Abstract
In Escherichia coli, ribosomes must interact with translocons on the membrane for the proper integration of newly synthesized membrane proteins, cotranslationally. Previous in vivo studies indicated that unlike the E. coli signal recognition particle (SRP), the SRP receptor FtsY is required for membrane targeting of ribosomes. Accordingly, a putative SRP-independent, FtsY-mediated ribosomal targeting pathway has been suggested (Herskovits, A.A., E.S. Bochkareva, and E. Bibi. 2000. Mol. Microbiol. 38:927–939). However, the nature of the early contact of ribosomes with the membrane, and the involvement of FtsY in this interaction are unknown. Here we show that in cells depleted of the SRP protein, Ffh or the translocon component SecE, the ribosomal targeting pathway is blocked downstream and unprecedented, membrane-bound FtsY–ribosomal complexes are captured. Concurrently, under these conditions, novel, ribosome-loaded intracellular membrane structures are formed. We propose that in the absence of a functional SRP or translocon, ribosomes remain jammed at their primary membrane docking site, whereas FtsY-dependent ribosomal targeting to the membrane continues. The accumulation of FtsY-ribosome complexes induces the formation of intracellular membranes needed for their quantitative accommodation. Our results with E. coli, in conjunction with recent observations made with the yeast Saccharomyces cerevisiae, raise the possibility that the SRP receptor–mediated formation of intracellular membrane networks is governed by evolutionarily conserved principles.
Keywords: E. coli; signal recognition particle; ribosome; protein targeting; membrane proteins
Introduction
Membrane-bound ribosomes in E. coli were extensively studied over 20 years ago. Despite clear evidence that this population of ribosomes is involved in protein synthesis (Randall and Hardy, 1975; Smith et al., 1978; Green and Inouye, 1983), this research venue decayed. Recently, it has been recognized that membrane-bound ribosomes are crucial for biogenesis of integral membrane proteins in E. coli, thus renewing interest in ribosome targeting to and association with the membrane in this organism. Similar to the mammalian protein targeting system (Walter and Johnson, 1994), E. coli also possesses signal recognition particle (SRP)* machinery (Rapoport, 1991; Luirink and Dobberstein, 1994), implicated in cotranslational membrane protein targeting (for review see Herskovits et al., 2000; de Gier and Luirink, 2001). This targeting system includes two proteins, Ffh (a homologue of the eukaryotic SRP54 protein) and FtsY (a homologue of the α-subunit of the eukaryotic SRP-receptor, SRα) (Bernstein et al., 1989; Romisch et al., 1989). The Ffh protein is important for proper assembly of integral membrane proteins (Macfarlane and Muller, 1995; de Gier et al., 1996; Ulbrandt et al., 1997), and FtsY is required for their expression (Seluanov and Bibi, 1997; unpublished data). Interestingly, only FtsY was shown to be essential for membrane targeting of ribosomes in vivo (Herskovits and Bibi, 2000), suggesting an alternative SRP-independent, FtsY-mediated targeting of ribosomes to the cytoplasmic membrane in E. coli (Herskovits et al., 2000). After targeting to the cytoplasmic membrane, ribosomes are transferred to the SecYEG site (the E. coli translocon) (Fig. 1, f) (Valent et al., 1998; Prinz et al., 2000a) or other translocation sites (Cristobal et al., 1999), but the molecular details of their initial interaction(s) with the membrane have remained unclear. Possibly, these interactions are transient and therefore attempts to isolate the relevant complexes in E. coli (other than the ribosome–translocon complex) have thus far been unsuccessful. We reasoned that by blocking essential targeting steps downstream, the putative transient ribosome-membrane contact(s) might be stabilized and prolonged. Our results show that under conditions of efficient depletion of the translocon or the SRP, membrane-bound ribosomal complexes containing FtsY are indeed accumulated, in accordance with the proposed role of FtsY in membrane targeting of ribosomes (Herskovits et al., 2000). Moreover, in both cases, novel endoplasmic membrane networks are synthesized possibly in response to the increase in the number of membrane-bound ribosomes. We propose that the FtsY-ribosome complexes represent primary membrane-docking sites for ribosomes in E. coli.
Figure 1.
