Abstract
The Aurora B kinase complex is a critical regulator of chromosome segregation and cytokinesis. In Caenorhabditis elegans, AIR-2 (Aurora B) function requires ICP-1 (Incenp) and BIR-1 (Survivin). In various systems, Aurora B binds to orthologues of these proteins. Through genetic analysis, we have identified a new subunit of the Aurora B kinase complex, CSC-1. C. elegans embryos depleted of CSC-1, AIR-2, ICP-1, or BIR-1 have identical phenotypes. CSC-1, BIR-1, and ICP-1 are interdependent for their localization, and all are required for AIR-2 localization. In vitro, CSC-1 binds directly to BIR-1. The CSC-1/BIR-1 complex, but not the individual subunits, associates with ICP-1. CSC-1 associates with ICP-1, BIR-1, and AIR-2 in vivo. ICP-1 dramatically stimulates AIR-2 kinase activity. This activity is not stimulated by CSC-1/BIR-1, suggesting that these two subunits function as targeting subunits for AIR-2 kinase.
Keywords: chromosome segregation; cytokinesis; central spindle; IAP; C. elegans
Introduction
Accurate duplication and segregation of the genome and cellular organelles are accomplished through an exquisitely coordinated series of events. Execution of several mitotic processes and cytokinesis requires members of the Aurora kinase family, which is subdivided into three subfamilies, Aurora A, B, and C, based on their subcellular localization (Nigg, 2001). Aurora B has essential functions in meiotic and mitotic chromosome segregation, and cytokinesis.
Aurora B kinase concentrates on the subcellular structures that it regulates. During meiosis in Caenorhabditis elegans, Aurora B kinase localizes to a discrete region between homologous chromosomes in diakinesis and metaphase, where it acts to control chromosome segregation (Kaitna et al., 2002; Rogers et al., 2002). In mammalian cells, during prometaphase and metaphase, Aurora B concentrates in the inner centromeric region, a localization that may underlie its function in mitotic chromosome segregation (Martineau-Thuillier et al., 1998). After anaphase onset, Aurora B kinase localizes to the central spindle where it promotes the stable localization of the centralspindlin complex and therefore probably regulates cytokinesis (Kaitna et al., 2002; Mishima et al., 2002). One outstanding question is how this kinase becomes concentrated on these distinct structures at different moments during meiosis and mitosis.
Although the mechanism of targeting Aurora B kinase is not fully understood, its localization in several systems requires at least two other proteins, Incenp and Survivin/BIR-1. Depletion of Aurora B kinase, Incenp, or Survivin/BIR-1 causes identical phenotypes, and biochemical interactions have been detected between these three proteins (for review see Adams et al., 2001). Thus, Aurora B, BIR-1 (Survivin), and Incenp appear to form a complex, hereafter referred to as the ABI* complex.
Here we describe the discovery and characterization of an additional subunit of the Aurora B kinase complex, CSC-1. C. elegans hermaphrodites mutant for csc-1 produce embryos that are inviable due to defects in meiotic and mitotic chromosome segregation and cytokinesis; identical phenotypes are caused by depletion of AIR-2 (Aurora B), ICP-1 (Incenp), or BIR-1 (Survivin). We provide genetic, cell biological, and biochemical evidence that CSC-1 is a subunit of the Aurora B kinase complex.
Results and discussion
CSC-1 is a novel 27-kD protein required for chromosome segregation and cytokinesis
During a large-scale screen for maternal effect embryonic lethal mutations, we isolated a mutant allele (t1171) that exhibited the set of phenotypes characteristic of the ABI complex members (Fig. 1 A). In particular, meiotic chromosome segregation and polar body extrusion are defective. During mitosis, spindle assembly appears normal, but chromosome segregation fails entirely and a single large nucleus reassembles in the subsequent interphase. After spindle elongation, a cleavage furrow forms and ingresses, but it fails to ingress to completion, thus producing a multinucleate embryo.
Figure 1.
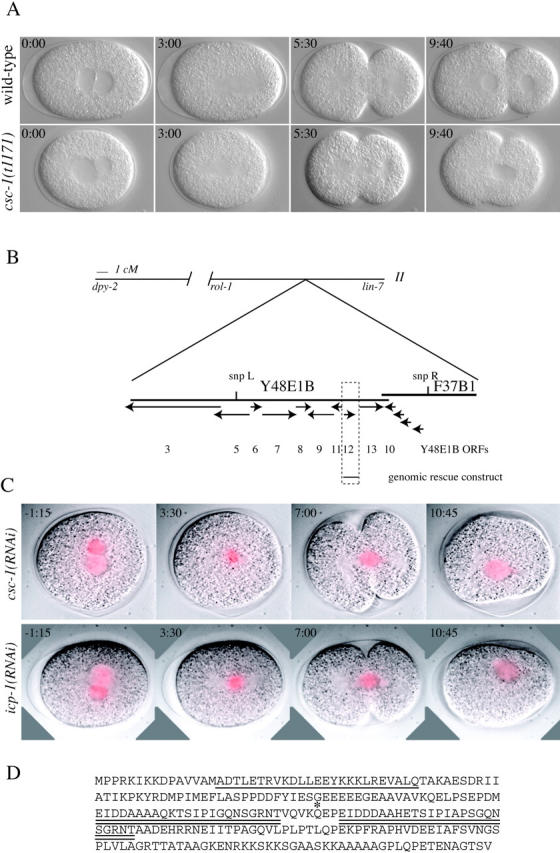
csc-1 encodes a 27-kD protein required for chromosome segregation and cytokinesis. (A) csc-1 mutants have defects in chromosome segregation and cytokinesis. Images shown are selected from a time-lapse Nomarski recording of a wild type and an embryo derived from a homozygous csc-1(t1171) mutant hermaphrodite. (B) Schematic depicting the position of the csc-1 locus. SNP mapping placed csc-1 between SNP L and SNP R. (C) csc-1(RNAi) and icp-1(RNAi) cause similar chromosome segregation phenotypes. Embryos dissected from worms expressing GFP–histone H2B and depleted of CSC-1 or ICP-1 by RNAi were imaged by Nomarski and fluorescence optics. Time shown is relative to nuclear envelope breakdown. The GFP signal is shown as a red overlay. (D) The sequence of the CSC-1 protein; the potential coiled-coil region is underlined and the imperfect repeat is double underlined. Glutamine 128, the residue mutated in csc-1(t1171), is indicated by an asterisk.
