Abstract
Identification of proteins that couple kinetochores to spindle microtubules is critical for understanding how accurate chromosome segregation is achieved in mitosis. Here we show that the protein hNuf2 specifically functions at kinetochores for stable microtubule attachment in HeLa cells. When hNuf2 is depleted by RNA interference, spindle formation occurs normally as cells enter mitosis, but kinetochores fail to form their attachments to spindle microtubules and cells block in prometaphase with an active spindle checkpoint. Kinetochores depleted of hNuf2 retain the microtubule motors CENP-E and cytoplasmic dynein, proteins previously implicated in recruiting kinetochore microtubules. Kinetochores also retain detectable levels of the spindle checkpoint proteins Mad2 and BubR1, as expected for activation of the spindle checkpoint by unattached kinetochores. In addition, the cell cycle block produced by hNuf2 depletion induces mitotic cells to undergo cell death. These data highlight a specific role for hNuf2 in kinetochore–microtubule attachment and suggest that hNuf2 is part of a molecular linker between the kinetochore attachment site and tubulin subunits within the lattice of attached plus ends.
Keywords: hNuf2; microtubules; kinetochores; mitosis; siRNA
Introduction
The kinetochore is a protein assembly at the centromere of each sister chromatid pair in mitosis. The inner core of the vertebrate kinetochore contains the proteins CENP-A, CENP-B, and CENP-C that tether the kinetochore to centromeric DNA (for review see Salmon and Rieder, 1998). The outer domain consists of an outer plate containing binding sites for ∼20 spindle microtubule plus ends in human cells (Rieder, 1982). A fine filamentous corona extending from the outer plate includes the microtubule motor protein CENP-E and the dynein/dynactin motor protein complex. These motors are involved with recruiting the plus ends of polar microtubules to attachment sites within the outer plate and with generating microtubule-dependent forces that pull kinetochores poleward (Salmon and Rieder, 1998). Attachment at the kinetochore–microtubule interface is dynamic because poleward movement of kinetochores is coupled to depolymerization at plus end attachment sites, whereas movement away from the pole is coupled to polymerization (Mitchison and Salmon, 1992). The kinetochore outer domain also contains spindle checkpoint proteins including Mps1, Mad1, Mad2, Bub1, BubR1, and Bub3, and the target of the spindle checkpoint, Cdc20 (for review see Milliband et al., 2002). The spindle checkpoint inhibits the ability of Cdc20 to activate the anaphase promoting complex/cyclosome until all kinetochores acquire a nearly full complement of kinetochore microtubules and achieve tension when sister pairs become bi-oriented on the spindle.
Identification of proteins that dynamically couple vertebrate kinetochores to microtubule plus ends is a critical issue that remains to be resolved in order to understand how chromosomes are accurately aligned on the spindle in prometaphase and then segregated to the poles in anaphase. The kinetochore-bound microtubule motors identified thus far are not sufficient because depletion of either CENP-E or dynein/dynactin from kinetochores suppresses, but does not block, kinetochore microtubule formation or chromosome movement in mammalian tissue cells (Howell et al., 2001; McEwen et al., 2001). Because the spindle checkpoint proteins were initially discovered in yeast and are conserved in humans, we suspected that key proteins at the kinetochore–microtubule interface might also be conserved. In budding yeast, a molecular complex of Nuf2p, Ndc80p, Spc24p, and Spc25p has been suggested to be essential for kinetochore microtubule formation (He et al., 2001; Wigge and Kilmartin, 2001). Human homologues of Nuf2 (hNuf2) and Ndc80 (HEC) have been identified and shown to localize to mitotic kinetochores (Chen et al., 1997; Nabetani et al., 2001; Wigge and Kilmartin, 2001). Because the fission yeast homologue of Nuf2 also plays a critical role in attaching chromosomes to spindles (Nabetani et al., 2001), and the Caenorhabditis elegans homologue, HIM-10, is essential for proper chromosome segregation (Howe et al., 2001), the vertebrate homologue of Nuf2 appeared to be an excellent candidate for a protein with key functions at the kinetochore–microtubule interface in human cells.