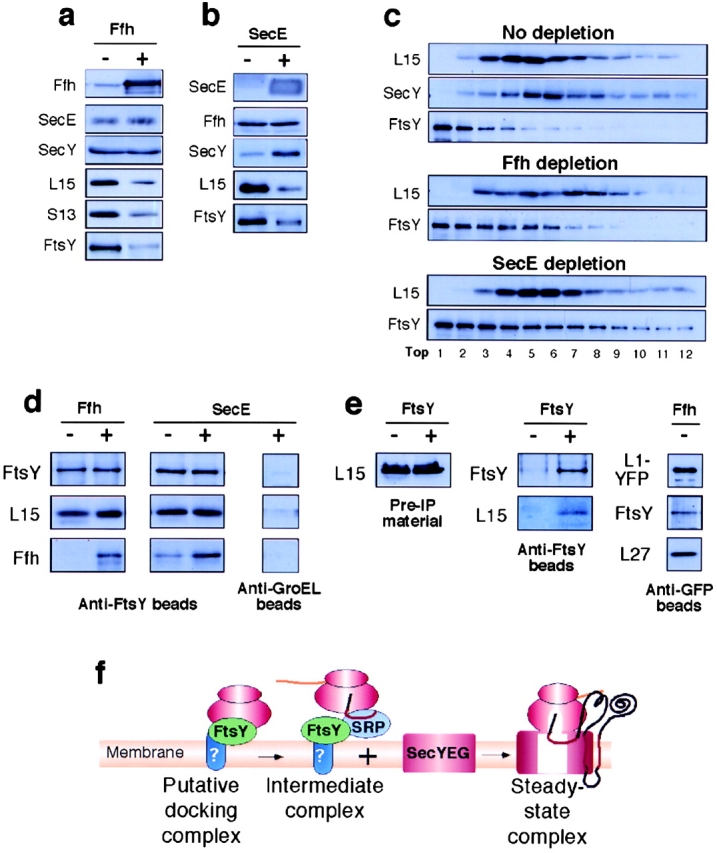
Accumulation of membrane-bound FtsY–ribosome complexes. Accumulation as analyzed by Western blotting (a, b, and c) and immunoprecipitation (d and e). (a and b) Accumulation of membrane-bound ribosomal proteins and FtsY in cells depleted of Ffh or SecE, respectively. (c) Sucrose density gradients of digitonin-solubilized membranes. (d) Coimmunoprecipitation of ribosomal proteins and Ffh with anti-FtsY antibodies. Purified membranes were solubilized by digitonin and the soluble material was incubated with anti-FtsY beads or anti-groEL beads as control. Immunoprecipitates were calibrated for equal amounts of FtsY by a separate semiquantitative Western blotting (not depicted) and then analyzed by Western blotting. (e) Control coimmunoprecipitation studies: total ribosomal fractions (left, membrane and cytosolic ribosomes, indicated as Pre-IP materials) from FtsY-depleted and nondepleted cells were coimmunoprecipitated with anti-FtsY beads and tested by Western blotting with anti-FtsY and anti-L15 antibodies (e, middle). (e, right) solubilized membranes from Ffh-depleted cells were immunoprecipitated with anti-YFP beads and the precipitates were analyzed by Western blotting with anti-GFP, anti-FtsY and anti-L27 antibodies. (f) Schematic representation of putative membrane-bound ribosomal complexes that form during and at the end of the ribosome targeting pathway.
Results and discussion
Accumulation of membrane-bound FtsY and ribosomes in cells depleted of Ffh or SecE
In mammalian cells (Gorlich et al., 1992) and in yeast (Prinz et al., 2000b) the translocon is the main ribosome receptor. Similarly, in E. coli the translocon also interacts with ribosomes with a high affinity (Prinz et al., 2000a). Therefore, at steady state, E. coli membrane preparations probably contain mainly ribosome-translocon complexes. In order to investigate earlier stages during membrane association of ribosomes, membranes were isolated from cells depleted of functional translocons (SecE-depleted) or functional SRP complexes (Ffh-depleted), and their content of ribosomes and additional components was analyzed. The results showed that Ffh-depleted membranes contain very little Ffh as expected (Fig. 1 a), and that membranes prepared from SecE-depleted cells contain almost no SecE and only a little SecY (Fig. 1 b), in agreement with the documented stabilizing effect of SecE on SecY (Nishiyama et al., 1992). In contrast to the observation that SecE or Ffh depletion did not affect each other's expression on the membrane, the amounts of membrane-bound ribosomes and FtsY were considerably affected. The Ffh-depleted membranes contained increased amounts of the ribosomal proteins L15 and S13, and also of FtsY, compared with nondepleted membranes (Fig. 1 a). Similar accumulation of ribosomal proteins and FtsY was observed in SecE-depleted membranes (Fig. 1 b). The accumulation of FtsY and ribosomes on the membrane requires neither Ffh nor translocon, as shown in Ffh- and SecE-depleted cells, respectively. Therefore, we hypothesized that the parallel accumulation of membrane-bound ribosomes and FtsY and the documented FtsY requirement for ribosomal targeting (Herskovits and Bibi, 2000) imply that FtsY plays functional or structural roles in the initial attachment of the ribosomes to the cytoplasmic membrane (Herskovits et al., 2001). Initial indication that FtsY might be associated with membrane-bound ribosomes in depleted cells arose from density gradient centrifugation experiments with solubilized membranes (Fig. 1 c). In samples from non-depleted cells, although some FtsY migrated with ribosomes, most FtsY was found in the top fractions, suggesting that at steady state, only some FtsY might be associated with membrane ribosomes. However, in samples from Ffh- or SecE-depleted cells, a marked shift was observed and considerable amount of FtsY was found comigrating with ribosomes, suggesting that more putative FtsY–ribosome complexes were trapped on the membrane under these conditions.