The csc-1 locus maps to 16 cM on chromosome II. As the known members of the ABI complex map to other regions of the genome, this locus must be distinct, and it was named csc-1(t1171) for chromosome segregation and cytokinesis defective. The map position was refined by single nucleotide polymorphism (SNP) mapping to a small region containing 12 predicted genes, four of which were predicted to encode glutathione transferases (Fig. 1 B). The eight unique genes in the region were depleted by RNA interference (RNAi); depletion of one predicted gene product, Y48E1B.12, caused chromosome segregation and cytokinesis defects identical to those seen in csc-1(t1171) mutant embryos (Fig. 1 C). Chromosome behavior was analyzed using GFP–histone H2B in csc-1(RNAi) embryos. These embryos have defects in chromosome congression to the metaphase plate and in segregation of the chromosomes to the spindle poles. The condensed chromosomes do not form a well-ordered metaphase plate, and subsequently the chromatin becomes stretched along the spindle axis, and cytokinesis initiates but ultimately fails (Fig. 1 C). These phenotypes are identical to those observed in embryos depleted for ICP-1 (Fig. 1 C) and AIR-2 (Kaitna et al., 2000).
The csc-1 locus is predicted to encode a 27-kD protein with a potential coiled coil in the NH2 terminus. No other conserved domains were detected. This protein also contains an imperfect direct repeat of 23–25 residues (Fig. 1 D). Surprisingly, with the exception of a sequence from the C. briggsae genome, no other sequences in the available databases have significant homology to CSC-1. To confirm the identity of csc-1, the gene was sequenced from animals homozygous for csc-1(t1171), and a single point mutation was identified that changes glutamine 128 into a stop codon. In addition, a 2.6-kb genomic fragment can rescue csc-1(t1171) to fertility. Worms homozygous for the strong loss of function allele of csc-1(t1171) are viable and semi-sterile, producing few embryos, all of which are inviable (average brood size of dpy-2[e8] is 51.2 [n = 13]; for dpy-2[e8];csc-1[t1171] the value is 7.2 [n = 105]). The viability and incomplete sterility of csc-1(t1171) animals may result from perdurance of the maternally provided protein.
CSC-1 localizes to meiotic and mitotic chromosomes and to the central spindle during anaphase
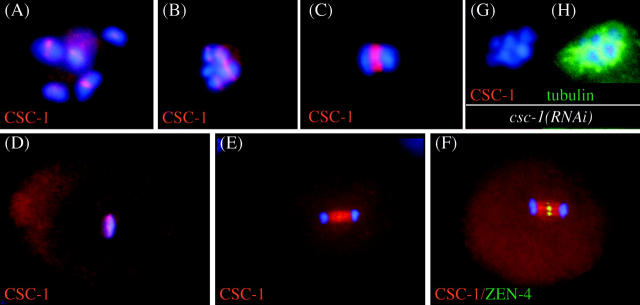
To examine whether CSC-1 shows the same subcellular location as other ABI complex members, a peptide-specific antibody was prepared and used to detect CSC-1 in wild-type embryos. In oocytes, during meiosis I, CSC-1 localizes to a discrete region of meiotic bivalents (Fig. 2, A and B) . During anaphase of meiosis I, CSC-1 localizes to the midzone of the meiotic spindle (Fig. 2 C). In mitosis, CSC-1 localizes to chromosomes during metaphase (Fig. 2 D) and to the spindle midzone during anaphase (Fig. 2 E) and telophase. The localization of CSC-1 on the central spindle is significantly broader than the localization of ZEN-4, the kinesin-like protein that is part of the centralspindlin complex (Fig. 2 F). The reactivity of anti–CSC-1 antibodies on chromatin (Fig. 2 G) and the central spindle is abolished in csc-1(RNAi) embryos. All features of the localization of CSC-1 are similar to those described for other members of the ABI complex (Schumacher et al., 1998; Severson et al., 2000; Oegema et al., 2001; Kaitna et al., 2002; Rogers et al., 2002).
Figure 2.
