Abstract
Astable cell line (GT2-LPk) derived from LLC-Pk was created in which endogenous DNA topoisomerase IIα (topoIIα) protein was downregulated and replaced by the expression of topoIIα fused with enhanced green fluorescent protein (EGFP–topoIIα). The EGFP–topoIIα faithfully mimicked the distribution of the endogenous protein in both interphase and mitosis. In early stages of mitosis, EGFP–topoIIα accumulated at kinetochores and in axial lines extending along the chromosome arms. During anaphase, EGFP–topoIIα diminished at kinetochores and increased in the cytoplasm with a portion accumulating into large circular foci that were mobile and appeared to fuse with the reforming nuclei. These cytoplasmic foci appearing at anaphase were coincident with precursor organelles of the reforming nucleolus called nucleolus-derived foci (NDF). Photobleaching of EGFP–topoIIα associated with kinetochores and chromosome arms showed that the majority of the protein rapidly exchanges (t1/2 of 16 s). Catalytic activity of topoIIα was essential for rapid dynamics, as ICRF-187, an inhibitor of topoIIα, blocked recovery after photobleaching. Although some topoIIα may be stably associated with chromosomes, these studies indicate that the majority undergoes rapid dynamic exchange. Rapid mobility of topoIIα in chromosomes may be essential to resolve strain imparted during chromosome condensation and segregation.
Keywords: DNA topoisomerase II; cell division; cell cycle proteins; kinetochore; nucleolus
Introduction
DNA topoisomerase II (topoII)* is essential in proliferating cells and is a major target for many anticancer drugs (Bakshi et al., 2001; Wang et al., 2001). In mammals, two genes code for isoforms of topoisomerase II, termed α and β, and alternative splicing confers further diversity (Petruti-Mot and Earnshaw, 2000). TopoII has been proposed to play both catalytic and structural roles in mitotic chromosome segregation. Genetic studies in yeast and inhibitor studies in mammalian cells suggest that topoII is required to resolve sister chromatid catenations during separation of the chromatids (Uemura et al., 1987; Holm et al., 1989; Gorbsky, 1994). In chromosomes that have been isolated from cells arrested in M phase, the bulk of topoII is tightly associated, suggesting that topoII forms a stable structural component or scaffold of chromosome architecture (Earnshaw et al., 1985). A study of rhodamine-labeled topoII in Drosophila embryos challenged this model with evidence that much topoII diffuses from the chromosomes at progressive stages of mitosis (Swedlow et al., 1993). The stability of topoIIα protein is cell cycle regulated, high during G2/M phases, and becoming unstable in the subsequent G1 phase (Heck et al., 1988).
Some immunolocalization and electron microscopy studies show a distribution of topoII in an axial core extending the length of the chromosome (for review see Warburton and Earnshaw, 1997). However, in both Drosophila embryos and condensed chromosomes formed in vitro in Xenopus extracts, topoII shows more uniform distribution on the chromosome arms (Hirano and Mitchison, 1993; Swedlow et al., 1993). In some immunolabeling studies, the enzyme was reported to be concentrated at kinetochores (Taagepera et al., 1993; Gorbsky, 1994; Rattner et al., 1996; Sumner, 1996). In interphase mammalian cells, various groups report differences in the subnuclear distribution of topoIIα with some but not all studies noting a concentration in the nucleoli (Meyer et al., 1997; Khelifa and Beck, 1999; Mo and Beck, 1999).
To investigate the dynamics of topoIIα in living cells, we produced a stable cell line expressing EGFP–topoIIα. FRAP experiments show that most topoIIα exchanges rapidly between cytoplasmic and kinetochore/chromosome-bound pools, and thus does not constitute a static structural chromosome scaffold. We also report a novel localization of topoIIα in late anaphase and telophase to discrete cytoplasmic foci that appear to incorporate into the reforming nucleoli during telophase.
Results and discussion
Establishment of a stable cell line expressing EGFP–topoIIα
To track the localization and dynamics of topoIIα in unperturbed living cells, we constructed a stable cell line, called GT2-LPk, that constitutively expresses EGFP–topoIIα. The overexpression of active topoII is cytotoxic, and although transient expression is possible, it has not previously been possible to obtain stable lines that constitutively express tagged forms of the enzyme (Mo et al., 1998; Mo and Beck, 1999). We compared the expression levels of endogenous topoIIα in the parental LLC-Pk and GT2-LPk lines. We found that expression of endogenous topoIIα was diminished in the GT2-LPk line, presumably accounting for the ability of the GT2-LPk line to escape cytotoxicity from topoIIα overexpression (Fig. 1 A). Immunoprecipitated EGFP–topoIIα efficiently catalyzed decatenation of kinetoplast DNA (Fig. 1 B). EGFP–topoIIα bound to protein A beads showed equivalent or greater catalytic activity compared with purified human topoIIα, a gift from Dr. Neal Osheroff (Vanderbilt University, Nashville, TN) (unpublished data).