FtsY forms a membrane-bound complex with the ribosome in the absence of Ffh
Next, we examined the interaction between FtsY and membrane-bound ribosomes in depleted and nondepleted cells using coimmunoprecipitation assays. Briefly, membranes purified from Ffh- and SecE-depleted cells were solubilized by digitonin, and the solubilized material was immunoprecipitated with immobilized, affinity-purified anti-FtsY antibodies, or anti-GroEL antibodies as a control. Samples containing equal amounts of immunoprecipitated FtsY (unpublished data) were subjected to Western blotting with antibodies to FtsY, L15, and Ffh. As shown in Fig. 1 d, anti-FtsY beads always precipitated in addition to FtsY also the ribosomal protein L15, indicating that FtsY interacts with membrane-bound ribosomes. In nondepleted samples, Ffh was also precipitated, as might be expected if some posttargeting intermediates exist (Murphy et al., 1997; Song et al., 2000; Fig. 1 f). However, a significantly lower amount of Ffh was coprecipitated with equal amounts of FtsY from the SecE-depleted samples, suggesting that some of the complexes do not contain Ffh. This was further confirmed by using saturating amounts of anti-FtsY beads (unpublished data). Furthermore, the surprising possibility that FtsY and ribosomes form complexes in the absence of SRP (Fig. 1 f) is strongly supported by the results obtained with Ffh-depleted samples, where in the absence of detectable amounts of Ffh, ribosomal proteins coprecipitated efficiently with the anti-FtsY beads (Fig. 1 d). In control experiments, the ability of anti-FtsY antibodies to immunoprecipitate ribosomes in the absence of FtsY was tested. Extracts of FtsY-depleted or nondepleted cells were ultracentrifuged, and the total ribosomal pellets (membrane and cytosolic ribosomes) were solubilized by digitonin. The solubilized material was subjected to immunoprecipitation with anti-FtsY beads. The results clearly show that despite the large amount of ribosomes, the anti-FtsY beads precipitated ribosomes only in the presence of FtsY (Fig. 1 e). Finally, in reverse coimmunoprecipitation experiments, we tested the ability of anti-ribosomal protein antibodies to precipitate FtsY from membranes of Ffh-depleted cells. For this purpose, a hybrid L1-YFP was constructed (Fig 5 a), and coimmunoprecipitation with anti-GFP beads showed that antibodies to the hybrid ribosomal protein efficiently precipitated other ribosomal proteins such as L27, and also FtsY, from solubilized Ffh-depleted membranes (Fig. 1 e). To our knowledge, this is the first demonstration of an interaction between the SRP receptor and membrane-bound ribosomes, in the absence of SRP. This complex may therefore represent the initial ribosome docking site on the E. coli membrane.
Figure 5.
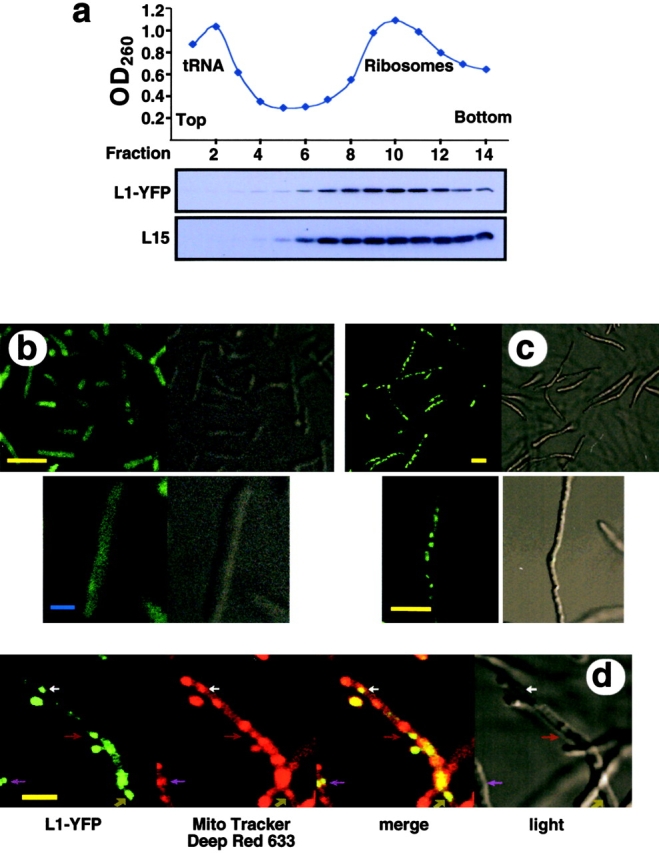
E. coli cells depleted of Ffh produce internal ribosomal clusters, as observed by CLSM. (a) Incorporation of L1-YFP into ribosomes as analyzed by density gradient separation of an E. coli extract and Western blotting with anti-L15 and anti-YFP antibodies. (b) CLSM of Nondepleted cells expressing L1-YFP. (c) CLSM of Ffh-depleted cells expressing L1-YFP. (d) Double labeling of Ffh- depleted cells expressing L1-YFP stained by Mito Tracker Deep Red 633. (b, c, and d, right) Light-microscope view. (d) Arrows point at colocalization of ribosomes and internal membranes. Bars, (yellow) 5 μm; (blue), 1 μm.