CSC-1 localizes in a similar manner as ABI complex members. CSC-1 localization in wild-type embryos was analyzed by immunofluorescence. Staining is observed in the central region of meiotic bivalents during diakinesis (A) and in prometaphase (B) of meiosis I. At anaphase I, CSC-1 localizes to the central region of the meiotic spindle (C). During mitotic metaphase, CSC-1 localizes to the chromosomes (D), and in anaphase, it localizes to the central spindle (E). The CSC-1 signal on the central spindle is wider than that of the centralspindlin component ZEN-4 (F). The above-mentioned staining patterns are absent in csc-1(RNAi) embryos (G); anti-tubulin staining demonstrates antibody accessibility (H).
CSC-1, BIR-1, and ICP-1 are mutually dependent for their localization and are all required for AIR-2 localization
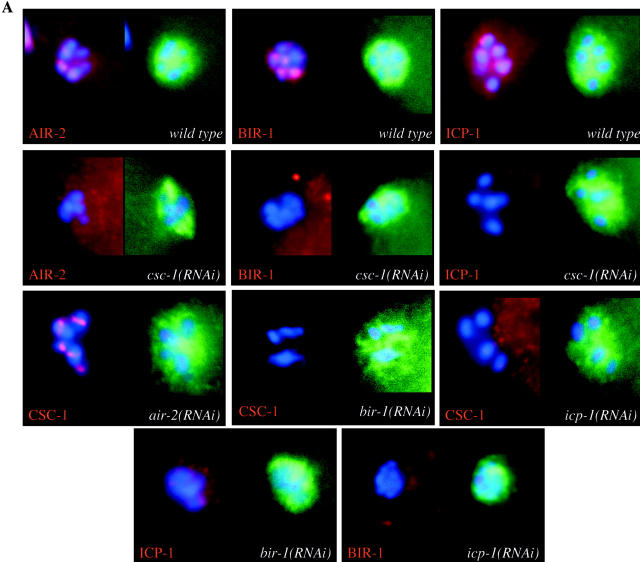
To examine whether CSC-1 is required for the localization of other ABI complex members, RNAi was used to deplete embryos of CSC-1, and the localization of AIR-2, ICP-1, and BIR-1 to meiotic chromosomes was examined by immunofluorescence, as this is the first instance where these proteins are functionally required. All three proteins require CSC-1 to localize to meiotic chromatin (Fig. 3 A, second row), mitotic chromosomes, and the spindle midzone in meiosis and mitosis (unpublished data). To determine whether CSC-1 localization requires ABI complex members, embryos were depleted of AIR-2, ICP-1, and BIR-1 by RNAi, and the localization of CSC-1 was examined by immunofluorescence. CSC-1 localization is dependent on BIR-1 and ICP-1, but independent of AIR-2 (Fig. 3 A, third row). Furthermore, BIR-1 and ICP-1 are interdependent, but AIR-2 independent, for their proper localization (Fig. 3 A, fourth row; Speliotes et al., 2000; unpublished data). Thus, CSC-1, BIR-1, and ICP-1 are mutually interdependent, but AIR-2 independent, for their localization to chromosomes. AIR-2 requires CSC-1, BIR-1, and ICP-1 to localize to chromosomes.
Figure 3.
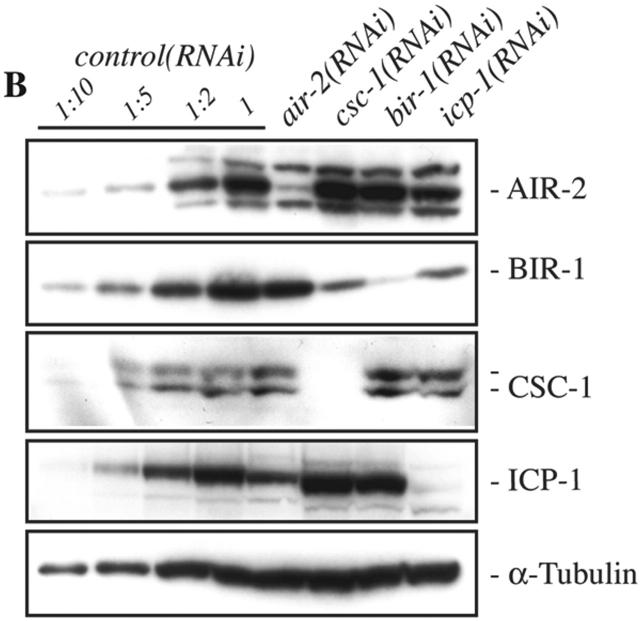
CSC-1, BIR-1, and ICP-1 are interdependent for their localization. (A) The localization of CSC-1 and known members of the ABI complex to meiotic chromatin was assayed by indirect immunofluorescence in fertilized oocytes of the indicated genotype. Embryos were costained with an anti-tubulin antibody to control for antibody accessibility. (B) Depletion of CSC-1 and ICP-1 reduces BIR-1 accumulation. Embryo extracts from RNAi-treated worms were analyzed by immunoblotting with antibodies directed against the indicated proteins. Immunoblotting with anti–α-tubulin (DM1α) antibodies demonstrates equal protein loading. Control embryo extracts were prepared using an RNAi construct that does not cause a phenotype. Dilutions of the control extracts were run in parallel to allow the degree of depletion to be estimated.
To examine whether these localization dependencies may also be due to an effect on protein stability, we performed Western blots on embryos isolated from RNAi-treated worms and assessed the levels of ICP-1, AIR-2, CSC-1, and BIR-1. Depletion of CSC-1 and ICP-1 reduced the levels of BIR-1 significantly, whereas depletion of BIR-1 and AIR-2 did not greatly affect the accumulation of the other subunits (Fig. 3 B). These results suggest a functional interaction between CSC-1 and BIR-1.
