Abstract
It was known that the uptake of tryptophan is reduced in the yeast erg6 mutant, which is defective in a late step of ergosterol biosynthesis. Here, we show that this is because the high affinity tryptophan permease Tat2p is not targeted to the plasma membrane. In wild-type cells, the plasma membrane localization of Tat2p is regulated by the external tryptophan concentration. Tat2p is transported from the Golgi apparatus to the vacuole at high tryptophan, and to the plasma membrane at low tryptophan. However, in the erg6 mutant, Tat2p is missorted to the vacuole at low tryptophan. The plasma membrane targeting of Tat2p is dependent on detergent-insoluble membrane domains, suggesting that sterol affects the sorting through the organization of lipid rafts. The erg6 mutation also caused missorting to the multivesicular body pathway in late endosomes. Thus, sterol composition is crucial for protein sorting late in the secretory pathway. Tat2p is subject to polyubiquitination, which acts as a vacuolar-targeting signal, and the inhibition of this process suppresses the Tat2p sorting defects of the erg6 mutant. The sorting mechanisms of Tat2p that depend on both sterol and ubiquitin will be discussed.
Keywords: ERG6; Tat2p; raft; multivesicular body; ubiquitin
Introduction
Understanding of protein sorting mechanisms in the secretory pathway is important to address how organelles establish their identities. Proteins are sent to their destinations along the pathway by being loaded onto particular kinds of transport vesicles. In the case of membrane proteins, this is often achieved through recognition of cytoplasmic sorting signals by vesicle coat or adaptor proteins. It is also known that some membrane proteins have their sorting signals in the membrane-spanning regions. For a typical example, yeast ER proteins Sec12p and Sec71p contain Golgi-to-ER retrieval signals in their transmembrane domains, and the Golgi protein Rer1p acts as the sorting receptor by recognizing these proteins as specific ligands and sending them back to the ER (Sato et al., 1996, 1997, 2001). In a different view, it is also plausible that lipid interacts with transmembrane domain signals, and thus the lipid composition affects the localization of proteins. In support of this idea, several mechanisms of sorting by lipids have been postulated. A well-known example is the sorting mediated by a sphingolipid- and cholesterol-rich membrane domain called “raft” (Simons and Ikonen, 1997). These lipids tend to cluster in the bilayer to form microdomains, which are not solubilized by detergents. A special set of proteins such as influenza virus HA and GPI-anchored proteins, which are targeted to the apical plasma membrane in epithelial cells, are associated with lipid rafts. This association appears to depend on protein–lipid interactions in the bilayer. HA requires both its transmembrane domain and cholesterol for segregation into rafts (Scheiffele et al., 1997). One of the roles of rafts is proposed to be serving as sorting platforms that emerge in the trans-Golgi and move to the apical surface.
For gaining further insights into the sterol-dependent sorting processes, the yeast Saccharomyces cerevisiae is an attractive organism. The structure of the major sterol in yeast, ergosterol, is slightly different from cholesterol, but its biosynthetic pathway has been almost completely understood (for review see Daum et al., 1998). In terms of membrane trafficking, the sterol composition has been shown to affect endocytosis in yeast (Heese-Peck et al., 2002). Evidence is also presented that yeast does have lipid rafts that are important for protein sorting (Bagnat et al., 2000, 2001). To further understand the role of sterols in traffic, we decided to start a study paying attention to yeast erg mutants, which are defective in the ergosterol biosynthesis. We examined phenotypes of the erg mutants to find potential defects in protein sorting. We were aware that the erg6 mutant was known to show reduced uptake of tryptophan from the medium (Gaber et al., 1989). The ERG6 gene encodes S-adenosylmethionine Δ24 methyltransferase, which acts at a late step of the ergosterol biosynthetic pathway by converting zymosterol to fecosterol. The tryptophan uptake defect raised the possibility that the high affinity tryptophan permease Tat2p (Schmidt et al., 1994) is not correctly targeted to the plasma membrane. In this article, we will show the results of our detailed analysis of the localization of Tat2p and its post-Golgi sorting, with a particular focus on the roles of ubiquitination and lipid raft association.
Results
Localization of Tat2p is regulated by the tryptophan concentration in the medium
Tryptophan-auxotrophic yeast trp1 strains were used in most experiments. Growth of trp1 cells depends on uptake of tryptophan from the medium, which is largely performed by Tat2p (Schmidt et al., 1994). Due to the reduced uptake of tryptophan (Gaber et al., 1989), Δerg6 cells were unable to grow at 20 μg/ml of tryptophan (Fig. 1), the standard concentration of this amino acid in synthetic media (Sherman, 1991). At a high concentration of tryptophan (200 μg/ml), the growth was restored. This severe tryptophan auxotrophy seemed to stem from the impaired function of Tat2p because overexpression of TAT2 by a multicopy YEp vector completely restored the growth of Δerg6 cells even at a low concentration of tryptophan (2 μg/ml). This result gave us a warning that we should be very cautious about the expression level of Tat2p.
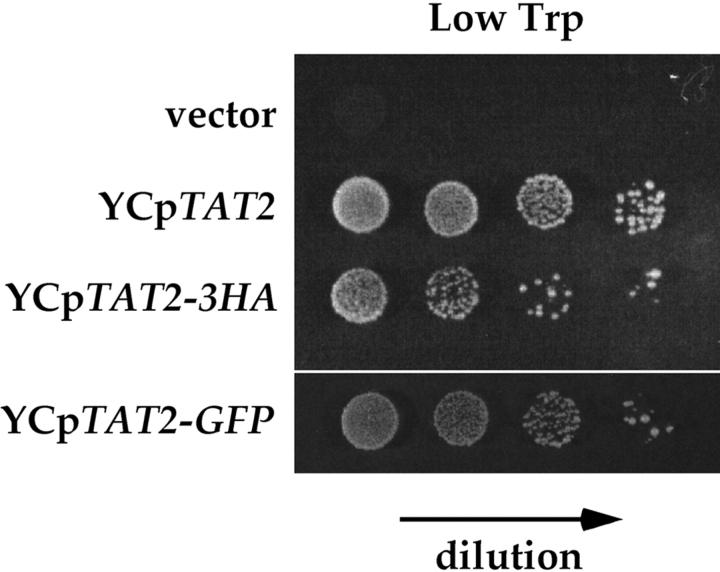
Figure 1.
Severe tryptophan auxotrophy of the Δerg6 mutant and its suppression by the overexpression of TAT2. YPH500 (WT, wild-type) and KUY136 (Δerg6) cells harboring the indicated plasmids were grown in the high tryptophan medium. Cells were washed and adjusted at the density of 107 cells/ml. 5-μl aliquots of 10-fold serial dilutions were spotted on MCD supplemented with adenine and the indicated concentrations of tryptophan.
Three copies of the HA epitope or the GFP was appended to the COOH terminus of Tat2p, which is predicted to orient to the cytoplasm (Beck et al., 1999). The TAT2 own promoter was chosen to express these variants. Either TAT2–3HA or TAT2-GFP on a single-copy YCp vector could support the growth of Δtat2 cells even at low tryptophan (Fig. 2), indicating that these fusion proteins are functional. In the following experiments, YCpTAT2–3HA or YCpTAT2-GFP was introduced into cells of the Δtat2 background.
Figure 2.
Epitope tagging of Tat2p. KUY121 (Δtat2) cells harboring the indicated plasmids were grown and spotted onto the low tryptophan medium as described in Fig. 1.
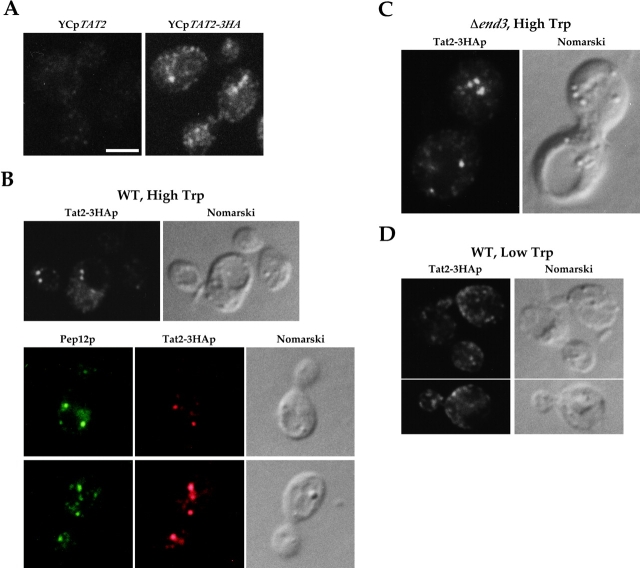
Localization of Tat2–3HAp was examined by indirect immunofluorescence microscopy with the anti-HA antibodies. Little staining was observed in Δtat2 cells harboring YCpTAT2 (Fig. 3 A), indicating that the nonspecific staining was negligible. Unexpectedly, plasma membrane staining of Tat2–3HAp was not evident at the standard concentration of tryptophan, and intracellular punctate structures were stained instead (Fig. 3 A). We reasoned that amino acids are sufficiently supplemented from the standard synthetic medium, and hence, high affinity amino acid permeases do not have to be localized to the plasma membrane under such a condition. Then, we tested the idea that the plasma membrane localization of permeases depends on the amino acid concentrations in the medium. TAT2 was dispensable for growth at high tryptophan (unpublished data), probably because other amino acid permeases, such as the low affinity tryptophan permease Tat1p (Schmidt et al., 1994), can incorporate sufficient tryptophan. Under this condition, Tat2–3HAp was found in the intracellular punctate structures, not on the plasma membrane (Fig. 3 B). Many of these spots located in the proximity of vacuoles. Such a perivacuolar localization pattern is known to be characteristic of the prevacuolar compartment, the yeast equivalent of late endosomes (Piper et al., 1995). Double labeling with Pep12p, the yeast syntaxin that marks late endosomes (Becherer et al., 1996), showed that Tat2–3HAp and Pep12p were clearly colocalized in the punctate structures (Fig. 3 B, bottom). These results indicate that Tat2–3HAp is localized to late endosomes when the tryptophan concentration is high in the medium. ER localization of HA-Tat2p was reported before (Beck et al., 1999), but we consider that this was due to overproduction.
