Abstract
Îndirect evidence suggests that p120-catenin (p120) can both positively and negatively affect cadherin adhesiveness. Here we show that the p120 gene is mutated in SW48 cells, and that the cadherin adhesion system is impaired as a direct consequence of p120 insufficiency. Restoring normal levels of p120 caused a striking reversion from poorly differentiated to cobblestone-like epithelial morphology, indicating a crucial role for p120 in reactivation of E-cadherin function. The rescue efficiency was enhanced by increased levels of p120, and reduced by the presence of the phosphorylation domain, a region previously postulated to confer negative regulation. Surprisingly, the rescue was associated with substantially increased levels of E-cadherin. E-cadherin mRNA levels were unaffected by p120 expression, but E-cadherin half-life was more than doubled. Direct p120–E-cadherin interaction was crucial, as p120 deletion analysis revealed a perfect correlation between E-cadherin binding and rescue of epithelial morphology. Interestingly, the epithelial morphology could also be rescued by forced expression of either WT E-cadherin or a p120-uncoupled mutant. Thus, the effects of uncoupling p120 from E-cadherin can be at least partially overcome by artificially maintaining high levels of cadherin expression. These data reveal a cooperative interaction between p120 and E-cadherin and a novel role for p120 that is likely indispensable in normal cells.
Keywords: p120ctn; p120; cadherin; catenin; SW48
Introduction
p120-catenin (p120)*is a member of the Armadillo (ARM)/β-catenin gene family (Reynolds et al., 1992; Peifer et al., 1994), and the prototypical member of the p120 subfamily (for review see Anastasiadis and Reynolds, 2000). Originally described as a substrate for the Src oncoprotein (Reynolds et al., 1989, 1992) and various receptor tyrosine kinases (Downing and Reynolds, 1991; Kanner et al., 1991), p120 interacts with the cytoplasmic domain of classical cadherins (Reynolds, 1994; Shibamoto et al., 1995; Staddon et al., 1995). Cadherins are both necessary and sufficient for the targeting of p120 to cell–cell junctions, indicating that cadherins are the only cellular proteins capable of recruiting p120 to membranes (Thoreson et al., 2000). The p120 binding site in the juxtamembrane domain (JMD) of cadherins is the most highly conserved region among members of the cadherin superfamily (Nollet et al., 2000), suggesting a general and indispensable role for the cadherin-p120 interaction.
Cadherins comprise a superfamily of transmembrane cell–cell adhesion receptors involved in many aspects of development, morphogenesis, and tumor malignancy (for reviews see Nollet et al., 1999; Takeichi, 1995; Yap, 1998). Extracellular domains of identical cadherins interact in a homophilic, Ca2+-dependent fashion to form adherens junctions between adjacent cells. The cytoplasmic domains interact with the catenins, which physically connect and/or regulate the interaction of the cadherin complex with the actin cytoskeleton. β-Catenin and p120 bind through their Armadillo repeat domains to the catenin binding domain and JMD of cadherins, respectively (Aberle et al., 1994; Hulsken et al., 1994; Funayama et al., 1995; Jou et al., 1995; Navarro et al., 1998; Yap et al., 1998). Compared with β-catenin, p120 is rather loosely associated with the cadherin complex (Thoreson et al., 2000), a property that may be important for its mechanism of action (Anastasiadis and Reynolds, 2001).
The role of p120 is controversial. Evidence suggests that p120 can both positively and negatively regulate cadherin activity, probably depending on signaling events that have yet to be clearly identified. JMD-deleted cadherins have been used to assay cadherin function in the absence of p120 binding. These studies suggest roles for the cadherin JMD, and by implication p120, in developmental events (Kintner, 1992; Horikawa and Takeichi, 2001), neuronal outgrowth (Riehl et al., 1996; Lilien et al., 1999), exclusion of one cadherin from junctions by another (Navarro et al., 1998), suppression of cell motility (Chen et al., 1997), cadherin clustering (Yap et al., 1998), and cadherin adhesiveness (Yap et al., 1998; Thoreson et al., 2000). Minimal p120-uncoupling mutations (e.g., triple alanine substitutions) have been used to minimize the risk of affecting non-p120 interactions (Thoreson et al., 2000). Nonetheless, novel JMD binding partners have been identified which are uncoupled by the same mutations that eliminate p120 binding. Both Hakai (Fujita et al., 2002) and presenilin (PS)-1 associate with the JMD (Baki et al., 2001; Marambaud et al., 2002), and have been implicated in mechanisms that down-regulate cadherin levels. The latter observations complicate the interpretation of p120's contribution to the results of JMD-deletion experiments.
p120 function has been more directly addressed in other systems. For example, in Colo205 cells, E-cadherin function is severely impaired despite the cells having normal levels of E-cadherin and catenins. Adhesion can be restored by p120 mutants lacking most of the NH2-terminal region, but not by full-length p120 (Aono et al., 1999). This result implies an aberrant signaling pathway in Colo205 cells that acts constitutively through an NH2-terminal region of p120 to block E-cadherin function. Although aspects of these data have been questioned recently (Horikawa and Takeichi, 2001), the ability of NH2-terminally truncated p120 to restore adhesiveness in these cells is clear. Thus, full-length p120 appears to negatively regulate adhesion in Colo205 cells.
p120 contains a 350-aa NH2-terminal region, followed by the ARM domain (i.e., ten ARM repeats), and a short COOH terminus. The NH2-terminal region immediately adjacent to the ARM domain encompasses the 100-aa phosphorylation domain which contains the majority of the p120 tyrosine phosphorylation sites (Mariner et al., 2001) and is probably an important regulator of p120 function. Most cells simultaneously express multiple p120 isoforms derived from alternative splicing. Splicing in the NH2-terminal region gives rise to four alternative start codons (Keirsebilck et al., 1998). Isoforms using start codons 1 (p120ctn1) and 3 (p120ctn3) are observed most frequently (Mo and Reynolds, 1996; Keirsebilck et al., 1998) and retain both the phosphorylation domain and the ARM domain. The fourth start codon gives rise to isoform 4 (p120ctn4), which retains the ARM domain but eliminates the NH2-terminal region, along with its phosphorylation domain. In contrast to other isoforms, p120ctn4 mRNA has been detected, but is rarely expressed as a protein. It is used in our study as a p120 mutant lacking the NH2 terminus.
E-cadherin is well established as a tumor and metastasis suppressor. Expression of E-cadherin is frequently down-regulated or extinguished in malignancy, an event strongly correlated with poor prognosis (for review see Birchmeier and Behrens, 1994; Nollet et al., 1999). Evidence both in vivo (Perl et al., 1998) and in vitro (Frixen et al., 1991; Vleminckx et al., 1991), link E-cadherin loss with the transition to metastasis during tumor progression (for review see Yap, 1998). Interestingly, a number of recent studies indicate p120 expression is also frequently lost in tumors of the breast, prostate, colon, stomach, and bladder (for review see Thoreson and Reynolds, 2002). In many cases, p120 loss is associated with poor prognosis, suggesting that alterations in p120 expression may be important in cancer.
Despite observations of p120 loss in tumors, mutation or loss of p120 in carcinoma cell lines has not been reported. Here, we show that the p120 gene is mutated in SW48 cells and that the cadherin adhesion system is impaired as a direct consequence of p120 insufficiency. p120 is apparently essential in normal epithelial cells to directly or indirectly stabilize E-cadherin, thereby explaining its positive role in regulating E-cadherin adhesiveness.
Results
Characterization of the cadherin complex in colorectal SW48 cells
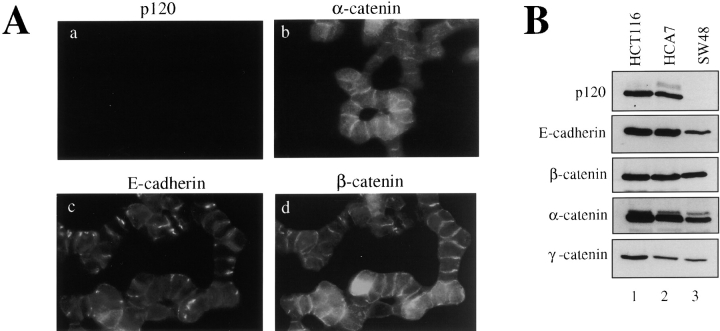
SW48 colon carcinoma cells have lost the ability to establish normal epithelial morphology. The cells form junctions, but line up end to end in loosely organized arrays that are unable to pull together into compact epithelial colonies (Fig. 1). To determine the cause of the defect, we examined the major components of the E-cadherin complex by immunofluorescence (Fig. 1 A) and by Western blotting (Fig. 1 B). Immunofluorescent analysis revealed E-cadherin staining that colocalized with α- and β-catenins at cell–cell contacts (Fig. 1 A, b–d). We initially attempted to localize p120 using well characterized monoclonal antibodies directed against the COOH-terminal epitopes (e.g., mAb's pp120, 15D2, 12F4) found on all known p120 isoforms (Wu et al., 1998). Interestingly, p120 staining was completely absent (Fig. 1 B, a). To date, this is the first observation of an adhesive cell line that does not stain with these antibodies. Consistent with these observations, analysis of SW48 cells by Western blotting showed normal levels of α-, β-, and γ-catenins (Fig. 1 B). E-cadherin levels were reduced, but significantly, p120 was not detected by mAb pp120. Although β-catenin levels are normally reduced in cells where E-cadherin expression is low, the β-catenin in SW48 cells contains a stabilizing mutation at codon 33 (Ilyas et al., 1997). This alteration probably accounts for the unexpectedly high β-catenin levels and the relatively high levels of cytoplasmic staining for both α- and β-catenins.
Figure 1.