CSC-1 associates with ABI complex members in vivo and binds to BIR-1 in vitro
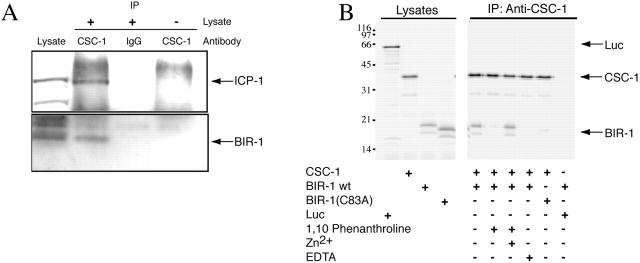
As CSC-1, BIR-1, and ICP-1 are interdependent for their localization, these proteins likely form a protein complex in vivo. To examine whether such a complex exists, we used Western blotting to determine if anti–CSC-1 immunoprecipitates (IPs) contain ABI complex members. Indeed, both BIR-1 and ICP-1 associate with CSC-1 in C. elegans embryo extracts (Fig. 4 A). We conclude that in vivo, CSC-1 is a member of the ABI complex.
Figure 4.
CSC-1 associates with ABI complex members in vivo and in vitro. (A) Whole cell extracts were prepared from wild-type embryos and immunoprecipitated with anti–CSC-1 antibodies. Western blots of the immunoprecipitates reveal that BIR-1 and ICP-1 associate with CSC-1. The coprecipitation of ICP-1 and BIR-1 is specific because it required anti–CSC-1 antibodies (lane 3) and embryonic lysates (lane 4). (B) CSC-1 binds to BIR-1 in vitro, and this interaction requires Zinc binding by BIR-1. Zinc chelation prevents the CSC-1/BIR-1 interaction. Mutation of one of the cysteine residues of BIR-1 (C83) predicted to coordinate zinc abolishes the interaction with CSC-1. CSC-1 and BIR-1 were translated in vitro in the presence of [35S]methionine and mixed in the presence of the indicated compounds. Immunoprecipitations were run on SDS-PAGE and the radioactive products detected with a phosphoimager. Luciferase (Luc) was included as a nonspecific control for the binding reactions.
The results above suggest that CSC-1 might directly bind to one of the members of this complex. To examine this possibility, binding assays were performed using in vitro–translated proteins. CSC-1 and BIR-1 associate in the absence of any other nematode proteins (Fig. 4 B). The baculovirus IAP repeat (BIR) domain of BIR-1 is stabilized by a Zn2+ atom that is coordinated by three cysteine residues and a histidine. The interaction between CSC-1 and BIR-1 is abolished when Zn2+ is chelated with 1,10 O-phenanthroline and restored by addition of excess zinc. In addition, mutation of BIR-1 at cysteine 83, one of the residues predicted to chelate zinc, to alanine, abolishes the interaction between CSC-1 and BIR-1 (Fig. 4 B). We conclude that the interaction between CSC-1 and BIR-1 requires a native, Zn2+-bound BIR domain. Furthermore, as several studies indicate that recombinant Survivin forms a stable dimeric structure (Shi, 2000), we investigated whether CSC-1 binds to a dimer of BIR-1 and whether the residues implicated in dimer formation are critical for the interaction between CSC-1 and BIR-1. We have not observed any indications that BIR-1 is a dimer when bound to CSC-1 (see Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200207117/DC1).
Efficient binding of the CSC-1/BIR-1 subcomplex to ICP-1 and formation of a multiprotein complex that activates AIR-2 kinase activity
We next investigated whether BIR-1, CSC-1, or the BIR-1/CSC-1 complex biochemically interacts with ICP-1 and AIR-2 kinase. Cell lysates were prepared from insect cells infected with baculoviruses expressing the individual proteins. Using chitin-binding domain (CBD)–tagged ICP-1, we examined whether a complex could be formed between ICP-1 and CSC-1 or BIR-1 by mixing these lysates and recovering the tagged ICP-1 on chitin beads. In these binary binding assays, no detectable interactions were found between ICP-1 and CSC-1; a weak but reproducible interaction was observed between ICP-1 and BIR-1. However when lysates containing all three subunits were mixed, robust formation of an ICP-1/BIR-1/CSC-1 complex was observed (Fig. 5 A). These data, combined with the previous observation of a robust BIR-1/CSC-1 complex, suggest that ICP-1 binds strongly to this subcomplex, but not to the individual subunits.
Figure 5.
