse Dsc3a and Dsc3b constructs were a gift from Drs. D. Garrod and C. Byrne (University of Manchester, Manchester, UK). Expression constructs (corresponding cytoplasmic domains depicted in Fig. 8 d) were made in pEF6Myc-HisB (Invitrogen) by amplifying the cDNA inserts with primer set 1 for Dsc3a (forward primer P5 with sequence 5′-aataggtaccgctgcactgtgcgatggtgg-3′, reverse primer 5′-aataactagtgcgcttagtgcaggtttttg-3′) and primer set 2 for Dsc3b (forward primer P5, reverse primer 5′-aataactagtaccagtgtgtcctctaatgg-3′). Dsc3bΔcyto1 and Dsc3bΔcyto2 inserts were amplified using primer sets 3 (forward primer P5, reverse primer 5′-aataactagtagaacacactctgtcatccc) and 4 (forward primer P5, reverse primer 5′-aataactagtcaaggtctggtgtcctttca), respectively. PCR products were KpnI/SpeI-cut and cloned into the KpnI/SpeI sites of the pEF6Myc-HisB vector.
Expression constructs encoding full-length myc-tagged human Dsg1, Dsg2, Dsc1a, and Dsc2a were described previously (Kowalczyk et al., 1994; Ishii et al., 2001); the human Dsg3 construct was a gift from Dr. J. Stanley (University of Pennsylvania, Philadelphia, PA). The FLAG-tagged DP and Myc-tagged Pg expression constructs were described previously (Kowalczyk et al., 1997; Bornslaeger et al., 2001).
Yeast two-hybrid plasmids
All PKP3 constructs were made by PCR using Pfu DNA polymerase (Stratagene) and primers containing appropriate additional restriction sites, which are underlined in the primer sequences. Silent mutations present in primers are in bold. All constructs (Fig. 3) were fully sequenced to ensure that mutations had not occurred during PCR. Constructs pGBKT7hPKP3, pGBKT7hPKP3arm, and pGBKT7hPKP3head were made using the following primers: forward primer 5′-atacgaattccaggacggtaacttcctg-3′ (P3) and reverse primer 5′-atacgtcgacacagccaacccccacctct-3′ (P4; full-length PKP3); forward primer 5′-atacgaattc ttgccggacgtgcatgggtt-3′ and reverse primer P4 (PKP3arm); forward primer P3 and reverse primer 5′-atacgtcgacgtgacccgagtcagccaggc-3′ (PKP3head). Amplified fragments were EcoRI/SalI-cut and cloned in the EcoRI/SalI sites of pGBKT7 (CLONTECH Laboratories, Inc.). Template DNA was the full-length human PKP3 cDNA cloned in pGEM11. The pGBKT7hPKP3ΔHR2 construct was made as follows: using DNA from plasmid p1912 as template, PCR was performed using forward primer P3 and reverse primer 5′-atacgaattc aggacccaggaagtcctcct-3′. The PCR product was EcoRI-cut and cloned in the EcoRI site of pGBKT7 to obtain pGBKT7hPKP3ΔHR2. To obtain pGBKT7hPKP3headΔHR2, the insert of p1912 was amplified with forward primer P3 and reverse primer 5′-atacgtcgacgtgacccgagtcagccaggc-3′, and the resulting PCR product was EcoRI/SalI-cut and cloned in the EcoRI/SalI sites of pGBKT7. The Dsg and Dsc constructs in pGAD424 were reported previously (Hatzfeld et al., 2000). Plasmids were EcoRI/SalI-cut and inserts were cloned in the EcoRI/XhoI sites of pGADT7, except for pGAD424Dsc2b. The pGAD424Dsc2b insert was EcoRI/MamI-cut and cloned in the EcoRI/SmaI sites of pGADT7. pGAD424 constructs containing the Dsg, CBS, IA, and CBS+Dsg domain of Dsg1 have been described previously (Hatzfeld et al., 2000). These plasmids were EcoRI/SalI-cut and cloned in the EcoRI/SalI sites of pGADT7. DNA from pGADT7Dsg1 was Tth111I-cut, blunted to generate a frameshift and self-ligated, resulting in pGADT7Dsg1 (519–789). Alternatively, the DNA was AvrII-cut, blunted to generate a frameshift and self-ligated, which resulted in pGADT7Dsg1 (IA+CBS) encoding aa 519–715 of Dsg1. Alternatively, DNA from pGADT7Dsg1 was BglII-cut and ligated, followed by digestion with Tth111I, blunting and ligation to generate pGADT7Dsg1 (aa 595–789). Plasmids were sequenced to ensure that the intended in-frame ligations or frameshifts had occurred correctly. pGADT7mEcad contains the cytoplasmic tail of mouse E-cadherin, and pGADT7hp1203AC and pGBKT7hp1203AC contain the human p120ctn isoform 3AC. The cytoplasmic domain of protocadherin-β15 was amplified via PCR using forward primer 5′-agcgaattcctgtgcaggaggagcagg-3′ and reverse primer 5′-atagtcgactacagatacctacattattctag-3′. The amplified fragment was EcoRI/SalI-cut and cloned in the EcoRI/SalI sites of pGBKT7 (CLONTECH Laboratories, Inc.). pACTIIDPNTP was described previously (Kowalczyk et al., 1997). In pACTIIDPNTPmut, a 1-bp deletion causes a frame shift in the DPNTP-coding sequence, resulting in translation of only the first 63 aa of DP.