Figure 1.
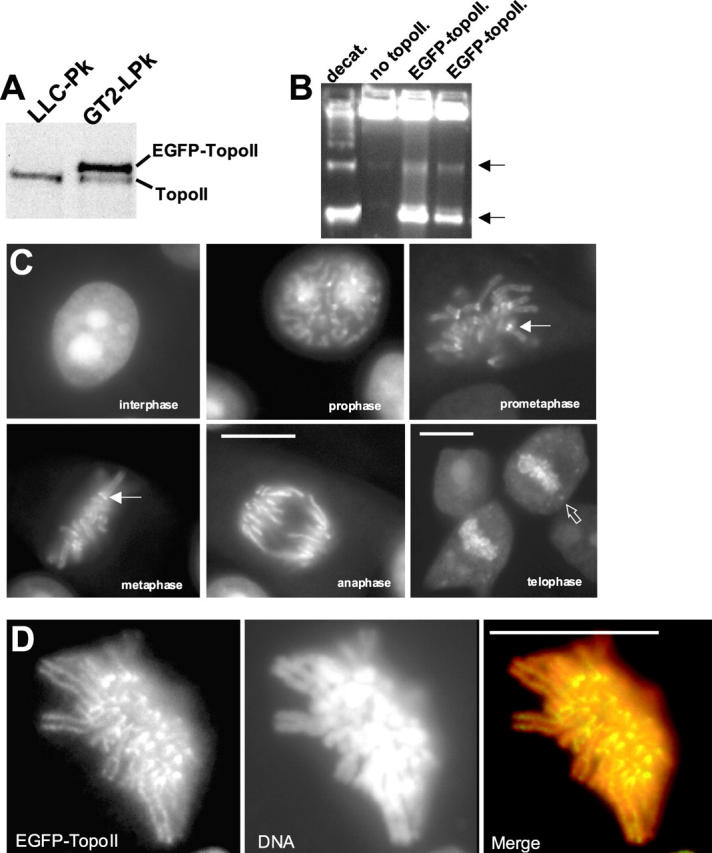
GT2-LPk cells express EGFP-topoIIα. (A) Immunoblotting with anti-topoIIα antibody of whole-cell extracts of parental LLC-Pk cells and GT2-LPk. GT2-LPk cells show downregulated expression of endogenous topoIIα. (B) EGFP–topoIIα immunoprecipitated from GT2-LPk cells catalyzes decatenation of kinetoplast DNA. Lanes contain respectively decatenated control, kinetoplast DNA with no topoisomerase II added, and two separate immunoprecipitations of GT2-LPk extracts with anti-GFP antibody. Arrows indicate the migration of the decatenated products (nicked and supercoiled). (C) EGFP– topoIIα localization in a panel of living cells at various stages of mitosis. EGFP–topoIIα associates with chromosomes in prophase, becomes concentrated at kinetochores (closed arrows) and along chromosome arms in prometaphase and metaphase, and associates with chromosome arms in anaphase. In late anaphase and telophase, EGFP–topoIIα increases in the cytoplasm with some becoming concentrated in cytoplasmic foci (open arrow). (D) Imaging of fixed GT2-LPk cell at late prometaphase after staining with DAPI shows localization of topoisomerase II to the kinetochore region and axes of chromosomes. Cells were simultaneously fixed and permeabilized by treatment with 2% formaldehyde, 0.5% TX-100 in microtubule stabilizing buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, 4 mM MgSO4, pH 6.95). The images represent a maximum projection of six 0.2-μm optical sections. In the merged image, EGFP–topoIIα is green and appears yellow due to overlap with the DNA in red. Bars, 10 μm.