Results and discussion
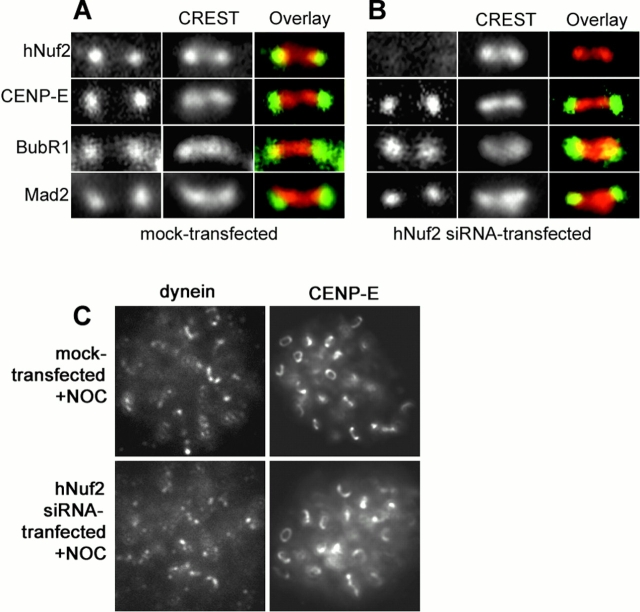
If hNuf2 plays a key role in microtubule plus end attachment within the kinetochore outer plate, we expected to see hNuf2 at kinetochores throughout mitosis and within the outer domain of the kinetochore. By immunofluorescence microscopy, we found that hNuf2 remains concentrated at kinetochores of HeLa (Fig. 1) , PtK1 (unpublished data), and CF-PAC cells (unpublished data) from prophase before nuclear envelope breakdown through the end of anaphase chromosome segregation. In addition, hNuf2 levels did not become significantly reduced at kinetochores by anaphase onset, as occurs for dynein/dynactin, CENP-E, and many of the spindle checkpoint proteins (Hoffman et al., 2001). Comparison of the immunofluorescence localization of hNuf2 to the inner core CENP-A, CENP-B, and CENP-C proteins recognized by CREST antibody and to the outer domain proteins CENP-E, cytoplasmic dynein, Mad2, or BubR1 showed that hNuf2 staining was peripheral to the CREST staining, and was similar to the localization of the other outer domain proteins (see Fig. 4; Wigge and Kilmartin, 2001).
Figure 1.
hNuf2 localizes to kinetochores throughout mitosis. HeLa cells were fixed and double stained with antibodies to hNuf2 and α-tubulin and observed using a spinning disc confocal fluorescence microscope.
Figure 4.
hNuf2-depleted kinetochores retain the spindle checkpoint proteins Mad2 and BubR1 and the motor proteins CENP-E and cytoplasmic dynein. Cells were mock transfected (A) or transfected with an hNuf2 siRNA (B) and fixed for immunofluorescence 48 h after transfection. The cells were stained using the indicated antibodies (left columns) and a kinetochore marker (CREST serum; center columns). Each image shows one kinetochore pair. (C) Kinetochore localization of CENP-E and dynein in nocodazole-treated cells. Cells were subjected to nocodazole treatment and immunofluorescence analysis.
To test how hNuf2 functions in mitosis, we turned to gene silencing by small interfering RNA (siRNA)* duplexes (Elbashir et al., 2001). HeLa cells were transfected with a 21-nucleotide duplex homologous to a portion of the hNuf2 sequence, and, as shown in Fig. 2 A, hNuf2 protein levels were strongly reduced by 24 h (by >80%), and levels remained low 48 h and 72 h after transfection. Depletion of hNuf2 was also confirmed by immunofluorescence microscopy (see Fig. 3) . Although most cells were depleted of hNuf2, a low level of kinetochore-bound hNuf2 could be detected at 48 h in ∼10% of the mitotic cells. To test how depletion of hNuf2 affects the cell cycle and mitosis, we recorded cell cultures by phase-contrast microscopy 24, 48, and 72 h after transfection. As seen in Fig. 2 B, by 24 h after hNuf2 siRNA transfection, several cells had progressed through the cell cycle and blocked in mitosis, exhibiting the rounded cell morphology and condensed chromosomes typical of mitosis (Fig. 2 B, 24 h, bottom). At 48 h, 22% of the cells were blocked in mitosis, and many cells (48%) also appeared to have undergone cell death (Fig. 2 B, 48 h, bottom; Fig. 2 C). By 72 h, few cells were left alive. In contrast, HeLa cells transfected with a control siRNA exhibited normal distribution of spread interphase cells and rounded mitotic cells at 24, 48, and 72 h (Fig. 2 B, top; Fig. 2 C).
Figure 2.