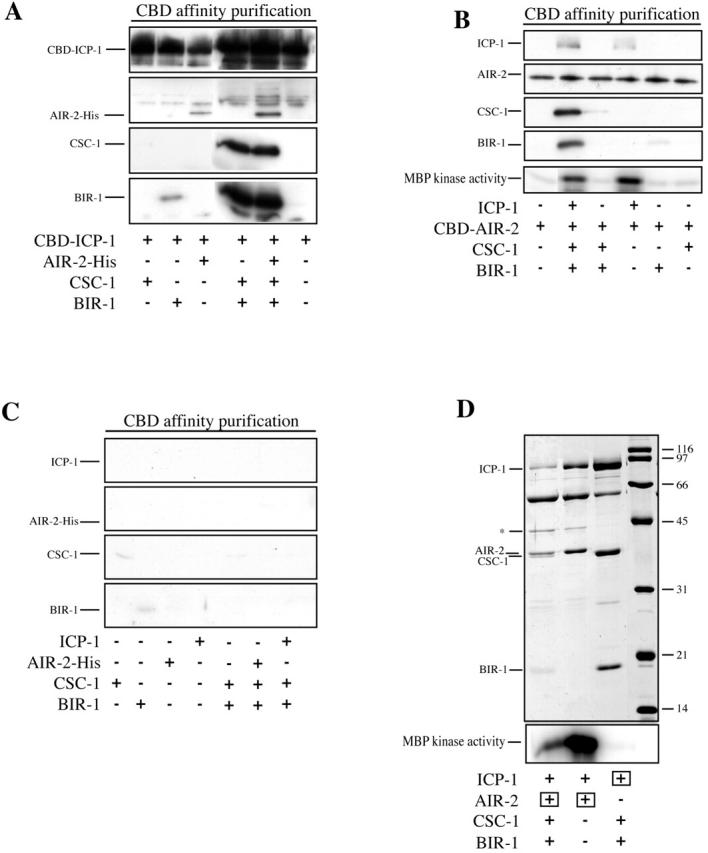
Recombinant AIR-2, ICP-1, CSC-1, and BIR-1 form complexes in vitro. AIR-2, ICP-1, CSC-1, and BIR-1 were individually expressed in insect cells, and lysates were mixed as indicated, purified using the CBD affinity tag, and analyzed by Western blotting to detect protein–protein interactions among ABI complex members. The Western blots shown in A, B, and C were processed in parallel. (A) CBD-tagged ICP-1 does not bind to CSC-1 and binds weakly to BIR-1. All three proteins together form a robust complex in vitro, CBD–ICP-1 binds to AIR-2–His, and the complex containing all four subunits can be reconstituted in vitro. (B) Using CBD-tagged AIR-2, similar complexes can be reconstituted. ICP-1 binds to CBD–AIR-2. CBD–AIR-2 does not bind to CSC-1 or BIR-1 individually, nor to the CSC-1/BIR-1 complex, but it does bind to the CSC-1/BIR-1/ICP-1 complex. The bottom panel shows the corresponding kinase activity using MBP as a substrate. ICP-1 is required for AIR-2 activity, and CSC-1 and BIR-1 do not enhance the MBP phosphorylation. (C) Control binding reactions for A and B show that little binding is observed in the absence of CBD-tagged subunits. (D) Coomassie staining of SDS-PAGE of purified soluble ABI complex. Insect cells were coinfected with the indicated combinations of viruses. The subunits containing the affinity tag used for the purification are indicated by a square. Uncleaved CBD–AIR-2 is indicated by an asterisk. The lower panel shows kinase activity toward MBP. The amount of kinase activity in lanes 1 and 2 correlates with the abundance of AIR-2 and ICP-1 (see also B).
Our previous studies showed that the COOH-terminal conserved domain of ICP-1 is sufficient to bind to AIR-2 (Kaitna et al., 2000). We therefore tested whether AIR-2 also binds directly to CSC-1 or BIR-1 or whether the presence of CSC-1/BIR-1 alters the binding of AIR-2 to ICP-1. Binary binding assays using epitope-tagged ICP-1 or AIR-2 confirmed the previously established interaction between ICP-1 and AIR-2, but no significant interactions were observed between AIR-2 and CSC-1 or BIR-1 (Fig. 5, A and B). Moreover, AIR-2 did not bind appreciably to the CSC-1/BIR-1 complex, except in the presence of ICP-1 (Fig. 5 B). In addition, the presence or absence of CSC-1/BIR-1 did not significantly affect the interaction between ICP-1 and AIR-2. The binding of AIR-2 to BIR-1 required CSC-1 (and ICP-1) and vice versa. Thus, AIR-2 can associate with the CSC-1/BIR-1 subcomplex, but this interaction is likely indirect via ICP-1.
The CSC-1/BIR-1/ICP-1 complex associates with AIR-2, raising the possibility that it regulates its activity. AIR-2 is not active above background levels, using myelin basic protein (MBP) as a substrate, when expressed alone in insect cells. Mixing of AIR-2–containing lysates with CSC-1, BIR-1, or both did not activate AIR-2 kinase. However, AIR-2 was significantly activated upon mixing with an ICP-1–containing lysate (Fig. 5 B). We next assessed whether CSC-1 or BIR-1, either alone or in combination, affected the ICP-1–activated kinase activity. We did not observe a significant, reproducible change in the ICP-1–stimulated AIR-2 kinase activity. These data confirm earlier reports that ICP-1 is an activator of AIR-2 (Kang et al., 2001; Bishop and Schumacher, 2002) and suggest that CSC-1 and BIR-1 do not function primarily by regulating AIR-2 kinase activity, at least toward the model substrate MBP.