EGFP–topoIIα localization in living cells
In living cells in interphase, EGFP-topoIIα was present diffusely in the nucleoplasm and concentrated in the nucleoli (Fig. 1 C). Nucleolar labeling persisted through early prophase when concentration of EGFP-topoIIα on chromosomes and at kinetochores became apparent. In living cells during prometaphase and metaphase topoIIα localized to thin lines running down the lengths of the chromosome arms (Fig. 1 C). Higher resolution images in fixed cells revealed that EGFP-topoIIα was concentrated along the central axis of each chromatid arm (Fig. 1 D). This distribution is consistent with the axial distribution of topoII observed in immunolabeling studies of fixed mammalian cells and isolated chromosomes (Earnshaw and Heck, 1985; Gorbsky, 1994; Rattner et al., 1996; Sumner, 1996). In anaphase, centromere concentration diminished while the localization to the chromosome arms persisted. In late anaphase and telophase, cytoplasmic levels of topoIIα increased similar to the behavior of rhodamine-labeled topoII in Drosophila (Swedlow et al., 1993). The anaphsase and telophase dynamics of topoIIα may also be preliminary to the decrease in topoIIα protein stability that occurs after mitosis (Heck et al., 1988). In contrast to a previous immunolabeling study (Barthelmes et al., 2000), we never observed concentration of EGFP-topoIIα at centrosomes at any stage of the cell cycle.
In late anaphase we observed a novel concentration of topoIIα in cytoplasmic foci (Figs. 1 C, telophase, and 2 A [Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200202053/DC1]). The foci were motile and appeared to move toward and fuse with the reforming nuclei in telophase. During telophase, most cytoplasmic topoIIα appeared to reenter the forming nuclei, and the concentration of EGFP–topoIIα in the nucleoli reappeared. The approximate size, behavior, and appearance of the cytoplasmic EGFP–topoIIα-containing foci were reminiscent of previously described dynamic structures termed nucleolus-derived foci (NDF) (Dundr et al., 2000). These cytoplasmic NDFs apparently travel to nuclei where they fuse with other nucleolar precursors within the nuclei, e.g., prenucleolar bodies, to reform the interphase nucleoli (Olson et al., 2000). We found that EGFP–topoIIα foci colocalized with antibody to B23, a known component of the NDFs (Fig. 2 B). In contrast, EGFP–topoIIα in anaphase and telophase GT2-LPk cells did not colocalize with nuclear pore antigens (Fig. 2 C).
Figure 2.
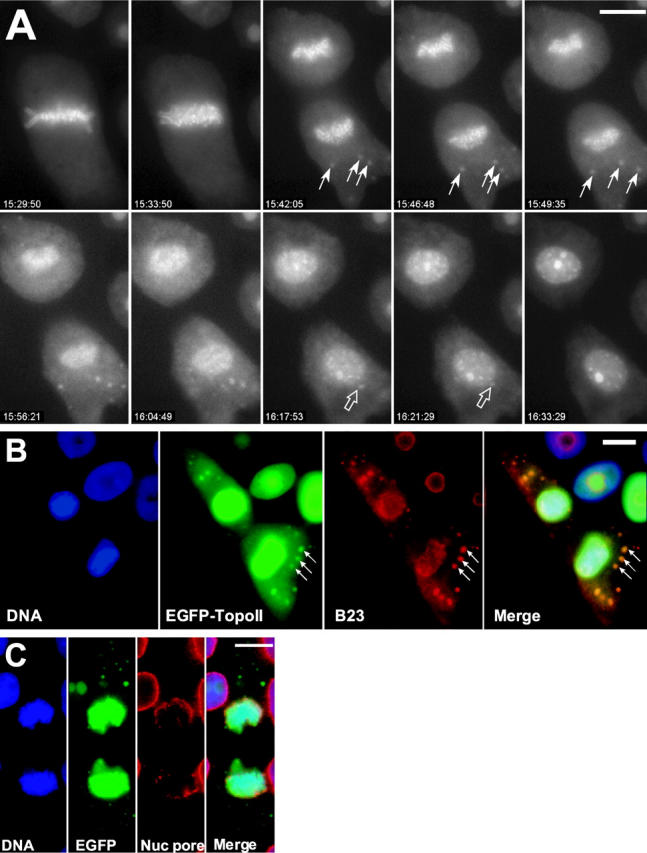
EGFP–topoIIα localization is dynamic at anaphase and telophase. (A) Selected frames from a time-lapse video of a GT2-LPk cell in late mitosis. In anaphase, some of the cytoplasmic EGFP–topoIIα concentrates into distinct foci (closed arrows) that are motile. At telophase, the foci appear to fuse with the reforming nucleus (open arrows), whereas nucleoli become more apparent. (Image frames in this sequence were individually adjusted for contrast to optimize visualization of the cyto- plasmic foci; Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200202053/DC1) (B) EGFP–topoIIα foci (arrows) in telophase cells colocalize with NDFs identified by labeling with antibody to nucleolar B23 protein, a gift from Dr. Mirek Dundr (National Cancer Institute, Bethesda, MD). (The EGFP– topoIIα image is oversaturated for the nuclear fluorescence to clearly visualize the cytoplasmic foci.) (C) EGFP–topoIIα foci do not colocalize with reforming nuclear envelope identified by RL1 antibody to nuclear pore proteins, a gift from Dr. Bryce Pascal (University of Virginia, Charlottesville, VA). Bars, 10 μm.