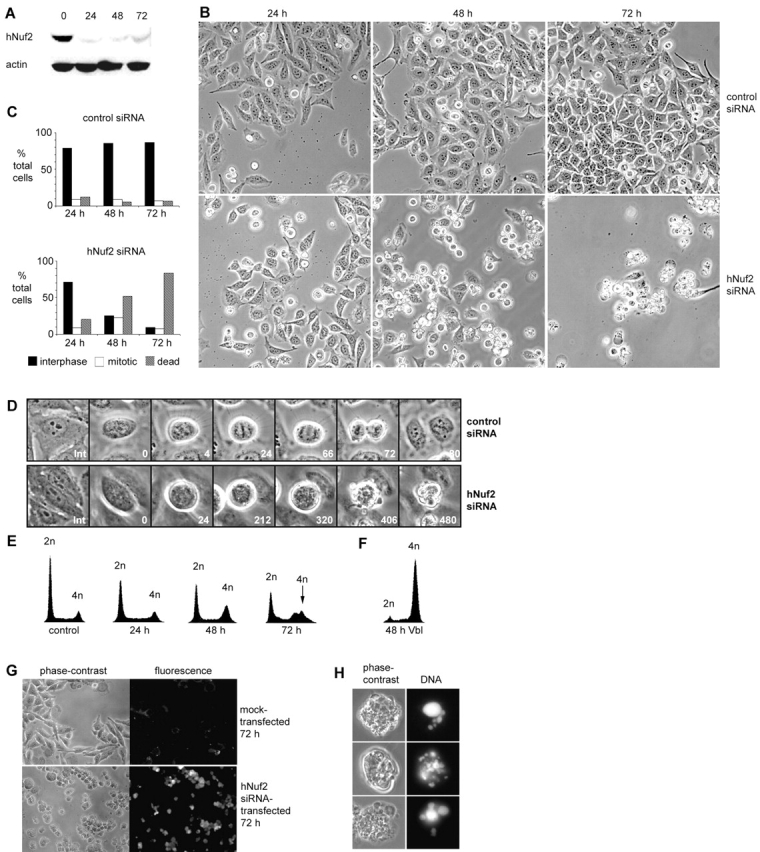
hNuf2-depleted cells exhibit a prolonged mitotic block and undergo cell death. (A) Reduction of hNuf2 levels after siRNA transfection. HeLa cells were transfected with hNuf2 siRNA, harvested 24, 48, and 72 h after transfection, and subjected to protein immunoblot analysis with hNuf2 antibodies and actin antibodies to control for gel loading. Time points are indicated above each lane, and time 0 indicates untransfected cells. (B) hNuf2 siRNA transfection results in a mitotic defect. Cells were transfected with either hNuf2 siRNA or a control 21-nucleotide siRNA duplex, and imaged 24, 48, and 72 h after transfection using a 10× phase objective. (C) Quantitation of control and hNuf2 siRNA–transfected cell phenotypes. Cells adherent to the culture dish with an intact nuclear envelope and decondensed chromatin were scored as interphase. Rounded cells with condensed chromosomes and smooth, uniform membranes were scored as mitotic. Cells that were multilobed, dense, and exhibited a nonuniform membrane were scored as dead (Mills et al., 1999; Zhang and Xu, 2002) (n = 6,000 cells per time point averaged from three independent experiments). (D) Mitotic progression of control siRNA– versus hNuf2 siRNA–transfected cells. 48 h after transfection, cells were transferred to live-cell chambers and time lapsed using a 40× phase objective. Top row, typical control siRNA–transfected cell; bottom row, typical hNuf2 siRNA–transfected cell. Time shown in minutes. (E) Time course of change in cellular DNA content after hNuf2 siRNA transfection. Time after transfection along the x axis is in hours, and untransfected cells were used as a control. Cells were analyzed by flow cytometry as described in the Materials and methods. (F) Cellular DNA content as analyzed by flow cytometry of cells treated for 48 h with 10 μM vinblastine. (G) Uptake of Trypan blue in hNuf2 siRNA–transfected cells. Mock-transfected and hNuf2 siRNA–transfected cells were incubated with Trypan blue 72 h after transfection and observed by phase-contrast and epifluorescence microscopy. (H) Nuclear fragmentation of hNuf2-depleted cells. Cells transfected with hNuf2 siRNA were fixed and stained with DAPI 48 h after transfection.
Figure 3.