Yeast two-hybrid analysis
Yeast two-hybrid interaction assays were performed using yeast strain AH109 (CLONTECH Laboratories, Inc.). Cotransformed yeast colonies were stamped on synthetic dropout medium (SD) lacking leucine and tryptophan for growth control, and SD lacking leucine, tryptophan, histidine, and adenine for interaction testing. SD lacking leucine, tryptophan, histidine, and adenine was supplemented with X-αgal, resulting in growth of blue yeast colonies when bait and prey fusion proteins interact.
CoIP experiments
HEK293T cells were transiently transfected with the constructs of interest and lysed 48 h later. The full-length PKP3-encoding plasmid used in these experiments was p1744. CoIP experiments were performed essentially as described by Kowalczyk et al. (1997), using the PKP3-specific antibody 23E3/4.
Acknowledgments
We are grateful to A. Haegeman for purification of antibodies, to J. Comijn and J. van Hengel for helpful discussions, and to W. Drijvers, E. Vanden Eynde, and C. Eichperger for technical assistance. We thank Dr. J. Stanley for the human Dsg3 expression construct, and Drs. D. Garrod and C. Byrne for the mouse Dsc3a and Dsc3b cDNA constructs
S. Bonné has been supported by the Instituut voor de Aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen (IWT). F. van Roy is supported by the Geconcerteerde Onderzoeksacties of Ghent University, the Fund for Scientific Research-Flanders, Fortis Verzekeringen (Belgium), and Interuniversitaire attractiepolen (Belgium). M. Hatzfeld is supported by the Deutsche Forschungsgemeinschaft, X. Chen is supported by an R.H. Lurie Baseball Charities Cancer Fellowship, and K.J. Green by NIH grants AR43380, AR41836, and project no. 4 of PO1 DE12328.
Address correspondence to Frans van Roy, Dept. for Molecular Biomedical Research, VIB-Ghent University, Ledeganckstraat 35, B-9000 Ghent, Belgium. Tel.: 32-9-264-50-17. Fax: 32-9-264-53-48. E-mail: F.Vanroy@dmb.rug.ac.be
Footnotes
Abbreviations used in this paper: CBS, catenin-binding segment; CK18, cytokeratin 18; CoIP, coimmunoprecipitation; DP, desmoplakin; DPNTP, desmoplakin amino-terminal fragment; DPNTPmut, mutated desmoplakin amino-terminal fragment; Dsc, desmocollin; Dsg, desmoglein; IA, intracellular anchor domain; Pg, plakoglobin; PKP, plakophilin.