Characterization of the E-cadherin complex in SW48 carcinoma cells. (A) Members of the cadherin complex were localized in SW48 cells by immunofluorescence. SW48 cells organize aberrantly into linear arrays and do not form compact epithelial colonies. p120 was localized with mAb pp120, which binds a COOH-terminal epitope present in all known p120 isoforms. (B) Cadherin complex proteins were analyzed by Western blotting of NP-40 cell lysates derived from the colon carcinoma cell lines HCT116, HCA7, and SW48. p120 is not detected in SW48 cells by mAb pp120, indicating that the COOH terminus is absent.
Mutations and splicing defects of p120 alleles in SW48 cells
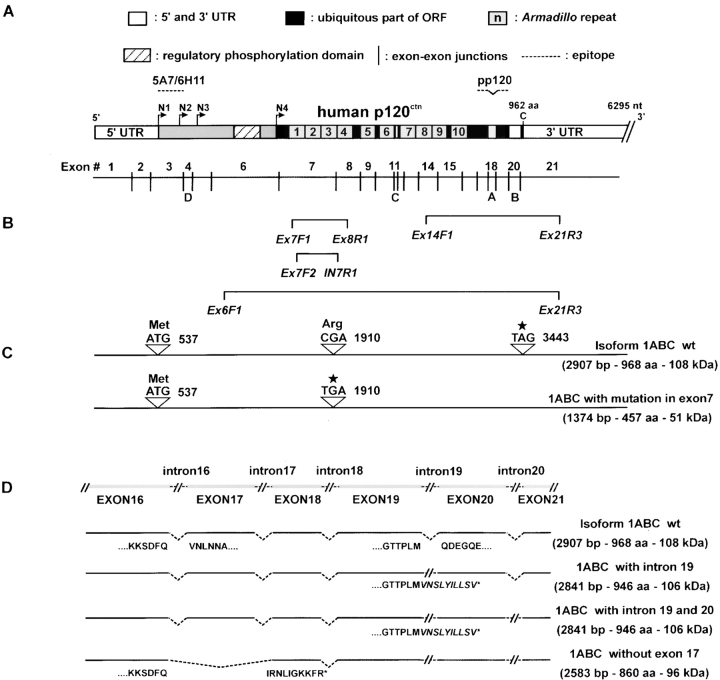
The failure to detect p120 with mAb pp120 suggested possible mutations in the p120 gene. However, no alterations in the genomic organization of p120 were detected by Southern blotting. To determine if mutations were present, p120 genomic DNA from SW48 cells was sequenced by a previously established method (Keirsebilck et al., 1998). In addition, p120 cDNA clones were generated by RT-PCR from SW48 RNA. Both genomic and cloned cDNA sequences revealed a heterozygous nonsense mutation in exon 7 at nucleotide 1908 (Fig. 2 C). This C to T mutation yields a premature stop codon that truncates the protein in the third ARM repeat.
Figure 2.
Characterization of mutated p120 alleles in SW48 cells. (A) Schematic diagram of human p120 protein and cDNA structure. N1–N4 are the four alternatively used start codons. A, B, C, and D, are alternatively spliced exons. Individual ARM repeats are numbered. Note the phosphorylation domain between N3 and N4, immediately NH2-terminal to the ARM domain. Approximate epitopes of mAbs are indicated. (B) Primer sets used for RT-PCR and genomic DNA analysis. (C) A nonsense mutation CGA to TGA in exon 7 is indicated by the C to T mutation detected in genomic DNA and cDNA clones. The normal ATG and TAG codons are indicated. (D) Aberrant mRNA forms detected by RT-PCR. The exon structure of p120 cDNA COOH-terminal end is shown across the top. Aberrant mRNAs, each resulting in premature stop codons, are located below. Aberrant splicing in the 3′-terminal part of p120 mRNA results in the indicated gene products, which include retention of intron 19, retention of both introns 19 and 20, or exclusion of exon 17.
An exact molecular defect associated with the other allele could not be identified. However, RT-PCR analysis of SW48 mRNA using p120-specific oligonucleotide primers (Fig. 2 B) revealed several cDNA abnormalities, including deletion of exon 17, retention of intron 19, or retention of both introns 19 and 20 (Fig. 2 D). Long-range RT-PCR indicated that these COOH-terminal changes existed in both the 1908 C/T and the 1908 WT alleles. All of these alterations result in premature stop codons that eliminate the normal p120 COOH terminus.
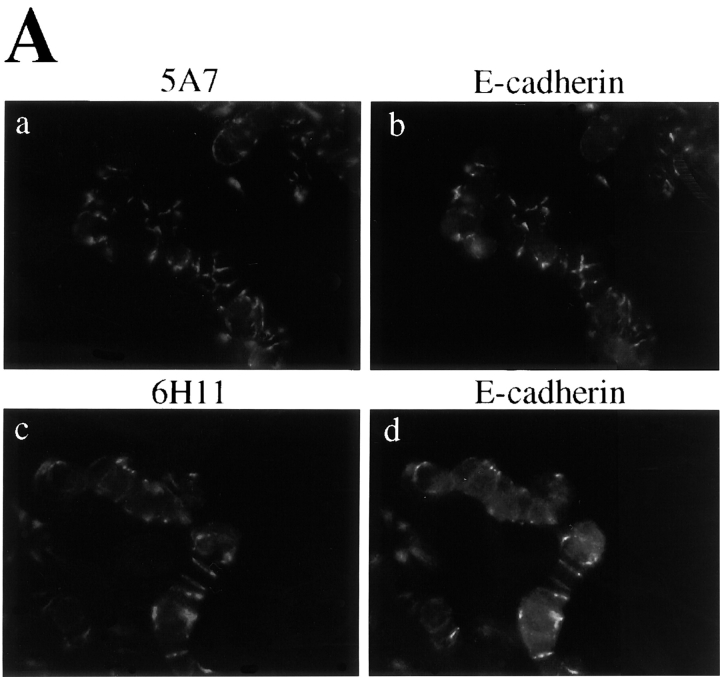
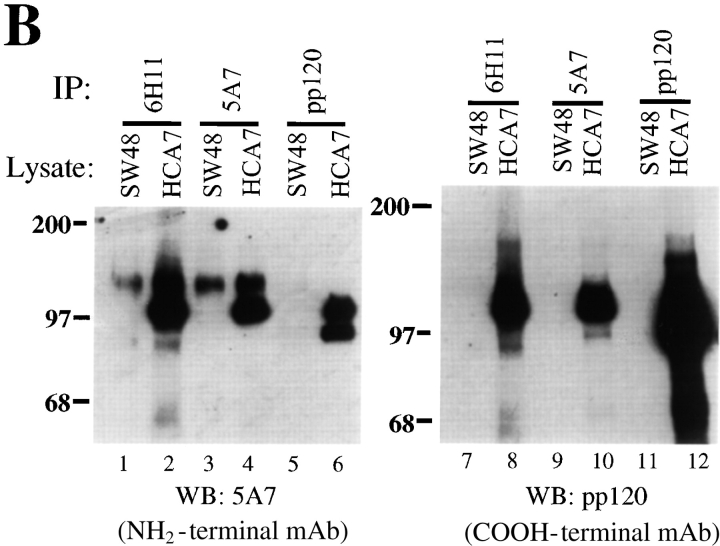
We then reevaluated p120 expression in the SW48 cells using monoclonal antibodies 5A7 and 6H11, which recognize different NH2-terminal epitopes (Fig. 3). Compared with other cell lines, these antibodies revealed weak and extremely punctate junctional staining (Fig. 3 A), indicating that p120 was not completely absent in SW48 cells. Immunoprecipitation and Western blotting with mAbs 5A7 and 6H11 revealed a near full-length p120 protein, and no other bands (Fig. 3 B, lanes 1 and 3). However, the very long exposure times required to detect the mutated p120 in these experiments indicate that it is grossly underexpressed. p120 remained undetectable by mAb pp120 (Fig. 3 B, lanes 7 and 9), confirming that the normal COOH terminus of p120 is absent.
Figure 3.
Antibodies against the NH2 terminus of p120 detect extremely low levels of p120 in SW48 cells. (A) p120 localization with NH2-terminally directed p120 mAbs. SW48 cells were costained with p120 mAb's 5A7 or 6H11 (a and c), and E-cadherin mAb C-20820 (b and d). (B) Quantitative analysis of p120 expression was performed by immunoprecipitating p120 from RIPA cell lysates with either NH2-terminally directed mAbs 5A7 or 6H11, or the COOH-terminally directed mAb pp120 (top). The immunoprecipitates were split and Western blotted with mAb 5A7 (lanes 1–6) or mAb pp120 (lanes 7–12). HCA7 cells expressing normal levels of p120 were used for comparison. The blots are overexposed to visualize the extreme low levels of p120.
Thus, in SW48 cells, one allele of the p120 gene contains a stop codon in exon 7 (ARM repeat 3), which predicts a short product that we have not been able to detect. In addition, long-range RT-PCR experiments indicate that both alleles appear to be sensitive to abnormal alternative splicing events that eliminate the normal COOH terminus. The data explain the initial failure to detect p120 in the parental SW48 cells by several p120 mAbs directed against the COOH-terminal end. We do not yet have an explanation for the very low expression level of p120 generated from the second allele.
