Abstract
The mechanisms that govern the assembly of nuclear pore complexes (NPCs) remain largely unknown. Here, we have established a role for karyopherins in this process. We show that the yeast karyopherin Kap121p functions in the targeting and assembly of the nucleoporin Nup53p into NPCs by recognizing a nuclear localization signal (NLS) in Nup53p. This karyopherin-mediated function can also be performed by the Kap95p–Kap60p complex if the Kap121p-binding domain of Nup53p is replaced by a classical NLS, suggesting a more general role for karyopherins in NPC assembly. At the NPC, neighboring nucleoporins bind to two regions in Nup53p. One nucleoporin, Nup170p, associates with a region of Nup53p that overlaps with the Kap121p binding site and we show that they compete for binding to Nup53p. We propose that once targeted to the NPC, dissociation of the Kap121p–Nup53p complex is driven by the interaction of Nup53p with Nup170p. At the NPC, Nup53p exists in two separate complexes, one of which is capable of interacting with Kap121p and another that is bound to Nup170p. We propose that fluctuations between these two states drive the binding and release of Kap121p from Nup53p, thus facilitating Kap121p's movement through the NPC.
Keywords: nuclear envelope; nucleoporins; NPC assembly; nuclear transport
Introduction
In eukaryotic cells, the constant flow of molecules between the nucleus and the cytoplasm is controlled by nuclear pore complexes (NPCs).* These structures decorate the surface of the nucleus and form aqueous channels through an otherwise impermeable double membrane nuclear envelope (NE). In yeast, the NPC has an estimated mass of ∼50 MD and is made up of ∼30 proteins termed nucleoporins or nups (for review see Rout and Aitchison, 2001). Nups can be divided into two broad categories depending on the presence or absence of Phe-Gly (FG) residues contained within repetitive peptide repeats generally of the type FXFG or GLFG (termed here FG-nups). Most yeast nups are distributed on both sides of the NPC (Nehrbass et al., 1996; Fahrenkrog et al., 1998; Marelli et al., 1998; Rout et al., 2000) and form a highly symmetrical core assembly, whereas others are asymmetrically distributed and make up filaments that extend into the nucleoplasm and the cytoplasm (for reviews see Allen et al., 2000; Rout and Aitchison, 2001).
To overcome the barrier imposed by the NE, cargo molecules are escorted to and through the NPC by carrier proteins referred to as karyopherins (importins and exportins are also widely used terms that designate direction of transport [for reviews see Wozniak et al., 1998; Macara, 2001; Strom and Weis, 2001]). Most karyopherins (kaps) can be grouped into a family of structurally related proteins collectively referred to as β-karyopherins (or β-kaps) of which there are 14 in yeast. A typical karyopherin-mediated transport process begins with the recognition of its cargo through discrete signal sequences broadly categorized as NLSs or nuclear export signals (NESs). An individual signal can be recognized by one or more karyopherins, with the latter scenario representing a level of functional redundancy.
The association of karyopherin–cargo complexes to peripherally localized nucleoporins is the initial step in their translocation through the NPC. These and subsequent interactions with the NPC are mediated largely by the FG-nups (for review see Ryan and Wente, 2000). The crystal structure of human karyopherin β1–importin β (Bayliss et al., 2000) in association with an FG-containing region of Nsp1p has demonstrated a direct interaction between the FG moiety and karyopherin β1–importin β. Similarly, two structurally distinct transport factors, Tap (Fribourg et al., 2001) and NTF2 (Bayliss et al., 2002), also directly interact with the FG repeats.
In addition to FG repeat–containing regions, other regions within nups have been shown to bind kaps. For example, the vertebrate Kap-β2–transportin can bind the FG repeat–containing nups Nup153 and Nup98 through a region similar to the M9 NLS recognized as cargo by Kap-β2 (Nakielny et al., 1999; Fontoura et al., 2000). Other nucleoporins, including the mammalian protein Tpr and the yeast protein Nic96p (Grandi et al., 1995; Bangs et al., 1998), also contain regions similar to known NLSs that function autonomously to target reporter proteins to the nucleus. The functions of these NLSs in the context of the nups are unclear. One possibility is that they function as docking sites for karyopherins at the NPC, analogous to the FG-containing domains. Other ideas also have been proposed. Fontoura et al. (2000) suggested that the M9 NLS of Nup98 assists in the release of cargo from Kap-β2. In addition, the M9 NLS of Nup153 has been suggested to play a role in the nuclear import of a shuttling pool of Nup153 (Nakielny et al., 1999).
Although most of the FG-nups appear to be capable of binding multiple karyopherins, it is clear, as suggested by the presence of specific NLSs, that certain nups exhibit preferences for members of the β-kap family. This is exemplified by the specific interaction of Nup53p with Kap121p (Marelli et al., 1998, 2001; Damelin and Silver, 2000). These and other observations have promoted the idea that there are specific docking sites within the NPC for certain kaps, and these serve to define different kap-mediated transport routes through the NPC (Rout et al., 1997; Marelli et al., 1998).
Despite our knowledge of the interactions between kaps and nups, the mechanism of translocation through the NPC remains largely undefined. Central to most current translocation models is the idea that the putative transport channel is lined by a high concentration of FG-nups. Rexach and Blobel (1995) proposed that kap–cargo complexes move stochastically through the NPC along a series of nups having increasing affinity for kaps, thereby lending directionality to the transport process. More recent data support the idea of such a gradient (Ben-Efraim and Gerace, 2001). Rout et al. (2000) have proposed that the narrow diameter of the central transporter and the random motion of the filaments formed by the FG-nups establishes a “virtual gate” that excludes most macromolecules except those that bind to these nups, i.e., the kaps. A seminal feature of this model is the idea that the vast majority of FG-nups lining the pore have a low affinity for the kaps, which allows them to rapidly exchange between nups until they encounter high affinity sites in the target compartment. Ribbeck and Gorlich (2001) envision a different model in which a network of FG-nups forms a hydrophobic barrier that is impermeable to most macromolecules. They propose that kaps, through their affinity for the FG-nups, can partition into this matrix and then diffuse across the NPC (Ribbeck and Gorlich, 2001, 2002).
Each of these models relies on the premise that the FG-nups are organized within a defined, potentially fluid matrix within the NPC. However, exactly how this network of nups is assembled and maintained in a functional state remains to be deciphered.
In yeast, the assembly of new NPCs occurs throughout the cell cycle until they are divided between the forming daughter nuclei during telophase. This would suggest that the machinery for the assembly of NPCs functions continuously (Winey et al., 1997). In this paper, we have examined the mechanism by which one FG-nup (Nup53p) is assembled into NPCs. A unique property of Nup53p is that it interacts specifically with Kap121p. Here, we show that Kap121p plays a role in the assembly of Nup53p into NPCs. We also provide evidence that once targeted to the NPC, Nup53p is released from Kap121p through a mechanism that is dependent on Nup170p. At the NPC, Nup53p exists in two states, one of which is capable of interacting with Kap121p.
Results
Defining the Kap121p-binding domain (KBD) of Nup53p
The specific interaction between Kap121p and the FG-nup Nup53p has led to the proposal that this nup plays a specific, perhaps regulatory, role in Kap121p-mediated import. Nup53p is slightly different from other FG-nups in that its FGs are not contained within the more common FXFG or GLFG peptide repeat patterns. In light of these observations, we have dissected the Kap121p–Nup53p interaction in an attempt to define the region of Nup53p responsible for Kap121p binding. To do this, NH2- and COOH-terminal deletions of Nup53p were synthesized as GST fusions in Escherichia coli (Fig. 1), and the binding of Kap121p to these fusion proteins was evaluated using a blot overlay assay previously used to detect interactions between Kap121p and a number of FG-nups including Nup53p (Marelli et al., 1998). As shown in Fig. 2 A, Kap121–protein A (pA) derived from yeast cytosol bound specifically to GST–Nup53p. Kap121-pA also bound GST fusions containing amino acid residues 203–475 and 374–475 of Nup53p, but failed to bind either of the COOH-terminal deletions containing residues 1–297 or 1–375. Further deletion analysis of the 374–475 region identified residues 401–448 as a minimal Kap121p-interacting region. A direct interaction between this region of Nup53p and Kap121p was confirmed by an in vitro solution binding assay using the GST-401–448 protein and recombinant Kap121p. As shown in Fig. 2 B, Kap121p bound specifically to GST-401–448, but not to GST alone.
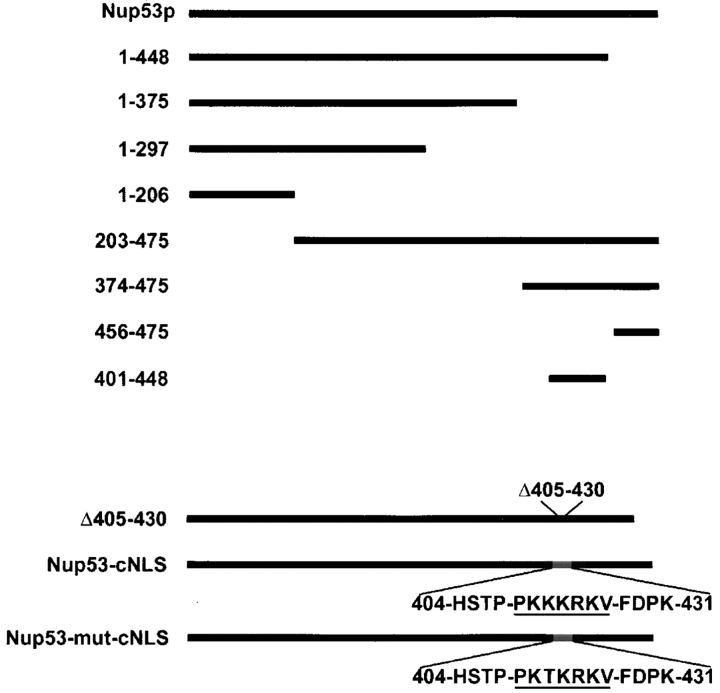
Figure 1.