TopoIIα exchanges rapidly in mitotic chromosomes
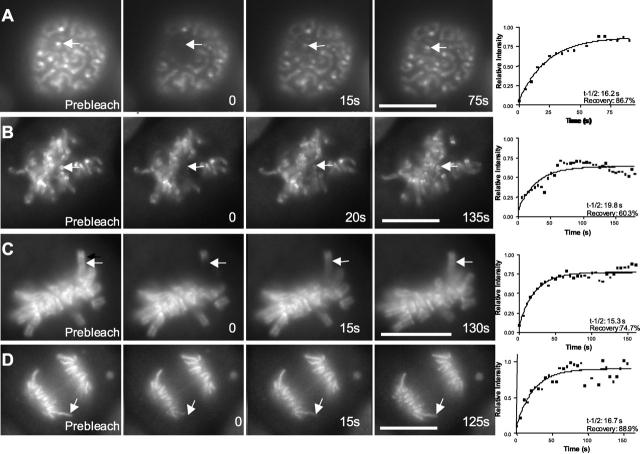
To quantify topoIIα exchange in living cells in mitosis, we used FRAP. If topoIIα served a static structural or nucleoskeletal role in maintaining the organization of the kinetochore or chromosome arm, then recovery should be slow. We found that recovery of the majority of the EGFP–topoIIα at kinetochores and chromosome arms occurs rapidly (Fig. 3 and Table I). Because the average recovery was <100%, there may be a subpopulation of kinetochore and chromosome-associated topoIIα that is more stably associated (for kinetochores an average of 38%; for chromosome arms an average of 24%). We found that the exchange of topoIIα occurred at all stages of mitosis and did not differ markedly from prophase through anaphase (Fig 3). Recovery of fluorescence likely reflects rapid exchange between the pools of topoIIα bound at chromosomes and kinetochores with the pool of cytoplasmic topoIIα.
Figure 3.
Recovery of EGFP–topoIIα fluorescence after photobleaching occurs rapidly at kinetochores and chromosome arms. Examples showing targeting of kinetochores (arrows) at prophase (A) and prometaphase (B) and targeting of chromosome arms (arrows) at metaphase (C) and anaphase (D) Images include a prebleach image, one taken just after photobleaching (0 time) and two images taken during recovery. Graphs at the end of each sequence show fitting of corrected intensity measurements in the photobleached regions to determine recovery half times (t-1/2) and total degree of recovery (recovery). Bars, 10 μm.
Table I.
Recovery of EGFP–topoIIα after photobleaching at kinetochores and chromosome arms
| Structure | t1/2 s (SD) | Range | % recovery (SD) | Range | n |
|---|---|---|---|---|---|
| Kinetochore | 15.7 (4.7) | 9.0–22.4 | 61.9 (14.7) | 42.8–86.7 | 8 |
| Chromosome arm | 15.5 (3.6) | 9.9–22.5 | 75.9 (13.0) | 52.4 – 90.0 | 10 |
Kinetochores and chromosome arms of GT2-LPk cells were photobleached with a nitrogen dye laser. Recovery of fluorescence was measured in the photobleached area from images taken at intervals after photobleaching. After correcting for background and loss of fluorescence due to image capture, measurements were used to calculate halftime recovery (t 1/2) and the percentage of recovery.
The rapid exchange of the majority of topoIIα in vivo contrasts to the tight association of topoIIα in chromosomes isolated from mitotic cells. In isolated chromosomes, topoII is resistant to DNAse treatment and extraction with high salt, and has thus been proposed to function as part of a stable structural scaffold for the mitotic chromosome. Although there may be a minor population of topoII that is stably associated with chromatin, we find the majority to exchange rapidly with topoIIα in the cytoplasm. Thus, in vivo, it appears unlikely that the bulk of topoIIα serves a static nucleoskeletal role in maintaining mitotic chromosome structure.