Restoring WT p120 induces normal epithelial morphology
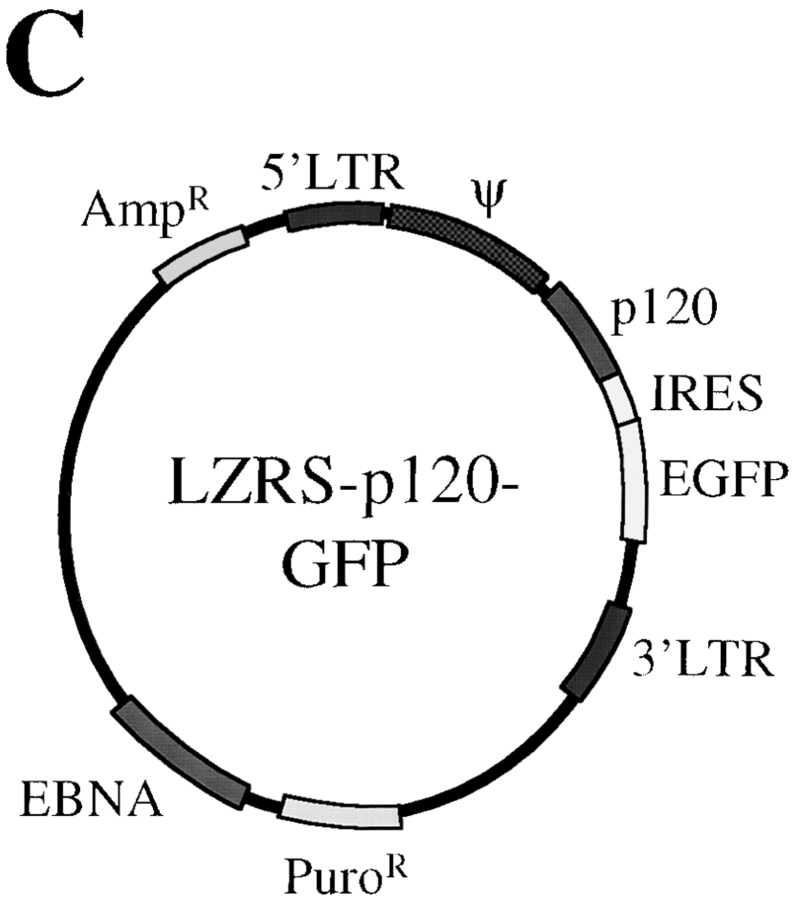
To determine whether the p120 loss or dysfunction was directly responsible for the aberrant SW48 morphology, WT-p120 expression was restored by infection with the retroviral vector pLZRS-p120-internal ribosomal entry site (IRES)-GFP. The structure of the vector is illustrated in Fig. 4 C. An IRES allows both p120 and GFP to be translated separately off the same mRNA transcript. p120 and GFP expression levels from this construct are directly proportional; cells containing high levels of GFP contain high levels of p120, and vice versa (Fig. 5 A).
Figure 4.
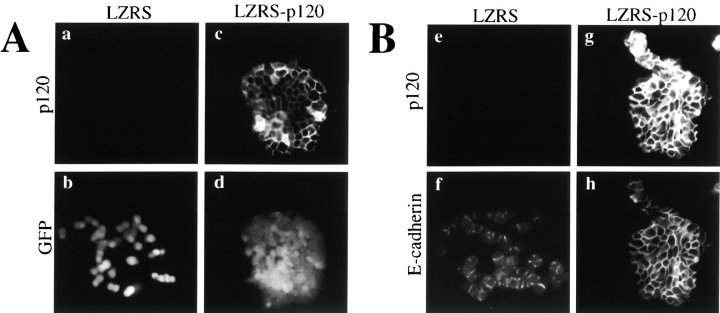
p120 expression in SW48 cells rescues epithelial colony morphology. (A and B) SW48 cells were infected with either LZRS- MS-IRES-GFP (LZRS; a, b, e, and f), or LZRS-p120-IRES-GFP (LZRS-p120; c, d, g, and h). Infected cells were sparsely plated and cultured until individual colonies appeared. (A) Rescue of epithelial morphology by p120. To visualize GFP, cells were fixed with paraformaldehyde to preserve the GFP signal (a–d). p120 was visualized with mAb 15D2 (a and c) and GFP with direct UV microscopy (b and d). GFP expression alone (b) had no effect on SW48 cell morphology. (B) Colocalization of p120 and E-cadherin in infected SW48 cells. To use the green wavelength for E-cadherin staining, cells were fixed in methanol to eliminate the GFP signal and then costained for p120 (e and g) and E-cadherin (f and h). (C) Organization of the LZRS-p120-GFP retroviral vector. An IRES is sandwiched between p120 and GFP cDNAs to allow for translation of both genes from a single mRNA transcript. Thus, the levels of GFP and p120 in infected cells are linked. Key functional elements of the vector are indicated.
Figure 5.
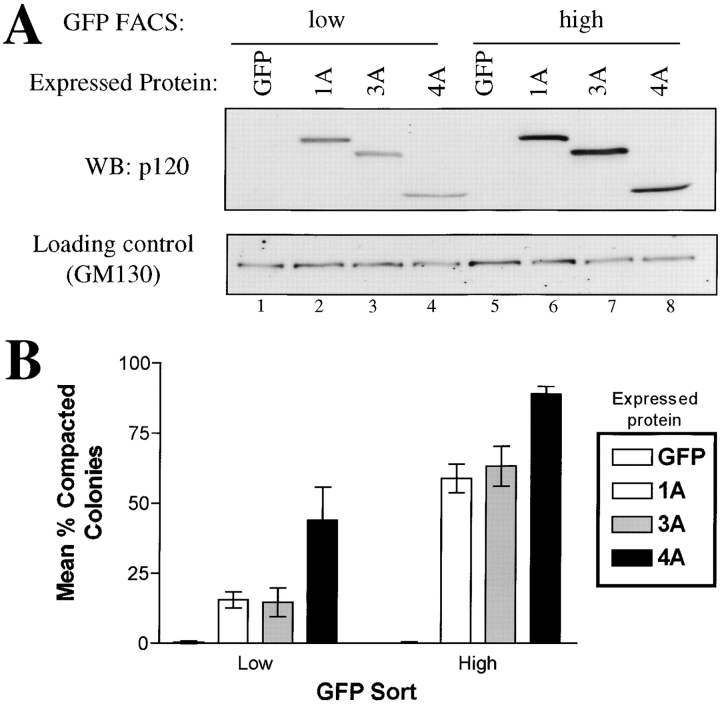
Epithelial rescue by p120 is both expression level and isoform dependent. (A) Isolation by GFP FACS of cell populations expressing low or high levels of p120 isoforms. Cells were infected with viruses containing GFP alone (lanes 1 and 5) or GFP linked by an IRES to p120 isoforms 1A (lanes 2 and 6), 3A (lanes 3 and 7), or 4A (lanes 4 and 8). Cells were sorted by FACS for low (lanes 1–4) and high (lanes 5–8) GFP expression. Protein normalization was confirmed by Western blotting with GM130 antibody. Note the tight correlation between GFP and p120 levels. Each isoform expression level is roughly equivalent within low and high level groups. (B) Quantification of the potency of p120 rescue as a function of expression level and isoform. Cells sorted as described above were plated at low density, grown into clonal colonies, and the colonies scored as compact or loose. The data reflect the ratio of compacted (rescued) colonies divided by the total number of colonies. Compacted colonies occurred more frequently when p120 is expressed at high levels. p120ctn4, which lacks the NH2-terminal phosphorylation domain, is more efficient at inducing compaction than p120ctn1 and 3, which retain this domain.
Interestingly, forced expression of WT-p120 (p120ctn3A) induced a striking rescue of normal epithelial morphology. The loose organization of the parental cells (Fig. 4, A and B, b and f) gave way to compacted epithelial colonies with strong staining of p120 at cell–cell junctions (Fig. 4, A and B, c and g). Infection with pLZRS-IRES-GFP, which expresses GFP but not p120, had no effect on morphology (Fig. 4 A, a and b). These data indicate that p120 expression by itself is sufficient to restore normal epithelial morphology in these cells. Unexpectedly, p120 expression was consistently associated with significantly brighter E-cadherin staining (Fig. 4 B, compare panels f and h).
p120 epithelial rescue is dose dependent and isoform specific
Observation of individual SW48 clones after p120 retroviral infection revealed significant heterogeneity in the degree of epithelial rescue. Cells containing high levels of GFP fluorescence were consistently epithelial, whereas low levels were generally associated with the parental SW48 phenotype. Moreover, initial characterization of various p120 isoforms suggested that isoforms might vary in their ability to rescue E-cadherin function. To quantify these effects, we used gated GFP FACS to isolate polyclonal cell lines expressing low or high levels of p120 isoforms 1A, 3A, and 4A (Fig. 5). Fig. 5 A shows that the low and high GFP sorted cells express low and high levels of p120, respectively. Moreover, the different p120 isoforms in each group were expressed at similar levels, enabling accurate comparisons between isoforms.
The effects of low and high p120 expression levels on the efficiency of epithelial rescue are shown in Fig. 5 B. For each of the isoforms tested, epithelial rescue was significantly more efficient in colonies expressing high levels of p120. Interestingly, epithelial rescue by isoform 4A was much more efficient than isoforms 1A and 3A. Isoform 4A lacks the NH2-terminal end, a region that contains the phosphorylation domain. Therefore, the sequences NH2-terminal to the ARM domain are not required for epithelial rescue, but confer a significant negative regulatory effect on the efficiency of this process.