AIR-2/ICP-1/CSC-1/BIR-1 form a robust complex, which was purified to obtain an indication of the relative abundance of each subunit in the complex. We coinfected insect cells with viruses that express CBD–AIR-2, ICP-1, CSC-1, and BIR-1 and purified AIR-2 and associated proteins by chitin affinity chromatography. The complex was eluted from chitin beads using the tobacco etch virus (TEV) protease to cleave a recognition site that was engineered between the CBD tag and AIR-2. These preparations contained approximately equal levels of CSC-1, BIR-1, and ICP-1 and slightly more AIR-2 (which carried the affinity tag) (Fig. 5 D). This complex has high kinase activity against MBP. We also purified a complex containing only AIR-2 and ICP-1. The kinase activity of both preparations paralleled the amount of AIR-2 and ICP-1, again indicating that CSC-1 and BIR-1 do not greatly affect AIR-2 kinase activity (Fig. 5 D).
Composition and function of the ABI complex
Genetic, cell biological, and biochemical assays indicate that CSC-1 functions as an essential subunit of the ABI kinase complex. This complex mediates chromosome segregation and cytokinesis in C. elegans and many other eukaryotic organisms. To understand these fundamental cellular events, it is critical to understand the function of this kinase complex and identify its important substrates. Furthermore, it is important to investigate the enzyme itself by characterizing its composition, establishing how it localizes, and deciphering how its activity is regulated. By genetic and cell biological criteria, CSC-1 is essential for all known functions of the Aurora B kinase, AIR-2. CSC-1 is required for AIR-2 localization, and the cellular consequences of CSC-1 depletion are identical to that of AIR-2. However, biochemical analysis indicates that AIR-2 kinase activity in vitro does not require CSC-1. Thus, we suggest that a complex containing CSC-1/BIR-1/ICP-1 functions to localize and activate AIR-2, the activation function being provided principally by ICP-1.
Conservation of CSC-1
Whereas AIR-2, BIR-1, and ICP-1 have well-defined orthologues in many other eukaryotes, we have been unable to detect proteins with significant similarity to CSC-1 in any other organism except for C. briggsae. One possibility is that CSC-1 is unique to nematodes. Alternatively, our failure to identify an orthologue of CSC-1 could be due to weak conservation at the primary sequence level. Finally, CSC-1 function might be provided by one of the previously characterized members of the ABI complex in other organisms. Vertebrate Survivin has been reported to bind directly to Incenp (Wheatley et al., 2001), the NH2 terminus of Incenp is sufficient to interact with Survivin in a cell extract system (Bolton et al., 2002), and affinity purification of Sli15p reveals coassociation of Ipl1p and Bir1p (the budding yeast orthologues of ICP-1, AIR-2, and BIR-1, respectively) (Cheeseman et al., 2002). In addition, interactions have been detected between Survivin and Aurora B (Wheatley et al., 2001; Bolton et al., 2002; Chen et al., 2003). In contrast, we have found that stable interaction of ICP-1 and BIR-1 as well as AIR-2 and BIR-1 requires CSC-1 (ICP-1 is also required in the latter case). A simple model is that the function of Incenp has diverged into two polypeptides, ICP-1 and CSC-1, at some point during evolution. However, we have not detected significant primary sequence conservation between CSC-1 and the NH2-terminal domain of vertebrate Incenp, though both proteins do contain regions predicted to form a coiled coil.
Although the split gene model for csc-1 is speculative, it is not without precedent. For example, in most organisms, separase is an inactive dimeric complex that is activated by degradation of its partner, securin. In Drosophila, however, separase is an inactive trimer that is activated by degradation of one subunit (Jager et al., 2001).
Subcellular targeting of the ABI complex
Our results provide some insight as to how the ABI complex concentrates on the structures it regulates in a cell cycle–dependent fashion, namely meiotic and mitotic chromosomes and the central spindle. The interdependence of BIR-1, ICP-1, and CSC-1 for centromeric localization (as well as to the central spindle) indicates that the binding sites for the ABI complex depend on all three (and perhaps additional) subunits. Binding of the ABI complex to chromosomes is dependent upon the integrity of the cohesin complex (Sonoda et al., 2001; Kaitna et al., 2002). This dependency is intriguing because one function that has been suggested for the ABI complex is to regulate microtubule–kinetochore interactions in response to tension exerted between associated sister chromatids (Kaitna et al., 2002; Tanaka et al., 2002).
The other prominent site of localization of the ABI complex is the central spindle. Notably, there are at least two distinct domains within the central spindle, a tightly restricted one that contains the centralspindlin complex and a somewhat broader domain in which the ABI complex localizes. Whereas stable localization of the centralspindlin complex requires the ABI complex (Kaitna et al., 2000), localization of the ABI complex is largely independent of centralspindlin (Jantsch-Plunger et al., 2000). Evidence is accumulating that posttranslational modification of the ABI complex regulates its redistribution to the spindle midzone. Aurora B kinase activity is required for the release of ABI complex members from chromosomes during anaphase and for association of the ABI complex with the central spindle (Severson et al., 2000; Murata-Hori and Wang, 2002; unpublished data).
Concluding remarks
The results presented here establish that the Aurora B kinase associates with a multisubunit targeting complex composed of CSC-1, BIR-1, and ICP-1. ICP-1 binds to the CSC-1/BIR-1 complex and to AIR-2; this latter interaction activates the catalytic activity of AIR-2 (Kang et al., 2001; Bishop and Schumacher, 2002; this paper). It remains to be determined how the BIR-1/CSC-1/ICP-1 subcomplex localizes to chromosomes and the central spindle, thereby recruiting Aurora B to these sites. CSC-1 and BIR-1 may have additional functions, such as regulating the substrate specificity of Aurora B.
