Abstract
The nucleolus is the site of ribosome biosynthesis, but is now known to have other functions as well. In the present study we have investigated how the distribution of signal recognition particle (SRP) RNA within the nucleolus relates to the known sites of ribosomal RNA synthesis, processing, and nascent ribosome assembly (i.e., the fibrillar centers, the dense fibrillar component (DFC), and the granular component). Very little SRP RNA was detected in fibrillar centers or the DFC of the nucleolus, as defined by the RNA polymerase I–specific upstream binding factor and the protein fibrillarin, respectively. Some SRP RNA was present in the granular component, as marked by the protein B23, indicating a possible interaction with ribosomal subunits at a later stage of maturation. However, a substantial portion of SRP RNA was also detected in regions of the nucleolus where neither B23, UBF, or fibrillarin were concentrated. Dual probe in situ hybridization experiments confirmed that a significant fraction of nucleolar SRP RNA was not spatially coincident with 28S ribosomal RNA. These results demonstrate that SRP RNA concentrates in an intranucleolar location other than the classical stations of ribosome biosynthesis, suggesting that there may be nucleolar regions that are specialized for other functions.
Keywords: nucleolus; signal recognition particle; SRP RNA; ribosome synthesis; peptide nucleic acids
Introduction
That the nucleolus is the site of ribosome biosynthesis was discovered over 35 years ago (Vincent and Miller, 1966), but recently it has emerged that additional processes are likely to be taking place in this region of the nucleus (Pederson, 1998a, 1998b; Carmo-Fonseca et al., 2000; Olson et al., 2000; Visintin and Amon, 2000). One initial clue to this idea was the discovery that signal recognition particle (SRP)* RNA injected into the nucleus of mammalian cells transiently passes through the nucleolus before its appearance in the cytoplasm (Jacobson and Pederson, 1998). The biological relevance of this observation was indicated by the finding that the nucleolar transit depends on particular sequence domains in the SRP RNA molecule (Jacobson and Pederson, 1998). These results were reinforced by the subsequent finding that endogenous SRP RNA as well as expressed GFP-labeled SRP protein components are present in the nucleolus of mammalian cells (Politz et al., 2000). Subsequent work in Saccharomyces cerevisiae has also revealed the nucleolar presence of SRP proteins (Ciufo and Brown, 2000; Grosshans et al., 2001).
The significance of SRP components in the nucleolus is not presently understood. A plausible explanation is that the SRP is at least partially assembled in the nucleolus. In addition, because mature SRP interacts with cytoplasmic ribosomes, SRP assembly may be coordinated with ribosome assembly within the nucleolus, perhaps as a mutual quality control mechanism assuring proper assembly of each particle (Pederson and Politz, 2000). Because the tripartite structural organization of the nucleolus has been so extensively studied and defined in relation to the steps of ribosome biosynthesis (Goessens, 1984; Hadjiolov, 1985; Hernandez-Verdun, 1991; Spector, 1993; Shaw and Jordan, 1995; Scheer and Hock, 1999; Huang, 2002), we reasoned that an important step toward defining the role of SRP RNA in the nucleolus, and any potential interaction with ribosomal components, would be to determine the precise sites within the nucleolus at which it is localized.
Results
Previously, we detected endogenous SRP RNA in the nucleoli of rat NRK fibroblasts by in situ nucleic acid hybridization using conventional phosphodiester (PO) backbone oligodeoxynucleotide probes (Politz et al., 2000), but the signal intensity was not high enough for high resolution intranucleolar mapping studies. Oligonucleosides linked by peptide bonds (peptide nucleic acid [PNA] oligos) form more stable DNA/RNA hybrids (Nielsen, 1999); therefore, we investigated the use of PNA oligos as a way to increase the detection of SRP RNA in the nucleolus. Fig. 1 shows that we detected an increase in both nucleolar and cytoplasmic signal of SRP RNA using PNA oligos as compared with PO backbone probes. The specificity of this result was demonstrated by the lack of detectable hybridization when a PNA oligo complementary to the evolutionary diverged yeast SRP RNA was used (Fig. 1 E). Signal was also reduced to background levels if cells were treated with RNase before in situ hybridization (unpublished data), demonstrating that the PNA probes were hybridizing to RNA as expected. It should be noted that the respective efficiencies with which PNAs and PO oligos hybridize to identical RNA target sequences cannot be directly compared because PNAs are typically much shorter than PO backbone probes and often must be designed to target different sequences because of PNA solubility limitations.
Figure 1.