SW48 adhesion defect results primarily from insufficient levels of p120
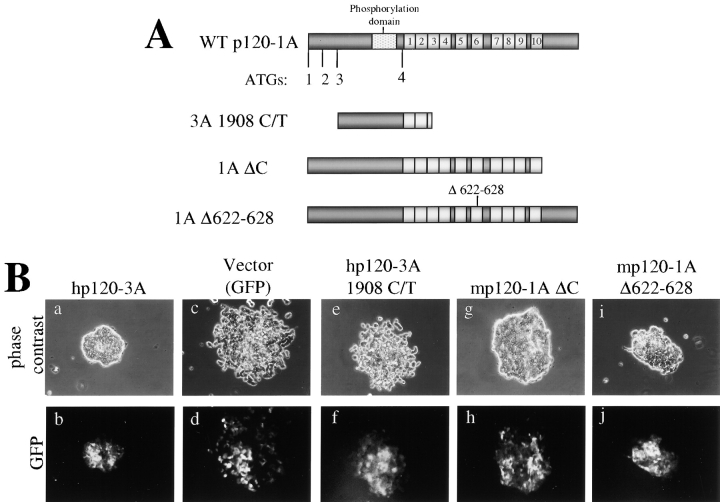
Because SW48 p120 is both mutated and underexpressed, it was initially unclear which alteration was the primary cause of the phenotypic aberrations. Therefore, we asked whether the putative endogenous mutant proteins in SW48 cells could rescue epithelial morphology if we forced their expression at significantly higher levels (Fig. 6). Mutant p120 cDNAs were expressed from the LZRS-p120-IRES-GFP virus, and high expressing cell populations were isolated by FACS as described previously. The constructs tested are illustrated in Fig. 6 A. The p120 cDNA containing the C/T mutation at nucleotide 1908 was directly cloned from SW48 cells. As expected from the fact that it lacks most of the ARM domain, the product of this cDNA was inactive (Fig. 6 B, e). We also overexpressed a murine p120 mutant lacking the COOH-terminal end (ΔC), a construct very similar to the three SW48 cDNAs containing COOH-terminal splicing defects. This mutant effectively rescued compaction (Fig. 6 B, g), indicating that the COOH terminus of p120 is dispensable, provided that the cDNA is adequately expressed.
Figure 6.
The p120 COOH terminus is dispensable for epithelial rescue. (A) Schematic of WT-p120 and p120 variants representing the mutants found in SW48 cells. Alternative ATG start sites are indicated. Light gray boxes represent ARM repeats. mp120–1A Δ622–628 contains a six-aa deletion in ARM 6 postulated to uncouple p120 from RhoA. (B) Effects of overexpression of p120 mutants in SW48 cells. Mutant constructs were cloned into pLZRS-p120-IRES-GFP and expressed in SW48 cells by retroviral infection. Infected cells were collected by gated GFP FACS, plated at low density, and colonies were photographed 1 wk later. Top panels are phase contrast images of colonies and bottom panels show GFP fluorescent images of the same cells. hp120–3A 1908 C/T does not affect the morphology (e and f), but both mp120–1A ΔC and mp120–1A Δ622–628 efficiently induce compaction (g–j).
Previous work shows that high level p120 overexpression induces extensive branching of cellular processes, an effect apparently induced by inhibition of RhoA, and/or activation of Rac and Cdc42 (Anastasiadis and Reynolds, 2001). The p120 deletion mutant Δ622–628 eliminates this effect, possibly by uncoupling p120 from Rho GTPases (Anastasiadis et al., 2000). Surprisingly, expression of this mutant in SW48 cells resulted in an efficient rescue of epithelial morphology (Fig. 6 B, i), suggesting that the p120 activity required for a branching morphology is not required for epithelial compaction.
Finally, the effect of human p120ctn3A was essentially identical to that of murine p120ctn3A (Fig. 6 B, a), thus eliminating the remote possibility that the murine p120-induced phenotype was the result of species specific effects.
Together, these data indicate that the aberrant SW48 phenotype results from insufficient p120 expression rather than loss of functional information in the p120 COOH terminus. Thus, the p120-deficiency makes SW48 cells an excellent model system for p120 structure–function analysis.
Epithelial colony rescue requires p120 binding to E-cadherin
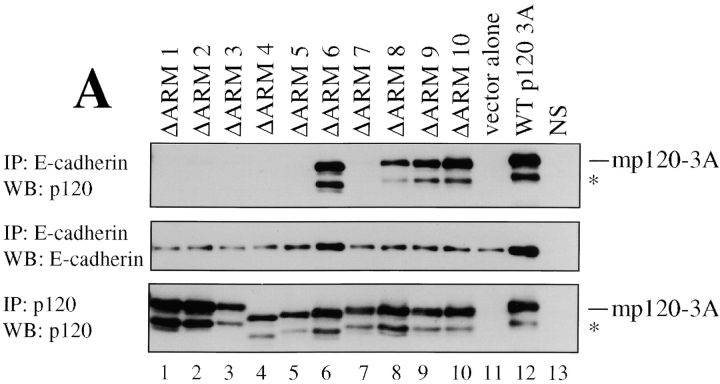
Apparently the sequences flanking the ARM repeat domain of p120 are dispensable for SW48 rescue, suggesting that the ARM domain likely contains the key functional information. To identify the critical sequences, we individually deleted the ten ARM repeats in the mouse p120ctn3A isoform and tested the constructs in SW48 cells. Infected cells were GFP sorted as described above to collect high expressing cell lines. Each of these lines expressed similar levels of p120 (Fig. 7 A, bottom). The mutant proteins were then assayed for both the ability to bind E-cadherin (Fig. 7 A) and the ability to restore epithelial morphology (Fig. 7 B). To assay the E-cadherin-p120 interaction, E-cadherin immunoprecipitates were split and Western blotted for p120 (Fig. 7 A, top) or E-cadherin (Fig. 7 A, middle). Deletion of ARM domains 1–5 and 7 disrupted the p120-E-cadherin interaction. Deletion of ARM domains 8, 9, and 10 showed a slight disruption, whereas deletion of ARM 6 had little effect.
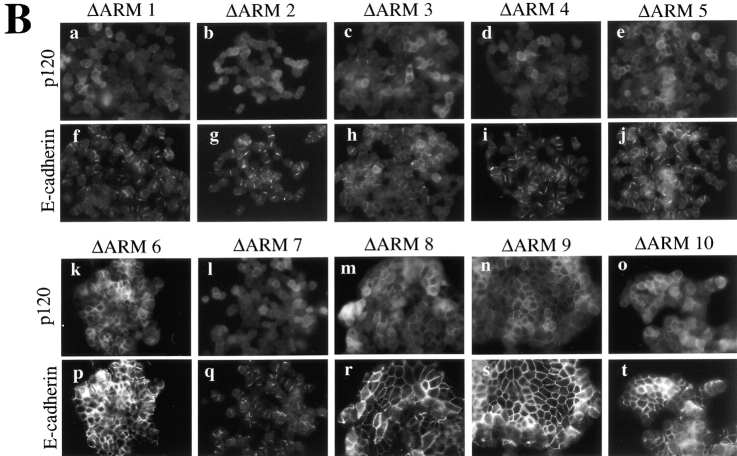
Figure 7.
p120-induced compaction is dependent on direct interaction between p120 and E-cadherin. (A) Structure-function analysis of p120 sequences necessary for interaction with E-cadherin. Each of the ten p120 ARM repeats was individually deleted, expressed in SW48 cells, and polyclonal cell lines derived by GFP FACS. E-cadherin immunoprecipitates were isolated, divided in half, and Western blotted for E-cadherin (middle) or p120 (top). p120 immunoprecipitation and Western blot with mAb p120 controlled for the exogenous expression of p120 mutants (bottom). (*) denotes p120 degradation products. Controls include cells expressing WT-p120 (lane 12), cells infected with empty vector (lane 11), and cells immunoprecipitated with an irrelevant nonspecific antibody (NS). (B) Effects of above constructs in SW48 cells assayed by immunofluorescence for p120 (top) and E-cadherin (bottom). Deletion of ARM repeats 1–5 and 7 block rescue of compaction. ARM repeats 6 and 8–10 are dispensable. E-cadherin binding (A) correlates perfectly with the ability to rescue epithelial morphology (B).
The ability of p120 to rescue epithelial morphology correlated precisely with its ability to bind E-cadherin (Fig. 7 B). Immunofluorescent staining revealed that p120-mediated rescue was abrogated by loss of ARM repeats 1–5 and 7, but repeat 6 and repeats 8–10 were dispensable. Thus, p120 ARM repeats required for E-cadherin interaction (Fig. 7 A) are also required for rescue of epithelial morphology (Fig. 7 B).
Expression of WT p120 increases E-cadherin levels in SW48 cells by posttranscriptionally stabilizing E-cadherin
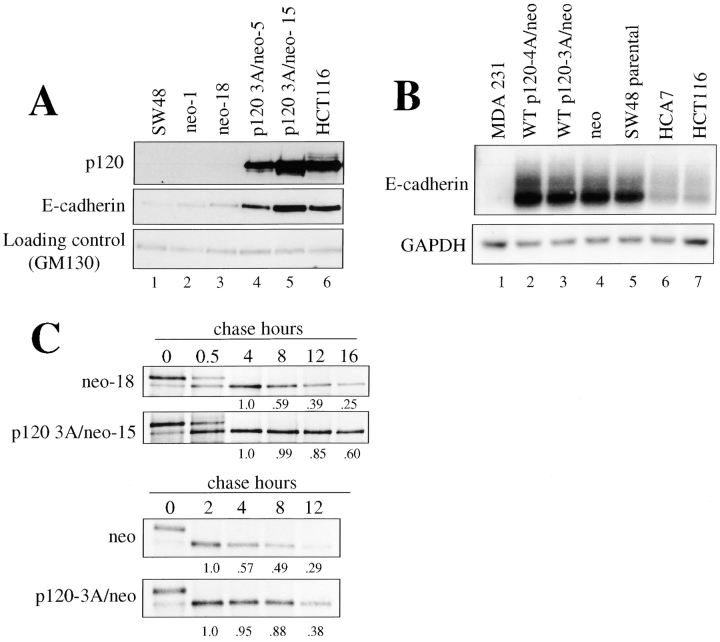
The data suggest a rescue mechanism whereby p120 stabilizes or increases E-cadherin levels in SW48 cells. To further characterize this effect, we replaced the GFP gene in LZRS-p120-IRES-GFP with the neomycin resistance gene (neo; LZRS-p120-IRES-neo) and generated clonal cell lines by G418 selection (Fig. 8 A). Lines expressing p120 levels equivalent to the control cell line HCT116 (Fig. 8 A, lane 6) were strongly compacted (unpublished data) and expressed levels of E-cadherin that were at least five fold higher than control cells (Fig. 8 A, compare lanes 4 and 5 to lanes 1–3). Thus, p120 expression significantly increases the expression or stability of E-cadherin in SW48 cells.