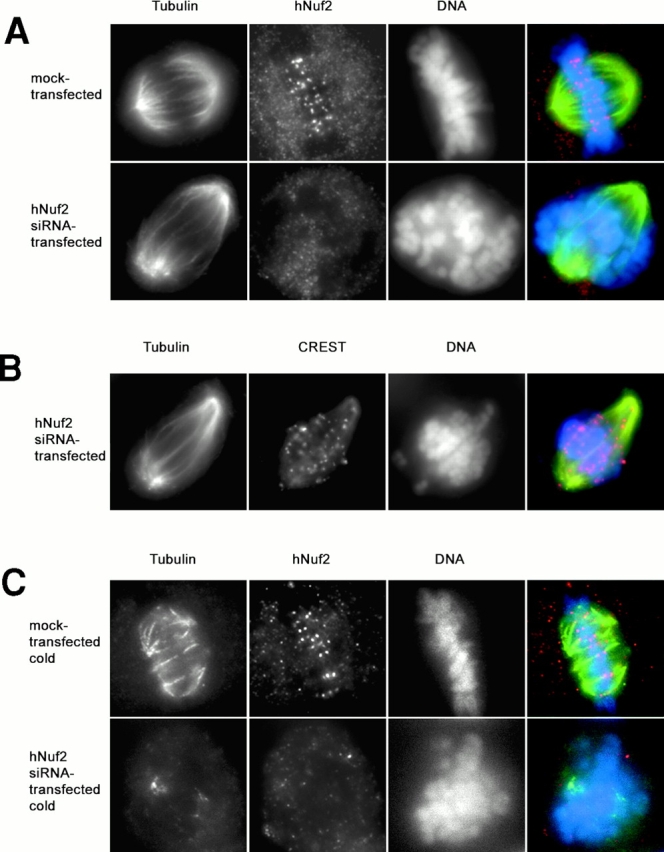
hNuf2-depleted cells lack stable kinetochore microtubules. (A and B) Immunofluorescent cell images of hNuf2 siRNA–transfected and mock-transfected cells. 48 h after transfection, cells were fixed for immunofluorescence and stained with the indicated antibodies. DAPI was used to visualize the DNA. (C) hNuf2 siRNA–transfected cells lack cold-stable kinetochore microtubules. Cells were incubated on ice for 10 min to induce complete microtubule disassembly of all nonkinetochore microtubules, and were subsequently fixed and processed for immunofluorescence using hNuf2 and tubulin antibodies and DAPI to stain DNA.
To better understand how depletion of hNuf2 affects mitotic progression, we performed long-term phase-contrast time-lapse imaging of multiple fields of live cells. We typically recorded images for 10 860 × 690-μm fields using a 10× objective at 90-s intervals over an 8-h duration to obtain statistical counts of the behavior of the cells, and we used 40× objective images of smaller regions to resolve chromosome movements during mitosis. As seen in Fig. 2 D (top), mitotic progression proceeded normally in control siRNA–transfected cells. The average length of mitosis and cell division (from onset of rounding to formation of two daughter cells) in control siRNA–transfected cells was 58.9 ± 17.4 min (n = 50 cells), not significantly different from the average length of mitosis in untransfected cells (68.5 ± 18.5 min; n = 70 cells). In hNuf2-depleted cells, however, cells entered mitosis normally, but then arrested without exhibiting any persistent alignment of their chromosomes into a metaphase plate (Fig. 2 D, bottom). Cells rarely exited mitosis and returned to their spread interphase morphology with an intact nuclear envelope and decondensed chromosomes (<5% of the total cells analyzed). Instead, after several hours blocked in mitosis, they suddenly developed multiple protrusions, membranes became nonuniform, and the cells eventually appeared as dense debris, typical of dying cells (Mills et al., 1999; Zhang and Xu, 2002). Statistical analysis of cells from the 10× time lapses indicated that the hNuf2 siRNA–transfected cells were blocked in mitosis on average at least 7.5 h before cell death. In a small population of the hNuf2 siRNA–transfected cells, chromosome alignment could be observed; in some cases, a metaphase plate persisted for several hours, whereas in others, the metaphase plate was transient. The majority of these cells, however, either remained blocked in mitosis throughout the duration of filming or eventually underwent cell death. We suspect that hNuf2 levels in these cells were not completely depleted, because when imaged by immunofluorescence, all cells that had achieved a metaphase plate (confirmed by DAPI staining) also had detectable levels of hNuf2 at kinetochores (see text below). It is important to note that the behavior of hNuf2-depleted cells differs from that of cells in previous reports in which depletion of a kinetochore protein resulted in a prolonged mitotic block. For example, HeLa cells depleted of CENP-E by antisense also arrest in mitosis (Yao et al., 2000), however, these cells do not undergo cell death, but maintain a persistent block for at least 20 h.