Figure 3.
Localization of Tat2–3HAp is controlled by the external tryptophan concentration. (A) KUY121 (Δtat2) cells harboring YCpTAT2 or YCpTAT2–3HA were grown in MCD with 20 μg/ml of tryptophan, and were processed for immunostaining with the anti-HA mAb. Bar, 2 μm. (B) KUY121 cells harboring YCpTAT2–3HA were grown in the high tryptophan medium, and were subjected to double staining using the anti-Pep12p mAb and the anti-HA pAb. (C) KUY137 (Δend3) cells harboring YCpTAT2–3HA were grown in the high tryptophan medium. (D) KUY121 cells harboring YCpTAT2–3HA were grown in the high tryptophan medium. Cells were washed and shifted to low tryptophan for 2 h.
The endosomal localization of Tat2–3HAp may result from plasma membrane targeting and rapid endocytosis. However, in the Δend3 mutant, which is defective in the endocytic internalization (Raths et al., 1993), Tat2–3HAp was still not detected on the plasma membrane at high tryptophan (Fig. 3 C), indicating that Tat2–3HAp is directed to late endosomes without detouring to the plasma membrane.
When wild-type cells were shifted from high to low tryptophan medium, staining of cell periphery became evident (Fig. 3 D), indicating that Tat2–3HAp is now targeted to the plasma membrane. This is reasonable because under the low tryptophan condition, Tat2–3HAp should be on the plasma membrane for efficient uptake of tryptophan.
These results demonstrate that the plasma membrane localization of Tat2–3HAp is regulated by the tryptophan concentration in the medium. Tat2–3HAp is targeted to late endosomes at high tryptophan, and to the plasma membrane at low tryptophan.
Tat2p is missorted to the vacuole in the Δerg6 mutant
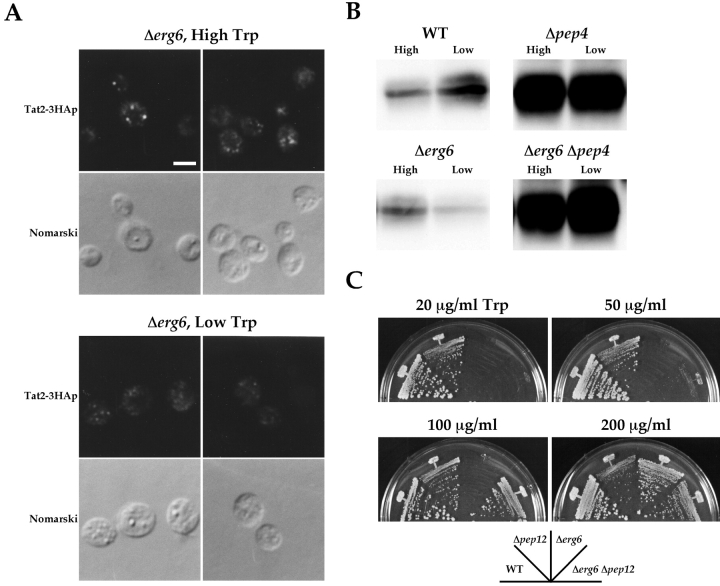
Next, we examined the localization of Tat2–3HAp in Δerg6 cells (Fig. 4 A). At high tryptophan, Tat2–3HAp was localized to punctate structures as in wild-type cells. However, when the Δerg6 cells were shifted to the low tryptophan medium, the staining of Tat2–3HAp did not change to the plasma membrane pattern, and the fluorescence within the cells became very faint.
Figure 4.
Tat2–3HAp is missorted to the vacuole in the Δerg6 mutant. (A) KUY153 (Δerg6) cells harboring YCpTAT2–3HA were grown in the high tryptophan medium. Cells were washed and shifted to high or low tryptophan for 2 h. Bar, 2 μm. (B) KUY121 (WT), KUY154 (Δpep4), KUY153 (Δerg6), and KUY156 (Δerg6 Δpep4) cells harboring YCpTAT2–3HA were grown as described in A. Cells were lysed and subjected to immunoblotting with the anti-HA antibody. (C) YPH500 (WT), KUY200 (Δpep12), KUY136 (Δerg6), and KUY204 (Δerg6 Δpep12) cells were grown on MCD supplemented with uracil, adenine, and the indicated concentrations of tryptophan at 26°C.
The amounts of Tat2–3HAp were examined by immunoblotting (Fig. 4 B). In wild-type cells, a larger amount of Tat2–3HAp was detected when incubated in the low tryptophan medium for 2 h than when kept in high tryptophan. In the vacuolar proteinase–deficient Δpep4 mutant, a high level of Tat2–3HAp was detected regardless of the tryptophan concentration. Together with the localization, these results indicate the regulated sorting of Tat2–3HAp in the secretory pathway. At high tryptophan, Tat2–3HAp is transported to the vacuole via late endosomes and eventually degraded. The degradation is slowed down at low tryptophan because Tat2–3HAp is rerouted to the plasma membrane.
In contrast, in Δerg6 cells, the amount of Tat2–3HAp was markedly reduced at low tryptophan (Fig. 4 B), consistent with the faint signal in the immunostaining. This reduction is due to vacuolar degradation because the disruption of PEP4 in Δerg6 prevented the loss of Tat2–3HAp at low tryptophan. Thus, in Δerg6 cells, Tat2–3HAp is missorted to the vacuole and quickly degraded under the low tryptophan condition.
The missorting of Tat2p implies that the severe tryptophan auxotrophy of Δerg6 can be suppressed if the vacuolar delivery is blocked. By using the pep12 mutation that inhibits the traffic to late endosomes and thereby redirects vacuolar proteins to the cell surface (Becherer et al., 1996), we show this is indeed the case. As shown in Fig. 4 C, the Δerg6 mutant did not grow below 100 μg/ml of tryptophan. However, this defect was clearly suppressed by Δpep12, although not completely. The Δerg6 Δpep12 double mutant grew well at 100 μg/ml and slowly at 50 μg/ml.
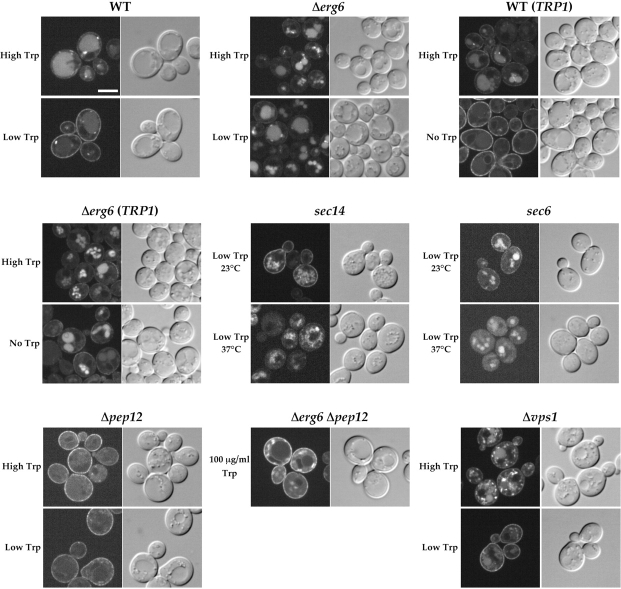
Sorting of Tat2-GFP late in the secretory pathway
To follow the route of Tat2p in more detail, the localization of another fusion construct, Tat2-GFP, was examined in various mutants defective in late steps of the secretory pathway. The results are shown in Fig. 5. In wild-type cells grown at high tryptophan, Tat2-GFP was localized to the vacuole as well as to perivacuolar late endosomes. When the cells were shifted to low tryptophan, Tat2-GFP was localized to the plasma membrane. The advantage using Tat2-GFP in living cells is that the plasma membrane was much more clearly visualized than in the fixed cells by immunostaining (compare with Fig. 3 D). This is probably because the enzymatic removal of the cell wall can be omitted if Tat2-GFP is used. Again, relocalization of Tat2-GFP to the plasma membrane in response to low tryptophan was not observed in Δerg6 cells, with prominent fluorescence in the vacuole.
Figure 5.
Sorting of Tat2-GFP late in the secretory pathway. KUY121 (WT), KUY153 (Δerg6), YPH259 (WT TRP1), KUY230 (Δerg6 TRP1), KUY177 (sec14), KUY196 (sec6), KUY202 (Δpep12), KUY209 (Δerg6 Δpep12), and KUY169 (Δvps1) cells harboring YCpTAT2-GFP were grown in the high tryptophan medium. GFP fluorescence was observed by confocal laser scanning microscopy. Alternatively, cells were washed, shifted to low tryptophan for 2 h, and then observed. In the case of TRP1 strains, cells were grown in the tryptophan-free medium. WT and Δerg6 were grown at 30°C. Others were at 23°C, except for Δerg6 Δpep12 at 26°C. To block secretion in sec14 and sec6, cells were shifted to the low tryptophan medium prewarmed to 37°C, and then incubated at 37°C for 2 h. For Δerg6 Δpep12, cells were grown in MCD with 100 μg/ml tryptophan. At this concentration of tryptophan, suppression of Δerg6 by Δpep12 was observed (see Fig. 4 C). Bar, 5 μm.
The localization of Tat2-GFP was also examined in TRP1 cells. The GFP fluorescence was clearly observed in these tryptophan prototrophs, indicating that Tat2p is expressed whether cells can synthesize tryptophan or not. Like in the trp1 cells, the localization of Tat2-GFP in TRP1 cells was regulated by external tryptophan. In wild-type TRP1 cells, Tat2-GFP was localized to the vacuole and perivacuolar late endosomes in the high tryptophan medium, and targeted to the plasma membrane in the tryptophan-free medium. In contrast, when Δerg6 TRP1 cells were grown in the tryptophan-free medium, Tat2-GFP was not localized to the plasma membrane.