Rapid exchange of topoIIα requires catalytic activity
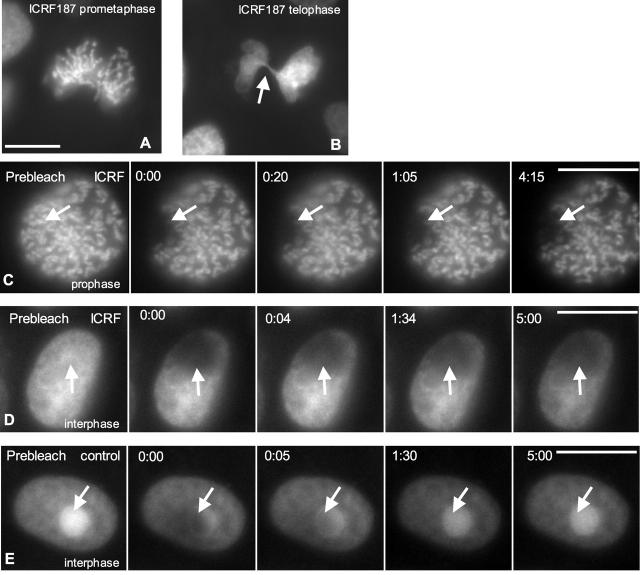
To determine whether catalytic activity of topoIIα was required for the rapid exchange observed in our FRAP analyses, we used a chemical topoIIα inhibitor, ICRF-187. ICRF-187 is an inhibitor of the bisdioxopiperazine class that blocks the enzyme by trapping it in the postcleavage closed clamp form (Roca et al., 1994). Applied to cells in mitosis, this inhibitor inhibits chromosome condensation and induces massive failure of chromosome segregation at anaphase in LLC-Pk cells, the parental line of the GT2-LPk cells (Gorbsky, 1994). Similarly, we found that ICRF-187 inhibits normal chromosome condensation and chromosome segregation in GT2-LPk cells (Fig. 4, A and B). In mitotic cells treated with ICRF-187, we detected only minimal recovery of EGFP–topoIIα into the bleached areas over a 10-min period (Fig. 4 C).
Figure 4.
Catalytic activity of topoIIα is required for rapid recovery after photobleaching. (A and B) GT2-LPk cells treated with ICRF-187, an inhibitor of topoIIα, showed impaired chromosome condensation in prometaphase (A) and mitotic catastrophe as the cleavage furrow has cut through the mass of unseparated chromatids in telophase (B, arrow). (C) Photobleaching of mitotic cells shows very limited recovery (arrow). (D) Photobleaching of interphase nucleus of ICRF-187–treated cell also shows limited recovery. ICRF-187 treatment reduces the concentration of EGFP-topoIIα at the nucleoli. (E) Photobleaching of nondrug treated interphase cell nucleus results in rapid recovery (arrow). Bars, 10 μm.
To corroborate the effects of ICRF-187 on the dynamics of topoIIα in mitotic cells, we also tested its effects in interphase nuclei. Treatment of cells with ICRF-187 greatly diminished the level of EGFP–topoIIα concentrated in the nucleolus. When tested by FRAP analysis of interphase cells, again ICRF-187 treatment was found to cause a marked decrease in the rate and extent of fluorescence recovery when compared with nondrug-treated interphase cells (Fig. 4, C and D). Together, these results suggest that the rapid exchange noted for EGFP–topoIIα in unperturbed cells requires catalytic activity. Moreover, ICRF-187 treatment slows the exchange rate of EGFP–topoIIα, lending further support to the idea that the EGFP–topoIIα protein is functional in vivo.
Rapid exchange of chromosome-associated and cytoplasmic pools of topoII
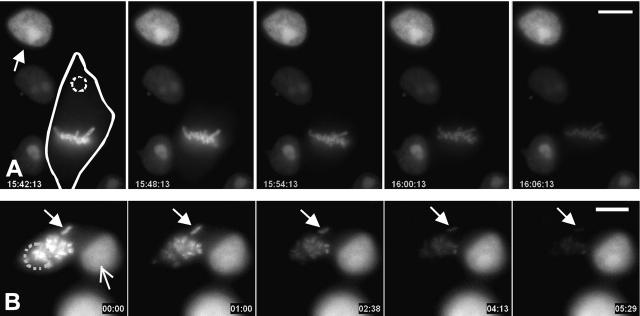
To further test whether the rapid exchange of topoIIα was somehow restricted to the protein associated with the chromosomes or occurred by exchange with a soluble cytoplasmic pool, we used fluorescence loss in photobleaching. We repeatedly photobleached a small area in the cytoplasm (Fig. 5 A) or on one region of the chromosomes of cells treated with a microtubule inhibitor to inhibit chromosome movement (Fig. 5 B; Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200202053/DC1). Loss of fluorescence of EGFP–topoIIα from unbleached mitotic chromosomes occurred upon repetitive bleaching at the distant area (Fig. 5). These data demonstrate that topoIIα mobility is not regionally confined. TopoIIα associated with chromosomes and kinetochores is mobile and exchanges with cytoplasmic pools of the enzyme.