Materials and methods
Worm strains and alleles
Strains used in this study include GE896 dpy-2(e8) csc-1(t1171)/mnC1 [II], him-3(e1147), MG303 ruIs32[pAZ132:pie-1/GFP/histoneH2B] III (derived from AZ212), Bristol N2, and CB4856, which were obtained from the Caenorhabditis Genetics Center.
Genetic mapping and genomic rescue
csc-1 was mapped to 16 cM on LGII. Fine mapping was performed using SNPs in crosses of GE896 with CB4856 (SNP data and primer sequences available on request). A 2.6-kb genomic fragment was coinjected with rol-6(su1006) and myo-2:GFP into GE896. Lines were established from Rol hermaphrodites. Dpy Rol worms were scored for fertility; 6/10 lines had rescue activity.
RNAi
Target sequences were PCR amplified from C. elegans cDNA, cloned into L4440 (provided by Andy Fire, Carnegie Institution of Washington, Baltimore, MD), transformed into HT115(DE3), and grown overnight. Nematode growth plates containing IPTG and ampicillin were seeded with these cultures for 24 h at 20°C. L4 animals were grown on these plates for 2 d at 20°C before analysis.
Immunostaining and microscopy
Immunostaining and microscopy were performed as previously described (Kaitna et al., 2002). Antibody sources and dilutions were as follows: Rb anti–phos-H3(S10) (Upstate Biotechnology), 1:500; Rb anti–CSC-1,1:750; Rb anti–AIR-2, 1:750; Rb anti–BIR-1 (Speliotes et al., 2000), 1:100; Rb anti–ICP-1 (Oegema et al., 2001), 1 μg/ml; Mo anti-GFP antibodies (Roche), 1:1,000; and rat YOL1/34 anti-tubulin monoclonal antibody, 1:300.
Anti–CSC-1 antibodies
CSC-1–specific antisera were produced in rabbits (Gramsch Laboratories) using a COOH-terminal peptide (CGPLQPETENAGTSV) coupled to keyhole limpet hemocyanin. An antibody raised against an NH2-terminal peptide provided essentially identical results in immunofluorescence and immunoprecipitation assays. Antibodies were affinity purified using immobilized peptides.
Embryo extract preparation
1 ml of C. elegans embryos obtained by alkaline hypochlorite treatment of gravid hermaphrodites was diluted in 1 ml buffer A (20 mM Hepes, 50 mM NaCl, 2 mM MgCl2, 5 mM EGTA, and 0.5% Triton X-100) and frozen in liquid nitrogen. The frozen pellets were ground to powder in a liquid nitrogen–cooled mortar. The powder was diluted in 1 ml buffer A and centrifuged at 20,000 g for 10 min at 4°C.
In vitro binding assays
Proteins were expressed in vitro with the TNT reticulocyte lysate system (Promega) in the presence of [35S]methionine and cysteine (Translabel; ICN Biomedicals). After translation, 10 μl of each translation product was diluted in 80 μl buffer A and incubated at 4°C for 30 min. Where indicated, EDTA was added to 10 mM; 1,10 O-phenanthroline 2 mM; and ZnCl2 5 mM. Proteins were immunoprecipitated or affinity purified with chitin beads. Proteins bound to the beads were analyzed by SDS-PAGE followed by autoradiography with a phosphoimager.
Expression of recombinant proteins and kinase assay
Recombinant baculoviruses were produced in Sf9 cells using the Bac-to-Bac system (Invitrogen). Recombinant proteins were expressed in High Five cells. Cells were harvested 48 h after viral infection and lysed in 20 mM Hepes, pH 7.5, 150 mM NaCl, 5 mM EGTA, 2 mM MgCl2, 1 mM PMSF, 0.1% Triton X-100, 5 mM β-mercaptoethanol, 10 μg/ml leupeptin, 10 μg/ml pepstatin. The lysates were centrifuged at 15 krpm for 10 min at 4°C. For binding experiments, equal volumes of the extracts were mixed in the indicated combinations (control extracts were substituted where appropriate) and recovered by overnight binding to chitin beads. Beads were washed in lysis buffer or kinase buffer and assayed for by Western blotting or MBP kinase activity.
Soluble purified recombinant protein complexes were obtained by coinfection of insect cells with the indicated viruses, and the purified complexes were eluted from chitin beads by incubation with tobacco etch virus (TEV) protease. Eluates were visualized by SDS-PAGE, and ∼20–60 ng of protein was assayed for kinase activity.
Kinase reactions were performed for 20 min at 20°C in kinase buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.5 μM okadaic acid, 50 μM ATP, 50 μCi/ml [γ-32P]ATP [NEN Life Science Products], 250 ng/μl MBP). Reactions were separated by SDS-PAGE, and radioactive phosphate incorporation was visualized by phosphoimaging.
Online supplemental material
A supplemental figure (Fig. S1) is available at http://www.jcb.org/cgi/content/full/jcb.200207117/DC1. This figure provides evidence that formation of the CSC-1/BIR-1 complex is insensitive to mutations of residues suggested to be involved in dimerization of the BIR-1 orhtologue, Survivin.
Supplemental Material
Acknowledgments
We thank Pierre Gönczy and Tony Hyman for help with the initial characterization of the t1171 allele, Liz Speliotes, H. Robert Horvitz, and Karen Oegema for providing antibodies, Tim Clausen for helpful discussions, and Susanne Kaitna and Jan-Michael Peters for comments on the manuscript.