References
- Armstrong, D.K.B., K.E. McKenna, P.E. Purkis, K.J. Green, R.A.J. Eady, I.M. Leigh, and A.E. Hughes. 1999. Haploinsufficiency of desmoplakin causes a striate subtype of palmoplantar keratoderma. Hum. Mol. Genet. 8:143–148. [DOI] [PubMed] [Google Scholar]
- Bannon, L.J., B.L. Cabrera, M.S. Stack, and K.J. Green. 2001. Isoform-specific differences in the size of desmosomal cadherin/catenin complexes. J. Invest. Dermatol. 117:1302–1306. [DOI] [PubMed] [Google Scholar]
- Basehoar, G., and M.W. Berns. 1973. Cloning of rat kangaroo (PTK2) cells following laser microirradiation of selected mitotic chromosomes. Science. 179:1333–1334. [DOI] [PubMed] [Google Scholar]
- Bierkamp, C., K.J. McLaughlin, H. Schwarz, O. Huber, and R. Kemler. 1996. Embryonic heart and skin defects in mice lacking plakoglobin. Dev. Biol. 180:780–785. [DOI] [PubMed] [Google Scholar]
- Bonné, S., J. van Hengel, F. Nollet, P. Kools, and F. van Roy. 1999. Plakophilin-3, a novel Armadillo-like protein present in nuclei and desmosomes of epithelial cells. J. Cell Sci. 112:2265–2276. [DOI] [PubMed] [Google Scholar]
- Bornslaeger, E.A., L.M. Godsel, C.M. Corcoran, J.K. Park, M. Hatzfeld, A.P. Kowalczyk, and K.J. Green. 2001. Plakophilin 1 interferes with plakoglobin binding to desmoplakin, yet together with plakoglobin promotes clustering of desmosomal plaque complexes at cell-cell borders. J. Cell Sci. 114:727–738. [DOI] [PubMed] [Google Scholar]
- Boukamp, P., R.T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N.E. Fusenig. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracke, M., B. Vyncke, G. Opdenakker, J.M. Foidart, G. De Pestel, and M. Mareel. 1991. Effect of catechins and citrus flavonoids on invasion in vitro. Clin. Exp. Metastasis. 9:13–25. [DOI] [PubMed] [Google Scholar]
- Chen, X., S. Bonné, M. Hatzfeld, F. van Roy, and K.J. Green. 2002. Protein binding and functional characterization of plakophilin 2: evidence for its diverse roles in desmosomes and beta-catenin signaling. J. Biol. Chem. 277:10512–10522. [DOI] [PubMed] [Google Scholar]
- Elias, P.M., N. Matsuyoshi, H. Wu, C. Lin, Z.H. Wang, B.E. Brown, and J.R. Stanley. 2001. Desmoglein isoform distribution affects stratum corneum structure and function. J. Cell Biol. 153:243–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto, F., U. Gluck, and B.M. Gumbiner. 1998. Nuclear localization signal-independent and importin karyopherin-independent nuclear import of beta-catenin. Curr. Biol. 8:181–190. [DOI] [PubMed] [Google Scholar]
- Funayama, N., F. Fagotto, P. McCrea, and B.M. Gumbiner. 1995. Embryonic axis induction by the armadillo repeat domain of beta-catenin: evidence for intracellular signaling. J. Cell Biol. 128:959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallicano, G.I., P. Kouklis, C. Bauer, M. Yin, V. Vasioukhin, L. Degenstein, and E. Fuchs. 1998. Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J. Cell Biol. 143:2009–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman, Y. 1981. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 23:175–182. [DOI] [PubMed] [Google Scholar]
- Green, K.J., and C.A. Gaudry. 2000. Are desmosomes more than tethers for intermediate filaments? Nat. Rev. Mol. Cell Biol. 1:208–216. [DOI] [PubMed] [Google Scholar]
- Hatzfeld, M. 1997. Band 6 protein and cytoskeletal organization. Cytoskeletal-Membrane Interactions and Signal Transduction. P. Cowin and M.W. Klymkowsky, editors. Landes Company and Chapman & Hall, Texas. 49–59.