Figure 5.
Fluorescence loss in photobleaching demonstrates exchange of topoIIα between chromosomal and cytoplasmic pools in mitotic cells. (A) Exchange of topoIIα occurs between the chromosomes and the cytoplasm of mitotic cells. A late prometaphase cell outlined by the solid line was targeted for photobleaching in the cytoplasm away from the chromosomes indicated (dotted circle). The region was photobleached every 20 s and images were captured every 2 min. Repeated photobleaching of the cytoplasm results in depletion of fluorescence in the chromosomes. The fluorescence intensity of the chromosomes was reduced by 79%, whereas that of the control interphase nucleus (arrow) in the adjacent interphase cell was reduced only 8%. (B) Exchange of topoIIα occurs through the cytoplasm among chromosomes. A cell treated with 0.1 μg/ml nocodazole was repeatedly bleached in the lower region of the mass of chromosomes (dotted circle). Fluorescence intensity of the isolated chromosome at top (closed arrows) diminished by 85%, whereas the intensity of the nucleus in the neighboring cell (open arrow) decreased by only 5% over the course of the observations (Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200202053/DC1). Bars, 10 μm.
Another study recently appeared analyzing GFP derivatives of topoIIα and topoIIβ in stable lines of 293 cells (Christensen et al., 2002). In general, the results from these authors and ours are consistent in describing the overall localization of topoIIα in interphase and mitotic cells. Both studies also agree that most topoIIα undergoes rapid exchange with the cytoplasm, and the studies concur on the necessity for topoIIα catalytic activity in rapid exchange. The studies do differ with regard to certain conclusions regarding topoIIα in mitotic chromosomes. Whereas Christensen et al. (2002) report that nearly 100% of topoIIα turns over rapidly, our results indicate that a population of topoIIα associated with the kinetochores and chromosome arms turns over more slowly. Moreover, whereas Christensen et al. (2002) conclude that in living cells topoIIα is uniformly distributed across the width of the chromosome arms, our evidence favors the idea that topoIIα concentrated toward the axes of the arms. There are several potential sources for the differences in the two studies. Apart from inherent differences between the two parental cell lines (LLC-Pk vs. 293), the GT2-LPk derivative line used in our work has downregulated levels of endogenous topoIIα that is replaced by EGFP–topoIIα. The 293 cell derivatives of Christensen et al. (2002) did not show evidence of down regulation of the endogenous enzyme. In addition, the photobleaching studies of mitotic chromosomes performed by Christensen et al. (2002) were carried out on cells arrested at M phase by treatment with the microtubule stabilizer paclitaxel. Apart from the cell shown in Fig. 5 B, our studies were done on cells not treated with drugs. In preliminary studies, we have noted greater recovery of EGFP–topoIIα after photobleaching GT2-LPk cells treated with either paclitaxel or nocodazole (unpublished data). Full resolution of the differences between the two studies will require further investigation.
Conclusions
Cell fractionation studies originally suggested that the bulk of topoIIα in mitotic cells was stably associated with the mitotic chromosomes and formed a structural scaffold. Although our evidence of rapid exchange of the bulk of chromosome-associated topoII does not rule out a structural role for the enzyme, any such role must accommodate the fact that most of the protein is highly dynamic, exchanging with the soluble pool in seconds. Consistent with the earlier findings on the single isoform of topoII in Drosophila embryos (Swedlow et al., 1993), we find in mammalian cells a net movement of topoIIα from the chromosomes to the cytoplasm in anaphase. However, two-way exchange between the chromosome arms and cytoplasm persists during anaphase. We report a novel association of topoIIα with NDFs in late mitosis. These organelles may serve to accumulate proteins to newly forming nucleoli in the daughter cells. The rapid dynamics of topoIIα throughout the mitotic chromosomes may be essential in allowing the enzyme to reach and rapidly relieve strain before torsional stresses imparted by chromosome condensation and chromosome movements cause breaks to the DNA.
Materials and methods
Cell culture and immunofluorescence
LLC-Pk were grown in DME supplemented with 10% FBS and antibiotics. To produce a stable cell line expressing EGFP–topoIIα, LLC-Pk cells were transfected using Lipofectamine Plus according to the manufacturer's instructions. Cells were selected for resistance to 2 μg/ml G418 and cloned by dilution in microwell plates. Cells were processed for immunofluorescence using methods described previously (Kallio et al., 1998).