This work has been supported by Boehringer Ingelheim, the Wiener Wirtschafts Förderungs Fonds, and an EMBO YIP award to M. Glotzer.
The online version of this article includes supplemental material.
H. Schnabel's present address is Max Planck Institut fur Neurobiologie, D-82152 Martinstried, Germany.
Footnotes
Abbreviations used in this paper: ABI, Aurora B, BIR-1, and Incenp; BIR, baculovirus IAP repeat; CBD, chitin-binding domain; MBP, myelin basic protein; RNAi, RNA interference; SNP, single nucleotide polymorphism.
References
- Adams, R.R., M. Carmena, and W.C. Earnshaw. 2001. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11:49–54. [DOI] [PubMed] [Google Scholar]
- Bishop, J.D., and J.M. Schumacher. 2002. Phosphorylation of the carboxy-terminus of INCENP by the Aurora B kinase stimulates Aurora B kinase activity. J. Biol. Chem. 277:27577–27580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton, M.A., W. Lan, S.E. Powers, M.L. McCleland, J. Kuang, and P.T. Stukenberg. 2002. Aurora B kinase exists in a complex with Survivin and INCENP and its kinase activity is stimulated by Survivin binding and phosphorylation. Mol. Biol. Cell. 13:3064–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Anderson, M. Jwa, E.M. Green, J. Kang, J.R. Yates III, C.S. Chan, D.G. Drubin, and G. Barnes. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172. [DOI] [PubMed] [Google Scholar]
- Chen, J., S. Jin, S.K. Tahir, H. Zhang, X. Liu, A.V. Sarthy, T.P. McGonigal, Z. Liu, S.H. Rosenberg, and S.C. Ng. 2003. Survivin enhances Aurora-B kinase activity and localizes Aurora-B in human cells. J. Biol. Chem. 278:486–490. [DOI] [PubMed] [Google Scholar]
- Jager, H., A. Herzig, C.F. Lehner, and S. Heidmann. 2001. Drosophila separase is required for sister chromatid separation and binds to PIM and THR. Genes Dev. 15:2572–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger, V., P. Gönczy, A. Romano, H. Schnabel, D. Hamill, R. Schnabel, A.A. Hyman, and M. Glotzer. 2000. CYK-4. A rho family GTPase activating protein (gap) required for central spindle formation and cytokinesis. J. Cell Biol. 149:1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitna, S., M. Mendoza, V. Jantsch-Plunger, and M. Glotzer. 2000. Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol. 10:1172–1181. [DOI] [PubMed] [Google Scholar]
- Kaitna, S., P. Pasierbek, M. Jantsch, J. Loidl, and M. Glotzer. 2002. The Aurora B Kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous chromosomes during meiosis. Curr. Biol. 12:798–812. [DOI] [PubMed] [Google Scholar]
- Kang, J.-S., I.M. Cheeseman, G. Kallstrom, S. Velmurugan, G. Barnes, and C.S. Chan. 2001. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 155:763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau-Thuillier, S., P.R. Andreassen, and R.L. Margolis. 1998. Colocalization of TD-60 and INCENP throughout G2 and mitosis: evidence for their possible interaction in signalling cytokinesis. Chromosoma. 107:461–470. [DOI] [PubMed] [Google Scholar]
- Mishima, M., S. Kaitna, and M. Glotzer. 2002. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell. 2:41–54. [DOI] [PubMed] [Google Scholar]
- Murata-Hori, M., and Y. Wang. 2002. The kinase activity of Aurora B is required for kinetochore-microtubule interactions during mitosis. Curr. Biol. 12:894–899. [DOI] [PubMed] [Google Scholar]
- Nigg, E.A. 2001. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2:21–32. [DOI] [PubMed] [Google Scholar]
- Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A.A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, E., J.D. Bishop, J.A. Waddle, J.M. Schumacher, and R. Lin. 2002. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J. Cell Biol. 157:219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, J.M., A. Golden, and P.J. Donovan. 1998. AIR-2: an Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 143:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson, A.F., D.R. Hamill, J.C. Carter, J. Schumacher, and B. Bowerman. 2000. The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr. Biol. 10:1162–1171. [DOI] [PubMed] [Google Scholar]
- Shi, Y. 2000. Survivin structure: crystal unclear. Nat. Struct. Biol. 7:620–623. [DOI] [PubMed] [Google Scholar]
- Sonoda, E., T. Matsusaka, C. Morrison, P. Vagnarelli, O. Hoshi, T. Ushiki, K. Nojima, T. Fukagawa, I.C. Waizenegger, J.M. Peters, et al. 2001. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell. 1:759–770. [DOI] [PubMed] [Google Scholar]
- Speliotes, E.K., A. Uren, D. Vaux, and H.R. Horvitz. 2000. The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell. 6:211–223. [DOI] [PubMed] [Google Scholar]
- Tanaka, T.U., N. Rachidi, C. Janke, G. Pereira, M. Galova, E. Schiebel, M.J. Stark, and K. Nasmyth. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 108:317–329. [DOI] [PubMed] [Google Scholar]
- Wheatley, S.P., A. Carvalho, P. Vagnarelli, and W.C. Earnshaw. 2001. INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr. Biol. 11:886–890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.