Figure 8.
p120 expression stabilizes E-cadherin by a post-transcriptional mechanism. (A) SW48 cells were infected with LZRS-p120-IRES-neo, and stable cell lines isolated. Levels of E-cadherin were assayed by Western blotting RIPA cell lysates. Parental (lane 1) or cell lines expressing neo only (lanes 2 and 3) were compared with two separately derived p120 expressing cell lines (lanes 4 and 5) and to HCT116 cells (which express normal levels of p120 and E-cadherin). GM130 blotting (bottom) was used as a loading control. p120 expression increased E-cadherin levels by more than fivefold in SW48 cells. (B) E-cadherin mRNA levels are unchanged by p120 expression. Total RNA was isolated and compared by Northern analysis using a human E-cadherin cDNA probe. To control for loading, the blot was stripped and reprobed with GAPDH cDNA. MDA-MB-231 cells (lane 1) do not express E-cadherin mRNA. SW48 cells stably expressing p120–4A (lane 2), p120–3A (lane 3), or neo alone (lane 4), are compared with SW48 parental (lane 5), HCA7 (lane 6), and HCT116 cells (lane 7). (C) Analysis of E-cadherin stability in p120-expressing cells. Clonal (p120 3A/neo-15) or polyclonal (p120–3A/neo) cell lines were generated by infection. E-cadherin half life was ascertained by pulse-chase analysis and compared with control clonal (neo-18) and polyclonal (neo) cell lines. Chase times are indicated across the top. Densitometric analyses are located below each lane and represent a normalized value where the 4 h chase lane (top) or the 2 h chase lane (bottom) represent the value 1.0. p120 expression essentially doubles the half life of E-cadherin.
To determine whether increased levels of E-cadherin resulted from effects of p120 on E-cadherin transcription, we assayed E-cadherin mRNA levels by Northern blotting samples obtained from parental SW48 cells and the same cells after rescue by p120 expression. Fig. 8 B shows that E-cadherin mRNA levels are unaltered by ectopic p120 expression, suggesting that p120 increases E-cadherin levels by a posttranscriptional mechanism. Interestingly, E-cadherin mRNA levels were relatively high in SW48 cells, suggesting a negative feedback loop to compensate for the low levels of E-cadherin protein. However, the increased E-cadherin levels associated with p120 overexpression did not reduce the levels of E-cadherin mRNA (Fig 8 B, lanes 2 and 3).
The effect of p120 on E-cadherin protein stability was assessed by pulse chase analysis. In clonal cell lines, ectopic expression of p120 increased the E-cadherin half-life dramatically from roughly 6 to more than 12 h (Fig. 8 C). In addition, a polyclonal population of SW48 cells, generated by LZRS-p120-IRES-neo infection and G418 selection, behaved similarly, although the stabilization was somewhat reduced relative to the clonal cell line, probably because the average p120 expression level in the population is lower. Thus, p120 expression in SW48 cells substantially increases the endogenous E-cadherin half life.
Forced E-cadherin expression induces epithelial colony morphology
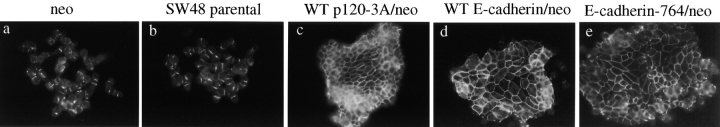
If p120 is acting essentially to stabilize or elevate E-cadherin levels, then forcing increased E-cadherin expression might also rescue epithelial morphology, even in the absence of p120. To test this hypothesis, we expressed WT-E-cadherin or p120-uncoupled E-cadherin (E-cad/764AAA) by infection of SW48 cells with LZRS-E-cad-IRES-neo or LZRS-E-cad/764AAA-IRES-neo followed by selection in G418 (Fig. 9). Both WT and p120 uncoupled E-cadherin rescued epithelial morphology. The rescue by E-cad-764AAA is particularly significant because the inability of this mutant to recruit p120 implies that p120 is not essential if E-cadherin levels are artificially maintained at high levels. Taken together, the data suggest that a crucial role of p120 is to regulate, stabilize, or maintain E-cadherin at levels that are high enough to generate strong adhesion.
Figure 9.
Epithelial rescue by overexpression of WT or p120-uncoupled E-cadherin in SW48 cells. SW48 cells were infected with LZRS-neo viruses containing p120, E-cadherin, or p120-uncoupled E-cadherin (E-cadherin-764), and colonies generated by G418 selection. Cells are stained for immunofluorescence using E-cadherin mAb C-20820. Expression of neo alone (a) had no effect on the parental SW48 phenotype (b). However, WT-E-cadherin (panel d) and p120-uncoupled E-cadherin (e) induced an epithelial morphology similar to that induced by p120 expression (c).
Discussion
We describe a novel role for p120 in regulating epithelial morphology. SW48 cells are the first example of p120 mutation in cancer. More importantly, p120 is grossly underexpressed in these cells, providing the first opportunity to examine the effects of p120 loss and reconstitution in vitro. The striking rescue of SW48 epithelial morphology by expression of WT-p120 argues strongly that p120 deficiency is the critical factor limiting adhesiveness and epithelial morphology in these cells. The ability of the endogenous E-cadherin to function efficiently in the presence of ectopic p120 indicates that the endogenous E-cadherin itself is fully functional, albeit underrepresented in the absence of p120. Thus, the low levels of E-cadherin in SW48 cells are likely secondary to the root problem of p120 insufficiency. A key observation is that E-cadherin levels increased substantially upon reconstitution of p120 due to a doubling of the E-cadherin half life. Moreover, the specific p120 ARM repeats required for interaction with E-cadherin were also necessary for reactivation of the cadherin system. Thus, p120 stabilizes E-cadherin and rescues its function via a mechanism dependent on their direct interaction.
The observations reported here are based on SW48 cells but are probably generally applicable. For example, significantly increased E-cadherin staining is observed in MDCK cells overexpressing either p120 (unpublished data), or δ-catenin (Lu et al., 1999), a close structural and functional relative of p120. However, in normal cells such as MDCK, the significance of this effect is not obvious because the cells already possess epithelial morphology and high levels of p120 and E-cadherin. In contrast, the effects of p120 overexpression in SW48 cells are striking due to the initial condition of p120 deficiency, the resulting low levels of endogenous E-cadherin, and the diffuse cellular morphology.
Evidence in the literature supports several possible explanations for these observations. The simplest is that the p120 interaction with E-cadherin is sufficient to physically stabilize the E-cadherin complex. Crystallographic studies indicate that the cadherin cytoplasmic domain is essentially unordered in the absence of binding partners (Huber et al., 2001). Alternatively, p120 may competitively block interactions of E-cadherin with other factors that promote E-cadherin recycling or degradation. Endocytosis has been proposed as a mechanism for down-regulating or recycling cadherins (Le et al., 1999) and is postulated to play a crucial role in the dynamic regulation of adhesiveness. The E3 ubiquitin-ligase Hakai interacts with the E-cadherin JMD in a tyrosine phosphorylation-dependent manner, thereby promoting ubiquitination and endocytosis of the complex (Fujita et al., 2002). Because multiple tyrosine kinases and phosphatases have been directly or indirectly linked with the cadherin complex (for review, see Daniel and Reynolds, 1997), the loss of p120 in SW48 cells may leave the E-cadherin JMD exposed to increased tyrosine phosphorylation resulting in Hakai-mediated endocytosis. PS1 also competes with p120 for binding the cadherin JMD (Baki et al., 2001), and although one report suggests that PS1 stabilizes the cadherin complex (Baki et al., 2001), a follow-up study suggests that PS1 may promote E-cadherin degradation by specific proteolytic cleavage events (Marambaud et al., 2002). Thus, p120 binding may block such interactions, and in the absence of p120, these proteins might act constitutively in SW48 cells to reduce the amount of E-cadherin.
Another possibility is that p120 may possess an activity that promotes stability of the cadherin complex. For example, recent evidence indicates that p120 can inhibit RhoA and activate Rac and Cdc42, known modulators of the actin cytoskeleton (for review see Anastasiadis and Reynolds, 2001). Although these reports focus on the regulation of GTPases by a soluble pool of p120, they also suggest the possibility that p120 recruits and coordinates the actions of these proteins in the cadherin complex. These GTPases may be necessary for cadherin clustering (Braga et al., 1997; Takaishi et al., 1997; Jou and Nelson, 1998). Thus, p120 binding to E-cadherin may promote the local assembly and/or organization of the underlying actin cytoskeleton resulting in stabilization of the adherens junction. On the other hand, our preliminary data (Fig. 6 B, i) show that a mutant (Δ622–628), presumably RhoA uncoupled and unable to induce cellular branching (Anastasiadis et al., 2000), effectively rescues epithelial morphology in SW48 cells. Thus, it is unclear whether p120's branching activity, and/or the p120/Rho GTPase connection is relevant to the observed epithelial rescue in SW48 cells.