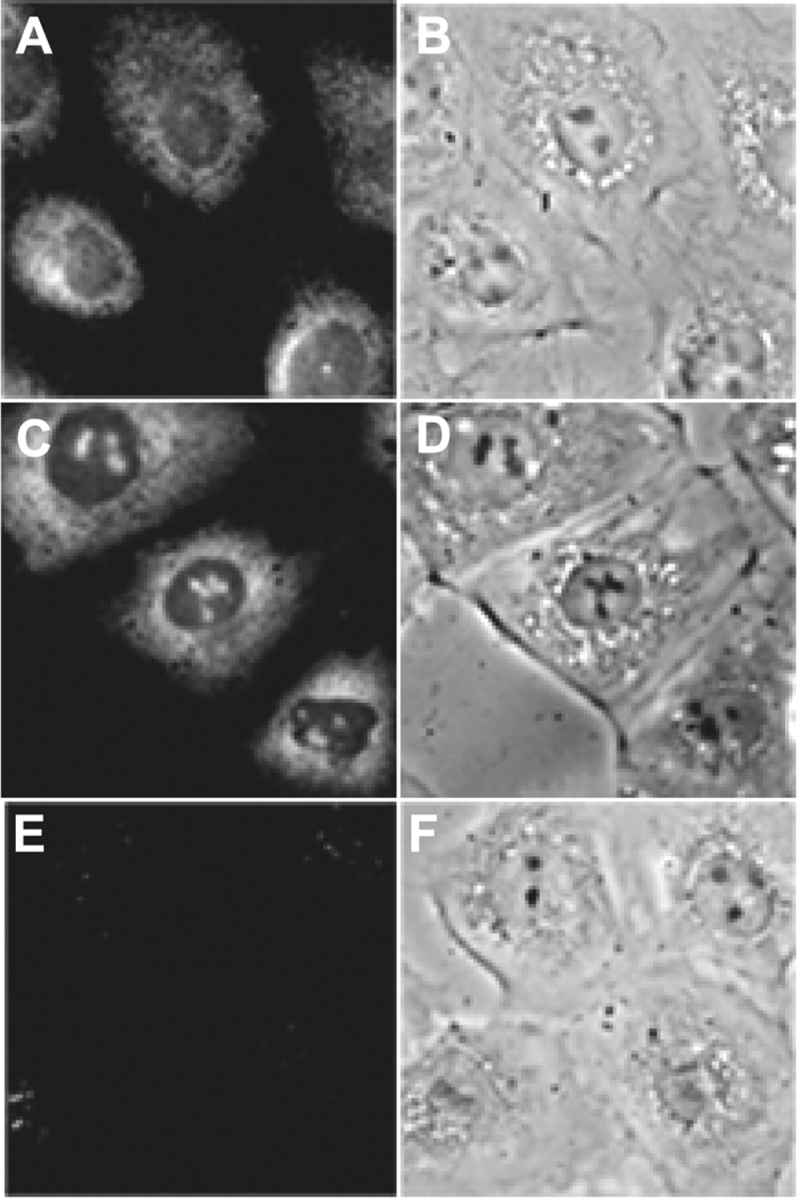
PNA probes enhance the detection of SRP RNA. In situ nucleic acid hybridization was performed on NRK cells using standard (PO backbone) oligodeoxynucleotides (A and B) or peptide nucleic acid (PNA) probes (C and D) complementary to human SRP RNA. A PNA probe complementary to S. pombe SRP RNA was used in parallel (E and F). (A, C, and E) Fluorescence images. (B, D, and F) Phase-contrast images.
The three domains of the nucleolus defined originally by electron microscopy and now as well by specifically localized proteins are (a) the fibrillar centers, in which the rDNA resides, for which upstream binding factor (UBF) is a marker; (b) the dense fibrillar component (DFC), into which nascent ribosomal RNA extends and rRNA processing and ribosome assembly commences, for which fibrillarin is a marker; and (c) the granular component, which contains partially processed rRNA and hosts the final stages of ribosome assembly, for which the protein B23 is a marker (Goessens, 1984; Hadjiolov, 1985; Hernandez-Verdun, 1991; Spector, 1993; Shaw and Jordan, 1995; Scheer and Hock, 1999).
UBF binds upstream of the rDNA promoters, and is thought to bind both actively transcribing genes as well as those open for transcription (Junera et al., 1997). As shown in Fig. 2 (A–F), SRP RNA only minimally overlapped with sites in NRK nucleoli that were immunostained for UBF. Similarly low levels of colocalization were also observed in HeLa cells expressing GFP-UBF to mark the fibrillar centers (Fig. 2, G–L). Next, three-dimensional optical stacks of immunostained, in situ–hybridized cells were processed using a constrained interactive deconvolution algorithm (Swedlow et al., 1997; Wallace et al., 2001) to obtain a higher resolution map of the intranucleolar space occupied by SRP RNA and UBF. Again, at this refined resolution, only a limited amount of overlap at the edges of a few fibrillar centers was observed (Fig. 3).
Figure 2.
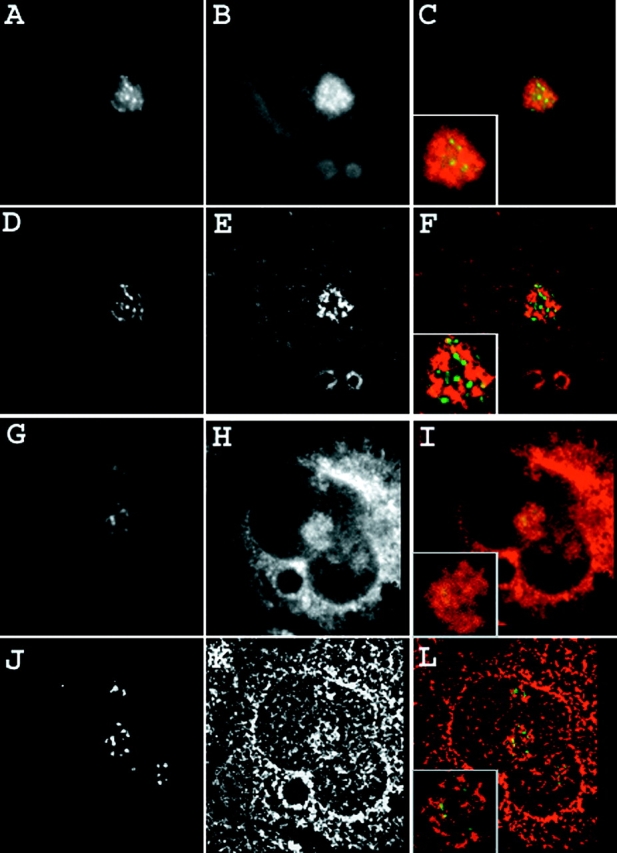
Combined detection of SRP RNA and nucleolar fibrillar centers. NRK cells were transfected with a GFP fusion of the RNA polymerase I–specific transcription factor UBF and, in parallel, untransfected NRK cells were immunostained for UBF. Both sets of cells were then subjected to in situ hybridization for SRP RNA with PNA probes (Materials and methods). (A–F) SRP RNA and UBF immunostaining. (A) Immunostained UBF. (B) SRP RNA. (C) Merged. (D–F) Nearest-neighbor deconvolutions of A–C, respectively. (G–L) SRP RNA and GFP-UBF. (G) GFP-UBF. (H) SRP RNA. (I) Merged. (J–L) Nearest-neighbor deconvolutions of G–I, respectively. (C, F, I, and L, insets) Nucleolus at higher magnification.