NUP53 mutants. NH2- and COOH-terminal truncations of Nup53p were designed as shown with amino acid residues listed on the left. Δ405–430 indicates the deletion of these residues. The Nup53-cNLS and Nup53-mut-cNLS constructs contain insertions encoding either the SV-40 T-antigen cNLS (underlined) or a mutant of the cNLS (mut-cNLS) in place of residues 405–430.
Figure 2.
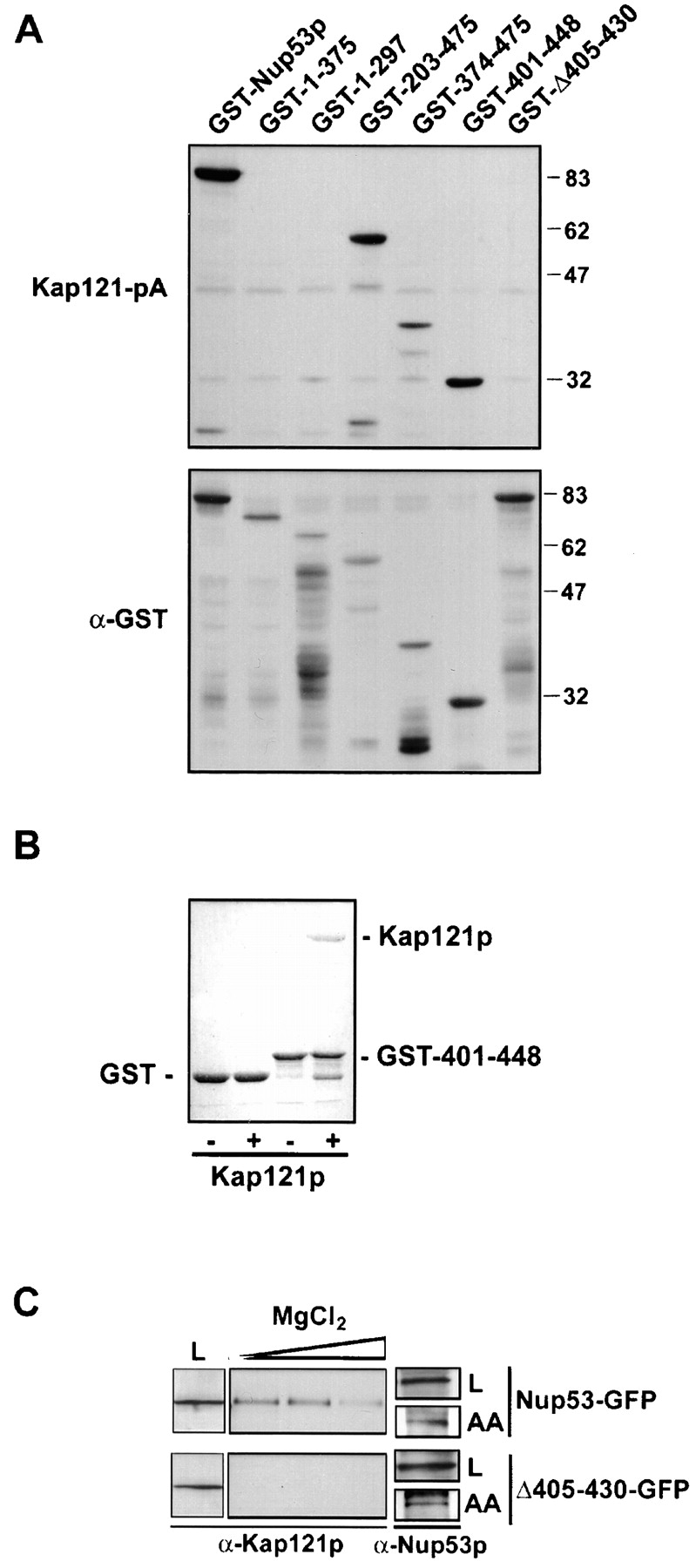
Defining the Kap121p-binding domain in Nup53p. (A) The truncations indicated were synthesized as GST fusions in E. coli, and total cell lysates were separated by SDS-PAGE and transferred to nitrocellulose. Membranes were probed with a cytosol prepared from a yeast strain (KP121pA) producing Kap121-pA or an anti-GST antibody (α-GST) to confirm the positions of the fusion proteins. Molecular mass markers are shown in kilodaltons. (B) GST and GST-401–448 were synthesized in E. coli and immobilized on Glutathione-Sepharose. Beads were incubated with buffer alone (−) or purified Kap121p (+), washed, and eluted with SDS-PAGE sample buffer. Proteins were separated by SDS-PAGE and visualized with Coomassie blue. (C) Immunoprecipitations of Nup53-GFP and Δ405–430-GFP were performed from a nup53Δ strain transformed with either pNP53-GFP or pΔ405–430-GFP. Both strains produced similar levels of the fusion proteins (not depicted). Anti-Nup53p antibodies were incubated with yeast extracts from these strains, followed by the addition of protein-G beads. The beads were collected, washed, and eluted with a gradient of 50, 200, and 500 mM MgCl2 and lastly with 0.5 M acetic acid, pH 3.4 (AA). Proteins in the eluted fractions were separated by SDS-PAGE, transferred to nitrocellulose, and probed with rabbit polyclonal anti-Kap121p (α-Kap121p) or anti-Nup53p (α-Nup53p) antibodies. L represents the load fractions.
Several other observations support the conclusion that residues 401–448 of Nup53p are necessary for binding Kap121p. First, a deletion mutant lacking residues 405–430 failed to bind Kap121-pA in overlay assays (Fig. 2 A). In addition, by two separate criteria, this deletion mutant failed to interact with Kap121p when expressed in yeast. First, when Nup53p or the 405–430 deletion mutant were affinity-purified from yeast lysates, Kap121p was associated with wild-type Nup53p, but not the 405–430 deletion mutant (Fig. 2 C). Second, we showed that the NE association of Kap121-GFP with Nup53p was dependent on residues 405–430. To do this, we took advantage of a previous observation that removal of Nup53p from the NPC caused a visible reduction in steady-state levels of Kap121p associated with NPCs (Marelli et al., 1998; Fig. 3 A). This phenotype was most striking in cells lacking both Nup53p and a structurally related protein Nup59p. Introduction of Nup53p into this strain reestablished NE binding of Kap121p to approximately wild-type levels (Fig. 3 B; Marelli et al., 1998). However, as shown in Fig. 3 C, the deletion of residues 405–430 from Nup53p eliminated its ability to recruit Kap121p to the NPC. As discussed below, a significant portion of this Nup53p mutant can associate with the NPC, suggesting that its inability to function as a binding site for Kap121p was not due to a complete mislocalization of the mutant protein. Together, these observations suggest that the KBD of Nup53p lies between residues 401 and 448 in a region lacking FG repeats.
Figure 3.
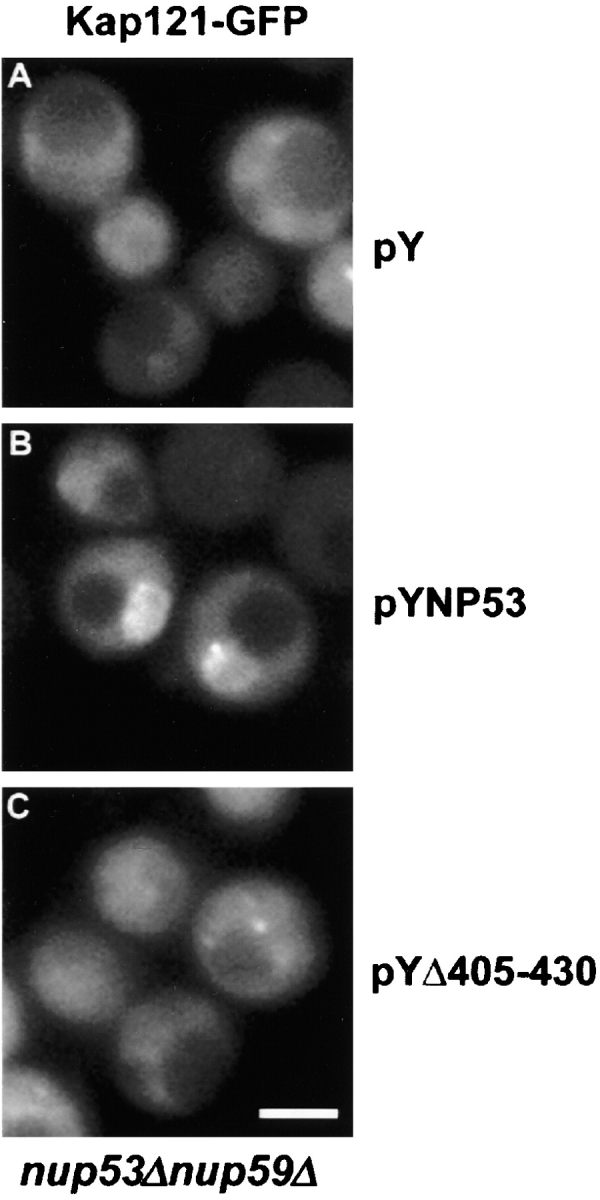
Kap121p interacts with the KBD at the NPC. A plasmid-born copy of KAP121-GFP was cointroduced with either (A) an empty plasmid (pY), (B) a plasmid containing NUP53 (pYNP53), or (C) a plasmid containing NUP53 lacking the coding region for the KBD, Δ405–430 (pYΔ405–430), into a haploid yeast strain lacking both NUP53 and NUP59 (NP53/NP59–2.1). The distribution of Kap121-GFP was determined by fluorescence microscopy. Bar, 5 μm.