To test whether Tat2-GFP is targeted to the plasma membrane by the exocytic pathway, temperature-sensitive sec mutants were examined. SEC14 is required for the exit from the Golgi (Stevens et al., 1982). When the sec14 mutant was shifted to the low tryptophan medium and grown at the permissive temperature 23°C, plasma membrane localization of Tat2-GFP was observed. However, when the low tryptophan medium was kept at the nonpermissive temperature 37°C, Tat2-GFP was not targeted to the plasma membrane and stayed in intracellular compartments. SEC6 encodes a component of the “exocyst” (TerBush et al., 1996), which is required for the fusion of Golgi-derived vesicles with the plasma membrane. At 37°C in the low tryptophan medium, Tat2-GFP was not targeted to the plasma membrane of the sec6 mutant. These results appear to indicate that Tat2-GFP follows the exocytic pathway to the plasma membrane at low tryptophan.
In Δpep12 cells, Tat2-GFP was localized to the plasma membrane irrespective of the tryptophan concentration. Thus, the inhibition of the vacuolar delivery by Δpep12 resulted in constitutive plasma membrane targeting of Tat2-GFP. Consistent with the suppression of Δerg6 by Δpep12 (Fig. 4 C), Tat2-GFP was also targeted to the plasma membrane in Δerg6 Δpep12 cells.
The vacuolar protein sorting (VPS)* pathway represents the direct vesicular traffic from the trans-Golgi to late endosomes. The VPS1 gene product is involved in this pathway and is considered to be necessary for the vesicle formation from the trans-Golgi (Nothwehr et al., 1995). Unlike in the Δpep12 mutant, Tat2-GFP was not missorted to the plasma membrane in Δvps1 cells at high tryptophan. Tat2-GFP was seen in the vacuole as well as perivacuolar dots. When the Δvps1 cells were shifted to low tryptophan, plasma membrane localization of Tat2-GFP was observed. Δvps1 did not suppress the severe tryptophan auxotrophy of Δerg6, either (unpublished data).
The result with Δvps1 indicates that Tat2-GFP does not follow the normal VPS pathway to reach late endosomes at high tryptophan. The Δpep12 mutant is defective not only in the VPS pathway, but also in the endocytic pathway. Traffic from early to late endosomes is blocked in Δpep12 (Gerrard et al., 2000). Thus, Tat2p must have taken the route from the trans-Golgi to late endosomes at high tryptophan via early endosomes. We would suggest that the tryptophan-dependent sorting of Tat2p occurs in early endosomes, and the Δerg6 mutant is defective in this sorting process (see Discussion and Fig. 9).
Figure 9.
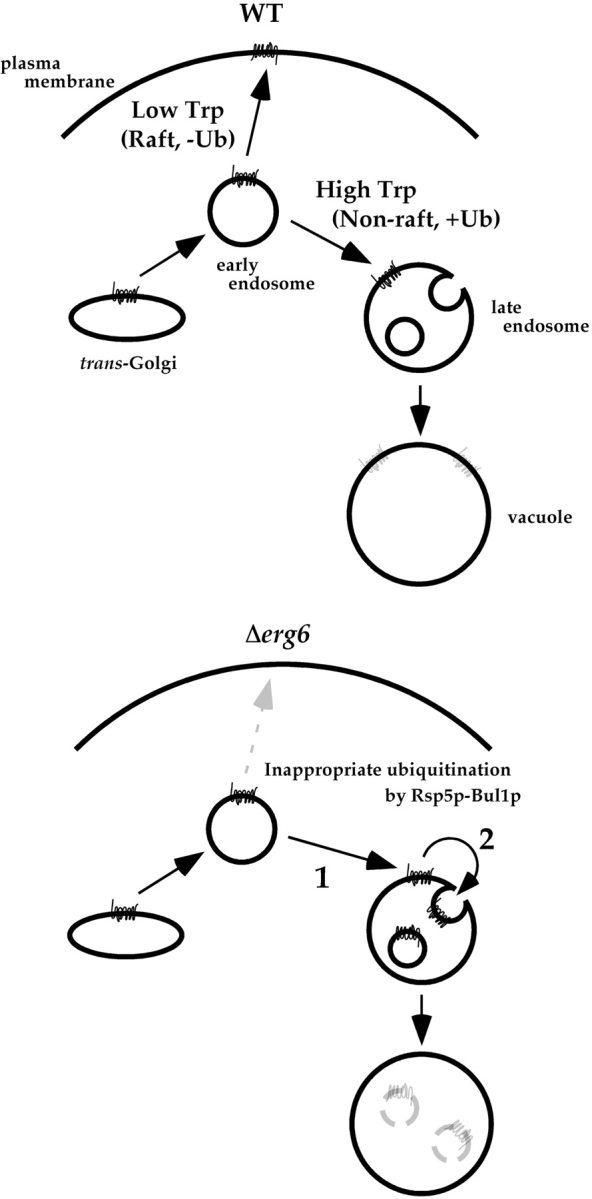
A model for the transport of Tat2p in wild-type and Δerg6 cells. See Discussion for details.
Ubiquitination and sorting of Tat2p
Evidence is rapidly accumulating that ubiquitin acts as a sorting signal at multiple steps in post-Golgi traffic. In the case of the yeast general amino acid permease Gap1p, sorting is affected by its ubiquitinated status (Helliwell et al., 2001; Soetens et al., 2001). Polyubiquitination of Gap1p by the Rsp5p ubiquitin ligase complex results in sorting to the vacuole instead of the plasma membrane. In other words, polyubiquitin is recognized as a vacuolar-targeting signal. This prompted us to examine the ubiquitination status of Tat2p. Tat2–3HAp was immunoprecipitated from cells expressing myc-tagged ubiquitin (Hochstrasser et al., 1991), and the precipitated materials were detected with the anti-myc or anti-HA antibody. To prevent degradation of Tat2–3HAp in the vacuole, Δpep4 strains were used. As shown in the right panel of Fig. 6 A, Tat2–3HAp was specifically precipitated (compare lane 1 with lanes 2–4). High mol wt myc–ubiquitin conjugates of Tat2–3HAp were detected in Δpep4 cells grown under the high tryptophan condition, (Fig. 6 A, left panel, lane 3), indicating that Tat2–3HAp is polyubiquitinated. It is known that ubiquitination of cargo proteins, such as the yeast pheromone receptor Ste2p, occurs in the plasma membrane on endocytic internalization (Hicke and Riezman, 1996). However, because Tat2–3HAp does not take the detour to the plasma membrane by exocytosis and endocytosis under the high tryptophan condition (Fig. 3), the place of its polyubiquitination must be somewhere else. On the other hand, the polyubiquitination of Tat2–3HAp was not detected in sec14 (Fig. 6 A, lane 5), indicating that the polyubiquitination reaction takes place after Tat2–3HAp has left the Golgi.
Figure 6.
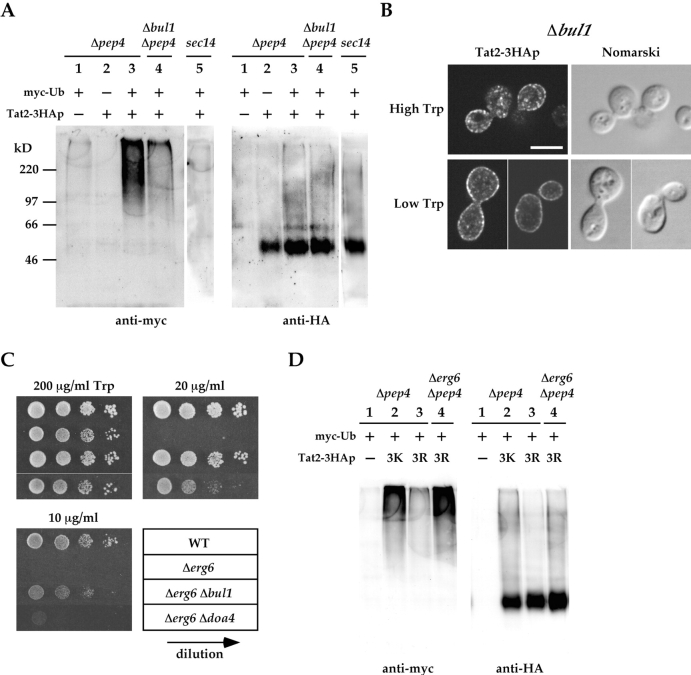
Ubiquitination and sorting of Tat2p. (A) KUY211 (Δpep4 TAT2; lane 1), KUY310 (Δpep4 TAT2–3HA; lane 2 and lane 3), and KUY314 (Δbul1 Δpep4 TAT2–3HA; lane 4) cells were grown in the high tryptophan medium. The strains harbored either pKU105 (Ub) or pKU106 (myc-Ub). In lane 5, KUY177 (sec14) cells harboring YCpTAT2–3HA and YEp105 (myc-Ub) were grown in the high tryptophan medium at 23°C, and then shifted to 37°C for 2 h. (B) KUY277 (Δbul1 Δtat2) cells harboring YCpTAT2–3HA were grown in the high tryptophan medium and shifted to low tryptophan for 2 h. Cells were processed for immunostaining with the anti-HA antibody. Bar, 5 μm. (C) YPH500 (WT), KUY136 (Δerg6), KUY266 (Δerg6 Δbul1), and KUY253 (Δerg6 Δdoa4) cells were grown in the high tryptophan medium. Cells were washed and adjusted at the density of 107 cells/ml. 5-μl aliquots of 10-fold serial dilutions were spotted on MCD supplemented with uracil, adenine, and the indicated concentrations of tryptophan. (D) KUY154 (Δpep4) and KUY156 (Δerg6 Δpep4) cells harboring YEp105 (myc-Ub) and a plasmid for the indicated variant of Tat2p were subjected to the detection of ubiquitination as described in A. Plasmids used to express Tat2p were YCpTAT2 in lane1, YCpTAT2–3HA in lane 2, and YCpTAT2 3K> R-3HA in lane 3 and lane 4.