Figure 3.
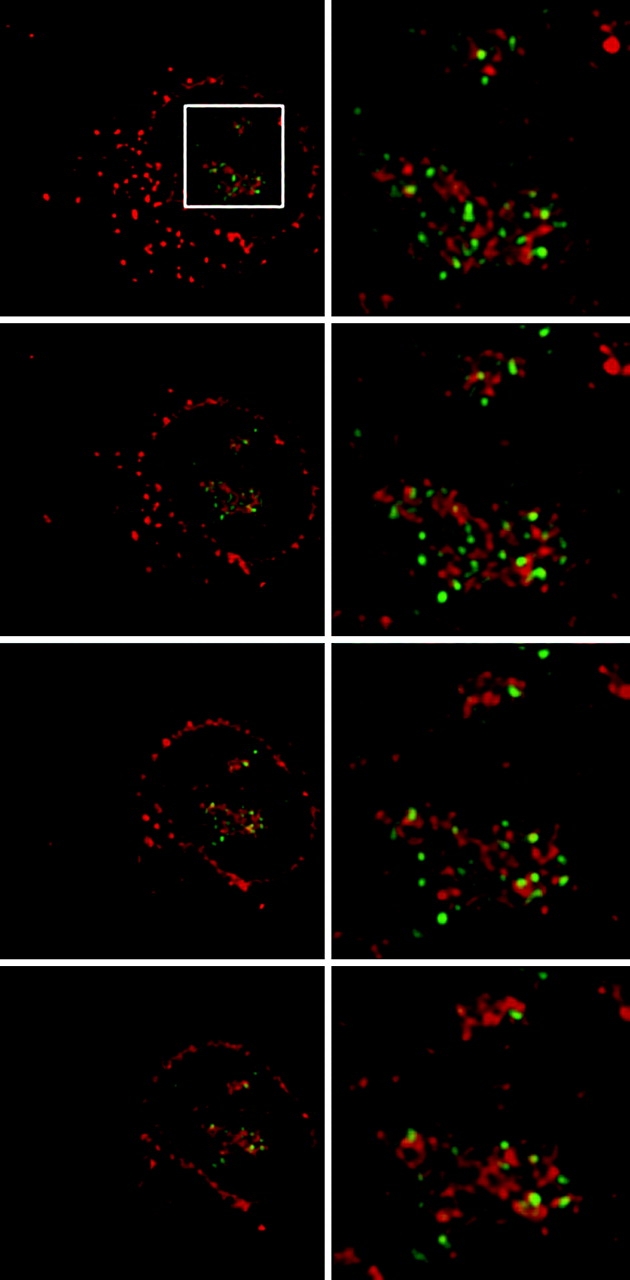
Three-dimensional deconvolution shows minimal overlap between UBF and SRP RNA. Image stacks were captured and subjected to constrained interative deconvolution as described in Materials and methods. (Left) Successive vertical midplanes of SRP RNA signal (red) in a NRK cell immunostained for UBF (green). (Right) Corresponding midplanes enlarged from the boxed region (top left). Yellow color indicates areas of overlap between UBF and SRP RNA.
SRP RNA was detectable in only low levels in the DFC, as defined by immunostaining of fibrillarin (Fig. 4) or by the localization of a yellow fluorescent protein fusion of fibrillarin (unpublished data). Colocalization between SRP RNA signal and fibrillarin protein was limited to a minimal spatial concordance of a minor fraction of SRP RNA along the edges of the DFC.
Figure 4.
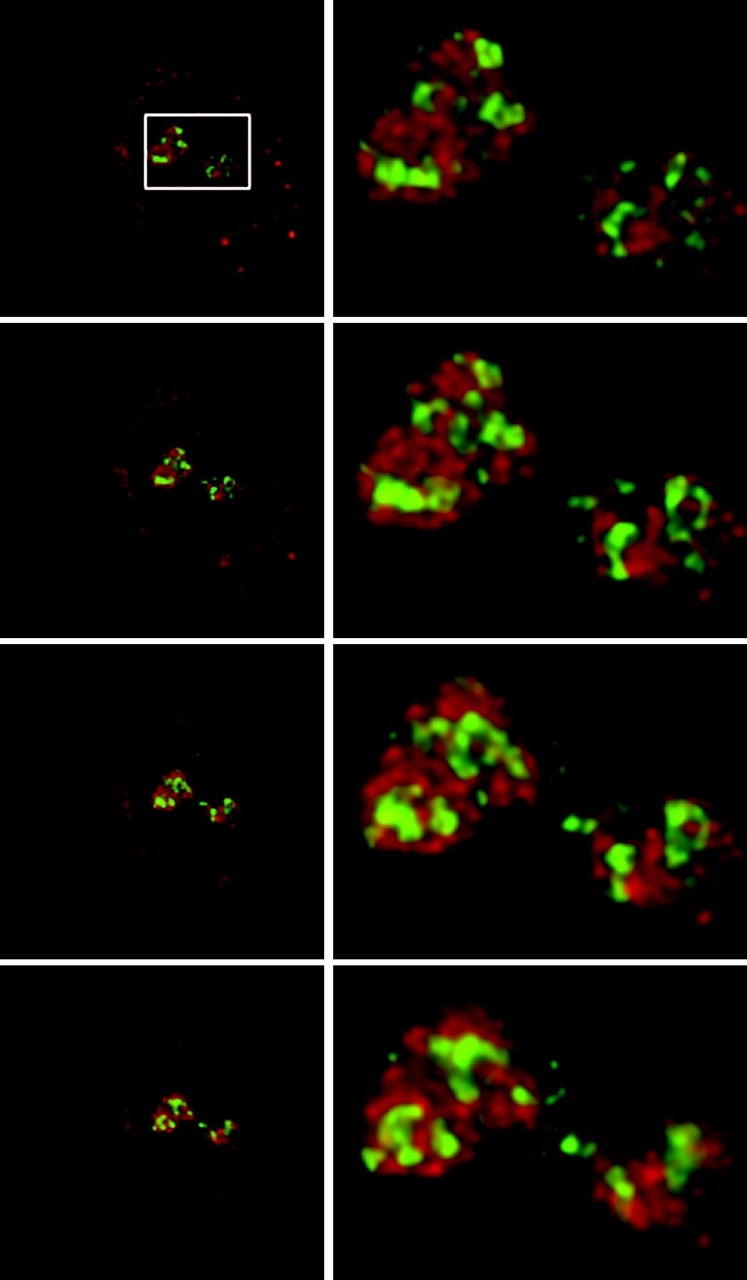
Three-dimensional deconvolution shows minimal overlap between fibrillarin and SRP RNA. NRK cells were subjected to immunostaining for fibrillarin and in situ hybridization for SRP RNA and image stacks were processed as in Fig. 5. (Left) Successive vertical midplanes of SRP RNA signal (red) and fibrillarin (green). (Right) Corresponding midplanes enlarged from the boxed region (top left). Yellow color indicates areas of overlap between fibrillarin and SRP RNA.