Kap121p assists in the targeting of Nup53p to the NPC
Analysis of the Nup53p KBD (Fig. 4 A) revealed that it appeared similar to known or potential NLSs in several Kap121p-specific cargoes including Pho4p (Kaffman et al., 1998), Spo12p (Chaves and Blobel, 2001), Ste12p (Leslie et al., 2002), and Yap1p (Isoyama et al., 2001). We tested if the KBD of Nup53p could function in such a capacity by constructing a reporter protein consisting of the Nup53p KBD (residues 401–448) fused to GFP (KBD–GFP). When expressed in wild-type DF5 cells, the KBD–GFP concentrated in the nucleus (Fig. 4 B). Moreover, the import of this reporter was specifically mediated by Kap121p, as cells harboring a KAP121 temperature-sensitive (ts) allele (kap121–41) failed to import the KBD–GFP fusion (Fig. 4 B), whereas import in a strain lacking KAP123 exhibited no import defect (unpublished data).
Figure 4.
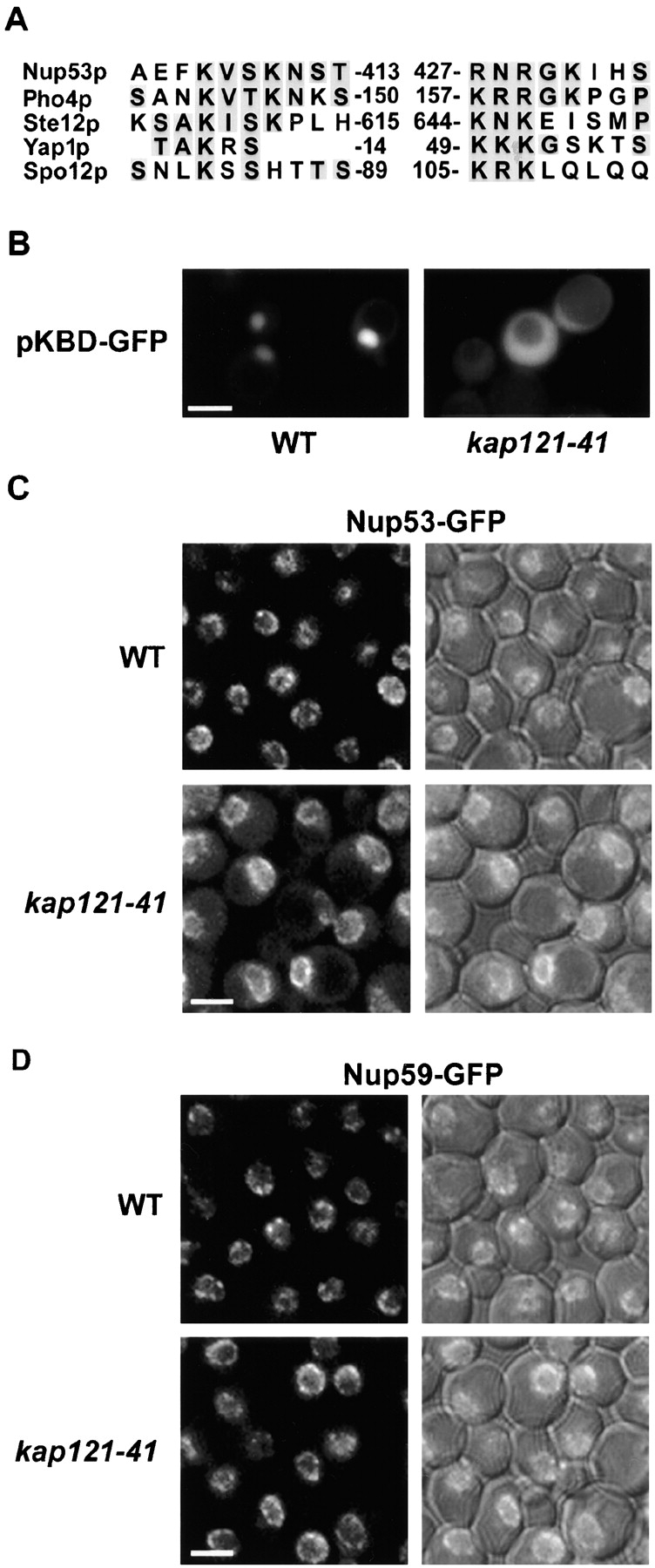
Kap121p functions in the efficient localization of Nup53p to the NPC. (A) Sequence alignment of the Nup53p KBD with NLS-containing regions of Pho4p, Ste12p, Yap1p, and Spo12p. Identical and similar amino acid residues in two or more sequences are shaded. Residue numbers are shown. Similarity between the NLSs was found to fall into two distinct regions with a spacer of variable length. (B) A plasmid expressing the coding region for the KBD of NUP53 fused to GFP (pKBD-GFP) was introduced into a haploid DF5 strain (WT) and the strain KP121–41 containing a temperature-sensitive allele of KAP121 (kap121–41). The distribution of KBD-GFP in these strains was monitored at 23°C by fluorescence microscopy. KP121–41 exhibits a constitutive import defect (Marelli et al., 1998). (C and D) The effect of the kap121–41 allele on the distribution of both Nup53-GFP and Nup59-GFP was examined in strains containing these fusion genes constructed by integrating the GFP ORF at the 3′ end of the chromosomal ORFs of the NUP53 and NUP59 genes in either DF5 (NP53GFP and NP59GFP) or the KP121–41 (NP53GFP/KP121–41 and NP59GFP/KP121–41) strains. 0.7-μm optical sections were acquired using a confocal microscope. Fluorescent and a combination of the fluorescent and differential interference contrast (DIC) images are shown. A cytoplasmic accumulation of Nup53-GFP is visible in the kap121–41–containing strain. Bars, 5 μm.
Our observation that the Nup53p KBD could be imported by Kap121p suggested that the interactions between these proteins were not restricted to events occurring at the NPC. Moreover, we have previously shown that overproduced Nup53p will enter the nucleus by a Kap121p-dependent mechanism (Marelli et al., 2001). We hypothesized that the Kap121p-mediated import machinery may play a role in the targeting of cytoplasmic Nup53p (arising either from newly synthesized Nup53p or by release from existing NPCs) to the NPC for its assembly into these structures. Several experiments were performed to test this potential mechanism. First, we tested whether a mutation in Kap121p would affect the ability of endogenous Nup53p to associate with NPCs. For these experiments, NUP53 was genomically tagged at the 3′ end of its ORF with the coding region of GFP in wild-type and kap121–41 ts strains. The Nup53-GFP in wild-type cells was visible in a defined punctate pattern along the nuclear periphery with no visible cytoplasmic staining (Fig. 4 C). However, in the strain lacking fully functional Kap121p (kap121–41) we observed, in addition to an NE signal, a clearly visible pool of cytoplasmic Nup53-GFP (Fig. 4 C). A similar result was also observed in the kap121–41 strain analyzed by immunofluorescence using antibodies specific for Nup53p (unpublished data). By comparison, a Nup59-GFP chimera was localized to the NPC in wild-type cells, and this localization was not altered in the kap121–41 strain (Fig. 4 D). The cytoplasmic pool of Nup53p in the kap121–41 strain could not be attributed to increased levels of Nup53p as determined by Western blot analysis (unpublished data). Rather, these results are consistent with the idea that Nup53p is inefficiently targeted to the NPC in the kap121–41 strain.
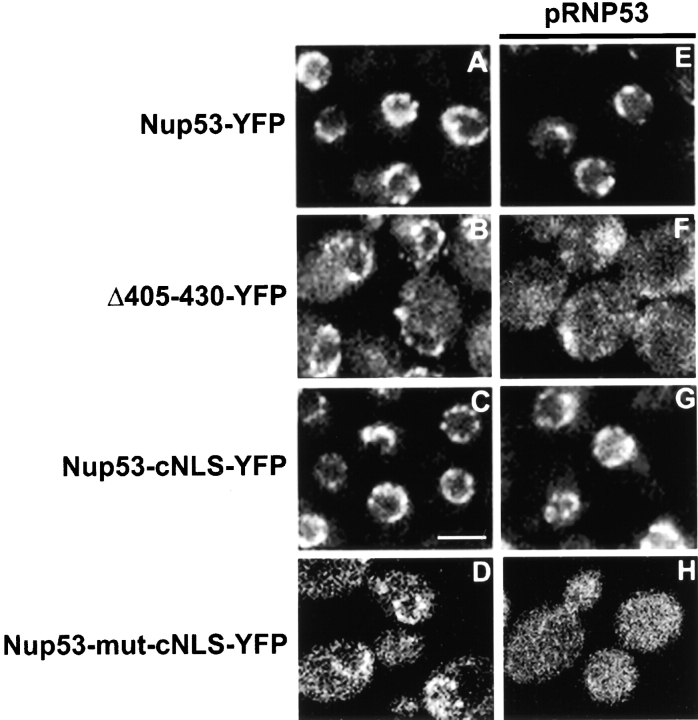
We tested what effects removing the KBD would have on the localization of Nup53p. To do this, an ORF encoding the Δ405–430 mutant COOH-terminally tagged with YFP (Δ405–430-YFP) was integrated at the NUP53 locus, replacing the wild-type ORF and placing the mutant construct under the control of the endogenous promoter. An examination of the localization of Δ405–430-YFP revealed that this protein was partially mislocalized to the cytoplasm in a pattern similar to that observed for Nup53p in the kap121–41 mutant (Fig. 5 B). By comparison, Nup53-YFP was visible exclusively at the nuclear rim in a punctate pattern (Fig. 5 A).
Figure 5.