BUL1 and BUL2 encode components of the Rsp5p ubiquitin ligase complex (Yashiroda et al., 1996, 1998). The deletion of these genes causes efficient plasma membrane delivery of Gap1p by decreasing its polyubiquitination (Helliwell et al., 2001). Similarly, we found that the myc–ubiquitin conjugation to Tat2–3HAp was markedly decreased by deletion of BUL1 (Fig. 6 A, lanes 3 and 4). We also examined the localization of Tat2–3HAp in Δbul1 cells and found that Tat2–3HAp was targeted to the plasma membrane even at high tryptophan (Fig. 6 B). At low tryptophan, Tat2–3HAp was localized to the plasma membrane very efficiently. Altogether, these results indicate that Tat2–3HAp is polyubiquitinated mostly by the Rsp5p–Bul1p ubiquitin ligase complex, and polyubiquitinated Tat2–3HAp is delivered to the vacuole without detouring to the plasma membrane. As has been reported for Gap1p (Soetens et al., 2001), the inhibition of polyubiquitination by Δbul1 would have dual roles for the marked accumulation of Tat2–3HAp in the plasma membrane; efficient targeting and inhibition of endocytic internalization.
Next, we examined the effect of Δbul1 on the tryptophan auxotrophy of Δerg6. Surprisingly, the Δerg6 Δbul1 double mutant could grow at as low as 10 μg/ml tryptophan (Fig. 6 C). Similarly, the deletion of DOA4, which reduces the efficiency of overall protein ubiquitination, also suppressed the severe tryptophan auxotrophy of Δerg6, although weakly. Δerg6 Δdoa4 cells could grow at 20 μg/ml of tryptophan (Fig. 6 C). These results led us to the hypothesis that Tat2p is inappropriately polyubiquitinated in Δerg6, resulting in the missorting to the vacuole.
There is another line of evidence that indicates aberrant polyubiquitination of Tat2p in Δerg6. Many lysine residues are present in the cytoplasmic domains of Tat2p, among which Beck et al. (1999) identified five lysine residues (10, 17, 20, 29, and 31) in the NH2-terminal domain as the ubiquitin acceptor sites on nutrient starvation. We confirmed that the three lysine residues (10, 17, and 20) are indeed the major ubiquitin acceptor sites of Tat2p. The variant of Tat2–3HAp, in which these three lysine residues were replaced by arginine (Tat23K>R-3HAp), was little ubiquitinated in the Δpep4 background (Fig. 6 D, compare lane 2 and lane 3). However, in Δerg6 Δpep4 cells, Tat23K>R-3HAp was again clearly ubiquitinated (Fig. 6 D, lane 4). The ubiquitination of Tat2p in Δerg6 must have occurred on improper lysine residues.
Missorting to the multivesicular body sorting pathway in the Δerg6 mutant
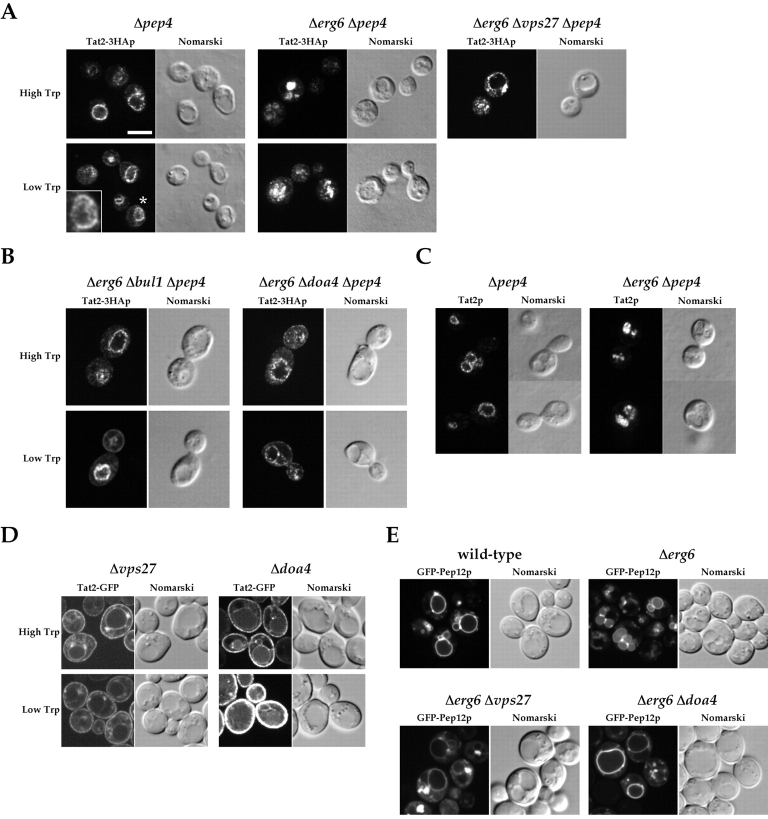
In immunofluorescence staining of Δpep4 strains to visualize vacuolar localization of Tat2–3HAp, we noticed an interesting difference between ERG6 and Δerg6 cells (Fig. 7 A). In Δpep4 cells, the vacuolar-limiting membrane was clearly stained, regardless of the tryptophan concentration. In contrast, in the Δerg6 Δpep4 cells, the fluorescence of Tat2–3HAp was not detected on the vacuole-limiting membrane, but almost exclusively in the lumen, either at high or low tryptophan. Such lumenal staining would indicate that Tat2–3HAp entered the multivesicular body (MVB)–sorting pathway, which transfers a subset of cargo proteins to the invaginating vesicles in yeast late endosomes (Odorizzi et al., 1998). To test this possibility, we examined the effect of VPS27 disruption. VPS27 is one of the class E VPS genes, all of which are required for MVB formation (Odorizzi et al., 1998). As shown in Fig. 7 A, Tat2–3HAp was localized to the vacuole-limiting membrane and the exaggerated class E compartment in Δerg6 Δvps27 Δpep4. These results indicate that Tat2–3HAp is efficiently sorted to the MVB pathway in Δerg6, regardless of the tryptophan concentration.
Figure 7.
Missorting of Tat2p to the MVB pathway in the Δerg6 mutant. (A) KUY154 (Δpep4), KUY156 (Δerg6 Δpep4), and KUY231 (Δerg6 Δvps27 Δpep4) cells harboring YCpTAT2–3HA were grown in the high or low tryptophan medium. Tat2–3HAp was stained with the anti-HA antibody. Inset on the bottom of Δpep4 shows the enlargement of the cell labeled with an asterisk, indicating that weak immunofluorescence is also seen in the vacuole lumen. Bar, 5 μm. (B) KUY268 (Δerg6 Δbul1 Δpep4) and KUY254 (Δerg6 Δdoa4 Δpep4) cells harboring YCpTAT2–3HA were stained with the anti-HA antibody. Note that cell surface staining was observed in cells exposed to low tryptophan. (C) The vacuolar localization of untagged Tat2p was detected in KUY211 (Δpep4) and KUY214 (Δerg6 Δpep4) cells harboring YEpTAT2, using the anti-Tat2p antibody. With this antibody, we could observe the signal in the cells expressing Tat2p from a multicopy plasmid. (D) GFP fluorescence was observed in KUY155 (Δvps27) and KUY251 (Δdoa4) cells harboring YCpTAT2-GFP. (E) Tryptophan-prototrophic YPH259 (wild-type) and KUY230 (Δerg6) cells harboring pKU84 (GFP-PEP12) were grown in MCD supplemented with uracil and fourfold excess adenine, and the GFP fluorescence was observed. KUY148 (Δerg6 Δvps27) and KUY253 (Δerg6 Δdoa4) were transformed with pKU144 (GFP-PEP12 TRP1) and converted into tryptophan prototrophs.
As shown in Fig. 6, inhibition of the ubiquitination by Δbul1 or Δdoa4 restored the tryptophan uptake in the Δerg6 mutant. Ubiquitin is also known to act as a sorting signal to the MVB (Katzmann et al., 2001), and the MVB sorting of cargo proteins is prevented by Δdoa4 (Losko et al., 2001; Reggiori and Pelham, 2001). We examined the effect of Δbul1 and Δdoa4 on the MVB sorting of Tat2–3HAp in Δerg6. The results are shown in Fig. 7 B. In contrast to Δerg6 Δpep4 cells, the vacuole-limiting membrane was clearly stained in Δerg6 Δbul1 Δpep4 and Δerg6 Δdoa4 Δpep4 cells, regardless of the tryptophan concentration. Plasma membrane staining at low tryptophan, which was not observed in Δerg6 Δpep4, was also appreciable in some Δerg6 Δbul1 Δpep4 and Δerg6 Δdoa4 Δpep4 cells, consistent with the suppression of the tryptophan auxotrophy (Fig. 6 C). Thus, the two defects of Tat2p sorting in the Δerg6 mutant, the cell surface–targeting defect and the MVB missorting, are simultaneously suppressed by the inhibition of ubiquitination.
In contrast to Tat2–3HAp, it may be noted that the fluorescence of Tat2-GFP is clearly seen in the vacuole lumen of wild-type cells (Fig. 5). We raised a specific antibody against Tat2p, and found that untagged Tat2p was detected on the vacuolar-limiting membrane in Δpep4, but in the vacuole lumen in Δerg6 Δpep4 (Fig. 7 C). This behavior is very similar to that of Tat2–3HAp, and therefore, the results with Tat2–3HAp may reflect the authentic nature of Tat2p. The sorting of Tat2-GFP into the vacuolar lumen was blocked in both Δvps27 and Δdoa4 cells (Fig. 7 D), indicating that it undergoes ubiquitin-dependent MVB sorting. Like in Δbul1 cells (Fig. 6 B), the plasma membrane signal of Tat2-GFP in Δdoa4 cells was obvious at high tryptophan, and became remarkable on the shift to low tryptophan.
Then why is Tat2-GFP efficiently sorted to the MVB in wild-type cells, even though it is functional and correctly targeted to the cell surface at low tryptophan? Tat2-GFP may be ubiquitinated more efficiently. Alternatively, vacuolar lumenal localization of the MVB vesicles could be detected more clearly by GFP fluorescence in living cells. Due to the fixation and subsequent permeabilization procedures, the MVB vesicles might look more obscure by immunofluorescence microscopy.