- Hatzfeld, M., G.I. Kristjansson, U. Plessman, and K. Weber. 1994. Band 6 protein, a major constituent of desmosomes from stratified epithelia, is a novel member of the armadillo multigene family. J. Cell Sci. 107:2259–2270. [DOI] [PubMed] [Google Scholar]
- Hatzfeld, M., C. Haffner, K. Schulze, and U. Vinzens. 2000. The function of plakophilin 1 in desmosome assembly and actin filament organization. J. Cell Biol. 149:209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid, H.W., A. Schmidt, R. Zimbelmann, S. Schäfer, S. Wintersimanowski, S. Stumpp, M. Keith, U. Figge, M. Schnolzer, and W.W. Franke. 1994. Cell type-specific desmosomal plaque proteins of the plakoglobin family: plakophilin 1 (band 6 protein). Differentiation. 58:113–131. [DOI] [PubMed] [Google Scholar]
- Hofmann, I., C. Mertens, M. Brettel, V. Nimmrich, M. Schnolzer, and H. Herrmann. 2000. Interaction of plakophilins with desmoplakin and intermediate filament proteins: an in vitro analysis. J. Cell Sci. 113:2471–2483. [DOI] [PubMed] [Google Scholar]
- Ishii, K., and K.J. Green. 2001. Cadherin function: breaking the barrier. Curr. Biol. 11:R569–R572. [DOI] [PubMed] [Google Scholar]
- Ishii, K., S.M. Norvell, L.J. Bannon, E.V. Amargo, L.T. Pascoe, and K.J. Green. 2001. Assembly of desmosomal cadherins into desmosomes is isoform dependent. J. Invest. Dermatol. 117:26–35. [DOI] [PubMed] [Google Scholar]
- King, I.A., A. Tabiowo, P. Purkis, I. Leigh, and A.I. Magee. 1993. Expression of distinct desmocollin isoforms in human epidermis. J. Invest. Dermatol. 100:373–379. [DOI] [PubMed] [Google Scholar]
- King, I.A., B.D. Angst, D.M. Hunt, M. Kruger, J. Arnemann, and R.S. Buxton. 1997. Hierarchical expression of desmosomal cadherins during stratified epithelial morphogenesis in the mouse. Differentiation. 62:83–96. [DOI] [PubMed] [Google Scholar]
- Klymkowsky, M.W. 1999. Plakophilin, armadillo repeats, and nuclear localization. Microsc. Res. Tech. 45:43–54. [DOI] [PubMed] [Google Scholar]
- Koch, P.J., M.D. Goldschmidt, R. Zimbelmann, R. Troyanovsky, and W.W. Franke. 1992. Complexity and expression patterns of the desmosomal cadherins. Proc. Natl. Acad. Sci. USA. 89:353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk, A.P., H.L. Palka, H.H. Luu, L.A. Nilles, J.E. Anderson, M.J. Wheelock, and K.J. Green. 1994. Posttranslational regulation of plakoglobin expression-influence of the desmosomal cadherins on plakoglobin metabolic stability. J. Biol. Chem. 269:31214–31223. [PubMed] [Google Scholar]
- Kowalczyk, A.P., E.A. Bornslaeger, J.E. Borgwardt, H.L. Palka, A.S. Dhaliwal, C.M. Corcoran, M.F. Denning, and K.J. Green. 1997. The amino-terminal domain of desmoplakin binds to plakoglobin and clusters desmosomal cadherin-plakoglobin complexes. J. Cell Biol. 139:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk, A.P., M. Hatzfeld, E.A. Bornslaeger, D.S. Kopp, J.E. Borgwardt, C.M. Corcoran, A. Settler, and K.J. Green. 1999. a. The head domain of plakophilin-1 binds to desmoplakin and enhances its recruitment to desmosomes. Implications for cutaneous disease. J. Biol. Chem. 274:18145–18148. [DOI] [PubMed] [Google Scholar]
- Kowalczyk, A.P., E.A. Bornslaeger, S.M. Norvell, H.L. Palka, and K.J. Green. 1999b. Desmosomes: intercellular adhesive junctions specialized for attachment of intermediate filaments. International Review of Cytology: A Survey of Cell Biology. Vol. 185. K.W. Jeon, editor. Academic Press, San Diego. 237–302. [DOI] [PubMed]
- McGrath, J.A. 1999. Hereditary diseases of desmosomes. J. Dermatol. Sci. 20:85–91. [DOI] [PubMed] [Google Scholar]
- McGrath, J.A., J.R. McMillan, C.S. Shemanko, S.K. Runswick, I.M. Leigh, E.B. Lane, D.R. Garrod, and R.A.J. Eady. 1997. Mutations in the plakophilin 1 gene result in ectodermal dysplasia skin fragility syndrome. Nat. Genet. 17:240–244. [DOI] [PubMed] [Google Scholar]
- McKoy, G., N. Protonotarios, A. Crosby, A. Tsatsopoulou, A. Anastasakis, A. Coonar, M. Norman, C. Baboonian, S. Jeffery, and W.J. McKenna. 2000. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease). Lancet. 355:2119–2124. [DOI] [PubMed] [Google Scholar]