Binding to a kap assists in the assembly of Nup53p into the NPC. (A–D) Strains were constructed where the ORF of one of the two NUP53 genes in the diploid DF5 strain was replaced with an ORF encoding either NUP53-YFP (NP53YFP), Δ405–430-YFP (Δ405–430-YFP), NUP53-cNLS-YFP (NP53-cNLS-YFP), or NUP53-mut-cNLS-YFP (NP53-mNLS-YFP). The distribution of the resulting fusion proteins was monitored by fluorescent confocal microscopy. (E–H) The effects of additional copies of wild-type NUP53 on the distribution of Nup53-YFP, Δ405–430-YFP, Nup53-cNLS-YFP, and Nup53-mut-cNLS-YFP were examined after introduction of the plasmid pRNP53 (CEN/URA3/NUP53). Strains were again examined using a confocal microscope. Increased levels of Nup53p abolish the binding of Δ405–430-YFP and Nup53-mut-cNLS-YFP to the NPC. Bar, 5 μm.
These data support the idea that the KBD of Nup53p and its recognition by Kap121p plays a role in the efficient localization of Nup53p to the NPC. However, it remained to be determined whether this reflects a unique property of the Kap121p–Nup53p complex or rather the general property of a kap targeting its cargo to the NPC. To address this question, we asked whether another kap could rescue the targeting of Nup53p to the NPC. To do this, we changed the specificity of the kap-binding site in Nup53p by replacing the KBD with an NLS (cNLS) recognized by the Kap60p–Kap95p heterodimer. Strikingly, the resulting construct was efficiently targeted to the NPC in a manner similar to wild-type Nup53p (Fig. 5 C). Moreover, the Nup53-cNLS-YFP construct was also efficiently targeted to the NPC in the kap121–41 ts strain (unpublished data). In contrast, a nonfunctional mutant cNLS did not rescue the targeting defect (Fig. 5 D).
It was also apparent from our results that even in the absence of the KBD, a portion of the Δ405–430-YFP associates with the NPC. Hence, the ability of Nup53p to associate with other nups in the NPC is not strictly dependent on its interactions with Kap121p. We predict that if its interaction with Kap121p facilitates Nup53p's association with other nups, the presence of a kap-binding domain should provide wild-type Nup53p with a competitive advantage over constructs lacking the KBD for assembly into the NPC. To determine whether this is the case, surplus copies of NUP53 were introduced by transformation with the plasmid pRNP53 into strains expressing NUP53-YFP, Δ405–430-YFP, NUP53-cNLS-YFP, and NUP53-mut-cNLS-YFP. As shown in Fig. 5 (E and G), the presence of wild-type Nup53p did not appreciably affect the association of Nup53-YFP or Nup53-cNLS-YFP with the NPC, but it blocked the binding of the Δ405–430-YFP and the Nup53-mut-cNLS-YFP constructs to the NPC (Fig. 5, F and H). These results demonstrated that the association of Nup53p with a kap played a distinct and advantageous role in the localization of Nup53p to the NPC.
The in vivo distribution of truncations of Nup53p
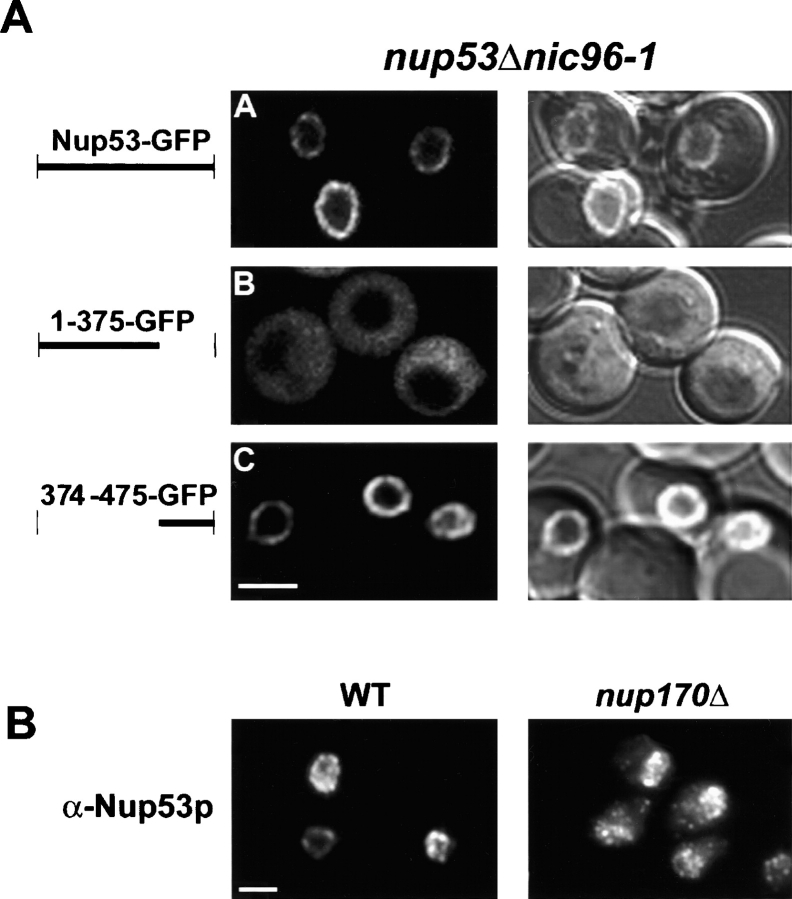
Once present within the context of the NPC, the steady-state association of Nup53p with this structure is determined by its association with nups. To define domains within Nup53p that mediate its association with other nups, we tested the ability of a series of Nup53p deletion mutants to associate with the NPC (Fig. 6). Each of the truncations and wild-type NUP53 were fused to the NH2 terminus of GFP in the plasmid pGFP-C-fus. Importantly, the localization of the chimeras was examined in strains lacking endogenous Nup53p (nup53Δ) to eliminate competition for binding sites at the NPC. In addition, these constructs were also examined in strains lacking the nucleoporin Nup120p. In the nup120Δ strain, NPCs cluster to focal points along the NE allowing NPC association to be distinguished from general NE localization. As shown in Fig. 6, Nup53-GFP was localized along the periphery of the nucleus in a punctate pattern in the nup53Δ strain and clustered to distinct foci in the nup53Δ nup120Δ strain (inset). A series of COOH-terminal deletions was examined, beginning with the removal of the last 27 amino acid residues of Nup53p. This 1–448-GFP construct, containing the KBD, exhibited an NPC-associated pattern similar to that of wild-type Nup53p (Fig. 6). A 1–375-GFP lacking the KBD also bound NPCs, albeit exhibiting a weaker NE signal and higher levels of cytoplasmic fluorescence. In contrast, the 1–206-GFP protein did not visibly associate with the nuclear periphery and was distributed throughout the cytoplasm. A construct containing residues 202–375 was also examined and did not associate with NPCs, suggesting this region was necessary, but not sufficient, for binding the NPC (unpublished data).
Figure 6.
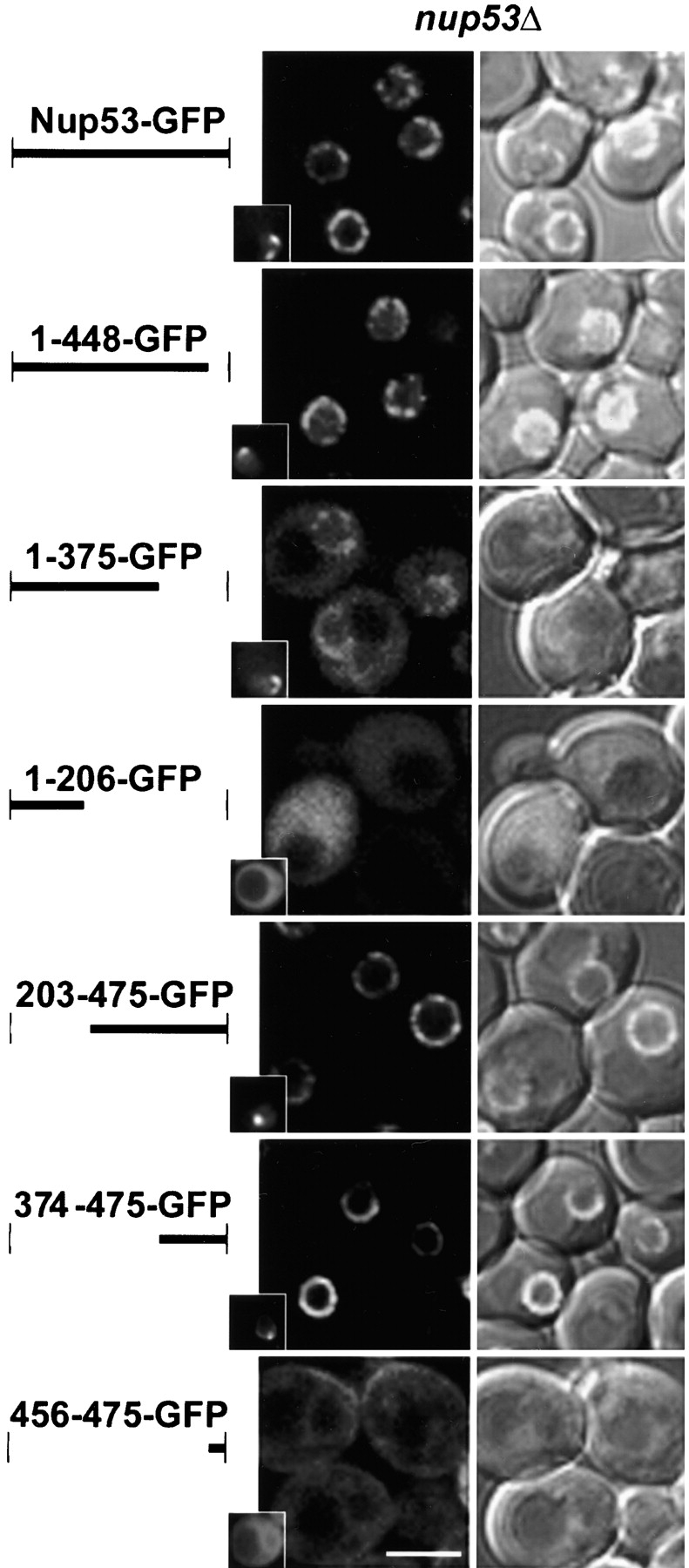
The in vivo localization of truncations of Nup53p. Plasmid-born copies of chimeric genes encoding full-length Nup53p and the indicated truncations fused to GFP were introduced into a haploid nup53Δ strain (NP53-A2) and a haploid nup53Δ nup120Δ strain where NPCs cluster (NP53/NP120–25–3). In each case, Western blot analysis showed that fusion proteins were of the predicted mass and were stable (not depicted). The cellular distribution of these chimeras was determined after growth to early-log phase at 23°C. Fluorescent and a combination of DIC and fluorescent confocal microscope images are shown as 0.7-μm sections for all constructs in the nup53Δ strain. Insets in panels in the left column show localization of the truncations in the nup53Δ nup120Δ strain imaged using a fluorescent microscope with a SPOT digital camera. Bar, 5 μm.