Cells depleted of Ffh or SecE produce endoplasmic membrane networks
In exponentially growing E. coli cells, ∼5–8% of all ribosomes are membrane-bound (Randall and Hardy, 1977; Herskovits and Bibi, 2000). Therefore, the accumulation of membrane-bound ribosomes in cells depleted of Ffh or SecE raised the question as to how the cytoplasmic membrane can accommodate such an increased amount of ribosomes. In order to investigate this issue, the morphology of Ffh- and SecE-depleted cells was examined by transmission EM (TEM). Although TEM sections from nondepleted cells (Fig. 2, a) or FtsY-depleted cells (Fig. 2 b) look similar, sections of Ffh- or SecE-depleted cells revealed tightly packed intracellular membrane structures (Fig. 2, c and e). At a higher magnification, the bilayers of these membranes (Fig. 2, d and f) are clearly apparent, as well as the ribosomes (Fig. 2, c, d, and f, arrowheads) that surround the membrane structures.
Figure 2.
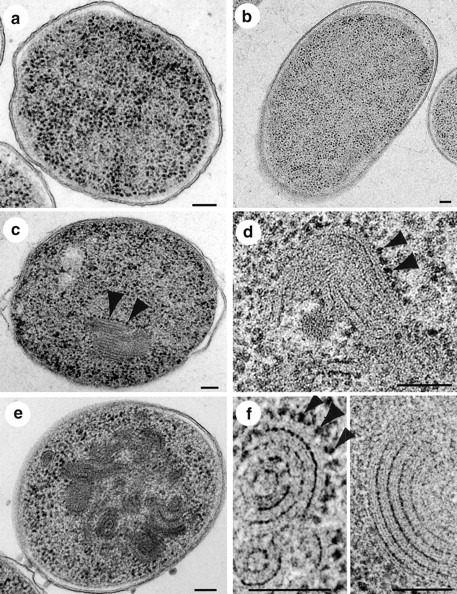
E. coli cells depleted of Ffh or SecE produce endoplasmic membrane networks as observed by electron microscopy. Representative micrographs are shown as follows: (a) Nondepleted cells; (b) FtsY-depleted cells; (c and d) Ffh-depleted cells; and (e and f) SecE-depleted cells. Bars, 100 nm. Arrowheads in c, d, and f indicate membrane-associated ribosomes.
Nondepleted and depleted cells stained with the carbocyanine fluorescent dye DiOC6, which stains intracellular membranes in eukaryotic cells (Sabnis et al., 1997), were examined by confocal laser scanning microscopy (CLSM). This dye produces a uniform background labeling of the cytoplasmic membranes in nondepleted cells (Fig. 3 a). Remarkably however, the laser scan across various sections (50 nm thick) revealed that all the Ffh- and SecE-depleted cells (Fig. 3, b and c) exhibited significant intracellular labeled structures. A closer look at individual cells (Fig. 3, b and c, bottom) showed that many of the intracellular labeled membranes did not occupy the entire cross section of the cell, and that they were located close to the cytoplasmic membrane. In contrast, as observed in TEM sections (Fig. 2 b), FtsY-depleted cells do not contain internal membrane structures (Fig. 3 d). This clear difference between SecE or Ffh depletion, and FtsY depletion provides further support for the proposed role of FtsY in membrane targeting of ribosomes (Herskovits et al., 2000). Furthermore, these results indicate that the formation of intracellular membrane structures is not a general response to defects in protein targeting, but rather specific to the inhibition of late stages during the process.
Figure 3.
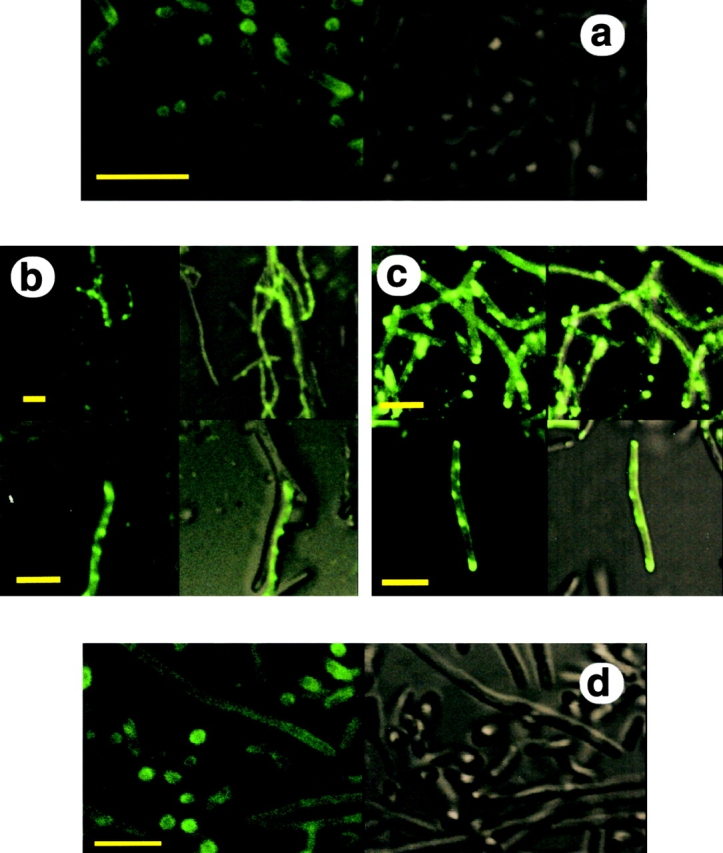
E. coli cells depleted of Ffh or SecE produce endoplasmic membrane networks as observed by CLSM. (a) Nondepleted cells stained by DiOC6. (b) Ffh-depleted cells stained by DiOC6. (c) SecE-depleted cells stained by DiOC6. (d) FtsY-depleted cells stained by DiOC6. Bars, 5 μm.