The same techniques were next used to map SRP RNA with respect to the granular component of the nucleolus. B23 is a multifunctional protein that is involved in nucleolar rRNA processing and ribosome assembly (Savkur and Olson, 1998; Szebeni and Olson, 1999; Okuwaki et al., 2002). It is primarily localized in the granular component in many cell types (Ochs et al., 1996). In agreement with these previous observations, we observed no overlap between concentrated regions of GFP-B23 and fibrillarin immunostaining in the NRK cells used in this study (unpublished data), demonstrating that B23 is a good marker for the granular component in these cells. A portion of SRP RNA overlapped with B23 in both immunostaining experiments (Fig. 5 F, arrow, yellow) and in the nucleoli of cells expressing GFP-B23 (Fig. 5 L, yellow). As can be seen in Fig. 5 (L), these areas of colocalization were distributed throughout the nucleolus, and were not limited to the edges of two adjoining signal regions. However, although colocalization was observed, a significant portion of SRP RNA was present in nucleolar regions where B23 was not concentrated (Fig. 5, F and L, red within nucleolus).
Figure 5.
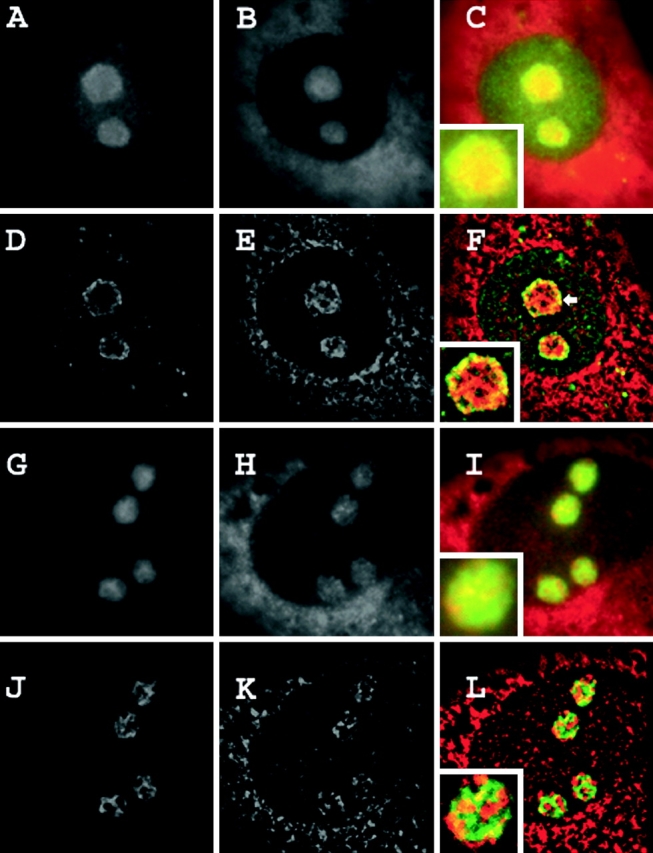
SRP RNA signal colocalizes with portions of the nucleolar granular component. (A–F) SRP RNA and B23 immunostaining. (A) B23. (B) SRP RNA. (C) Merged. (D–F) Nearest neighbor deconvolutions of A–C, respectively. (G–L) SRP RNA and GFP-B23. (G) GFP-B23. (H) SRP RNA. (I) Merged. (J–L) Nearest-neighbor deconvolutions of G–I, respectively. (C, F, I, and L, insets) Nucleolus at higher magnification.
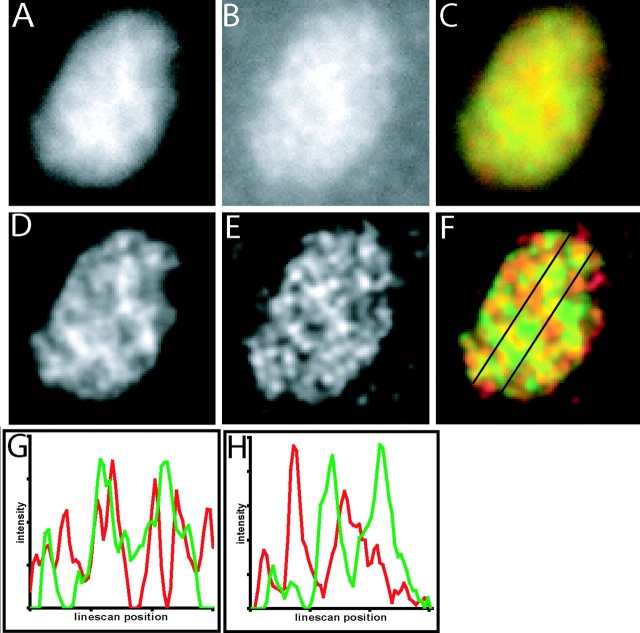
Using constrained iterative deconvolution to increase the resolution of subnucleolar regions, the intensity distribution of SRP RNA signal (Fig. 6, red) was found to clearly differ from that of B23 (Fig. 6 F, green). Specifically, the most concentrated regions of SRP RNA (Fig. 6, G and H, red peaks in linescans) often did not overlap with the most concentrated regions of B23 (Fig. 6, G and H, green peaks in linescans). Although the fraction of SRP RNA that colocalized with B23 varied among nucleoli in the same cell and between cells, all nucleoli contained some SRP RNA signal that was concentrated in intranucleolar regions where B23, fibrillarin, and UBF were least concentrated.