As another way to assess the MVB missorting in the Δerg6 mutant, we looked at a different GFP marker, GFP-Pep12p. The results are shown in Fig. 7 E. As reported previously (Reggiori et al., 2000), GFP-Pep12p resides mostly on the vacuolar-limiting membrane in wild-type cells when overexpressed. However, in Δerg6 cells, the GFP fluorescence was now evident in the vacuole lumen. The fluorescence on the limiting membrane still remained, indicating that GFP-Pep12p is not completely relocated to the vacuole lumen. The lumenal signal in Δerg6 cells disappeared by either Δvps27 or Δdoa4. Thus, GFP-Pep12p in Δerg6 cells is also missorted to the MVB in a ubiquitin-dependent manner, indicating that a subset of cargo proteins is inappropriately ubiquitinated and then sorted to the MVB in Δerg6.
Transport of Tat2p to the plasma membrane depends on detergent-insoluble membrane domains
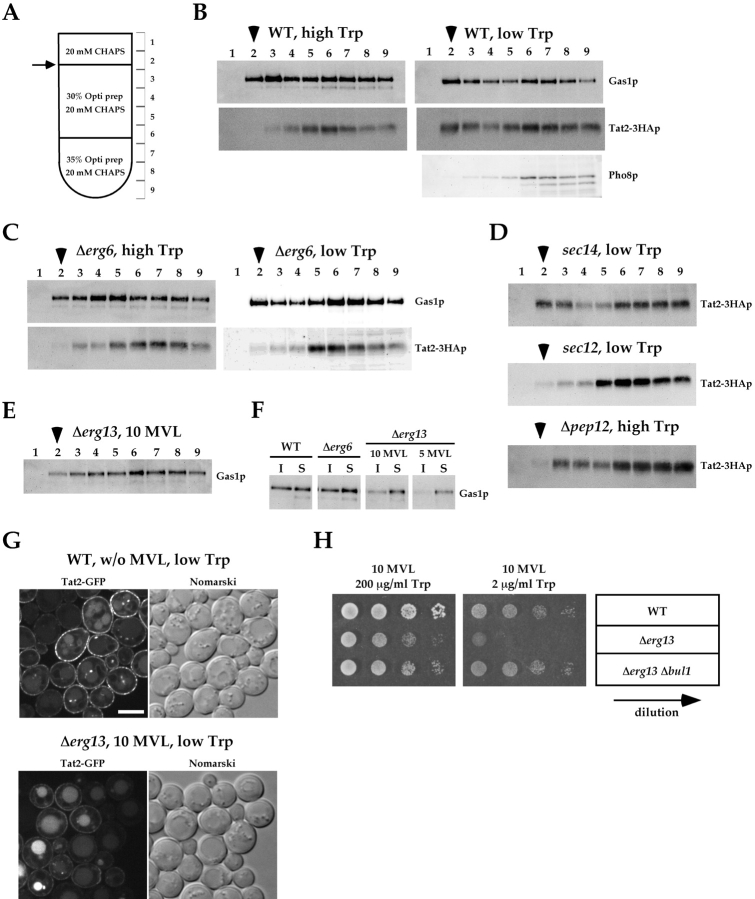
Because the deficiency of normal sterol in Δerg6 affected the sorting of Tat2p, we further investigated whether sterol-rich, detergent-insoluble membrane domains (so-called rafts) are involved in the plasma membrane delivery of Tat2p. Detergent insolubility of Tat2p was examined by treatment with CHAPS followed by a flotation analysis as diagrammed in Fig. 8 A. In wild-type cells (Fig. 8 B), a fraction of the GPI-anchored protein Gas1p floated to the interphase between 0 and 30% of OptiPrep™ (Fig. 8 B, fraction 2, arrowhead), whereas the vacuolar alkaline phosphatase Pho8p did not, as reported previously (Bagnat et al., 2000). Tat2–3HAp did not float to fraction 2 under the high tryptophan condition where it is sorted to late endosomes. However, Tat2–3HAp was clearly detected in fraction 2 under the low tryptophan condition where it is targeted to the plasma membrane. In the Δerg6 mutant (Fig. 8 C), Gas1p was still detected in the floating fraction 2. This is consistent with a recent report (Sievi et al., 2001), and indicates that the sterol intermediates accumulating in the Δerg6 mutant can replace ergosterol in the context of detergent-insoluble membrane domain formation. However, Tat2–3HAp in Δerg6 cells failed to float to fraction 2 even at low tryptophan (Fig. 8 C).
Figure 8.
Detergent-insoluble membrane domain is involved in the plasma membrane transport of Tat2p. (A) The flotation procedure to monitor the detergent insolubility. Detergent-insoluble membrane domain is floated to the interface (arrow) that corresponds to fraction 2. (B) KUY121 (WT) cells harboring YCpTAT2–3HA were grown in the high tryptophan medium, washed, and shifted to low tryptophan for 2 h. Cells were subjected to the flotation analysis. In B–D, arrowheads indicate fraction 2, which contains detergent-insoluble membrane domains. (C) KUY153 (Δerg6) cells harboring YCpTAT2–3HA were grown and analyzed as described in B. The amount of Tat2–3HAp at low tryptophan is significantly decreased in this mutant (see Fig. 4 B), but an enhanced image is shown here. (D) KUY177 (sec14), KUY197 (sec12), and KUY202 (Δpep12) cells harboring YCpTAT2–3HA were grown in the high tryptophan medium at 23°C. The sec mutant cells were washed, shifted to the prewarmed (37°C) low tryptophan medium, and incubated for 2 h. (E) KUY256 (Δerg13) cells harboring YCpTAT2–3HA were grown in the high tryptophan medium supplemented with 10 mg/ml MVL. (F) Distribution of Gas1p between the detergent-insoluble (I; mixture of the fractions 2 and 3) and soluble (S; mixture of the fractions 7–9) fractions. The high Trp samples of B–D were used. The Δerg13 cells were initially grown at 10 mg/ml MVL, and then incubated with 5 mg/ml MVL for 2 h. (G) KUY121 (WT) and KUY256 (Δerg13) cells harboring YCpTAT2-GFP were grown in the high tryptophan medium, shifted to low tryptophan, and incubated for 4 h. For Δerg13 cells, MVL was included in the medium at 10 mg/ml. Bar, 5 μm. (H) YPH500 (WT), KUY255 (Δerg13), and KUY257 (Δerg13 Δbul1) cells were grown in the high tryptophan medium supplemented with 50 mg/ml MVL. Cells were washed and adjusted at the density of 107 cells/ml. 5-μl aliquots of 10-fold serial dilutions were spotted on the high or low tryptophan medium supplemented with 10 mg/ml MVL, uracil, and adenine.
These results suggest that the association of Tat2p with the detergent-insoluble membrane rafts is required for plasma membrane delivery. Alternatively, the detergent insolubility of Tat2p could simply reflect the fact that rafts are the major lipid phase in the plasma membrane (Bagnat et al., 2001). We prefer the former possibility. First, Tat2–3HAp acquired the detergent insolubility even when its exit from the Golgi was blocked in the sec14 mutant (Fig. 8 D). When ER export was blocked by the sec12 mutation (Nakano et al., 1988), the detergent insolubility of Tat2–3HAp was not observed (Fig. 8 D), indicating that Tat2–3HAp is partitioned into lipid rafts after it reached the Golgi apparatus. Second and more importantly, when Tat2–3HAp was forced to localize to the plasma membrane in Δpep12 cells grown at high tryptophan, Tat2–3HAp was not found in the detergent-insoluble fraction (Fig. 8 D).
To confirm the role of rafts, we also attempted to disrupt the detergent-insoluble domain by inhibiting the initial step of the ergosterol biosynthesis. The ERG13 gene encodes HMG-CoA synthase. Δerg13 cells require mevalonate in the medium for growth. As reported before (Dimster-Denk et al., 1994), the growth of Δerg13 cells was arrested at a low concentration (5 mg/ml) of mevalonic acid lactone (MVL). At 10 mg/ml of MVL, Δerg13 cells were able to grow slowly. As shown in Fig. 8 E, only a small amount of Gas1p was found in the floating fraction 2 when MVL was supplied at 10 mg/ml. For simplicity, the distribution of Gas1p was compared between the detergent-insoluble (I) fractions (Fig. 8 A, defined as the mixture of fractions 2 and 3) and the detergent-soluble (S) fractions (Fig. 8 A, the mixture of fractions 7–9). As shown in Fig. 8 F, Gas1p gradually disappeared from the I fractions of Δerg13 cells according to the decrease of the supplementing MVL, indicating that the detergent-insoluble domains were significantly depleted when the flux of sterol synthesis was reduced. Exactly under the same condition, Tat2-GFP was inefficiently routed to the plasma membrane, and the vacuolar staining remained prominent (Fig. 8 G). In addition, Δerg13 cells were unable to grow at low tryptophan (Fig. 8 H), implying that the plasma membrane targeting of Tat2p is critical for growth under this condition. Strikingly, Δbul1 suppressed such severe tryptophan auxotrophy of the Δerg13 mutant (Fig. 8 H), indicating that the raft requirement for the plasma membrane targeting of Tat2p can be bypassed by the inhibition of polyubiquitination.
All these results strongly suggest that the partitioning of Tat2p into the rafts at low tryptophan is not the indirect consequence of the plasma membrane targeting, but is rather the cause of the sorting into the plasma membrane route. In other words, lipid raft sorting is very important for the cell surface delivery of Tat2p. However, it should be remembered that Tat2p can also be delivered to the plasma membrane in the absence of raft association under some conditions (for example, when the vacuolar trafficking pathway is blocked or the polyubiquitination is inhibited).
Discussion
In this paper, we present clear evidence that sterol is crucial for the correct sorting of the high affinity tryptophan permease Tat2p in yeast cells. The routes that we propose Tat2p follows are illustrated in Fig. 9. In the wild-type cells, Tat2p goes to the plasma membrane at low tryptophan, and to the vacuole via late endosomes at high tryptophan. The localization analysis of Tat2p in mutants defective in post-Golgi traffic indicates that Tat2p is delivered to late endosomes at high tryptophan not directly from the trans-Golgi nor via endocytosis from the plasma membrane. It is probably once delivered from the trans-Golgi to early endosomes, and there the tryptophan-dependent sorting occurs. These elaborate trafficking regulations of Tat2p are compromised in the Δerg6 mutant. Tat2p is not delivered to the plasma membrane, but to the vacuole at low tryptophan (Fig. 9, Δerg6, arrow 1). This is why Δerg6 cells are unable to take up tryptophan efficiently. In addition, Tat2p is missorted to the MVB pathway in late endosomes (Fig. 9, Δerg6, arrow 2).