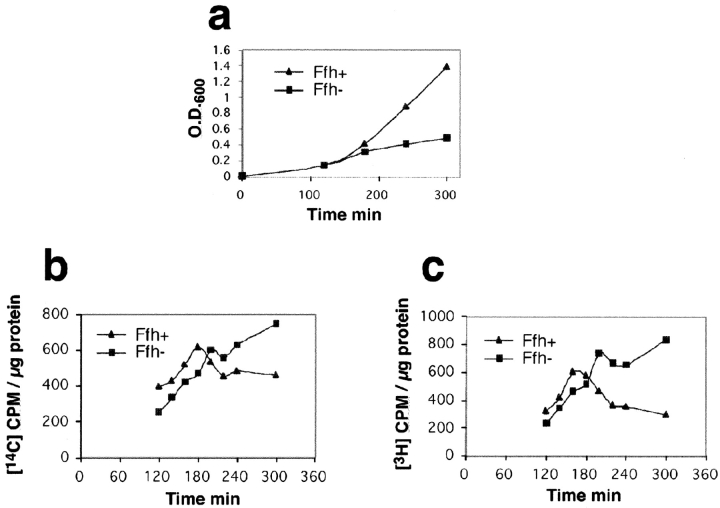
The rate of lipid synthesis in depleted cells versus nondepleted cells was then compared by in vivo labeling with [2-14C]-acetate and L-[3-3H]-serine, and the results suggest that membrane proliferation was induced in the depleted cells (Fig. 4, b and c). Notably, similar reports on an increased lipid biosynthesis and formation of internal membrane networks have been observed previously, under conditions of a defective translocon (de Cock et al., 1989), or overexpression of certain membrane proteins (Arechaga et al., 2000). In both cases, it seems likely that the ribosomal targeting pathway was blocked at the level of the translocon.
Figure 4.
Lipid synthesis by Ffh-depleted or nondepleted cells. (a) Growth as monitored by measuring the optical densities of the cultures at the indicated times. (b) Incorporation of [2-14C]-acetate in samples withdrawn at the indicated times. (c) Incorporation of L-[3-3H]-serine in samples withdrawn at the indicated times. In both b and c, the incorporated radioactivity is expressed as CPM per mg of proteins. The data presented in b and c represent the averages of two independent experiments and the standard error for each point was within 10%.
Colocalization of ribosomes and endoplasmic membrane networks in Ffh-depleted cells
To assess the notion that the heavily stained particles observed in the electron micrographs are ribosomes (Fig. 2, d and f), ribosomes were labeled with the yellow fluorescence protein YFP. A hybrid protein containing YFP fused to the COOH terminus of the ribosomal protein L1 was expressed in Ffh-depleted or nondepleted cells, and was successfully incorporated into ribosomes (Fig. 5 a). Using CLSM, a diffuse pattern of the YFP fluorescence was observed in nondepleted cells (Fig. 5 b), whereas heavily labeled ribosomal clusters, similar to those obtained with the internal membrane-specific dye DiOC6 (Fig. 3), were detected in depleted cells (Fig. 5 c). To test by CLSM whether ribosomes co-localize with the internal membranes, we implemented another hydrophobic dye (Mito Tracker Deep Red 633), which enables usage of a set of distinct CLSM filters for double labeling (see Materials and methods). The results indicate overlap between the YFP fluorescence and some of the internal Mito Tracker Deep Red 633 labeled spots (indicated by arrows in Fig. 5 d). Although the dye labels also the poles, the septa, and the cytoplasmic membrane (unpublished data), it is likely that the colabeled regions represent the internal membranes with associating ribosomes.
In summary, we have described the identification of unprecedented membrane-bound complexes, which contain ribosomes and FtsY, but not Ffh. Such ribosome–FtsY complexes presumably accumulate when the final steps of ribosomal targeting to the translocon are blocked, and therefore may represent an early ribosome-docking step during the biosynthesis of membrane proteins. These results lend further support to our previous proposal that in addition to the suggested SRP-dependent pathway for membrane targeting of ribosomes in E. coli, an alternative, Ffh-independent pathway exist, in which FtsY plays a central role (Herskovits and Bibi, 2000; Herskovits et al., 2000, 2001). The later pathway implies that FtsY mediates constant supply of essential membrane bound ribosomes. According to this model, the transfer of these ribosomes to the translocon requires recognition of newly translated hydrophobic nascent peptides by SRP. This scenario implies that the SRP can also function downstream of FtsY during the biosynthesis of SRP substrates (Fig. 1 f). This model is currently being examined by additional studies of putative steps along the pathway of membrane protein biogenesis in E. coli. Future studies will also determine whether the interaction between the receptor and the ribosome is direct or mediated by additional components, such as a functional analogue of the β-subunit of the mammalian SRP-receptor (Fig. 1 f; Bacher et al., 1999; Fulga et al., 2001).