Several NH2-terminal deletions were also examined (Fig. 6). Most strikingly, both the 203–475-GFP and 374–475-GFP proteins containing the KBD showed the same localization patterns described above for wild-type Nup53p. However, when we further deleted the KBD, the resulting protein, 456–475-GFP, was observed in a diffuse staining pattern throughout the cytoplasm. In addition, 456–475-GFP also associated with the NE and plasma membrane, suggesting it was capable of binding multiple membrane systems in the cell. However, the NE membrane association does not appear to involve a stable interaction with the NPC, as the protein failed to cluster with NPCs in nup53Δ nup120Δ cells. Together, the deletion analysis suggests that two regions within Nup53p play a predominant role in its association with the NPC: one region lying between amino acid residues 1 and 375 and a second region, containing the KBD, between residues 374 and 475.
Mapping interactions between Nup53p and other nups
To investigate which nups interact with the two NPC binding regions of Nup53p, we examined the localization of both 1–375-GFP and 374–475-GFP in different nup mutants. A potential candidate was Nic96p because several pieces of data supported the idea that these two nups interact. Work from Fahrenkrog et al. (2000), as well as our unpublished data, showed that Nic96p is bound to Nup53p affinity purified from yeast extracts. Moreover, an interaction between these proteins was detected by two-hybrid analysis (Fahrenkrog et al., 2000; Uetz et al., 2000; unpublished data). On the basis of these observations, we examined the localization of Nup53-GFP, 1–375-GFP, and 374–475-GFP in a strain harboring a ts allele of NIC96, nic96–1 (Grandi et al., 1995), and lacking NUP53 (nup53Δ nic96–1). In this strain, the NPC association of 1–375-GFP was clearly reduced as compared with the nup53Δ strain (compare Fig. 7 A, panel B, with Fig. 6). In contrast, both Nup53-GFP and 374–475-GFP exhibited a strong nuclear peripheral signal indicating that the distribution of these proteins were not affected by the nic96–1 allele. These results suggest that Nic96p, either directly or indirectly, mediates the association of the 1–375 region of Nup53p with the NPC.
Figure 7.
Nic96p is required to properly localize the NH 2 terminus of Nup53p to the NPC. (A) The subcellular distributions of Nup53-GFP and two truncations of Nup53p (1–375-GFP and 374–475-GFP) were examined in a strain containing a ts allele of NIC96, nic96–1, and lacking endogenous NUP53 (NP53/nic96–1). Shown are fluorescent and a combination of fluorescent and DIC images obtained from 0.7-μm confocal sections of cells grown to mid-log phase at 23°C. Compare the distribution of 1–375-GFP to that of 1–375-GFP in the nup53Δ strain (Fig. 6). (B) Immunofluorescence microscopy using affinity-purified α-Nup53p antibodies was performed on W303 (WT) and nup170Δ (NP170–11.1) strains. Antibody binding was detected using Cy3-conjugated anti–rabbit antibodies. Bars, 5 μm.
We have postulated that Nup170p is also an important binding partner for Nup53p at the NPC. This was based on several observations, including the association of Nup170p with an isolated, Nup53p-containing subcomplex (Marelli et al., 1998) and the requirement for Nup170p in the targeting of overproduced Nup53p to the NPC (Marelli et al., 2001). Moreover, we have also observed that endogenous Nup53p is partially mislocalized in a nup170-null mutant (Fig. 7 B) in a pattern reminiscent of that observed in the kap121–41 ts mutant (Fig. 4 C). These results suggest that Nup170p plays a role in the deposition of Nup53p at the NPC. To further investigate the interactions between these nups, we mapped the region of Nup53p that binds to Nup170p using overlay assays. As shown in Fig. 8 A, Nup170-pA derived from yeast cell lysates bound to a COOH-terminal region of Nup53p (amino acid residues 374–475) containing the KBD, but did not bind to residues 1–375 and showed attenuated reactivity with the Δ405–430 construct. Consistent with these data, when the 374–475-GFP fusion protein was produced in cells lacking Nup170p (nup170Δ), it no longer associated with the NPC, but instead was mislocalized to the cytoplasm (Fig. 8 B). Together, these results suggest that Nup170p, either directly or indirectly, mediates the association of residues 374–475 of Nup53p with the NPC.
Figure 8.
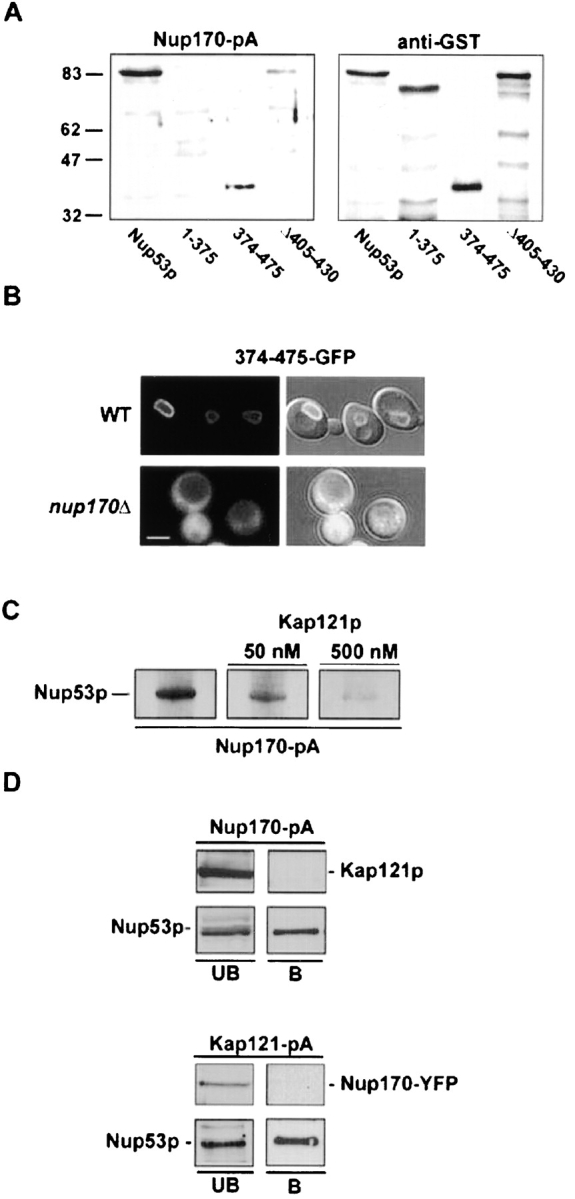
Nup170p and Kap121p interact with an overlapping region of Nup53p. (A) Nitrocellulose membranes containing SDS-PAGE–separated E. coli total cell lysates containing GST fusions of Nup53p and the indicated mutants were probed with either a cell lysate derived from a yeast strain expressing NUP170-pA (NP170pA) or an anti-GST antibody (α-GST). The positions of molecular mass markers of the indicated size in kilodaltons are shown on the left. (B) The subcellular distribution of the Nup170p binding domain of Nup53p was examined in WT (W303) and nup170Δ (NP170–11.1) strains using a 374–475-GFP fusion protein. Shown are confocal microscopy images acquired in both the fluorescent channel and a combination of the fluorescent and DIC images. (C) Equivalent amounts of GST–Nup53p in E. coli cell lysates were separated by SDS-PAGE, transferred to nitrocellulose and probed separately with an equal amount of a yeast total cell lysate containing Nup170-pA and supplemented with 0-, 50-, or 500-nM concentrations of recombinant Kap121p. The Nup170-pA fusion was detected using HRP-conjugated IgG antibodies and ECL. (D) Total cell lysates derived from yeast strains synthesizing Nup170-pA (NP170pA) or Kap121-pA (KP121pA/NP170YFP) were immunopurified on IgG–Sepharose columns. Shown are the unbound fraction (UB) and a fraction eluted from the beads with 1 M MgCl2 (B) analyzed by immunoblotting with α-Nup53p, α-GFP, or α-Kap121p antibodies to detect the indicated protein. Bar, 5 μm.
These results led us to investigate the possibility that Nup170p and Kap121p competed for the same binding site on Nup53p, and that this could provide a mechanism for releasing Nup53p from Kap121p at the NPC. To test this idea, overlay assays examining the binding of Nup170-pA to Nup53p were performed in the presence of recombinant Kap121p. As shown in Fig. 8 C, increasing concentrations of Kap121p (beginning at 50 nM) competed for the binding of Nup170-pA to Nup53p.
We concluded from these data that in vivo Nup53p might exist in two separate complexes, bound either to Nup170p or to Kap121p. In light of these results, our previous data showing that Nup170p and Kap121p copurified with Nup53-pA (Marelli et al., 1998) could represent a combination of separate Kap121p–Nup53p and Nup53p–Nup170p complexes. To test this, Nup170-pA and Kap121-pA were separately affinity-purified from yeast cells, and eluted fractions were examined by Western blotting (Fig. 8 D). Nup53p was detected in fractions eluted from both the Nup170-pA and Kap121-pA. In both cases, Nup53p was also detected in the unbound fraction. However, Nup170p was not bound to Kap121-pA, nor was Kap121p detected in fractions eluted from Nup170-pA.