Immunoprecipitation and decantenation assays
GT2-LPk cells were lysed in high salt buffer (20 mM Tris, pH 8.0, 0.75 M KCl, 0.75% NP40) and immunoprecipitated with anti-GFP antibody using previously described methods (Kallio et al., 1998). Decatenation assays were performed using a kit supplied by Topogen according to the manufacturer's directions.
Western blotting
Proteins were separated by SDS polyacrylamide gel electrophoresis using 4–20% gradient acrylamide gels. Proteins were transferred to Immobilon-P membrane. Anti-topoIIα was used at 1:1,000. Blots were then incubated with HRP-conjugated goat anti–rabbit and proteins visualized by chemiluminescence and imaged with a digital camera.
Laser photobleaching
Cells were observed on a warm stage using a Zeiss Axioplan II microscope equipped with the ORCA II CCD camera (Hamamastu) and controlled with MetaMorph software (Universal Imaging Corporation). A nitrogen laser-pumped dye laser (Photonic Instruments) delivered 4 ns laser pulses at 440 nm attenuated with a neutral density slider. Typically, we bleached for 1 s at a repetition rate of 30 Hz. For each time-lapse series in Figs. 3, 4, and 5, the individual frames were imaged and adjusted for contrast using identical conditions.
Data analysis
Metamorph software was used for data collection and analysis. Data were analyzed essentially as described (Gorbsky et al., 1990; Howell et al., 2000; Phair and Misteli, 2000). Briefly, an area slightly larger than the target area was used for local cytoplasmic background subtraction. Integrated intensity of the target and background regions and their associated area measurements were logged to Excel spreadsheets. Acquisitions were normalized to adjust for reduction of total cellular fluorescence due to photobleaching and image acquisition (Phair and Misteli, 2000). Normalized relative fluorescence intensity values were exported to GraphPad Prism software and analyzed by nonlinear regression. The rate constant k was determined using the perturbation-relaxation equation and half time of recovery was determined using t1/2 = ln2/k. Percent recovery was calculated according to the method described by Howell et al. (2000) using normalized relative intensity values.
Online supplemental material
Videos 1 and 2, corresponding to Figs. 2 A and Fig. 5 B, respectively, are available online as Quicktime movies at http://www.jcb.org/cgi/content/full/jcb.200202053/DC1.
Supplemental Material
Acknowledgments
We thank Dr. Mirek Dundr for helpful advice and for supplying anti-B23 antibody. We thank Dr. Neil Osheroff for supplying purified topoIIα enzyme. We thank Dr. Bryce Paschal for the gift of RL1 antibody.
P.A. Tavormina was supported by a National Research Service Award provided by the National Institute of General Medical Sciences. M.-G. Come was supported in part by a fellowship from the Association pour le Reserche sur le Cancer.
The online version of this article contains supplemental material.
Footnotes
Abbreviations used in this paper: EGFP, enhanced green fluorescent protein; NDF, nucleolus-derived foci; topoIIα, topoisomerase IIα.
References
- Bakshi, R.P., S. Galande, and K. Muniyappa. 2001. Functional and regulatory characteristics of eukaryotic type II DNA topoisomerase. Crit. Rev. Biochem. Mol. Biol. 36:1–37. [DOI] [PubMed] [Google Scholar]
- Barthelmes, H.U., P. Grue, S. Feineis, T. Straub, and F. Boege. 2000. Active DNA topoisomerase IIα is a component of the salt-stable centrosome core. J. Biol. Chem. 275:38823–38830. [DOI] [PubMed] [Google Scholar]
- Christensen, M.O., M.K. Larsen, H.U. Barthelmes, R. Hock, C.L. Andersen, E. Kjeldsen, B.R. Knudsen, O. Westergaard, F. Boege, and C. Mielke. 2002. Dynamics of human DNA topoisomerases IIα and IIβ in living cells. J. Cell Biol. 157:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr, M., T. Misteli, and M.O. Olson. 2000. The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 150:433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw, W.C., B. Halligan, C.A. Cooke, M.M. Heck, and L.F. Liu. 1985. Topoisomerase II is a structural component of mitotic chromosome scaffolds. J. Cell Biol. 100:1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw, W.C., and M.M. Heck. 1985. Localization of topoisomerase II in mitotic chromosomes. J. Cell Biol. 100:1716–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky, G.J. 1994. Cell cycle progression and chromosome segregation in mammalian cells cultured in the presence of the topoisomerase II inhibitors ICRF-187 ([+]-1,2-bis[3,5-dioxopiperazinyl-1-yl]propane; ADR-529) and ICRF-159 (razoxane). Cancer Res. 54:1042–1048. [PubMed] [Google Scholar]