Finally, the accumulation of membrane-bound ribosome–FtsY complexes is intriguingly linked to the production of novel endoplasmic membrane networks, suggesting that these two consequences of Ffh- or SecE-depletion are related. In this regard, it has been shown recently that in the yeast Sacharomyces cerevisiae, the ER morphology is disrupted by conditional mutations in SP101 and SP102, encoding the respective α- and β-subunits of the SRP receptor (Prinz et al., 2000c). Accordingly, it was suggested that the role of the SRP receptor in maintaining the ER structure is related to its known function in targeting ribosomes to the ER membrane. Thus, our results in E. coli bring about a possibility that the principles of ribosome targeting and intracellular membrane network formation are evolutionarily conserved.
Materials and methods
Bacterial strains and growth conditions
E. coli WAM113 was used for depletion of Ffh (Phillips and Silhavy, 1992), E. coli CM124 was used for depletion of SecE (Traxler and Murphy, 1996) and E. coli FJP10 was used for depletion of FtsY (Herskovits et al., 2001). Cultures were grown at 37°C in LB medium supplemented with the required antibiotics and arabinose (0.2%). For depletion, cells were grown overnight, washed three times in LB broth and suspended in LB (O.D.600 = 0.01) without arabinose. In all experiments, efficient depletion was achieved after 4.5–5 h of growth in the absence of arabinose.
Cell fractionation
Membranes were purified as described previously (Herskovits and Bibi, 2000) with some modifications. Usually, a 10 liter-cultures was harvested, and washed in buffer R (25 mM K-Hepes, pH 7.6, 150 mM KOAc, 10 mM Mg(OAc)2, 1 mM PMSF) supplemented with 0.25 M sucrose and 20 μg/ml RNAse-free DNase. After French Press treatment (four times at 8000 psi), and removal of cell debris, membranes were collected by ultracentrifugation (1.5 h, 53,000 rpm) and resuspended in buffer R. The membranes were further purified by flotation in a step-wise sucrose gradient (8 h, 39,000 rpm, 4°C) using SW41 rotor (Beckman Coulter). The isolated membranes were washed in buffer R supplemented with 0.25 M sucrose, and aliquots of 2 mg membrane proteins were frozen in liquid nitrogen and stored at −80°C.
Membrane solubilization, density gradient separation, and immunoprecipitation
1 mg/ml of membrane proteins was solubilized in buffer R containing 3% digitonin (Calbiochem), and 1 mM dithiotreithol. After a 40-min incubation at room temperature and 2 h at 10°C the mixture was separated in 10 ml of 5–20% sucrose gradients prepared in buffer R containing 0.1% digitonin, by ultracentrifugation (1.5 h, 39,000 rpm, 4°C) using SW41 rotor (Beckman Coulter). The migration of ribosomes in the gradients was followed by measuring the absorbance of RNA at O.D.260. Fractions were then precipitated in 10% TCA, separated by SDS-PAGE and analyzed by Western blotting with antibodies to ribosomal proteins. Immunoprecipitation of the solubilized membranes was performed using 4 Fast Flow protein A-Sepharose beads (Amersham Biosciences) covalently coated with purified anti-FtsY antibodies, anti-GFP antibodies (BAbCO) or anti-GroEL antibodies as a control. For cross-linking we used dimethyl pimalidate (Packman and Perhan, 1982). Initially, nonsolubilized material was removed from the digitonin-solubilized membranes (see previous section) by centrifugation through a 20% sucrose cushion (10 min, 67,000 rpm, 4°C) using a TLA100.3 rotor (Beckman Coulter). Samples were incubated with 40 μl of anti-FtsY, anti-GFP or anti-GroEL beads as a control, washed thoroughly and eluted in SDS sample buffer.
Transmission EM
Nondepleted or depleted cells were drawn into cellulose capillary tubes and frozen in a Bal-Tec HPM010 high-pressure freezing machine (Hohenberg et al., 1994). Cells were subsequently freeze-substituted in a Leica AFS device in anhydrous acetone containing 0.1% osmium tetroxide for 3 d at −90°C and then warmed up to 0°C over 24 h. Samples were embedded in Epon; 60–80-nm sections, stained with uranyl acetate and lead citrate and examined in an FEI Tecnai T12 electron microscope operating at 120 kV.
Membrane staining and confocal microscopy
For staining, cells were incubated with 2.5 μg/ml DiOC6 (3,3′-dihexyloxacarbocyanine iodide) or 0.5 μM Mito Tracker Deep Red 633 (Molecular Probes) for 5 and 15 min, respectively, on ice. The cells were washed three times with 10 mM potassium phosphate buffer, pH 7.4, 0.14 M NaCl, fixed in 0.25% glutaraldehyde for 10 min and washed again. The stained cells were observed using the FV500 laser scanning confocal microscope and the Fluoview software (Olympus). The following filters were used (excitation–emission): for DiOC6 and L1-YFP fluorescence, 488–525 nm; Mito Tracker Deep Red 633, 633–665 nm.