Figure 6.
Constrained iterative deconvolution of SRP RNA and B23 protein signal in the nucleolus. Raw (A) and deconvolved (D) midplane of a NRK cell nucleolus containing GFP-B23. Raw (B) and deconvolved (E) midplane of same nucleolus showing SRP RNA signal. (C and F) Overlays of B23 (green) and SRP RNA (red) images showing regions of similar intensities in yellow. (G) Linescan of left vertical line in F. (H) Linescan of right vertical line in F. Each linescan shows, from left to right, the intensities (arbitrary units) of B23 (green) and SRP RNA (red) along the line indicated in F, proceeding downward from top to bottom. Linescans are displayed with the minimal linescan intensity at the origin of the y-axis.
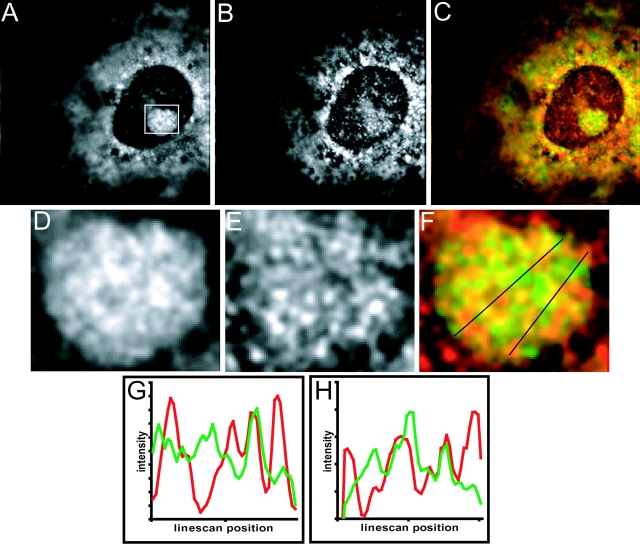
Because these results so far indicated that a considerable portion of SRP RNA resides in intranucleolar regions that do not correspond with any of the three classically defined sites of ribosome synthesis, we sought to confirm this conclusion by determining the spatial relationship of SRP RNA to 28S ribosomal RNA in the nucleolus by performing double in situ hybridization experiments. As shown in the deconvolved images in Fig. 7, although some SRP RNA signal was coincident with 28S rRNA (Fig. 7 F, yellow signal; overlapping green and red peaks in linescans), a significant portion was not (Fig. 7 C, red; red peaks in linescans). Thus, all our results taken together reveal a region extending throughout the nucleolus from which the markers characteristic of the three classical stations of ribosomal biosynthesis are absent (or present in low concentration), and in which 28S rRNA is also deficient. SRP RNA is concentrated within this previously unidentified region of the nucleolus.
Figure 7.
SRP RNA and 28S rRNA distribution in the nucleolus revealed by double in situ hybridization. Deconvolved midplane of NRK cell containing 28S rRNA signal (A), SRP RNA signal (B), and these two images combined (C) where 28S rRNA is green and SRP RNA is red. Overlapping regions of similar intensities are shown in yellow. (D–F) Deconvolved images of nucleolus shown in boxed area in A. (D) 28S rRNA signal. (E) SRP RNA signal. (F) Color combined images of D and E coded as in the C above. (G) Linescan of leftmost line in F. (H) Linescan of rightmost line in F. Each linescan shows, from left to right, the intensities (arbitrary units) of 28S rRNA (green) and SRP RNA (red) along the line indicated in F, proceeding downward from top to bottom. Linescans are displayed with the minimal linescan intensity at the origin of the y-axis.
Discussion
The discovery that there is a connection between the nucleolus and SRP RNA was first made when fluorescent SRP RNA was microinjected into the nucleus of mammalian cells and observed to transiently pass through the nucleolus (Jacobson and Pederson, 1998). It was subsequently found that endogenous SRP RNA is also present in the nucleolus of mammalian cells as are some of the SRP's protein components (Politz et al., 2000). Our goal in the present study was to determine the intranucleolar distribution of SRP RNA at higher spatial resolution than in our earlier work, and this was made possible by our use of PNA probes for in situ hybridization. This resulted in a substantial increase in the detection of SRP RNA in the nucleolus. Indeed, the levels of SRP RNA detected in the nucleolus with PNA probes were similar to or greater than, on an average intensity per pixel basis, that observed in the most SRP RNA–rich regions of the cytoplasm. Although we do not know the efficiency with which our PNA probes detect SRP RNA, it is to be noted that SRP RNA was readily observed biochemically in previous studies with purified rat hepatoma and HeLa cell nucleoli (Reddy et al., 1981; Mitchell et al., 1999). Our results confirm these cell fractionation studies and further suggest that, at least as determined by in situ nucleic acid hybridization, the level of SRP RNA in the nucleolus is quantitatively substantial.