Because p120-uncoupled cadherins can restore epithelial morphology in several systems (Aono et al., 1999; Thoreson et al., 2000; this work, Fig. 9), it has been suggested that cadherins can function efficiently in the absence of p120 (Aono et al., 1999). Our data provide a cautionary note on the interpretation of experiments using p120-uncoupled cadherins based on the fact that these experiments rely on ectopic cadherin expression driven by constitutively active promoters. If stabilizing E-cadherin is an important role for p120 under normal circumstances, then constitutive overexpression of p120-uncoupled cadherins may at least partially compensate for p120 function despite the absence of p120 binding. Indeed, overexpression of p120-uncoupled E-cadherin can rescue epithelial morphology in SW48 cells, but the absence of p120 in the parental cell line clearly leads to malfunction of the cadherin system. Moreover, the known p120-uncoupling mutations simultaneously eliminate binding sites for p120, Hakai, and PS1, thereby potentially abolishing both cadherin stabilizing and destabilizing mechanisms. These possibilities highlight the significance of our observations in SW48 cells, which directly address the consequences of p120 loss and reconstitution.
Based on experiments in Colo205 cells, it was suggested that p120 negatively regulates cadherin adhesiveness (Aono et al., 1999). These cells normally express all members of the cadherin complex, including p120, but are nonetheless poorly adhesive (Aono et al., 1999). An NH2-terminally truncated p120 mutant (similar to our p120ctn4 isoform) strongly reactivates the cadherin system in these cells, whereas WT-p120 has no effect (Aono et al., 1999). Thus, the cadherin system in Colo205 cells appears to be constitutively inactivated through a signaling mechanism requiring the phosphorylation domain of p120. In contrast, we find that both WT and NH2-terminally truncated p120 (p120ctn4) reactivate cadherin function in SW48 cells. These contrasting scenarios suggest that the principle defect in SW48 cells is p120 insufficiency, whereas the principle defect in Colo205 cells involves aberrant signaling through p120. Although these cell lines represent extreme cases, it is possible that both scenarios exist to various degrees in many tumors and may contribute to cadherin malfunction in malignancy. It is worth noting that the p120 NH2-terminal domain had a negative effect on adhesion in SW48 cells (as also reported in Colo205), but the effect was considerably less prominent than the inhibition observed in Colo205 cells. In light of our data, we suggest that p120 is principally a positive regulator of adhesion. The regulatory region containing the phosphorylation domain is likely to act like a dimmer switch and provides a mechanism for signaling pathways to modulate cadherin functions, such as adhesion and motility, through posttranslational modification of p120.
The data support a basic model whereby the positive role of p120 in adhesion requires its interaction with E-cadherin and results in subsequent stabilization of the complex. The regulatory region then acts as a dimmer switch, potentially controlled by various signaling events resulting in phosphorylation and/or dephosphorylation of specific p120 residues. These actions may control the on/off rate of p120, whose affinity for cadherins appears to be relatively weak (Reynolds et al., 1994; Shibamoto et al., 1995; Staddon et al., 1995). Conditions that promote p120 binding would therefore be expected to increase adhesiveness. Cytoplasmic p120 may promote cell motility through action on Rho GTPases (Noren et al., 2000; Grosheva et al., 2001). Thus, p120 translocation to the cytoplasm could be important for other events typically associated with E-cadherin downregulation, as suggested previously (Anastasiadis and Reynolds, 2001). Alternatively, p120 phosphorylation may control a specific p120 activity that functions within the cadherin complex (e.g., recruitment of Rho GTPases or as yet unidentified binding partners), which in turn may be important for dynamic modulation of adhesion as occurs in motile cells. The various mechanisms proposed are not mutually exclusive and the end result is likely influenced by several events that act together.
Accumulating evidence indicates that p120 expression is frequently altered in human carcinomas. Quite striking are reports in which p120 staining is completely absent from certain tumors of the breast, stomach, prostate, bladder, and colon (for review see Thoreson and Reynolds, 2002). In light of these data, it is unclear why p120-deficient epithelial cell lines are not common. Nonetheless, these observations suggest that p120 function may be important in cancer and raise new questions as to how p120 expression is regulated.
E-cadherin downregulation is common in tumors and occurs through several mechanisms, including mutation (for review see Berx et al., 1998; Berx and van Roy, 2001), promoter methylation (Matsumura et al., 2001), and regulatory alterations mediated by various transcription factors (Batlle et al., 2000; Cano et al., 2000; Comijn et al., 2001). We suggest a new mechanism based on the ability of p120 to posttranscriptionally stabilize E-cadherin. In some tumors, p120 loss may be an initial event leading to reduced levels of E-cadherin. Since E-cadherin stabilizes α- and β-catenin (De Leeuw et al., 1997), one consequence of p120 downregulation may be reduced levels of all members of the cadherin complex, and thus an overall reduction in cadherin activity. Ironically, β-catenin is not downregulated in SW48 cells because of a stabilizing mutation at codon 33 (Ilyas et al., 1997).
In conclusion, we describe the first example of a p120-deficient carcinoma cell line and a novel role for p120. p120 insufficiency in SW48 cells appears to be directly responsible for a significant reduction in the amount of E-cadherin and for the associated defects in epithelial morphology. The residual p120 levels in these cells may very well be essential for the fact that even limited junctions were observed. Restoring p120 substantially increased E-cadherin levels and rescued epithelial morphology by mechanisms that were completely dependent on the direct interaction between these proteins. We believe this activity to be responsible for the positive role of p120 in adhesion and necessary to maintain E-cadherin abundance and activity at levels sufficient to mediate strong cell–cell adhesion. Our data also suggests that the NH2-terminal domain contains regulatory sequences necessary for dynamic modulation of this otherwise positive action. Based on these observations, we also suggest that p120 loss in tumors may induce the subsequent downregulation of E-cadherin to levels that cannot support normal epithelial morphology.
Materials and methods
Cell culture
SW48 cells were grown in DME/F-12 with 1% l-glutamine. All other cells were grown in DME. All media was supplemented with 10% FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin (GIBCO BRL). Phoenix 293 cell media contained heat-inactivated FCS.
Immunofluorescence
5 × 103 cells per coverslip were grown 7–10 d before immunofluorescent labeling as previously described (Mariner et al., 2000). To eliminate GFP, cells were fixed in 100% methanol for 7 min at −20°C. To visualize GFP expression, cells were fixed in 3% paraformaldehyde for 30 min and permeablized with PBS/0.2% Triton-X for 5 min. Primary antibodies were used at: p120, 0.5 μg/ml (Transduction Labs); anti–β-catenin, 1/1,000 (C-2206; Sigma-Aldrich); anti–α-catenin, 1/1,000 (C-2081; Sigma-Aldrich); anti–E-cadherin, 1/2,500 (C-20820; Transduction Labs); p120 mAbs 5A7 and 6H11, 2 μg/ml (Wu et al., 1998); and hECD1, 1 μg/ml. Secondary antibodies goat anti–mouse IgG2a Alexa 488, goat anti–mouse IgG1 Alexa 594, goat anti–mouse Alexa 488, and goat anti–rabbit Alexa 594 (Molecular Probes) were diluted 1:600.
Immunoprecipitation and Western blotting
Immunoprecipitation and Western blot procedures have been described previously (Reynolds et al., 1994). For coimmunoprecipitation, cells were lysed in CSK buffer (10 mM Pipes, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 0.5% NP-40). RIPA and NP-40 whole cell lysates were prepared as in (Mariner et al., 2000). Protein was quantitated by BCA Protein Assay (Pierce Chemical Co.). Antibodies pp120, hECD1, anti–β-catenin, anti–α-catenin, and peroxidase-conjugated donkey anti–mouse and mouse anti–rabbit secondaries (Jackson ImmunoResearch Laboratories) were used as described in (Thoreson et al., 2000). Other antibody concentrations were: Plakoglobin mAb I5F11, 2.5 μg/ml; p120 mAb 5A7, 2 μg/ml; and GM130, 1:250 (610823; Transduction Labs).
Viral production, infection, and production of stable cell lines
Phoenix 293 cell transfection (Grignani et al., 1998) was performed as described on the Nolan lab website (http://www.stanford.edu/group/nolan/), incubating for 7–9 h at 37°C. Cells were selected with 5 μg/ml puromycin (Sigma-Aldrich) and passaged once before viral harvest. To harvest virus, cells at 75% confluence were incubated for 16 h at 37°C. Collected media was passed through a 0.45-μm syringe filter (Pall Corporation) and immediately used or stored at −70°C. To infect, 4 μg/ml polybrene (Sigma-Aldrich) was added to virus stock before overlaying cells plated at 6 × 104 cells/60-mm dish or 1.8 × 105 cells/100-mm dish. Cells were incubated overnight and replated in SW48 media. To produce clonal stable cell lines, cells were infected with the LZRS-MS-IRES-neo virus containing the genes of interest, sparsely plated, selected with 875 active units/mL Geneticin (GIBCO BRL) for 7–10 d, and isolated using cloning cylinders.
FACS
Cells infected with LZRS-MS-IRES-GFP virus containing the genes of interest were split 1:1, grown 24 h at 37°C, and suspended at 2 × 106 cells/ml in PBS (15 mM NaCl, 10 mM NaH2PO4). GFP gates were set to collect the cells in the bottom third (low expressors) or top third (high expressors) of the GFP range (488 nm) using a FACStar Plus cell sorter. Gating parameters were predetermined by observing GFP levels in pilot experiments.
Quantitative analysis of epithelial rescue
Cells expressing p120 isoforms were sorted into high and low GFP expressing populations. 5 × 103 cells/90-mm dish were grown for 7 d. The number of colonies out of 100 that exhibited the compacted colony phenotype were counted. GraphPad Prism software was used for statistical (Bonferroni's Multiple Comparison Test) and graphical analysis. Results reflect pooled data from three independent sorting experiments and show standard deviation of the means.