Regulated transport of Tat2p
The activity of the Tat2p permease is controlled by the regulated sorting of membrane traffic rather than by synthesis. When and where the commitment is made as to whether Tat2p is transported to the vacuole or to the plasma membrane is a very important question, which turned out to be not an easy one. The experiments using Δend3, Δvps1, and Δpep12 mutants indicated that Tat2p takes the unconventional route, trans-Golgi to early endosomes to late endosomes under the high tryptophan condition. On the other hand, it is not clear how Tat2p is routed to the plasma membrane at low tryptophan. Both SEC14 and SEC6 are required for the plasma membrane targeting of Tat2p, but this does not necessarily mean that it follows the conventional secretory pathway because one branch of the yeast exocytic pathway transits through endosomes (Harsay and Schekman, 2002). TLG1 and RCY1, which encode the early endosomal syntaxin and the F-box protein, respectively, are required for recycling from endosomes to the plasma membrane (Wiederkehr et al., 2000). However, their deletion mutants did not give clear-cut results, perhaps because their recycling defects were only partial (unpublished data). At present, the simplest model is that Tat2p always travels via early endosomes, which is the place of sorting, and the final destination either to the plasma membrane or to the vacuole is determined there.
Ubiquitin-dependent sorting of amino acid permeases
It has been known that intracellular sorting of yeast nutrient transporters, such as Gap1p and the ferrichrome transporter Arn1p, is regulated late in the secretory pathway (Roberg et al., 1997; Kim et al., 2002). In the case of Gap1p, the sorting depends on the nitrogen source in the medium. Gap1p is targeted to the plasma membrane when cells are grown on urea, but to the vacuole in the glutamate medium. The similarity between Tat2p and Gap1p regarding nutrient-dependent regulation of sorting led us to suspect the presence of a common mechanism (i.e., ubiquitination).
Kaiser's group has shown that polyubiquitination by the Rsp5p ubiquitin ligase complex directs Gap1p to the vacuole instead of the plasma membrane (Helliwell et al., 2001). This also turns out to be the case with Tat2p. Bul1p, known as a component of the Rsp5p complex required for elongation of polyubiquitin chains (Yashiroda et al., 1996; Helliwell et al., 2001), plays a critical role in the regulation of ubiquitination status of Tat2p. Polyubiquitination of Tat2p is little detected in the Δbul1 mutant. Surprisingly, almost all the defects of Δerg6 in Tat2p sorting are simultaneously suppressed by the knockout of BUL1. Presumably, aberrant polyubiquitination in the Δerg6 mutant is alleviated by the Δbul1 mutation. The anomaly of ubiquitination in Δerg6 is also seen on the acceptor sites of ubiquitin. Tat2p is polyubiquitinated on inappropriate lysine residues in Δerg6.
MVB sorting of Tat2p
Δerg6 cells also show a peculiar behavior in the MVB sorting. Although Tat2p remains on the limiting membrane when it is finally targeted to the vacuole in wild-type cells, Tat2p is almost completely segregated into the lumen of the vacuole in Δerg6. Similarly, Pep12p is also missorted into the MVB in Δerg6 cells. This MVB mistargeting is blocked by the class E Δvps27 mutation, suggesting that normal mechanisms of MVB sorting by the ESCRT complexes (Katzmann et al., 2001; Babst et al., 2002a, 2002b) are operating in this process. Interestingly, a CHO cell mutant defective in cholesterol biosynthesis also shows MVB missorting of the cation-independent mannnose 6-phosphate receptor (Miwako et al., 2001). This kind of missorting may be a general outcome caused by defects of normal sterol synthesis.
Our finding that the MVB missorting of Tat2p and Pep12p in Δerg6 is suppressed by either Δbul1 or Δdoa4 indicates that it occurs in a ubiquitin-dependent manner. The sequential sorting defects of Tat2p in Δerg6, namely in early and late endosomes, could be explained solely by ubiquitination. That is, Tat2p is inappropriately ubiquitinated in Δerg6, delivered from early to late endosomes, and then sequestered into the MVB by being caught by the ESCRT-1 complex, a putative sorting receptor for ubiquitinated cargoes (Katzmann et al., 2001).
In several cases, monoubiquitination has been shown sufficient for the entry of cargo into the MVB. Tat2p is polyubiquitinated even under the low tryptophan condition in Δpep4 cells (unpublished data). Beck et al. (1999) have also shown that Tat2p is polyubiquitinated under the starvation condition and found on the vacuolar-limiting membrane. Then the question is why Tat2p is not always sorted to the MVB pathway. Mono- vs. polyubiquitination could explain the difference. Alternatively, the position of ubiquitination may be important. For example, ubiquitin signals near the membrane could be recognized by the MVB-sorting machinery more easily than the distal ones (Reggiori and Pelham, 2002). For Tat2p, the major ubiquitin acceptor sites for the wild-type are Lys10, Lys17, and Lys20, all near the NH2 terminus. Ubiquitination of Tat2p in the Δerg6 mutant might occur on lysine residues proximal to the membrane, resulting in more efficient MVB sorting.
Lipid raft–dependent sorting of Tat2p
We present two lines of evidence indicating that association with the detergent-insoluble membrane domain is required for the plasma membrane delivery of Tat2p. First, Tat2p became detergent-insoluble under the low tryptophan condition. Second, depletion of sterols by using the mevalonate auxotroph Δerg13 mutant disrupted the detergent-insoluble membrane domain and simultaneously blocked the plasma membrane targeting of Tat2p. The detergent insolubility and sterol dependence of this membrane domain fit well with the concept of the lipid raft (Simons and Ikonen, 1997). Our results with Δerg13 indicate that Tat2p is missorted to the vacuole in the absence of lipid rafts. Similarly, the proton ATPase Pma1p is delivered to the plasma membrane in association with rafts, and missorted to the vacuole when rafts are disrupted (Bagnat et al., 2001). It appears that raft and nonraft domains are segregated for the plasma membrane and late endosomal delivery, respectively.
The raft association is not obligatory for the plasma membrane targeting of Tat2p if its vacuolar sorting is inhibited by Δpep12 or Δbul1. The fact that the severe tryptophan auxotrophy of the raft-deficient Δerg13 was suppressed by Δbul1 indicates that raft association and polyubiquitination have counteracting effects in the sorting of Tat2p. On aberrant polyubiquitination, Tat2p is probably diverted from the raft-dependent plasma membrane targeting pathway to the nonraft pathway to late endosomes. Because ERG6 is involved in a late step of the ergosterol biosynthetic pathway, sterols are not depleted in Δerg6, but intermediates such as zymosterol accumulate (Munn et al., 1999). These intermediates are still capable of forming rafts, judging from the detergent insolubility of the GPI-anchored protein in Δerg6. However, Tat2p cannot be associated with such altered rafts any more and missorted to the vacuole. Tat2p may be just unable to reside stably in the rafts with the unusual sterol composition, or could be excluded from the rafts due to its inappropriate polyubiquitination.
Experiments with the sec mutants grown at low tryptophan indicated that Tat2p becomes associated with rafts in the Golgi. Tat2p associates with rafts even at high tryptophan in the sec14 mutant (unpublished data). Although the altered phospholipid composition of this mutant (McGee et al., 1994) might indirectly affect the raft organization, this observation suggests that Tat2p can gain access to rafts in the Golgi. On the other hand, polyubiquitination of Tat2p was not detected in the sec14 mutant, indicating that the polyubiquitination occurs after the exit from the Golgi. Thus, Tat2p would be first partitioned into rafts, and then be subjected to the ubiquitin-dependent sorting, presumably in early endosomes.
How polyubiquitin acts as a sorting signal to the nonraft, vacuolar trafficking pathway remains to be resolved. Lafont and Simons (2001) have shown that the ubiquitin ligases Cbl and Nedd4 are partitioned into rafts. Interestingly, the yeast Nedd4 homologue Rsp5p is partially resistant to detergent extraction (Wang et al., 2001), implying that polyubiquitination of Tat2p by the Rsp5p–Bul1p complex could occur in the rafts. Sorting receptors such as Hrs (Raiborg et al., 2002) may bind to polyubiquitin and divert cargo proteins to the nonraft membrane domains. Alternatively, Tat2p might dissociate from rafts independently of ubiquitin. The dissociation could change the environment around the molecule and would then trigger its polyubiquitination and sorting to late endosomes.
That the slight alteration in sterol structure or composition can dramatically change the destination of a plasma membrane protein raises a possibility that similar regulation could be used for differentiation of the cell surface, for example, during the development in higher eukaryotes. Our future work will aim at understanding how sterols might be involved in such higher order regulations and how they are linked to ubiquitin, a key player in the post-Golgi traffic.
While this manuscript was in preparation, Bagnat and Simons (2002) reported that Fus1p, a plasma membrane protein required for yeast mating, is largely excluded from rafts and mislocalized to the vacuole in the Δerg6 mutant. This behavior of Fus1p in Δerg6 is similar to that of Tat2p, and supports the view that a subset of plasma membrane proteins are missorted in Δerg6 to cause pleiotropic phenotypes. Indeed, the mating deficiency (Gaber et al., 1989) and the drug hypersensitivity (Kaur and Bachhawat, 1999) of Δerg6 might all be explained by the missorting of plasma membrane proteins due to impaired raft association and inappropriate ubiquitination.
Materials and methods
Yeast strains and media
Yeast strains used in this study are listed in Table I.