The major goal of the present study was to resolve the intranucleolar distribution of SRP RNA within the nucleolus, particularly in relation to the stations of ribosome synthesis as defined by the classical tripartite organization of the nucleolus. Because we have hypothesized that SRP RNA in the nucleolus is related to the regulated construction of the overall translation machinery (Pederson and Politz, 2000), we wished to determine whether SRP RNA might be uniquely present in one, and only one, of the three nucleolar compartments, as a clue to its possible role at a discrete step in ribosome biosynthesis. In the present study we have used the RNA polymerase I–specific transcription factor UBF to demarcate the fibrillar centers, the protein fibrillarin as a fiduciary landmark of the DFC of the nucleolus, and the protein B23 to identify the rRNA containing regions of the granular component. In all three cases, we used both specific antibodies as well as fluorescent protein expression to identify these components of the nucleolus, combined with the highly sensitive detection of SRP RNA. Additionally, we performed dual in situ hybridization experiments to resolve the spatial relationship between 28S rRNA and SRP RNA. We found that very little SRP RNA was present in the fibrillar centers and the DFC. Rather, a portion of the SRP RNA was localized with B23 in the granular component and, surprisingly, the remainder was concentrated in rRNA deficient regions of the nucleolus.
The intranucleolar distribution pattern of SRP RNA observed in the present investigation renders unlikely a model where the SRP RNA is intimately associated with precursor ribosomes throughout their synthesis and assembly. SRP RNA is not concentrated in either fibrillar centers or the DFC and therefore it probably does not play a role during rRNA transcription or early processing in any direct, interactive fashion, although it remains possible that very low concentrations of SRP RNA might carry out functional interactions with formative ribosomes at these sites.
Rather, our results indicate that SRP RNA might interact with nucleolar ribosomes much later during assembly, perhaps within the B23-rich portion of the granular component. However, the SRP RNA that was localized within the granular component displayed a heterogeneous distribution and was not closely correlated with the abundance of B23 protein. The most concentrated regions of SRP RNA were often not coincident with concentrated regions of B23 protein, indicating that portions of SRP RNA and B23 protein have different locations within the granular component.
Studies at the electron microscopic level have generally conveyed the granular component as having a fairly compact and generally homogenous particulate appearance (for review see Hadjiolov, 1985). A protein termed No55 identified by Ochs et al. (1996) using a rare human patient autoimmune serum, and not presently known to be involved in ribosome biosynthesis, was found to be distributed fairly homogeneously throughout the granular component. However, B23, which is involved in a rRNA processing step (Savkur and Olson, 1998; Okuwaki et al., 2002), exhibits a somewhat uneven distribution within the granular component (Ochs et al., 1996; unpublished data). Our finding that a substantial portion of the SRP RNA is present in regions at which B23 is not concentrated (and yet are not DFC) may indicate that within the granular component itself, particular activities are spatially organized into different functional domains. Such activities may require different levels of SRP RNA as compared with B23. Indeed, it is not known whether all of the “granules” of the granular component that are visualized by electron microscopy actually represent nascent ribosomal subunits, or whether there are other particles present also. Our results raise the possibility that a portion of the particulate texture of the granular component might in part represent nascent SRPs, and that regions containing these particles are identifiable at the fluorescent light microscopy level.
Consistent with our results showing that SRP RNA is concentrated in regions other than fibrillar centers, the DFC or B23-rich regions of the nucleolus, we also found that SRP RNA was most concentrated in regions of the nucleolus that were deficient in 28S rRNA. This suggests that this portion of SRP RNA may not directly interact with ribosomes during assembly, but instead is sequestered from those areas. To the best of our knowledge, this intranucleolar distribution pattern has not been previously observed, and it raises a number of intriguing issues. Do these SRP RNA–rich areas have different structural characteristics that can be detected using electron microscopy, now that we are alerted to their existence? What else might colocalize within this domain of the nucleolus? More and more factors apparently unrelated to ribosome biogenesis are being found in the nucleolus, including several growth factors, components of gene silencing machinery, anaphase exit equipment and telomerase (Pederson, 1998a, 1998b; Mitchell et al., 1999; Carmo-Fonseca et al., 2000; Olson et al., 2000; Visintin and Amon, 2000; Wong et al., 2002). These various factors would not necessarily be expected to associate with the nucleolar regions dedicated to ribosome biosynthesis (and indeed might well disrupt ribosome synthesis if they did). Thus, it is conceivable that the SRP RNA-rich areas of the nucleolus discovered in the present study may define territories in which activities other than ribosome biosynthesis predominate. An intriguing possibility, among others, is that the functions taking place in this compartment may nonetheless be coordinated with the ribosome synthesis pathway, perhaps in a cell cycle regulated way.
Materials and methods
Cells
NRK (rat kidney–derived) fibroblasts were cultured as previously described (Wang et al., 1991). HeLa cells were grown in Eagle's minimum essential medium containing 10% fetal bovine serum.
PNA probes and in situ hybridization
PNA probes complementary to nucleotides 88–102, 208–221, and 231–245 of rat (conserved in human) SRP RNA were synthesized by PE Biosystems. Each PNA was labeled with a rhodamine group at its 5′ end. A PNA probe complementary to nucleotides 231–245 of Schizosaccharomyces pombe SRP RNA was also synthesized for use as a control (Results). Cy3-labeled PO backbone oligodeoxynucleotides complementary to SRP RNA were as described earlier (oligo 1 is complementary to nucleotides 208–240, oligo 2 to 118–151, oligo 3 to 249–284; Politz et al., 2000).
PO oligos complementary to rat 28S rRNA were as follows (see De Rijk et al., 1999 for database and nomenclature information): Oligo 1 in loop E11_1 (D7b), G*TACCGGCAC*GGACGCC*CGCGGCGCCCA*C; Oligo 2 in loop E9_1 (D-7a), C*GAGGGCAACGGAGGCCA*CGCCCG*CCCT*C; Oligo 3 in loop B13_1 (D1), G*ACGCCACAT*TCCCGCGCC*CGGCGCGCG*C; Oligo 4 in loop C1_1 (D2), C*CGCGCCGCCGGG*TCAATCC*CCGGGCGG*C; and Oligo 5 in loops H1_2, H1_3 (D12), A*GGCTC*CCGCACCGGACCCCGG*CCCGAC*C. *Indicates positions of aminohexyl-modified thymidine residues added during synthesis (Integrated DNA Technologies) and subsequently labeled with fluorescein (Politz and Singer, 1999). These five oligos were used together to detect 28S rRNA.