- Gorbsky, G.J., C. Simerly, G. Schatten, and G.G. Borisy. 1990. Microtubules in the metaphase-arrested mouse oocyte turn over rapidly. Proc. Natl. Acad. Sci. USA. 87:6049–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck, M.M., W.N. Hittelman, and W.C. Earnshaw. 1988. Differential expression of DNA topoisomerases I and II during the eukaryotic cell cycle. Proc. Natl. Acad. Sci. USA. 85:1086–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, T., and T.J. Mitchison. 1993. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J. Cell Biol. 120:601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B.J., D.B. Hoffman, G. Fang, A.W. Murray, and E.D. Salmon. 2000. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 150:1233–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, C., T. Stearns, and D. Botstein. 1989. DNA Topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol. Cell. Biol. 9:159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio, M., J. Weinstein, J.R. Daum, D.J. Burke, and G.J. Gorbsky. 1998. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J. Cell Biol. 141:1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelifa, T., and W.T. Beck. 1999. Merbarone, a catalytic inhibitor of DNA topoisomerase II, induces apoptosis in CEM cells through activation of ICE/CED-3-like protease. Mol. Pharmacol. 55:548–556. [PubMed] [Google Scholar]
- Meyer, K.N., E. Kjeldsen, T. Straub, B.R. Knudsen, I.D. Hickson, A. Kikuchi, H. Kreipe, and F. Boege. 1997. Cell cycle-coupled relocation of types I and II topoisomerases and modulation of catalytic enzyme activities. J. Cell Biol. 136:775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo, Y.Y., and W.T. Beck. 1999. Association of human DNA topoisomerase IIα with mitotic chromosomes in mammalian cells is independent of its catalytic activity. Exp. Cell Res. 252:50–62. [DOI] [PubMed] [Google Scholar]
- Mo, Y.Y., K.A. Ameiss, and W.T. Beck. 1998. Overexpression of human DNA topoisomerase II alpha by fusion to enhanced green fluorescent protein. Biotechniques. 25:1052–1057. [DOI] [PubMed] [Google Scholar]
- Olson, M.O.J., M. Dundr, and A. Szebeni. 2000. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10:189–196. [DOI] [PubMed] [Google Scholar]
- Petruti-Mot, A.S., and W.C. Earnshaw. 2000. Two differentially spliced forms of topoisomerase IIα and β mRNAs are conserved between birds and humans. Gene. 258:183–192. [DOI] [PubMed] [Google Scholar]
- Phair, R.D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature. 404:604–609. [DOI] [PubMed] [Google Scholar]
- Rattner, J.B., M.J. Hendzel, C.S. Furbee, M.T. Muller, and D.P. Bazett-Jones. 1996. Topoisomerase IIα is associated with the mammalian centromere in a cell cycle- and species-specific manner and is required for proper centromere/kinetochore structure. J. Cell Biol. 134:1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca, J., R. Ishida, J.M. Berger, T. Andoh, and J.C. Wang. 1994. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc. Natl. Acad. Sci. USA. 91:1781–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner, A.T. 1996. The distribution of topoisomerase II on mammalian chromosomes. Chromosome Res. 4:5–14. [DOI] [PubMed] [Google Scholar]
- Swedlow, J.R., J.W. Sedat, and D.A. Agard. 1993. Multiple chromosomal populations of topoisomerase II detected in vivo by time-lapse, three dimensional wide-field microscopy. Cell. 73:97–108. [DOI] [PubMed] [Google Scholar]
- Taagepera, S., P.N. Rao, F.H. Drake, and G.J. Gorbsky. 1993. DNA topoisomerase IIα is the major chromosome protein recognized by the mitotic phosphoprotein antibody MPM-2. Proc. Natl. Acad. Sci. USA. 90:8407–8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura, T., H. Ohkura, Y. Adachi, K. Morino, K. Shiozaki, and M. Yanagida. 1987. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 50:917–925. [DOI] [PubMed] [Google Scholar]
- Wang, H., Y. Mao, N. Zhou, T. Hu, T.S. Hsieh, and L.F. Liu. 2001. ATP-bound topoisomerase II as a target for antitumor drugs. J. Biol. Chem. 276:15990–15995. [DOI] [PubMed] [Google Scholar]
- Warburton, P.E., and W.C. Earnshaw. 1997. Untangling the role of DNA topoisomerase II in mitotic chromosome structure and function. Bioessays. 19:97–99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.