In vivo lipid radiolabeling
E. coli WAM113 cells were grown in LB broth with and without arabinose for 5 h at 37°C. After 1 h, 1 μCi/ml of [2-14C]-acetate and 10 μCi/ml of L-[3-3H]-serine (Amersham Biosciences) were added to the cultures. At various time points samples were withdrawn and subjected to phospholipids extraction (Folch et al., 1957) and total protein determination. Lipids were extracted as follow: cells (1 ml samples) were washed three times in LB and transferred to a tube containing 3 ml chloroform/methanol (2:1). After 2 h incubation at room temperature, the mixture was diluted by addition of 1 ml of water, mixed and centrifuged for 5 min at 1,400 rpm. After removal of the water-methanol phase, the chloroform phase was washed twice by 2 ml of water/methanol (1:1) and centrifuged for 5 min at 1,400 rpm. Finally, ethanol (0.5 ml) was added to the chloroform phase and the samples were dried under nitrogen. For determination of the incorporated radioactivity, the dried pellets were resuspended in 2 ml of benzene:methanol (1:1) and 0.5 ml samples were transferred to scintillation vials containing scintillation fluid Ultima Gold (Sigma-Aldrich).
Ribosome labeling
For the construction of L1-YFP, the chromosomal gene for L1, rplA was amplified by PCR and cloned upstream to the YFP encoding gene using the pEYFP plasmid (CLONTECH Laboratories, Inc.). E. coli WAM113 expressing the L1-YFP hybrid were grown with or without arabinose, and subjected to confocal microscopy.
Acknowledgments
We thank M. Cohen for assistance in L1-YFP construction. We are grateful to V. Kiss for excellent guidance with the confocal microscopy, N. Pepo, D. Pelled and J. Bodennec for their invaluable help in staining of membranes and radiolabeling of phospholipids, and B. Traxler (University of Washington, Seattle, WA) for E. coli CM124. We thank E.S. Bochkareva for critical discussions and reading of the manuscript.
This work was supported by the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities, and in part by the German-Israeli Foundation for Scientific Research and Development.
Footnotes
Abbreviations used in this paper: CLSM, confocal laser scanning microscopy; SRP, signal recognition particle; TEM, transmission EM.
References
- Arechaga, I., B. Miroux, S. Karrasch, R. Huijbregts, B. de Kruijff, M.J. Runswick, and J.E. Walker. 2000. Characterisation of new intracellular membranes in Escherichia coli accompanying large scale over-production of the b subunit of F(1)F(o) ATP synthase. FEBS Lett. 482:215–219. [DOI] [PubMed] [Google Scholar]
- Bacher, G., M. Pool, and B. Dobberstein. 1999. The ribosome regulates the GTPase of the beta-subunit of the signal recognition particle receptor. J. Cell Biol. 146:723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, H.D., M.A. Poritz, K. Strub, P.J. Hoben, S. Brenner, and P. Walter. 1989. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 340:482–486. [DOI] [PubMed] [Google Scholar]
- Cristobal, S., P. Scotti, J. Luirink, G. von Heijne, and J.W. de Gier. 1999. The signal recognition particle-targeting pathway does not necessarily deliver proteins to the sec-translocase in Escherichia coli. J. Biol. Chem. 274:20068–20070. [DOI] [PubMed] [Google Scholar]
- de Cock, H., J. Meeldijk, P. Overduin, A. Verkleij, and J. Tommassen. 1989. Membrane biogenesis in Escherichia coli: effects of a secA mutation. Biochim. Biophys. Acta. 985:313–319. [DOI] [PubMed] [Google Scholar]
- de Gier, J.W., and J. Luirink. 2001. Biogenesis of inner membrane proteins in Escherichia coli. Mol. Microbiol. 40:314–322. [DOI] [PubMed] [Google Scholar]
- de Gier, J.W., P. Mansournia, Q.A. Valent, G.J. Phillips, J. Luirink, and G. von Heijne. 1996. Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett. 399:307–309. [DOI] [PubMed] [Google Scholar]
- Fulga, T.A., I. Sinning, B. Dobberstein, and M.R. Pool. 2001. SRbeta coordinates signal sequence release from SRP with ribosome binding to the translocon. EMBO J. 20:2338–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D., S. Prehn, E. Hartmann, K.U. Kalies, and T.A. Rapoport. 1992. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 71:489–503. [DOI] [PubMed] [Google Scholar]
- Green, P., and M. Inouye. 1983. In vivo and in vitro systems for studying bacterial membrane biogenesis. Methods Enzymol. 96:74–84. [DOI] [PubMed] [Google Scholar]
- Herskovits, A.A., and E. Bibi. 2000. Association of Escherichia coli ribosomes with the inner membrane requires the signal recognition particle receptor but is independent of the signal recognition particle. Proc. Natl. Acad. Sci. USA. 97:4621–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits, A.A., E.S. Bochkareva, and E. Bibi. 2000. New prospects in studying the prokaryotic signal recognition particle pathway. Mol. Microbiol. 38:927–939. [DOI] [PubMed] [Google Scholar]