The methods used for cell fixation, permeabilization and in situ hybridization were as previously described (Politz et al., 2000), except that 3.8 ng of each of the three SRP PNAs (or 11.5 ng of yeast control PNA) in 25 μl total hybridization mixture was used per coverslip. Coverslips were then mounted in Prolong Antifade mounting medium (Molecular Probes) and dried overnight at room temperature before viewing.
Antibodies and immunofluorescence
UBF, a RNA polymerase I–specific transcription factor specifically localized in fibrillar centers of the nucleolus, was detected with an antibody (Chan et al., 1991; Roussel et al., 1993; Dousset et al., 2000) provided by Daniele Hernandez-Verdun (Institut Jacques Monod, Paris, France). Fibrillarin, a protein specific to the DFC of the nucleolus, was detected with a monoclonal antibody we have previously used (Jacobson et al., 1995), provided by Edward Chan and Eng Tan (Scripps Research Institute, La Jolla, CA). B23, which identifies the granular component, was detected with a monoclonal antibody (Ochs et al., 1983) provided by Pui-Kwong Chan (Baylor College of Medicine, Houston, TX). Immunostaining and sequential in situ hybridization was performed using a minor modification of a protocol provided by Sui Huang (Northwestern University School of Medicine, Chicago, IL). Cells were grown on coverslips and fixed for 12 min in 4% (vol/vol) formaldehyde in PBS, containing 5 mM MgCl2. Fixation and all subsequent steps were performed at room temperature unless otherwise noted. Coverslips were washed three times in PBS containing 1% bovine serum albumin (PBSB) for 5–10 min each, incubated for 5 min in 0.5% Triton X-100 in PBSB, and then again washed three times in PBSB for 5–10 min each time. Coverslips were then incubated with the desired primary antibody (1:2,000 for anti-B23, 1:75 for anti-fibrillarin and 1:100 for anti-UBF) in PBSB for 1 h in a humidified chamber, washed three times in PBSB (10 min each), incubated with the appropriate secondary antibody (1:200 anti–mouse IgG for B23, 1:80 anti–mouse IgG for fibrillarin and 1:750 anti–human IgG for UBF; all secondary antibodies were from Sigma-Aldrich) in PBSB in humidified chambers for one hour and then washed three times in PBSB (10 min each). Cells were then refixed by immersing the coverslips in 2% (vol/vol) formaldehyde in PBS containing 5 mM MgCl2, for 5 min, followed by three rinses in 70% (vol/vol) ethanol. Coverslips were stored in absolute ethanol at 4°C for 18–24 h before performing in situ hybridization as described above.
Fluorescent fusion proteins
Plasmids encoding green fluorescent protein fusions to human UBF and human B23 were obtained from Sui Huang (Northwestern University School of Medicine; Chen and Huang, 2001). A plasmid encoding a yellow fluorescent protein fusion to human fibrillarin was obtained from Angus Lamond (University of Dundee, Dundee, Scotland). These fluorescent protein-encoding plasmids were transfected into NRK cells with Lipofectamine 2000 (Invitrogen Life Technologies) following manufacturer's instructions. In the case of YFP-fibrillarin, a stable cell line was constructed. HeLa cells were transfected as above and 18 h later the medium was replaced with fresh medium containing 800 μg/ml Geneticin (G-418; GIBCO BRL). The medium was changed twice weekly and the cells were cultured for 6–8 wk before subcloning and further selection.
Microscopy and image processing
Results were analyzed with a Leica DMIRB microscope equipped with a 100× objective (N.A. 1.4) and appropriate filter sets, and images were captured using a Quantix 57 CCD camera (Roper Scientific Photometrics). For high resolution spatial mapping, three-dimensional optical stacks (containing 21 consecutive 0.25 micron slices) were captured using a PIFOC microscope focusing drive (Polytec PI). Images were dark current subtracted, intensity scaled, and in some cases, subjected to two-dimensional nearest-neighbor deconvolution using Metamorph software. Alternatively, image stacks were processed by constrained iterative deconvolution (Applied Precision) using an empirical point-spread function (Swedlow et al., 1997; Wallace et al., 2001).
Acknowledgments
We thank Christina N. Alavian in our laboratory for her important role in the initial application of PNA oligos for detection of SRP RNA in the nucleolus. We are very grateful to Jason Swedlow (University of Dundee) for his most generous assistance in carrying out the constrained iterative deconvolution of some of our images. We also thank Craig Brumwell in our laboratory for constructing the YFP-fibrillarin stable cell line, and Christine Powers for help with the B23 experiments. We also gratefully acknowledge the following scientists, who provided key materials: Pui-Kwong Chan, Edward Chan, Daniele Hernandez-Verdun, Sui Huang, Angus Lamond, and Eng Tan.
This work was supported by National Institutes of Health grant GM-21595, which requires us to state that the content of our paper is not the official position of the U.S. government.
Footnotes
Abbreviations used in this paper: DFC, dense fibrillar component; PNA, peptide nucleic acid; PO, phosphodiester; SRP, signal recognition particle; UBF, upstream binding factor.
References
- Carmo-Fonseca, M., L. Mendes-Soares, and I. Campos. 2000. To be or not to be in the nucleolus. Nat. Cell Biol. 2:E107–E112. [DOI] [PubMed] [Google Scholar]
- Chan, E.K.L., H. Imai, J.C. Hamel, and E.M. Tan. 1991. Human autoantibody to RNA polymerase I transcription factor hUBF: molecular identity of nucleolus organizer region autoantigen NOR-90 and ribosomal RNA transcription upstream binding factor. J. Exp. Med. 174:1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., and S. Huang. 2001. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153:169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciufo, L.F., and J.D. Brown. 2000. Nuclear export of yeast signal recognition particle lacking Srp54p by the Xpo1p/Crm1p NES-dependent pathway. Curr. Biol. 10:1256–1264. [DOI] [PubMed] [Google Scholar]
- De Rijk, P., E. Robbrecht, S. De Hoog, A. Caers, Y. Van de Peer, and R. De Wachter. 1999. Database on the structure of the large subunit ribosomal RNA. Nucleic Acid Res. 27:174–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dousset, T., C. Wang, C. Verheggen, D. Chen, D. Hernandez-Verdun, and S. Huang. 2000. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol. Biol. Cell. 11:2705–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goessens, G. 1984. Nucleolar structure. Int. Rev. Cytol. 87:107–158. [DOI] [PubMed] [Google Scholar]
- Grosshans, H., K. Deinert, E. Hurt, and G. Simos. 2001. Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J. Cell Biol. 153:745–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolov, A.A. 1985. Cell Biology Monographs. Vol. 12. Springer-Verlag, Vienna/Berlin. 263 pp.