Discussion
Our studies examining the specific interaction of Nup53p with Kap121p have uncovered a role for karyopherins in the targeting and assembly of nucleoporins into NPCs. We propose a model in which Kap121p, in addition to binding Nup53p at the NPC, can bind to a soluble, cytoplasmic pool of Nup53p, potentially derived from either newly synthesized Nup53p or from that released from existing NPCs, and target it to NPCs where it associates with other nups. The binding of Kap121p to Nup53p occurs through a region of the nup that lacks FG repeats (amino acid residues 401–448), but can function autonomously as an NLS (Fig. 4 B). However, it remains to be determined whether Nup53p shares the same binding site on Kap121p as the prototypical cargo NLSs or if it binds to a unique site.
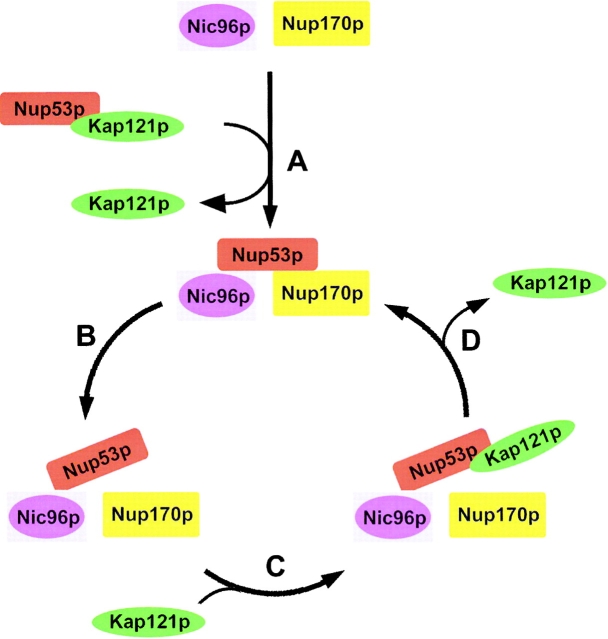
We propose that once bound to free Nup53p, Kap121p assists in its assembly into NPCs (see model in Fig. 9, step A). Although Kap121p is not strictly necessary for Nup53p to bind NPCs, it is clear that the efficient targeting of Nup53p to the NPC requires both functional Kap121p and the KBD of Nup53p. Moreover, the KBD imparts a distinct competitive advantage on Nup53p for integration into the NPC over mutant constructs lacking only the KBD (Fig. 5). One interpretation of these results is that Kap121p can act as a chaperone that maintains Nup53p in a conformation capable of binding more efficiently to its neighboring nups. Consistent with this idea, the competitive advantage provided by the KBD is not solely a property of this peptide, but rather appears to be related to its ability to bind a karyopherin. We show that the KBD can be replaced with a cNLS recognized by the Kap95p–Kap60p complex, and the resulting construct is assembled into NPCs as efficiently as wild-type Nup53p (Fig. 5).
Figure 9.
A model for the interactions of Nup53p with Kap121p at the NPC. (A) Kap121p interacts with a soluble, cytoplasmic pool of Nup53p and assists in its direct or indirect association with Nup170p and Nic96p at the NPC. This is accompanied by the release of Kap121p from Nup53p. (B) Nup53p's association with Nup170p is thought to be transient and it can dissociate from Nup170p. (C) Dissociation from Nup170p exposes the KBD of Nup53p, allowing it to act as a binding site for Kap121p. (D) Kap121p bound to Nup53p at the NPC could then be released by stochastic changes that allow Nup53p to bind Nup170p.
On the basis of these results, it seems likely that other kaps may perform similar assembly functions for other nups. One can envision that nups containing NLSs, and possibly other kap-binding sites such as FG repeats, could be recognized by kaps after their synthesis, and that these interactions play a role in their assembly into NPCs. However, this hypothesis is inherently difficult to test because unlike Nup53p, most of the kap-binding nups bind multiple kaps, and thus individual kap mutants are unlikely to cause assembly defects. This is the case for mammalian Nup153, which contains both FG repeats and an M9 NLS-like region (Nakielny et al., 1999). Nup153 appears to represent a class of mobile nups (Daigle et al., 2001), and it has been suggested that Kap-β2–transportin 1, by binding the M9 NLS-like region, plays a role in mediating its movement to the NPC (Nakielny et al., 1999). However, this potential function for Kap-β2–transportin 1 remains to be established. Other nups, including mammalian Nup98 (Fontoura et al., 2000), Tpr (Bangs et al., 1998), and yeast Nic96p (Grandi et al., 1995) also contain functional NLSs, and it will be of interest to determine their roles in NPC assembly and dynamics.
At the NPC, two domains of Nup53p are required for its steady-state association with the NPC. Deletion analysis mapped these binding sites within amino acid residues 1–375 and a region encompassing the KBD between residues 374 and 475. Residues 1–375 appear to interact either directly or indirectly with Nic96p. We observed that the 1–375 deletion construct failed to bind the NPC and was mislocalized to the cytoplasm in a strain containing a ts mutation in NIC96 (Fig. 7 A). This is in contrast to the 374–475 segment of Nup53p, whose localization to the NPC was not affected in the nic96–1 strain. The NPC localization of the 374–475 fragment was, however, strictly dependent on Nup170p (Fig. 8 B). This observation is consistent with in vitro mapping data showing that Nup170p binds to this region of Nup53p (Fig. 8 A). However, due to the nature of the overlay assays used to define these interactions, we cannot exclude the possibility that the association of Nup170p with this region of Nup53p is bridged by another nup. This could theoretically include a number of nups that interact with Nup53p including, potentially, itself. In the latter case, a Nup53p dimer would be predicted to form through the KBD-containing region; however, this seems unlikely because we have been unable to observe a specific interaction between Nup53-pA and the Nup53p truncations using the overlay assay (unpublished data). It also appears unlikely that two other nups known to interact with Nup53p (Nup59p and Nup157p) mediate this interaction, as mutants lacking these nups have no effect on the localization of Nup53p or its truncations (unpublished data).
Whether direct or indirect, the association of Nup170p with Nup53p occurs through a region that overlaps with the KBD. This overlap likely reveals an important step in the incorporation of Nup53p into the NPC. We propose that once the Kap121p–Nup53p complex enters the confines of the NPC, Nup53p's association with Nup170p drives the release of Nup53p from Kap121p (Fig. 9, step A). Consistent with this idea, Nup170p and Kap121p compete with one another for binding to Nup53p (Fig. 8 C). Moreover, the binding of Kap121p or Nup170p to Nup53p appears to be mutually exclusive. When purified from yeast extracts, Nup53p is bound to either Kap121p or Nup170p but not simultaneously to both (Fig. 8 D). We envision two possible scenarios that could explain the release of Nup53p from Kap121p and its association with Nup170p. First, the binding of the Kap121p–Nup53p complex to the NPC through other FG-containing nups could stimulate its dissociation, perhaps by a mechanism analogous to the accelerated release of import cargo from kaps observed upon binding to Nup1p, Nup2p, and Nup98 (Rexach and Blobel, 1995; Fontoura et al., 2000; Solsbacher et al., 2000; Gilchrist et al., 2002). Once released, Nup53p could then bind to multiple Nup170p sites within the NPC. Alternatively, the direct binding of Nup170p to Nup53p could alter its conformation and reduce its affinity for Kap121p, leading to the release of Kap121p. By either mechanism, once bound to Nup170p, Nup53p is predicted to exhibit a lower affinity for Kap121p.
What must be reconciled with this model is that NPC-associated Nup53p, through its KBD, acts as a binding site for Kap121p (Fig. 3), and thus some portion of NPC-bound Nup53p is predicted to be free from Nup170p. A clue to solving this conundrum may come from the results of previous work showing that Nup53p's association with Nup170p may be transient (Marelli et al., 2001). This is suggested by the observation that when overproduced, Nup53p is targeted to the NPC from where it is released, and then associates with the inner nuclear membrane. This process is dependent on Nup170p, but Nup170p does not accompany the excess Nup53p to the inner nuclear membrane. Our interpretation of these results is that Nup53p can readily dissociate from Nup170p (Fig. 9, step B). However, what would drive their dissociation is unclear. If the off rate is sufficient, normal stochastic changes in the NPC may allow Nup53p to fluctuate between two conformational states within the NPC. In one conformation, Nup53p would be free from Nup170p and capable of binding Kap121p. In the other, Nup53p would be bound to Nup170p and either unable to bind to, or exhibit a reduced affinity for, Kap121p. This again is consistent with our observation that Nup53p can be isolated in separate complexes with Kap121p or Nup170p (Fig. 8 D). In either state, we propose that Nup53p is also tethered to the NPC, likely through a mechanism requiring Nic96p. This interaction is unlikely to interfere with Nup53p's interaction with either Nup170p or Kap121p (Fig. 9, step C). Consistent with this idea, it is interesting to note that the same NIC96 mutant allele (nic96–1) that fails to interact with Nup53p also abolishes the nuclear rim localization of Kap121p (Damelin and Silver, 2000).
To assume different conformations while positioned within the NPC, a necessary property of Nup53p would likely be a flexible tertiary structure. This concept is supported by recent observations by Denning and co-workers (2002) showing that Nup2p, and in particular its FG repeat region, exists in a native unfolded structure. These data also lead the authors to propose that this may be a general property of the FG repeat nups.