Northern analysis
Total RNA was isolated with TRIzol Reagent (Invitrogen) according to the manufacturer's protocol. 20 μg of each RNA sample was analyzed by Northern blotting with an E-cadherin probe generated by PCR-amplification of a 1,326-bp fragment of human E-cadherin using 5′-TGATGTGAACACCTACAATGCCG-3′ and 5′-CCTCCGAAGAAACAGCAAGAGC-3′ as primers. Probing with full-length murine GAPDH controlled for loading. Probes were α-P32 dCTP-labelled by primer extention (Prime-It II Kit; Stratagene). Nylon membranes were hybridized using Express Hyb hybridization solution (CLONTECH Laboratories, Inc.) following the manufacturer's protocol. Results were visualized by autoradiography using Phosphor Screens (Molecular Dynamics) and a PhosphoImager 445-SI (Molecular Dynamics).
Cloning
pLZRS-MS-IRES-GFP and pLZRS-MS-IRES-neo retroviral vectors were generated by replacing the lacZ gene of pLZRS-lacZ with EGFP or neo genes (from pIRES2-EGFP and pIRES-neo; CLONTECH Laboratories. Inc.). A shuttle vector (pMS) was created by replacing the pBluescript (Stratagene) polylinker with a polylinker containing SgfI, EcoR1, AfeI, Kpn1, and SfiI restriction sites. The same polylinker was inserted into LZRS-IRES-MS-GFP (or neo). All constructs were made by cloning cDNA into pMS, then moving it to LZRS by a SgfI-SfiI ligation. LZRS-hp120-3A-1908-C/T-GFP was generated from cDNA cloned directly from SW48 mRNA. All other deletion mutants were created using ExSite Mutagenesis (Stratagene). The deleted sequences in the LZRS-mp120-GFP constructs are based on the aa numbering system for p120-1A (Anastasiadis and Reynolds, 2000). Listed aa are inclusive of the deletion. ΔC, (aa 831–932); ΔARM1, (aa 358–400); ΔARM2, (aa 401–443); ΔARM3, (aa 445–486); ΔARM4, (aa 487–545); ΔARM5, (aa 546–591); ΔARM6, (aa 592–649); ΔARM7, (aa 650–696); ΔARM8, (aa 697–737); ΔARM9, (aa 738–787); ΔARM10, (aa 788–824). All ARM repeat domain mutants were made in mp120 3A. mp120-1A Δ622-628 and p120 uncoupled E-cadherin (E-cad-764AAA) have been described previously (Anastasiadis et al., 2000; Thoreson et al., 2000). The p120-4A isoform was generated from p120-3A by using PCR to eliminate the first three start codons and starts at aa 324.
RT-PCR and genomic sequencing
For RT-PCR analysis, RNA was isolated from human tumor cell lines using the RNeasy kit (QIAGEN). cDNA was synthesized using SuperscriptTM II Rnase H− reverse transcriptase (GIBCO BRL), incubated with 0.5 U Rnase H (GIBCO BRL), and PCR amplified using Taq DNA polymerase (GIBCO BRL). Products were isolated using a Concert Rapid Gel extraction kit (GIBCO BRL) and directly sequenced or cloned in the pGEMR-T cloning system (Promega). Primers EX7F1, EX8R1, EX14F1, EX21R3, and EX6F1 were used and have been previously described (Keirsebilck et al., 1998). Intron-exon boundaries were determined by partial sequence analysis of subcloned fragments containing protein-encoding sequences (Keirsebilck et al., 1998). Intron-specific amplimers were used to screen all the p120-specific exons for relevant mutations. As exon 7 contains 464 nucleotides, two primer pairs were used. The mutation was detected using primers EX7F2 (5′-GTCAAGTCCAATGCAGCTGCATA-3′) and IN7R1 (5′-ATCCCTCTAGTTCTCAACATCAC-3′) by PCR using Goldstar DNA polymerase (Eurogentec) followed by sequencing.
Pulse chase
Pulse chase analysis was performed as described (Fujita et al., 2002). Cells were lysed in RIPA buffer and E-cadherin immunoprecipitated with 2 μg of hECD-1 mAb. Prior to drying, gels were fixed and treated with Amplify (Amersham Biosciences) according to manufacturer's protocol. Radioactive bands were visualized by autoradiography with phosphorscreens as described above and quantified using ImageQuant (Molecular Dynamics).
Acknowledgments
This work was supported by the NIH grant CA55724, the Vanderbilt-Ingram Cancer Center Support grant CA68684-06, and grants from the FWO, the Geconcerteerde Onderzoeksacties of Ghent University, and the Fortis Bank-Verzekeringen, Brussels, Belgium. We thank Dr. Gary Nolan for LZRS retroviral constructs, Dr. Bob Coffey for use of the fluorescent microscope, and Dr.'s Margaret Wheelock and Masatoshi Takeichi for mAbs I5F11 and hECD-1, respectively. Gregg Wildenberg, Sarah Brockway, Kim Brown, and Alyssa Bonine generated several constructs and viruses. Jolanda van Hengel is a postdoctoral fellow with the FWO (Fund for Scientific Research, Flanders).
Footnotes
Abbreviations used in this paper: ARM, armadillo; IRES, internal ribosomal entry site; JMD, juxtamembrane domain; neo, neomycin resistance gene; p120, p120-catenin; PS, presenilin.
References
- Aberle, H., S. Butz, J. Stappert, H. Weissig, R. Kemler, and H. Hoschuetzky. 1994. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J. Cell Sci. 107:3655–3663. [DOI] [PubMed] [Google Scholar]
- Anastasiadis, P.Z., and A.B. Reynolds. 2000. The p120 catenin family: complex roles in adhesion, signaling and cancer. J. Cell Sci. 113:1319–1334. [DOI] [PubMed] [Google Scholar]
- Anastasiadis, P.Z., and A.B. Reynolds. 2001. Regulation of Rho GTPases by p120-catenin. Curr. Opin. Cell Biol. 13:604–610. [DOI] [PubMed] [Google Scholar]
- Anastasiadis, P.Z., S.Y. Moon, M.A. Thoreson, D.J. Mariner, H.C. Crawford, Y. Zheng, and A.B. Reynolds. 2000. Inhibition of RhoA by p120 catenin. Nat. Cell Biol. 2:637–644. [DOI] [PubMed] [Google Scholar]
- Aono, S., S. Nakagawa, A.B. Reynolds, and M. Takeichi. 1999. p120(ctn) acts as an inhibitory regulator of cadherin function in colon carcinoma cells. J. Cell Biol. 145:551–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baki, L., P. Marambaud, S. Efthimiopoulos, A. Georgakopoulos, P. Wen, W. Cui, J. Shioi, E. Koo, M. Ozawa, V.L. Friedrich, Jr., and N.K. Robakis. 2001. Presenilin-1 binds cytoplasmic epithelial cadherin, inhibits cadherin/p120 association, and regulates stability and function of the cadherin/catenin adhesion complex. Proc. Natl. Acad. Sci. USA. 98:2381–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle, E., E. Sancho, C. Franci, D. Dominguez, M. Monfar, J. Baulida, and A. Garcia De Herreros. 2000. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2:84–89. [DOI] [PubMed] [Google Scholar]
- Berx, G., and F. van Roy. 2001. The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res. 3:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berx, G., F. Nollet, and F. van Roy. 1998. Dysregulation of the E-cadherin/catenin complex by irreversible mutations in human carcinomas. Cell Adhes. Commun. 6:171–184. [DOI] [PubMed] [Google Scholar]
- Birchmeier, W., and J. Behrens. 1994. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim. Biophys. Acta. 1198:11–26 [DOI] [PubMed] [Google Scholar]
- Braga, V.M., L.M. Machesky, A. Hall, and N.A. Hotchin. 1997. The small GTPases Rho and Rac are required for the establishment of cadherin-dependent cell–cell contacts. J. Cell Biol. 137:1421–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano, A., M.A. Perez-Moreno, I. Rodrigo, A. Locascio, M.J. Blanco, M.G. del Barrio, F. Portillo, and M.A. Nieto. 2000. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2:76–83. [DOI] [PubMed] [Google Scholar]
- Chen, H., N.E. Paradies, M. Fedor-Chaiken, and R. Brackenbury. 1997. E-cadherin mediates adhesion and suppresses cell motility via distinct mechanisms. J. Cell Sci. 110:345–356. [DOI] [PubMed] [Google Scholar]
- Comijn, J., G. Berx, P. Vermassen, K. Verschueren, L. van Grunsven, E. Bruyneel, M. Mareel, D. Huylebroeck, and F. van Roy. 2001. The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion. Mol. Cell. 7:1267–1278. [DOI] [PubMed] [Google Scholar]
- Daniel, J.M., and A.B. Reynolds. 1997. Tyrosine phosphorylation and cadherin/catenin function. Bioessays. 19:883–891. [DOI] [PubMed] [Google Scholar]
- De Leeuw, W.J., G. Berx, C.B. Vos, J.L. Peterse, M.J. Van de Vijver, S. Litvinov, F. Van Roy, C.J. Cornelisse, and A.M. Cleton-Jansen. 1997. Simultaneous loss of E-cadherin and catenins in invasive lobular breast cancer and lobular carcinoma in situ. J. Pathol. 183:404–411. [DOI] [PubMed] [Google Scholar]
- Downing, J.R., and A.B. Reynolds. 1991. PDGF, CSF-1, and EGF induce tyrosine phosphorylation of p120, a pp60src transformation-associated substrate. Oncogene. 6:607–613. [PubMed] [Google Scholar]
- Frixen, U.H., J. Behrens, M. Sachs, G. Eberle, B. Voss, A. Warda, D. Lochner, and W. Birchmeier. 1991. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J. Cell Biol. 113:173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, Y., G. Krause, M. Scheffner, D. Zechner, H.E. Leddy, J. Behrens, T. Sommer, and W. Birchmeier. 2002. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 4:222–231. [DOI] [PubMed] [Google Scholar]