Table 1. Yeast strains used in this work.
| Strain | Genotype | Reference/Source |
|---|---|---|
| YPH259 | MATα ura3 lys2 ade2 his3 leu2 | Sikorski and Hieter (1989) |
| YPH499 | MAT a ura3 lys2 ade2 trp1 his3 leu2 | Sikorski and Hieter (1989) |
| YPH500 | MATα ura3 lys2 ade2 trp1 his3 leu2 | Sikorski and Hieter (1989) |
| YPH501 | MAT a/MATα ura3/ura3 lys2/lys2 ade2/ade2 trp1/trp1 his3/his3 lys2/lys2 | Sikorski and Hieter (1989) |
| KUY121 | MATα Δtat2::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From YPH500 |
| KUY136 | MATα Δerg6::LEU2 ura3 lys2 ade2 trp1 his3 leu2 | From YPH500 |
| KUY137 | MATα Δend3::HIS3 Δtat2::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY121 |
| KUY148 | MATα Δerg6::LEU2 Δvps27::HIS3 ura3 lys2 ade2 trp1 his3 leu2 | From YPH501 |
| KUY153 | MATα Δerg6::LEU2 Δtat2::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY121 |
| KUY154 | MATα Δpep4::HIS3 Δtat2::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY121 |
| KUY155 | MATα Δvps27::HIS3 Δtat2::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY121 |
| KUY156 | MATα Δerg6::LEU2 Δpep4::HIS3 Δtat2::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY154 |
| KUY169 | MATα Δvps1::HIS3 Δtat2::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY121 |
| KUY177 | MATα Δtat2::hisG sec14-3 ura3 leu2 trp1 his3/4 | From ANY21xANS14-2C |
| KUY196 | MATα Δtat2::hisG sec6-4 ura3 leu2 trp1 his3/4 | From ANS6-2D |
| KUY197 | MAT a Δtat2::hisG sec12-4 ura3 leu2 trp1 his3 his4 suc gal2 | From MBY10-7A |
| KUY200 | MATα Δpep12::HIS3 ura3 lys2 ade2 trp1 his3 leu2 | From YPH500 |
| KUY202 | MATα Δpep12::HIS3 Δtat2::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY121 |
| KUY204 | MATα Δpep12::HIS3 Δerg6::LEU2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY136 |
| KUY209 | MATα Δpep12::HIS3 Δerg6::LEU2 Δtat2::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY202 |
| KUY211 | MATα Δpep4::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From YPH500 |
| KUY214 | MATα Δerg6::LEU2 Δpep4::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY211 |
| KUY230 | MATα Δerg6::LEU2 ura3 lys2 ade2 his3 leu2 | From YPH259 |
| KUY231 | MATα Δerg6::LEU2 Δvps27::HIS3 Δpep4::ADE2 Δtat2::hisG ura3 lys2 ade2 trp1 his3 leu2 | From YPH500 |
| KUY251 | MATα Δdoa4::HIS3 Δtat2::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY121 |
| KUY253 | MATα Δdoa4::HIS3 Δerg6::LEU2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY136 |
| KUY254 | MATα Δerg6::LEU2 Δdoa4::HIS3 Δpep4::ADE2 Δtat2::hisG ura3 lys2 ade2 trp1 his3 leu2 | From YPH500 |
| KUY255 | MATα Δerg13::HIS3 ura3 lys2 ade2 trp1 his3 leu2 | From YPH500 |
| KUY256 | MATα Δerg13::HIS3 Δtat2::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY121 |
| KUY257 | MATα Δerg13::HIS3 Δbul1::LEU2 ura3 lys2 ade2 trp1 his3 leu2 | From YPH500 |
| KUY266 | MATα Δbul1::HIS3 Δerg6::LEU2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY136 |
| KUY268 | MATα Δerg6::LEU2 Δbul1::HIS3 Δpep4::ADE2 Δtat2::hisG ura3 lys2 ade2 trp1 his3 leu2 | From YPH500 |
| KUY277 | MATα Δbul1::LEU2 Δtat2::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY121 |
| KUY310 | MATα TAT2-3HA Δpep4::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY211 |
| KUY314 | MATα TAT2-3HA Δbul1::HIS3 Δpep4::ADE2 ura3 lys2 ade2 trp1 his3 leu2 | From KUY310 |
| ANY21 | MAT a ura3 leu2 trp1 his3 his4 suc gal2 | Lab collection |
| ANS14-2C | MATα sec14-3 ura3 leu2 his3/4 | Lab collection |
| ANS6-2D | MATα sec6-4 ura3 leu2 trp1 his3/4 | Lab collection |
| MBY10-7A | MAT a sec12-4 ura3 leu2 trp1 his3 his4 suc gal2 | Lab collection |
Yeast cells were grown in MCD medium, composed of 0.67% yeast nitrogen base without amino acids (Difco Laboratories), 0.5% casamino acids (Difco Laboratories), and 2% glucose. Casamino acid is the mixture of amino acids lacking tryptophan. Adenine and uracil were supplemented at 20 μg/ml. Tryptophan was supplemented at a high (200 μg/ml), standard (20 μg/ml), or low (2 μg/ml) concentration. Unless otherwise indicated, yeast cells were grown at 30°C.
Plasmids and antibodies
Details of the various plasmid constructions and antibodies are described in the supplemental materials and methods section (available at http://www.jcb.org/cgi/content/full/jcb.200303088/DC1).
Fluorescence microscopy
Immunofluorescence microscopy was performed essentially as described before (Nishikawa and Nakano, 1991), except that permeabilization of fixed cells was performed by spheroplasting buffer containing 1% (wt/vol) BSA and 0.1% (vol/vol) Triton X-100 for 10 min at RT. Cells were observed and photographed using a photomicroscope (model BX-60; Olympus). Alternatively, the same microscope equipped with a confocal laser scanner unit (model CSU10; Yokogawa Electronic Corp.) was used. Images were acquired by a high resolution digital charge-coupled device camera (model C4742–95; Hamamatsu Photonics) and processed by IPLab software (Scanalytics).
Detection of the ubiquitinated forms of Tat2–3HAp
Detection of ubiquitinated Tat2–3HAp was performed basically according to the method used for the case of Gap1p (Helliwell et al., 2001). To enhance the detection, the myc-tagged ubiquitin was exogenously expressed. The expression of myc-Ub was under the control of the CUP1 promoter, which was inducible by addition of CuSO4 to the medium (Ellison and Hochstrasser, 1991). However, myc-Ub conjugates were detectable even when the promoter was uninduced, as was reported previously (Hochstrasser et al., 1991). In this work, cells were grown at the basal expression level of the CUP1 promoter. 5 × 108 cells were collected and treated with NaN3 and potassium flouride at the final concentration of 20 mM each. The cells were resuspended in 125 μl lysis buffer (20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 1.6% SDS, 6 M urea, 5 mM N-ethylmaleimide, and 0.02% NaN3) containing a protease inhibitor mixture (1 mM PMSF, 5 μg/ml chymostatin, leupeptin, antipain, and pepstatin A, and 2.5 μg/ml aprotinin), lysed by agitation with glass beads, and incubated at 37°C for 20 min. 875 μl IP dilution buffer (1.1% Triton X-100, 170 mM NaCl, 6 mM EDTA, 60 mM Tris-HCl, pH 7.4, 5 mM N-ethylmaleimide, and 0.02% NaN3, the protease inhibitor mixture) was added to the cell lysates, and insoluble material was removed by centrifugation. 750 μl supernatant was mixed with 40 μl protein G Sepharose 4 Fast Flow (Amersham Biosciences), and precleared by rotation at RT for 30 min. The samples were centrifuged, and 700 μl supernatant was incubated overnight with 10 μl rat anti-HA antibody (3F10; Roche Diagnostics) and 15 μl protein G Sepharose suspension, with rotation at 4°C. The beads were washed twice with IP buffer (1% Triton X-100, 0.2% SDS, 150 mM NaCl, 5 mM EDTA, and 50 mM Tris-HCl, pH 7.4), twice with urea wash buffer (1% Triton X-100, 0.2% SDS, 2 M urea, 250 mM NaCl, 5 mM EDTA, and 50 mM Tris-HCl, pH 7.4) and once with high salt wash buffer (1% Triton X-100, 0.2% SDS, 50 mM NaCl, 5 mM EDTA, and 50 mM Tris-HCl, pH 7.4). The beads were suspended with SDS-PAGE sample buffer (2% SDS, 5% β-mercaptoethanol, 10% glycerol, 50 mM Tris-HCl, pH 6.8, and 0.025% bromophenol blue) containing 6 M urea, and incubated at 37°C for 20 min. 24 μl of the sample was subjected to SDS-PAGE and immunoblotting with the anti-myc antibody (9E10) to detect myc-Ub conjugates. To detect Tat2–3HAp, the sample was diluted 15-fold and 20 μl was loaded. Anti-HA antibody (16B12) was used for immunoblotting.
Analysis of lipid rafts
5 × 108 cells were collected, treated with NaN3 and potassium flouride at a final concentration of 20 mM each, and resuspended in 275 μl TNE buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 5 mM EDTA) containing a protease inhibitor mixture (1 mM PMSF and 5 μg/ml chymostatin, leupeptin, antipain, and pepstatin A). After adding glass beads, the suspension was vortexed for 30 s and was then chilled on ice for 30 s, repeating six times. Unbroken cells and debris were removed by centrifugation at 500 rpm for 5 min. The cleared lysate (175 μl) was mixed with equal volume of TNE containing 40 mM CHAPS (Sigma-Aldrich), and was then incubated at 4°C for 30 min. The tube was centrifuged at 5,000 rpm for 5 min, and 330 μl supernatant was mixed with 770 μl 50% OptiPrep™ (Nycomed Pharma)/TNE/20 mM CHAPS to give the final concentration of 35% OptiPrep™. The solution was set on the bottom of a 3 PC tube (Hitachi Koki Co., Ltd.), and overlaid with 1.4 ml 30% OptiPrep™/TNE/20 mM CHAPS and 0.5 ml TNE/20 mM CHAPS. The gradients were centrifuged at 4°C for 7.5 h using a rotor (model RPS65T; Hitachi Koki Co., Ltd.) at 35,000 rpm, and nine fractions (320 μl each) were collected from the top. Each fraction was mixed with 288 μl 110 mM Tris-HCl, pH 6.8/4.4% SDS/22% glycerol and 32 μl β-mercaptoethanol, incubated at 37°C for 5 min, and subjected to SDS-PAGE.