The ability of Nup53p to exist in separate conformations has important implications for its role in Kap121p-mediated transport. A prediction of this model is that the release of Kap121p from its Nup53p binding site in the NPC can be facilitated by the transition of Nup53p to a Nup170p-bound state, allowing Kap121p to proceed to other nups or exit the NPC (Fig. 9, step D). This mechanism could occur rapidly and independently of RanGTP. However, it is also possible that RanGTP could facilitate the release of Kap121p. RanGTP can dissociate the Kap121p–Nup53p complex in vitro (Marelli et al., 1998), an effect that RanGTP has on a number of nup–import karyopherin complexes (for an extensive list see Allen et al., 2001). This includes nups that are positioned asymmetrically or symmetrically (such as Nup53p) on the cytoplasmic and nucleoplasmic faces of the NPC. However, it is unclear whether RanGTP performs this function throughout the NPC or whether it is restricted to the nucleoplasmic face.
A mechanism by which dynamic interactions between nups can modulate their affinity for kaps may also extend to other kap-binding nups including the FG-nups. In principle, once bound to a receptor nup, the kap could then be released by the interaction of the receptor nup with specific neighboring nups. This could be accomplished by the neighboring nups inducing a conformational change in the receptor nup that promotes a change in its affinity for the kap, or by direct competition for the kap-binding site. This idea may also partially explain the apparent discrepancy between the higher affinity interactions observed between nups and kaps in vitro (Allen et al., 2001; Ben-Efraim and Gerace, 2001) and the apparent requirement for low affinity interactions necessary to accommodate the rapid flux of kaps through the NPC (Ribbeck and Gorlich, 2001). Taken a step further, one can envision that similar types of binding sites (e.g., FG repeats) could exhibit different release rates depending on their position in the NPC and their neighboring nups.
Materials and methods
Yeast strains and media
All strains were grown at 30°C unless otherwise indicated in YPD (1% yeast extract, 2% bactopeptone, and 2% glucose) or in synthetic media (SM) supplemented with the necessary nutrients containing 2% glucose (Sherman et al., 1983). Yeast transformations were performed as described by Delorme (1989) and tetrad dissections as described in Sherman et al. (1983). The following strains were used in this study. DF5 (Mata ura3–52 his3-Δ200 trp1–1 leu2–3,112 lys2–801), W303 (Mata ade2–1 ura3–1 his3–11,15 trp1–1 leu2–3,112 can1–100), NP53-A2 and NP53/NP59–2.1 (Marelli et al., 1998), NP170pA (Aitchison et al., 1995b), KP121pA (Rout et al., 1997), KP121–41 (Leslie et al., 2002), NP170–11.1 (Aitchison et al., 1995b), NP120–25–3 (Aitchison et al., 1995a), and nic96–1 (Grandi et al., 1995; provided by Ed Hurt, University of Heidelberg, Heidelberg, Germany). NP53/nic96–1, NP53/NP120, and KP121pA/NP170YFP are segregants from crosses between NP53-A2 and either nic96–1 or NP120–25–3 and KP121pA with NP170YFP. All genomic integrations of the GFP and YFP ORFs were performed in a diploid DF5 strain or the strain KP121–41 to produce NP53GFP, NP59GFP, NP53GFP/KP121–41, NP59GFP/KP121–41, NP53YFP, Δ405–430-YFP, NP53-cNLS-YFP, NP53-mNLS-YFP, and NP170YFP. The genomic integrations of the GFP ORF were performed as described previously (Aitchison et al., 1995b) using the plasmid pGFP/HIS5 (Dilworth et al., 2001). A similar approach was used to produce strains containing YFP-tagged constructs using the plasmids pRNP53-YFP, pRΔ405–430-YFP, pRNP53-cNLS-YFP, pRNP53-mNLS-YFP, and pYFP/URA4 (a gift from Rick Rachubinski, University of Alberta, Edmonton, Alberta, Canada).
The synthesis of each of the GFP and YFP fusions was confirmed by immunoblotting total cell extracts using either anti-Nup53p, anti-Nup59p (Marelli et al., 1998), or anti-GFP antibodies (a gift of Mike Rout, The Rockefeller University, New York, NY).
Plasmids
All of the following plasmids contain PCR products amplified with the Expand High Fidelity PCR System (Roche) and primers containing BamHI linkers. The PCR products were inserted into the BamHI site of the indicated plasmid. The following NUP53 cDNAs, where 1 is the A of the initiation codon, were inserted into pGEX-4T-1 (Amersham Biosciences): pGNUP53 (1–1428), pG203–475 (607–1428), pG374–475 (1120–1428), and pG401–448 (1201–1344). The following NUP53 cDNAs containing a 5′ BamHI site and a 3′ blunt end were inserted into pGEX-3X (Amersham Biosciences) cut with BamHI and filled in EcoRI site: pG1–375 (1–1126), pG1–297 (1–892), and pG1–201 (1–604).
All of the following NUP53 cDNAs containing a 5′ BamHI site were subcloned into the BamHI site in pGFP-C-fus (Niedenthal et al., 1996): pNP53-GFP (−8 to 1428), p203–475-GFP (−3 to 3 and 607–1428), p374–475-GFP (−3 to 3 and 1120–1428), p456–475-GFP (−3 to 3 and 1366–1428), p1–448-GFP (−8 to 1343), p1–375-GFP (−8 to 1119), p1–206-GFP (−8 to 617), and pKBD-GFP (−3 to 3 and 1201–1343).
The deletion constructs of the NUP53 ORF lacking the KBD and containing an insertion of a cNLS and a mutant cNLS were made as follows. Antisense and sense oligonucleotides corresponding to nucleotides +1199–1215 and +1299–1318, respectively, of NUP53 were designed with 5′ BglII linkers. These primers were used in a PCR to synthesize a linear copy of the pNP53-GFP plasmid lacking the coding region for aa residues 405–430. This cDNA was digested with BglII and ligated to produce the pΔ405–430-GFP plasmid. The plasmids pNP53-cNLS-GFP and pNP53-mNLS-GFP were constructed by inserting the coding region of the SV-40 large T antigen NLS (cNLS) or a mutant of the cNLS (aa residues HSTPPKKKRKVEDPK or HSTPPKTKRKVEDPK; signal in bold) into the BglII site of pΔ405–430-GFP.
The plasmids pRNP53-YFP, pRΔ405–430-YFP, pRNP53-cNLS-YFP, and pRNP53-mNLS-YFP were constructed as follows: The EYFP ORF was amplified by the PCR from pEYFP (CLONTECH Laboratories, Inc.) with primers containing BamHI and NotI linkers and directionally cloned into pRS306 (URA3; Sikorski and Hieter, 1989) to produce pRYFP. The regions coding for NUP53, Δ405–430, NUP53-cNLS, and NUP53-mut-cNLS were subsequently inserted into pRYFP creating pRNP53-YFP, pRΔ405–430-YFP, pRNP53-cNLS-YFP, and pRNP53-mNLS-YFP.
pYNP53 is pCUP1-NUP53 from Marelli et al., 2001). pΔ405–430 is subcloned into pYEX-BX (CLONTECH Laboratories, Inc.).
Overlay and in vitro binding assays
Total cell lysates were prepared for SDS-PAGE from E. coli BLR 21 cells synthesizing the indicated GST fusion proteins. Cells were grown to an OD600 of 0.5 and induced with 1 mM IPTG for 3 h at 37°C. Cell pellets were solubilized in sample buffer and proteins were separated by SDS-PAGE and transferred to nitrocellulose. The membranes were then blocked and probed with a cytosolic fraction isolated from yeast cells synthesizing a Kap121-pA fusion as described previously (Marelli et al., 1998). The bound pA was detected with HRP-conjugated donkey anti–rabbit antibodies and ECL (Amersham Biosciences).
Overlay assays using Nup170-pA derived from yeast lysates were performed using a similar procedure. A yeast strain synthesizing Nup170-pA (NP170pA) was grown to an OD600 of 1.0, harvested by centrifugation, and lysed in 15 ml of 50 mM Hepes, pH 7.5, 110 mM potassium acetate, 2 mM MgCl2, 0.2 mM PMSF, 2 μg/ml leupeptin, 2 μg/ml aprotinin, and 0.4 μg/ml pepstatin A using a French Press. Triton X-100 was then added to a final concentration of 1% and incubated on ice for 10 min. The lysate was then clarified by centrifugation at 11,300 g for 15 min at 4°C. The supernatant fraction was diluted twofold in lysis buffer and used to probe nitrocellulose membranes. For Nup170-pA overlay experiments performed in the presence of recombinant Kap121p, clarified Nup170-pA lysates were supplemented with purified Kap121p (Marelli et al., 1998) to a final concentration of 0, 50, or 500 nM before probing membranes.
Solution binding assays, performed using recombinant Kap121p and the KBD-containing region of Nup53p, were conducted as follows. GST and GST-401–448 were synthesized in E. coli and purified as described by the manufacturer (Amersham Biosciences). 5 μg of purified GST or GST-401–448 was bound to 10 μl of Glutathione Sepharose 4B beads (Amersham Biosciences). Beads were then incubated with ∼10 μg of purified Kap121p in 50 μl PBS for 1 h at 4°C. The beads were washed extensively with PBS and proteins were eluted with SDS-PAGE sample buffer.
Fluorescence microscopy
GFP and YFP fusion proteins were visualized in live cells with either a fluorescence microscope (BX-50; Olympus) equipped with a SPOT digital camera or a confocal microscope (LSM 510; Carl Zeiss MicroImaging, Inc.). Before viewing, yeast strains were grown under the following conditions: strains harboring the plasmid pPS1069 (KAP121-GFP [TRP1]; Seedorf and Silver, 1997; provided by M. Seedorf and P. Silver, Dana Farber Cancer Institute, Boston, MA) were grown in SM lacking tryptophan to early-log phase. Genomically tagged strains were grown in YPD to mid-log phase. Truncations of Nup53p in the pGFP-C-fus vectors were grown in SM lacking uracil to early-log phase. It should be noted that the ORFs cloned into the pGFP-C-fus vector are under the control of the MET25 promoter. However, for this work, all media contained methionine because the promoter's basal level of expression was close to that observed from NUP53's endogenous promoter.