- Herskovits, A.A., A. Seluanov, R. Rajsbaum, C.M. ten Hagen-Jongman, T. Henrichs, E.S. Bochkareva, G.J. Phillips, F.J. Probst, T. Nakae, M. Ehrmann, et al. 2001. Evidence for coupling of membrane-targeting and function of the SRP-receptor FtsY. EMBO Rep. 2:1040–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch, J., M. Lees, and G.H.S. Stanley. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226:497–509. [PubMed] [Google Scholar]
- Hohenberg, H., K. Mannweiler, and M. Muller. 1994. High-pressure freezing of cell suspensions in cellulose capillary tubes. J. Microsc. 175:34–43. [DOI] [PubMed] [Google Scholar]
- Luirink, J., and B. Dobberstein. 1994. Mammalian and Escherichia coli signal recognition particles. Mol. Microbiol. 11:9–13. [DOI] [PubMed] [Google Scholar]
- Macfarlane, J., and M. Muller. 1995. The functional integration of a polytopic membrane protein of Escherichia coli is dependent on the bacterial signal-recognition particle. Eur. J. Biochem. 233:766–771. [DOI] [PubMed] [Google Scholar]
- Murphy, E.C., III, T. Zheng, and C.V. Nicchitta. 1997. Identification of a novel stage of ribosome/nascent chain association with the endoplasmic reticulum membrane. J. Cell Biol. 136:1213–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, K., S. Mizushima, and H. Tokuda. 1992. The carboxyl-terminal region of SecE interacts with SecY and is functional in the reconstitution of protein translocation activity in Escherichia coli. J. Biol. Chem. 267:7170–7176. [PubMed] [Google Scholar]
- Packman, L.C., and R.N. Perhan. 1982. Quaternary structure of the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus studied by a new reversible cross-linking procedure with bis(imidoesters). Biochemistry. 21:5171–5175. [DOI] [PubMed] [Google Scholar]
- Phillips, G.J., and T.J. Silhavy. 1992. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 359:744–746. [DOI] [PubMed] [Google Scholar]
- Prinz, A., C. Behrens, T.A. Rapoport, E. Hartmann, and K.U. Kalies. 2000. a. Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J. 19:1900–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz, A., E. Hartmann, and K.U. Kalies. 2000. b. Sec61p is the main ribosome receptor in the endoplasmic reticulum of Saccharomyces cerevisiae. Biol. Chem. 381:1025–1029. [DOI] [PubMed] [Google Scholar]
- Prinz, W.A., L. Grzyb, M. Veenhuis, J.A. Kahana, P.A. Silver, and T.A. Rapoport. 2000. c. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150:461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall, L.L., and A.J. Hardy. 1975. Analysis of the ribosomes engaged in the synthesis of the outer membrane proteins of Escherichia coli. Mol. Gen. Genet. 137:151–160. [DOI] [PubMed] [Google Scholar]
- Randall, L.L., and S.J. Hardy. 1977. Synthesis of exported proteins by membrane-bound polysomes from Escherichia coli. Eur. J. Biochem. 75:43–53. [DOI] [PubMed] [Google Scholar]
- Rapoport, T.A. 1991. Protein translocation. A bacterium catches up. Nature. 349:107–108. [DOI] [PubMed] [Google Scholar]
- Romisch, K., J. Webb, J. Herz, S. Prehn, R. Frank, M. Vingron, and B. Dobberstein. 1989. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 340:478–482. [DOI] [PubMed] [Google Scholar]
- Sabnis, R.W., T.G. Deligeorgiev, M.N. Jachak, and T.S. Dalvi. 1997. DiOC6 (3): a useful dye for staining the endoplasmic reticulum. Biotech. Histochem. 72:253–258. [DOI] [PubMed] [Google Scholar]
- Seluanov, A., and E. Bibi. 1997. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J. Biol. Chem. 272:2053–2055. [DOI] [PubMed] [Google Scholar]
- Smith, W.P., P.C. Tai, and B.D. Davis. 1978. Nascent peptide as sole attachment of polysomes to membranes in bacteria. Proc. Natl. Acad. Sci. USA. 75:814–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W., D. Raden, E.C. Mandon, and R. Gilmore. 2000. Role of Sec61alpha in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell. 100:333–343. [DOI] [PubMed] [Google Scholar]
- Traxler, B., and C. Murphy. 1996. Insertion of the polytopic membrane protein MalF is dependent on the bacterial secretion machinery. J. Biol. Chem. 271:12394–12400. [DOI] [PubMed] [Google Scholar]
- Ulbrandt, N.D., J.A. Newitt, and H.D. Bernstein. 1997. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 88:187–196. [DOI] [PubMed] [Google Scholar]
- Valent, Q.A., P.A. Scotti, S. High, J.W. de Gier, G. von Heijne, G. Lentzen, W. Wintermeyer, B. Oudega, and J. Luirink. 1998. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 17:2504–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, P., and A.E. Johnson. 1994. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 10:87–119. [DOI] [PubMed] [Google Scholar]