Online supplemental materials
Plasmid construction, antibodies, and immunoblotting procedures are included in the online supplemental materials, available at http://www.jcb.org/cgi/content/full/jcb.200303088/DC1.
Supplemental Material
Acknowledgments
We are grateful to Kunihiko Iwamoto, Hidemitsu Nakamura, and Akinori Ohta (University of Tokyo, Tokyo, Japan) for plasmids and advice during the initial planning of this work, Ryogo Hirata (RIKEN, Japan) for antibodies and valuable suggestions, Marcus Lee (University of California, Berkeley, Berkeley, CA) and Tamotsu Yoshimori (National Institute of Genetics, Mishima, Japan) for advice on the raft isolation method, Yoshiko Kikuchi (University of Tokyo, Tokyo, Japan) for plasmids and stimulating discussion, Mark Hochstrasser (Yale University, New Haven, CT) for the myc-Ub expression plasmid, and the members of the Nakano laboratory for helpful discussions throughout this work.
This work was supported by grants from the Biodesign and Bioarchitect Research Projects of RIKEN and by a President's Special Research Grant of RIKEN. K. Umebayashi is a recipient of the Special Postdoctoral Researcher fellowship of RIKEN.
The online version of this article includes supplemental material.
K. Umebayashi's present address is Department of Cell Genetics, National Institute of Genetics, 1111 Yata, Mishima, Shizuoka 411-8540, Japan.
Footnotes
Abbreviations used in this paper: MVB, multivesicular body; MVL, mevalonic acid lactone; VPS, vacuolar protein sorting.
References
- Babst, M., D.J. Katzmann, E.J. Estepa-Sabal, T. Meerloo, and S.D. Emr. 2002. a. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell. 3:271–282. [DOI] [PubMed] [Google Scholar]
- Babst, M., D.J. Katzmann, W.B. Snyder, B. Wendland, and S.D. Emr. 2002. b. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 3:283–289. [DOI] [PubMed] [Google Scholar]
- Bagnat, M., and K. Simons. 2002. Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA. 99:14183–14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat, M., S. Keränen, A. Shevchenko, A. Shevchenko, and K. Simons. 2000. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc. Natl. Acad. Sci. USA. 97:3254–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnat, M., A. Chang, and K. Simons. 2001. Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Mol. Biol. Cell. 12:4129–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becherer, K.A., S.E. Rieder, S.D. Emr, and E.W. Jones. 1996. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol. Biol. Cell. 7:579–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, T., A. Schmidt, and M.N. Hall. 1999. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 146:1227–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum, G., N.D. Lees, M. Bard, and R. Dickson. 1998. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 14:1471–1510. [DOI] [PubMed] [Google Scholar]
- Dimster-Denk, D., M.K. Thorsness, and J. Rine. 1994. Feedback regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase in Saccharomyces cerevisiae. Mol. Biol. Cell. 5:655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison, M.J., and M. Hochstrasser. 1991. Epitope-tagged ubiquitin. J. Biol. Chem. 266:21150–21157. [PubMed] [Google Scholar]
- Gaber, R.F., D.M. Copple, B.K. Kennedy, M. Vidal, and M. Bard. 1989. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol. Cell. Biol. 9:3447–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard, S.R., B.P. Levi, and T.H. Stevens. 2000. Pep12p is a multifunctional yeast syntaxin that controls entry of biosynthetic, endocytic and retrograde traffic into the prevacuolar compartment. Traffic. 1:259–269. [DOI] [PubMed] [Google Scholar]
- Harsay, E., and R. Schekman. 2002. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J. Cell Biol. 156:271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese-Peck, A., H. Pichler, B. Zanolari, R. Watanabe, G. Daum, and H. Riezman. 2002. Multiple functions of sterols in yeast endocytosis. Mol. Biol. Cell. 13:2664–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, S.B., S. Losko, and C.A. Kaiser. 2001. Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J. Cell Biol. 153:649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke, L., and H. Riezman. 1996. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 84:277–287. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M., M.J. Ellison, V. Chau, and A. Varshavsky. 1991. The short-lived MATα2 transcriptional regulator is ubiquitinated in vivo. Proc. Natl. Acad. Sci. USA. 88:4606–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann, D.J., M. Babst, and S.D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-1. Cell. 106:145–155. [DOI] [PubMed] [Google Scholar]
- Kaur, R., and A.K. Bachhawat. 1999. The yeast multidrug resistance pump, Pdr5p, confers reduced drug resistance in erg mutants of Saccharomyces cerevisiae. Microbiology. 145:809–818. [DOI] [PubMed] [Google Scholar]
- Kim, Y., C.-W. Yun, and C.C. Philpott. 2002. Ferrichrome induces endosome to plasma membrane cycling of the ferrichrome transporter, Arn1p, in Saccharomyces cerevisiae. EMBO J. 21:3632–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafont, F., and K. Simons. 2001. Raft-partitioning of the ubiquitin ligases Cbl and Nedd4 upon IgE-triggered cell signaling. Proc. Natl. Acad. Sci. USA. 98:3180–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losko, S., F. Kopp, A. Kranz, and R. Kölling. 2001. Uptake of the ATP-binding cassette (ABC) transporter Ste6 into the yeast vacuole is blocked in the doa4 mutant. Mol. Biol. Cell. 12:1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee, T.P., H.B. Skinner, E.A. Whitters, S.A. Henry, and V.A. Bankaitis. 1994. A phosphatidylinositol transfer protein controls the phosphatidylcholine content of yeast Golgi membranes. J. Cell Biol. 124:273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwako, I., A. Yamamoto, T. Kitamura, K. Nagayama, and M. Ohashi. 2001. Cholesterol requirement for cation-independent mannose 6-phosphate receptor exit from multivesicular late endosomes to the Golgi. J. Cell Sci. 114:1765–1776. [DOI] [PubMed] [Google Scholar]
- Munn, A.L., A. Heese-Peck, B.J. Stevenson, H. Pichler, and H. Riezman. 1999. Specific sterols required for the internalization step of endocytosis in yeast. Mol. Biol. Cell. 10:3943–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, A., D. Brada, and R. Schekman. 1988. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J. Cell Biol. 107:851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa, S., and A. Nakano. 1991. The GTP-binding Sar1 protein is localized to the early compartment of the yeast secretory pathway. Biochim. Biophys. Acta. 1093:135–143. [DOI] [PubMed] [Google Scholar]
- Nothwehr, S.F., E. Conibear, and T.H. Stevens. 1995. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J. Cell Biol. 129:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi, G., M. Babst, and S.D. Emr. 1998. Fab1p PtIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 95:847–858. [DOI] [PubMed] [Google Scholar]
- Piper, R.C., A.A. Cooper, H. Yang, and T.H. Stevens. 1995. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131:603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg, C., K.G. Bache, D.J. Gillooly, I.H. Madshus, E. Stang, and H. Stenmark. 2002. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 4:394–398. [DOI] [PubMed] [Google Scholar]
- Raths, S., J. Rohrer, F. Crausaz, and H. Riezman. 1993. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 120:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., and H.R.B. Pelham. 2001. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 20:5176–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., and H.R.B. Pelham. 2002. A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat. Cell Biol. 4:117–123. [DOI] [PubMed] [Google Scholar]
- Reggiori, F., M.W. Black, and H.R.B. Pelham. 2000. Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol. Biol. Cell. 11:3737–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberg, K.J., N. Rowley, and C.A. Kaiser. 1997. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J. Cell Biol. 137:1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M., K. Sato, and A. Nakano. 1996. Endoplasmic reticulum localization of Sec12p is achieved by two mechanisms: Rer1p-dependent retrieval that requires the transmembrane domain and Rer1p-independent retention that involves the cytoplasmic domain. J. Cell Biol. 134:279–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, K., M. Sato, and A. Nakano. 1997. Rer1p as common machinery for the endoplasmic reticulum localization of membrane proteins. Proc. Natl. Acad. Sci. USA. 94:9693–9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, K., M. Sato, and A. Nakano. 2001. Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatmer. J. Cell Biol. 152:935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele, P., M.G. Roth, and K. Simons. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, A., M.N. Hall, and A. Koller. 1994. Two FK506 resistance-conferring genes in Saccharomyces cerevisiae, TAT1 and TAT2, encode amino acid permeases mediating tyrosine and tryptophan uptake. Mol. Cell. Biol. 14:6597–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21. [DOI] [PubMed] [Google Scholar]
- Sievi, E., T. Suntio, and M. Makarow. 2001. Proteolytic function of GPI-anchored plasma membrane protease Yps1p in the yeast vacuole and Golgi. Traffic. 2:896–907. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature. 387:569–572. [DOI] [PubMed] [Google Scholar]
- Soetens, O., J.-O. De Craene, and B. André. 2001. Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J. Biol. Chem. 276:43949–43957. [DOI] [PubMed] [Google Scholar]
- Stevens, T., B. Esmon, and R. Schekman. 1982. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 30:439–448. [DOI] [PubMed] [Google Scholar]
- TerBush, D.R., T. Maurice, D. Roth, and P. Novick. 1996. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Wang, G., J.M. McCaffery, B. Wendland, S. Dupré, R. Haguenauer-Tsapis, and J.M. Huibregtse. 2001. Localization of the Rsp5p ubiquitin-protein ligase at multiple sites within the endocytic pathway. Mol. Cell. Biol. 21:3564–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr, A., S. Avaro, C. Prescianotto-Baschong, R. Haguenauer-Tsapis, and H. Riezman. 2000. The F-box protein Rcy1p is involved in endocytic membrane traffic and recycling out of an early endosome in Saccharomyces cerevisiae. J. Cell Biol. 149:397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiroda, H., T. Oguchi, Y. Yasuda, A. Toh-e, and Y. Kikuchi. 1996. Bul1, a new protein that binds to the Rsp5 ubiquitin ligase in Saccharomyces cerevisiae. Mol. Cell Biol. 16:3255–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiroda, H., D. Kaida, A. Toh-e, and Y. Kikuchi. 1998. The PY-motif of Bul1 protein is essential for growth of Saccharomyces cerevisiae under various stress conditions. Gene. 225:39–46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.