- Mertens, C., C. Kuhn, and W.W. Franke. 1996. Plakophilins 2a and 2b: constitutive proteins of dual location in the karyoplasm and the desmosomal plaque. J. Cell Biol. 135:1009–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens, C., C. Kuhm, R. Moll, I. Schwetlick, and W.W. Franke. 1999. Desmosomal plakophilin 2 as a differentiation marker in normal and malignant tissues. Differentiation. 64:277–290. [DOI] [PubMed] [Google Scholar]
- Messent, A.J., M.J. Blissett, G.L. Smith, A.J. North, A. Magee, D. Foreman, D.R. Garrod, and M. Boulton. 2000. Expression of a single pair of desmosomal glycoproteins renders the corneal epithelium unique amongst stratified epithelia. Invest. Ophthalmol. Vis. Sci. 41:8–15. [PubMed] [Google Scholar]
- Nollet, F., P. Kools, and F. van Roy. 2000. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J. Mol. Biol. 299:551–572. [DOI] [PubMed] [Google Scholar]
- Norgett, E.E., S.J. Hatsell, L. Carvajal-Huerta, J.C. Ruiz Cabezas, J. Common, P.E. Purkis, N. Whittock, I.M. Leigh, H.P. Stevens, and D.P. Kelsell. 2000. Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum. Mol. Genet. 9:2761–2766. [DOI] [PubMed] [Google Scholar]
- North, A.J., M.A.J. Chidgey, J.P. Clarke, W.G. Bardsley, and D.R. Garrod. 1996. Distinct desmocollin isoforms occur in the same desmosomes and show reciprocally graded distributions in bovine nasal epidermis. Proc. Natl. Acad. Sci. USA. 93:7701–7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North, A.J., W.G. Bardsley, J. Hyam, E.A. Bornslaeger, H.C. Cordingley, B. Trinnaman, M. Hatzfeld, K.J. Green, A.I. Magee, and D.R. Garrod. 1999. Molecular map of the desmosomal plaque. J. Cell Sci. 112:4325–4336. [DOI] [PubMed] [Google Scholar]
- Ruiz, P., V. Brinkmann, B. Ledermann, M. Behrend, C. Grund, C. Thalhammer, F. Vogel, C. Birchmeier, U. Gunthert, W.W. Franke, and W. Birchmeier. 1996. Targeted mutation of plakoglobin in mice reveals essential functions of desmosomes in the embryonic heart. J. Cell Biol. 135:215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runswick, S.K., M.J. O'Hare, L. Jones, C.H. Streuli, and D.R. Garrod. 2001. Desmosomal adhesion regulates epithelial morphogenesis and cell positioning. Nat. Cell Biol. 3:823–830. [DOI] [PubMed] [Google Scholar]
- Schäfer, S., S. Stumpp, and W.W. Franke. 1996. Immunological identification and characterization of the desmosomal cadherin Dsg2 in coupled and uncoupled epithelial cells and in human tissues. Differentiation. 60:99–108. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., L. Langbein, M. Rode, S. Pratzel, R. Zimbelmann, and W.W. Franke. 1997. Plakophilins 1a and 1b: widespread nuclear proteins recruited in specific epithelial cells as desmosomal plaque components. Cell Tissue Res. 290:481–499. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., L. Langbein, S. Pratzel, M. Rode, H.R. Rackwitz, and W.W. Franke. 1999. Plakophilin 3 -- a novel cell-type-specific desmosomal plaque protein. Differentiation. 64:291–306. [DOI] [PubMed] [Google Scholar]
- Simmons, N.L. 1990. A cultured human renal epithelioid cell line responsive to vasoactive intestinal peptide. Exp. Physiol. 75:309–319. [DOI] [PubMed] [Google Scholar]
- Smith, E.A., and E. Fuchs. 1998. Defining the interactions between intermediate filaments and desmosomes. J. Cell Biol. 141:1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin, V., E. Bowers, C. Bauer, L. Degenstein, and E. Fuchs. 2001. Desmoplakin is essential in epidermal sheet formation. Nat. Cell Biol. 3:1076–1085. [DOI] [PubMed] [Google Scholar]
- Vermeulen, S.J., E.A. Bruyneel, M.E. Bracke, G.K. De Bruyne, K.M. Vennekens, K.L. Vleminckx, G.J. Berx, F.M. van Roy, and M.M. Mareel. 1995. Transition from the noninvasive to the invasive phenotype and loss of alpha-catenin in human colon cancer cells. Cancer Res. 55:4722–4728. [PubMed] [Google Scholar]