Immunofluorescence microscopy was performed as described in Marelli et al. (2001) using affinity-purified polyclonal rabbit anti-Nup53p antibodies (Marelli et al., 1998) and Cy3-conjugated, donkey anti–rabbit secondary antibodies (Jackson ImmunoResearch Laboratories).
Immunoprecipitations and immunoblotting
250-ml cultures of yeast strains synthesizing either Kap121-pA (KP121pA/NP170YFP) or Nup170-pA (NP170pA) were grown to an OD600 of 1.0. All steps were performed at 4°C. The cells were harvested by centrifugation and resuspended in 12 ml of lysis buffer (20 mM Na2HPO4, pH 7.5, 150 mM NaCl, 0.1 mM MgCl2, 0.2 mM PMSF, 2 μg/ml leupeptin, 2 μg/ml aprotinin, and 0.4 μg/ml pepstatin A). Cells were lysed with a French Press followed by the addition of 12 ml lysis buffer containing 40% DMSO and 2% Triton X-100. Lysates were cleared by consecutive centrifugations at 11,300 g for 20 min and 311,000 g for 45 min. The supernatant was incubated overnight with 50-μl IgG Sepharose beads (Amersham Biosciences), washed extensively with wash buffer, and eluted with 1 M MgCl2.
Immunoprecipitations of Nup53-GFP and Δ405–430-GFP were performed as follows. A nup53Δ strain containing either pNP53-GFP or pΔ405–430-GFP was grown and lysates prepared as described above. Supernatants were incubated overnight with anti–Nup53p pAbs, followed by a 1-h incubation with 50 μl of Protein G–Sepharose beads (Amersham Biosciences). The beads were collected, washed, and eluted with a step gradient of 50, 200, and 500 mM MgCl2, followed by 0.5 M acetic acid, pH 3.4. Proteins were analyzed by Western blotting.
Western blots were performed as follows. Post-transfer nitrocellulose membranes were blocked with 5% skim milk powder in PBS containing 0.1% Tween 20. GST, Nup53p, Nup170-YFP, and Kap121p were detected using specific rabbit pAbs. The anti-GFP antibody was used to detect Nup170-YFP. Secondary antibodies were HRP-conjugated, donkey anti–rabbit (Amersham Biosciences). Protein A chimeras were detected using a rabbit IgG (ICN Biomedicals) followed by the HRP-conjugated, donkey anti–rabbit antibodies. Binding was visualized using ECL as described by the manufacturer (Amersham Biosciences).
Sequence alignments
Alignments of the various NLS sequences indicated were performed online at the European Bioinformatics Institute (http://www.ebi.ac.uk/) using ClustalW.
Acknowledgments
We thank David Goldfarb for valuable criticisms of the manuscript and Deena Leslie and members of the Wozniak laboratory for important discussions. We are also grateful for all those cited in the text for reagents, and Honey Chan for assistance with the confocal microscope.
Funding for this work was provided by the Canadian Institute for Health Research and the Alberta Heritage Foundation for Medical Research.
Footnotes
Abbreviations used in this paper: FG, Phe-Gly; kap, karyopherin; KBD, Kap121p binding domain; NE, nuclear envelope; NES, nuclear export signal; NPC, nuclear pore complex; pA, protein A; ts, temperature sensitive.
References
- Aitchison, J.D., G. Blobel, and M.P. Rout. 1995. a. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J. Cell Biol. 131:1659–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison, J.D., M.P. Rout, M. Marelli, G. Blobel, and R.W. Wozniak. 1995. b. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131:1133–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, N.P., L. Huang, A. Burlingame, and M. Rexach. 2001. Proteomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276:29268–29274. [DOI] [PubMed] [Google Scholar]
- Allen, T.D., J.M. Cronshaw, S. Bagley, E. Kiseleva, and M.W. Goldberg. 2000. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J. Cell Sci. 113:1651–1659. [DOI] [PubMed] [Google Scholar]
- Bangs, P., B. Burke, C. Powers, R. Craig, A. Purohit, and S. Doxsey. 1998. Functional analysis of Tpr: identification of nuclear pore complex association and nuclear localization domains and a role in mRNA export. J. Cell Biol. 143:1801–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss, R., T. Littlewood, and M. Stewart. 2000. Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell. 102:99–108. [DOI] [PubMed] [Google Scholar]
- Bayliss, R., S.W. Leung, R.P. Baker, B.B. Quimby, A.H. Corbett, and M. Stewart. 2002. Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J. 21:2843–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim, I., and L. Gerace. 2001. Gradient of increasing affinity of importin β for nucleoporins along the pathway of nuclear import. J. Cell Biol. 152:411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves, S.R., and G. Blobel. 2001. Nuclear import of Spo12p, a protein essential for meiosis. J. Biol. Chem. 276:17712–17717. [DOI] [PubMed] [Google Scholar]
- Daigle, N., J. Beaudouin, L. Hartnell, G. Imreh, E. Hallberg, J. Lippincott-Schwartz, and J. Ellenberg. 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damelin, M., and P.A. Silver. 2000. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol. Cell. 5:133–140. [DOI] [PubMed] [Google Scholar]
- Delorme, E. 1989. Transformation of Saccharomyces cerevisiae by electroporation. Appl. Environ. Microbiol. 55:2242–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning, D.P., V. Uversky, S.S. Patel, A.L. Fink, and M. Rexach. 2002. The Saccharomyces cerevisiae nucleoporin Nup2p is a natively unfolded protein. J. Biol. Chem. 277:33447–33455. [DOI] [PubMed] [Google Scholar]
- Dilworth, D.J., A. Suprapto, J.C. Padovan, B.T. Chait, R.W. Wozniak, M.P. Rout, and J.D. Aitchison. 2001. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 153:1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog, B., E.C. Hurt, U. Aebi, and N. Pante. 1998. Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes. J. Cell Biol. 143:577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog, B., W. Hubner, A. Mandinova, N. Pante, W. Keller, and U. Aebi. 2000. The yeast nucleoporin Nup53p specifically interacts with Nic96p and is directly involved in nuclear protein import. Mol. Biol. Cell. 11:3885–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura, B.M., G. Blobel, and N.R. Yaseen. 2000. The nucleoporin Nup98 is a site for GDP/GTP exchange on ran and termination of karyopherin β2-mediated nuclear import. J. Biol. Chem. 275:31289–31296. [DOI] [PubMed] [Google Scholar]
- Fribourg, S., I.C. Braun, E. Izaurralde, and E. Conti. 2001. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell. 8:645–656. [DOI] [PubMed] [Google Scholar]
- Gilchrist, D., B. Mykytka, and M. Rexach. 2002. Accelerating the rate of disassembly of karyopherin-cargo complexes. J. Biol. Chem. 277:18161–18172. [DOI] [PubMed] [Google Scholar]
- Grandi, P., N. Schlaich, H. Tekotte, and E.C. Hurt. 1995. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J. 14:76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoyama, T., A. Murayama, A. Nomoto, and S. Kuge. 2001. Nuclear import of the yeast AP-1-like transcription factor Yap1p is mediated by transport receptor Pse1p, and this import step is not affected by oxidative stress. J. Biol. Chem. 276:21863–21869. [DOI] [PubMed] [Google Scholar]
- Kaffman, A., N.M. Rank, and E.K. O'Shea. 1998. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 12:2673–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, D.M., B. Grill, M.P. Rout, R.W. Wozniak, and J.D. Aitchison. 2002. Kap121p-mediated nuclear import is required for mating and cellular differentiation in yeast. Mol. Cell. Biol. 22:2544–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara, I.G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli, M., J.D. Aitchison, and R.W. Wozniak. 1998. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143:1813–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli, M., C.P. Lusk, H. Chan, J.D. Aitchison, and R.W. Wozniak. 2001. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 153:709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny, S., S. Shaikh, B. Burke, and G. Dreyfuss. 1999. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 18:1982–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass, U., M.P. Rout, S. Maguire, G. Blobel, and R.W. Wozniak. 1996. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J. Cell Biol. 133:1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedenthal, R.K., L. Riles, M. Johnston, and J.H. Hegemann. 1996. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 12:773–786. [DOI] [PubMed] [Google Scholar]
- Rexach, M., and G. Blobel. 1995. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 83:683–692. [DOI] [PubMed] [Google Scholar]
- Ribbeck, K., and D. Gorlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck, K., and D. Gorlich. 2002. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21:2664–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., and J.D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593–16596. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., G. Blobel, and J.D. Aitchison. 1997. A distinct nuclear import pathway used by ribosomal proteins. Cell. 89:715–725. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., J.D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B.T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, K.J., and S.R. Wente. 2000. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12:361–371. [DOI] [PubMed] [Google Scholar]
- Seedorf, M., and P.A. Silver. 1997. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc. Natl. Acad. Sci. USA. 94:8590–8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1983. Methods In Yeast Genetics: Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 120 pp.
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher, J., P. Maurer, F. Vogel, and G. Schlenstedt. 2000. Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin α. Mol. Cell. Biol. 20:8468–8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom, A.C., and K. Weis. 2001. Importin-beta-like nuclear transport receptors. Genome Biol. 2:REVIEWS3008. [DOI] [PMC free article] [PubMed]
- Uetz, P., L. Giot, G. Cagney, T.A. Mansfield, R.S. Judson, J.R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 403:623–627. [DOI] [PubMed] [Google Scholar]
- Winey, M., D. Yarar, T.H. Giddings, Jr., and D.N. Mastronarde. 1997. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes. Mol. Biol. Cell. 8:2119–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak, R.W., M.P. Rout, and J.D. Aitchison. 1998. Karyopherins and kissing cousins. Trends Cell Biol. 8:184–188. [DOI] [PubMed] [Google Scholar]