- Hernandez-Verdun, D. 1991. The nucleolus today. J. Cell Sci. 99:465–471. [DOI] [PubMed] [Google Scholar]
- Huang, S. 2002. Building an efficient factory: where is pre-rRNA synthesized in the nucleolus? J. Cell Biol. 157:739–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, M.R., L.-G. Cao, Y.-L. Wang, and T. Pederson. 1995. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J. Cell Biol. 131:1649–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, M.R., and T. Pederson. 1998. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc. Natl. Acad. Sci. USA. 95:7981–7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junera, H.R., C. Masson, G. Geraud, J. Suja, and D. Hernandez-Verdun. 1997. Involvement of in situ conformation of ribosomal genes and selective distribution of upstream binding factor in rRNA transcription. Mol. Biol. Cell. 8:145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J.R., J. Cheng, and K. Collins. 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 19:567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, P.E. 1999. Applications of peptide nucleic acids. Curr. Opin. Biotechnol. 10:71–75. [DOI] [PubMed] [Google Scholar]
- Ochs, R., M. Lischwe, P. O'Leary, and H. Busch. 1983. Localization of nucleolar phosphoproteins B23 and C23 during mitosis. Exp. Cell Res. 146:139–149. [DOI] [PubMed] [Google Scholar]
- Ochs, R.L., T.W. Stein, Jr., E.K. Chan, M. Ruutu, and E.M. Tan. 1996. cDNA cloning and characterization of a novel nucleolar protein. Mol. Biol. Cell. 7:1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuwaki, M., M. Tsujimoto, and K. Nagata. 2002. The RNA binding activity of a ribosome biogenesis factor, nucleophosmim/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol. Biol. Cell. 13:2016–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, M.O.J., M. Dundr, and A. Szebeni. 2000. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10:189–196. [DOI] [PubMed] [Google Scholar]
- Pederson, T. 1998. a. Growth factors in the nucleolus? J. Cell Biol. 143:279–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson, T. 1998. b. The plurifunctional nucleolus. Nucleic Acids Res. 26:3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson, T., and J.C. Politz. 2000. The nucleolus and the four ribonucleoproteins of translation. J. Cell Biol. 148:1091–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz, J.C., and R.H. Singer. 1999. In situ reverse transcription for detection of hybridization between oligonucleotides and their intracellular targets. Methods. 18:281–285. [DOI] [PubMed] [Google Scholar]
- Politz, J.C., S. Yarovoi, S. Kilroy, K. Gowda, C. Zwieb, and T. Pederson. 2000. Signal recognition particle components in the nucleolus. Proc. Natl. Acad. Sci. USA. 97:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, R., W.-Y. Li, D. Henning, Y.C. Choi, K. Nohga, and H. Busch. 1981. Characterization and subcellular localization of 7-8 S RNAs of Novikoff hepatoma. J. Biol. Chem. 25:8452–8457. [PubMed] [Google Scholar]
- Roussel, P., C. Andre, C. Masson, G. Geraud, and D. Hernandez-Verdun. 1993. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J. Cell Sci. 104:327–337. [DOI] [PubMed] [Google Scholar]
- Savkur, R., and M.O.J. Olson. 1998. Preferential cleavage in pre-ribosomal RNA by protein B23 endoribonuclease. Nucleic Acids Res. 26:4508–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, U., and R. Hock. 1999. Structure and function of the nucleolus. Curr. Opin. Cell Biol. 11:385–390. [DOI] [PubMed] [Google Scholar]
- Shaw, P.J., and E.G. Jordan. 1995. The nucleolus. Annu. Rev. Cell Dev. Biol. 11:93–121. [DOI] [PubMed] [Google Scholar]
- Spector, D.L. 1993. Macromolecular domains within the cell nucleus. Annu. Rev. Biochem. 9:265–315. [DOI] [PubMed] [Google Scholar]
- Swedlow, J.R., J.W. Sedat, and D.A. Agard. 1997. Deconvolution in optical microscopy. In Deconvolution of Images and Spectra. P.A. Jansson, editor. Academic Press, New York. 284–309.
- Szebeni, A., and M.O.J. Olson. 1999. Nucleolar protein B23 has molecular chaperone activity. Protein Sci. 8:905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, W.S., and O.L. Miller, Jr. 1966. International Symposium on the Nucleolus. Its Structure and Function. Montevideo, Uruguay. J. Natl. Cancer Inst. Monograph. 23:1–610.
- Visintin, R., and A. Amon. 2000. The nucleolus: the magician's hat for cell cycle tricks. Curr. Opin. Cell Biol. 12:372–377. [DOI] [PubMed] [Google Scholar]
- Wallace, W., L.H. Schaefer, and J.R. Swedlow. 2001. A working person's guide to deconvolution in light microscopy. Biotechniques. 31:1076–1097. [DOI] [PubMed] [Google Scholar]
- Wang, J., L.G. Cao, Y.L. Wang, and T. Pederson. 1991. Localization of pre-messenger RNA at discrete nuclear sites. Proc. Natl. Acad. Sci. USA. 88:7391–7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, J.M.Y., L. Kusdra, and K. Collins. 2002. Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat. Cell Biol. 4:731–736. [DOI] [PubMed] [Google Scholar]