- Funayama, N., F. Fagotto, P. McCrea, and B.M. Gumbiner. 1995. Embryonic axis induction by the armadillo repeat domain of beta-catenin: evidence for intracellular signaling. J. Cell Biol. 128:959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grignani, F., T. Kinsella, A. Mencarelli, M. Valtieri, D. Riganelli, L. Lanfrancone, C. Peschle, G.P. Nolan, and P.G. Pelicci. 1998. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 58:14–19. [PubMed] [Google Scholar]
- Grosheva, I., M. Shtutman, M. Elbaum, and A.D. Bershadsky. 2001. p120 catenin affects cell motility via modulation of activity of Rho - family GTPases: a link between cell-cell contact formation and regulation of cell locomotion. J. Cell Sci. 114:695–707. [DOI] [PubMed] [Google Scholar]
- Horikawa, K., and M. Takeichi. 2001. Requirement of the juxtamembrane domain of the cadherin cytoplasmic tail for morphogenetic cell rearrangement during myotome development. J. Cell Biol. 155:1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, A.H., D.B. Stewart, D.V. Laurents, W.J. Nelson, and W.I. Weis. 2001. The cadherin cytoplasmic domain is unstructured in the absence of beta-catenin. A possible mechanism for regulating cadherin turnover. J. Biol. Chem. 276:12301–12309. [DOI] [PubMed] [Google Scholar]
- Hulsken, J., W. Birchmeier, and J. Behrens. 1994. E-cadherin and APC compete for the interaction with beta-catenin and the cytoskeleton. J. Cell Biol. 127:2061–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyas, M., I. Tomlinson, A. Rowan, M. Pignatelli, and W. Bodmer. 1997. Beta-catenin mutations in cell lines established from human colorectal cancers. Proc. Natl. Acad. Sci. USA. 94:10330–10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jou, T.S., and W.J. Nelson. 1998. Effects of regulated expression of mutant RhoA and Rac1 small GTPases on the development of epithelial (MDCK) cell polarity. J. Cell Biol. 142:85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jou, T.S., D.B. Stewart, J. Stappert, W.J. Nelson, and J.A. Marrs. 1995. Genetic and biochemical dissection of protein linkages in the cadherin-catenin complex. Proc. Natl. Acad. Sci. USA. 92:5067–5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanner, S.B., A.B. Reynolds, and J.T. Parsons. 1991. Tyrosine phosphorylation of a 120-kilodalton pp60src substrate upon epidermal growth factor and platelet-derived growth factor receptor stimulation and in polyomavirus middle-T-antigen-transformed cells. Mol. Cell. Biol. 11:713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keirsebilck, A., S. Bonne, K. Staes, J. van Hengel, F. Nollet, A. Reynolds, and F. van Roy. 1998. Molecular cloning of the human p120ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics. 50:129–146. [DOI] [PubMed] [Google Scholar]
- Kintner, C. 1992. Regulation of embryonic cell adhesion by the cadherin cytoplasmic domain. Cell. 69:225–236. [DOI] [PubMed] [Google Scholar]
- Le, T.L., A.S. Yap, and J.L. Stow. 1999. Recycling of E-cadherin: a potential mechanism for regulating cadherin dynamics. J. Cell Biol. 146:219–232. [PMC free article] [PubMed] [Google Scholar]
- Lilien, J., C. Arregui, H. Li, and J. Balsamo. 1999. The juxtamembrane domain of cadherin regulates integrin-mediated adhesion and neurite outgrowth. J. Neurosci. Res. 58:727–734. [DOI] [PubMed] [Google Scholar]
- Lu, Q., M. Paredes, M. Medina, J. Zhou, R. Cavallo, M. Peifer, L. Orecchio, and K.S. Kosik. 1999. delta-catenin, an adhesive junction-associated protein which promotes cell scattering. J. Cell Biol. 144:519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marambaud, P., J. Shioi, G. Serban, A. Georgakopoulos, S. Sarner, V. Nagy, L. Baki, P. Wen, S. Efthimiopoulos, Z. Shao, et al. 2002. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 21:1948–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariner, D.J., J. Wang, and A.B. Reynolds. 2000. ARVCF localizes to the nucleus and adherens junction and is mutually exclusive with p120(ctn) in E-cadherin complexes. J. Cell Sci. 113:1481–1490. [DOI] [PubMed] [Google Scholar]
- Mariner, D.J., P. Anastasiadis, H. Keilhack, F.-D. Böhmer, J. Wang, and A.B. Reynolds. 2001. Identification of Src phosphorylation sites in the catenin p120ctn. J. Biol. Chem. In press. [DOI] [PubMed] [Google Scholar]
- Matsumura, T., R. Makino, and K. Mitamura. 2001. Frequent down-regulation of E-cadherin by genetic and epigenetic changes in the malignant progression of hepatocellular carcinomas. Clin. Cancer Res. 7:594–599. [PubMed] [Google Scholar]
- Mo, Y.Y., and A.B. Reynolds. 1996. Identification of murine p120 isoforms and heterogeneous expression of p120cas isoforms in human tumor cell lines. Cancer Res. 56:2633–2640. [PubMed] [Google Scholar]
- Navarro, P., L. Ruco, and E. Dejana. 1998. Differential localization of VE- and N-cadherins in human endothelial cells: VE-cadherin competes with N-cadherin for junctional localization. J. Cell Biol. 140:1475–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollet, F., G. Berx, and F. van Roy. 1999. The role of the E-cadherin/catenin adhesion complex in the development and progression of cancer. Mol. Cell Biol. Res. Comm. 2:77–85. [DOI] [PubMed] [Google Scholar]
- Nollet, F., P. Kools, and F. van Roy. 2000. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. Journal of Molecular Biology. 299:551–572. [DOI] [PubMed] [Google Scholar]
- Noren, N.K., B.P. Liu, K. Burridge, and B. Kreft. 2000. p120 catenin regulates the actin cytoskeleton via Rho family GTPases. J. Cell Biol. 150:567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer, M., S. Berg, and A.B. Reynolds. 1994. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 76:789–791. [DOI] [PubMed] [Google Scholar]
- Perl, A.K., P. Wilgenbus, U. Dahl, H. Semb, and G. Christofori. 1998. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 392:190–193. [DOI] [PubMed] [Google Scholar]
- Reynolds, T. 1994. Prostate cancer genetics research picks up pace. J. Natl. Cancer Inst. 86:1112–1115. [DOI] [PubMed] [Google Scholar]
- Reynolds, A.B., D.J. Roesel, S.B. Kanner, and J.T. Parsons. 1989. Transformation-specific tyrosine phosphorylation of a novel cellular protein in chicken cells expressing oncogenic variants of the avian cellular src gene. Mol. Cell. Biol. 9:629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, A.B., L. Herbert, J.L. Cleveland, S.T. Berg, and J.R. Gaut. 1992. p120, a novel substrate of protein tyrosine kinase receptors and of p60v-src, is related to cadherin-binding factors beta-catenin, plakoglobin and armadillo. Oncogene. 7:2439–2445. [PubMed] [Google Scholar]
- Reynolds, A.B., J. Daniel, P.D. McCrea, M.J. Wheelock, J. Wu, and Z. Zhang. 1994. Identification of a new catenin: the tyrosine kinase substrate p120cas associates with E-cadherin complexes. Mol. Cell. Biol. 14:8333–8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehl, R., K. Johnson, R. Bradley, G.B. Grunwald, E. Cornel, A. Lilienbaum, and C.E. Holt. 1996. Cadherin function is required for axon outgrowth in retinal ganglion cells in vivo. Neuron. 17:837–848. [DOI] [PubMed] [Google Scholar]
- Shibamoto, S., M. Hayakawa, K. Takeuchi, T. Hori, K. Miyazawa, N. Kitamura, K.R. Johnson, M.J. Wheelock, N. Matsuyoshi, M. Takeichi, et al. 1995. Association of p120, a tyrosine kinase substrate, with E-cadherin/catenin complexes. J. Cell Biol. 128:949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staddon, J.M., C. Smales, C. Schulze, F.S. Esch, and L.L. Rubin. 1995. p120, a p120-related protein (p100), and the cadherin/catenin complex. J. Cell Biol. 130:369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi, K., T. Sasaki, H. Kotani, H. Nishioka, and Y. Takai. 1997. Regulation of cell-cell adhesion by rac and rho small G proteins in MDCK cells. J. Cell Biol. 139:1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi, M. 1995. Morphogenetic roles of classic cadherins. Curr. Opin. in Cell Biol. 7:619–627. [DOI] [PubMed] [Google Scholar]
- Thoreson, M.A., P.Z. Anastasiadis, J.M. Daniel, R.C. Ireton, M.J. Wheelock, K.R. Johnson, D.K. Hummingbird, and A.B. Reynolds. 2000. Selective uncoupling of p120(ctn) from E-cadherin disrupts strong adhesion. J. Cell Biol. 148:189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreson, M.A., and A.B. Reynolds. 2002. Altered expression of the catenin p120 in human cancer: implications for tumor progression. Differentiation. In press. [DOI] [PubMed] [Google Scholar]
- Vleminckx, K., L. Vakaet, Jr., M. Mareel, W. Fiers, and F. van Roy. 1991. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 66:107–119. [DOI] [PubMed] [Google Scholar]
- Wu, J., D.J. Mariner, M.A. Thoreson, and A.B. Reynolds. 1998. Production and characterization of monoclonal antibodies to the catenin p120ctn. Hybridoma. 17:175–183. [DOI] [PubMed] [Google Scholar]
- Yap, A.S. 1998. The morphogenetic role of cadherin cell adhesion molecules in human cancer: a thematic review. Cancer Invest. 16:252–261. [DOI] [PubMed] [Google Scholar]
- Yap, A.S., C.M. Niessen, and B.M. Gumbiner. 1998. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J. Cell Biol. 141:779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]