To further examine the pathway of mitotic exit by cell death resulting from depletion of hNuf2, we determined the DNA content by flow cytometry (Fig. 2 E). 48 h after hNuf2 siRNA transfection, a significant population of the cells had accumulated with a 4n DNA content, and by 72 h, the large population of cells containing 4n DNA had shifted to a state in which they contained a variable amount of sub-4n DNA. For comparison, HeLa cells treated with vinblastine to depolymerize microtubules remained blocked in mitosis with a 4n DNA content, even after 48 h (Fig. 2 F). These results suggest that the hNuf2 siRNA–transfected cells persist in mitosis for some time and subsequently undergo cell death. To test this directly, we incubated cells with a Trypan blue solution 72 h after transfection for 10 min, and confirmed that cells transfected with hNuf2 siRNA had undergone cell death, as only these cells were permeable to the dye (Fig. 2 G). Furthermore, we stained cells at 48 and 72 h after hNuf2 siRNA transfection with DAPI to directly image cellular DNA (Fig. 2 H) and observed the DNA to be very dense, opaque, and present in globules of varying sizes and shapes, characteristic of cells undergoing cell death (Mills et al., 1999; Zhang and Xu, 2002). We conclude from these data that depletion of hNuf2 from HeLa cells results in a prolonged mitotic block followed by cell death. This aberrant exit from mitosis has characteristics of both apoptosis and mitotic catastrophe (also known as mitotic cell death) (Nabha et al., 2002). Although researchers are beginning to understand the differences in these processes, there remains much controversy regarding distinctions between their biochemical pathways. It will be interesting for both cancer and cell biology fields to determine the specific suicidal mechanisms that are executed in hNuf2 depletion–induced cell death.
To test how depletion of hNuf2 blocks chromosome alignment into a metaphase plate, we used immunofluorescence microscopy. Fig. 3 A (top) shows the typical distribution of spindle microtubules and hNuf2 localization for a control cell. The formation of bundles of kinetochore microtubules and their subsequent pulling forces make the spindle short and oblate, on average 11.3 μm from pole to pole (±1.2 μm; n = 8). In hNuf2-depleted cells (Fig. 3 A, bottom), robust microtubule arrays extended from the poles and penetrated the chromosomes, but these spindles were ∼60% longer (17.0 ± 2.2 μm; n = 18) than control spindles, consistent with the absence of pulling forces from kinetochore fibers, which were not detectable (Fig. 3 A). Furthermore, in cells stained for CREST to clearly mark the kinetochores, we could detect no kinetochore fibers (Fig. 3 B). We next measured the distance between sister kinetochores to determine if hNuf2 kinetochores were stretched, which would suggest tension produced by attached microtubules. The average interkinetochore distance in hNuf2-depleted cells was 0.88 μm (±0.19 μm; n = 152 kinetochores), compared with the control metaphase distance of 1.56 μm (±0.36 μm; n = 138 kinetochores). Cells treated with vinblastine to depolymerize all microtubules had an average interkinetochore distance of 0.88 μm (±0.14 μm; n = 258 kinetochores). These results suggest that kinetochores in hNuf2-depleted cells are not under tension due to microtubule forces. It is known that kinetochore microtubules are differentially stable to cooling at 4°C in comparison to nonkinetochore microtubules (Rieder, 1981). When we cooled cells for 10 min before fixation, kinetochore fibers were abundant in control cells (Fig. 3 C, top), and the spindle length was reduced to 4.8 μm (±1.1 μm; n = 9). However, in hNuf2 siRNA–transfected cells, only a few microtubules were observed (Fig. 3 C, bottom), and in cells with detectable microtubules, the spindles remained elongated with an average spindle length of 13.6 ± 1.7 μm (n = 17). Those cells that did exhibit a low level of hNuf2 staining at kinetochores contained more prominent fluorescence bundles of kinetochore microtubules in the cold-treated preparations (unpublished data). Thus, the formation of stable kinetochore microtubules depends critically on hNuf2.
Consistent with our time-lapse data, we observed by DAPI staining that although most hNuf2 siRNA–treated mitotic cells were unable to properly align their chromosomes (90%), ∼10% of the treated cells that entered mitosis did achieve a metaphase alignment of chromosomes. The kinetochores of these cells contained hNuf2, although the levels were decreased to an average of 28% of control metaphase levels (unpublished data). Our time-lapse data indicate that such cells also block in mitosis and subsequently undergo cell death, suggesting that reduced levels of hNuf2 at kinetochores can inhibit chromosome segregation and induce mitotic cell death, but permit sufficient kinetochore fiber formation for metaphase chromosome alignment.
In both budding and fission yeast, depletion of Nuf2 blocks microtubule attachment to chromosomes and also inactivates the spindle checkpoint, a result that suggests that Nuf2 depletion produces major disruption of the assembly of many proteins at kinetochores rather than a specific effect on kinetochore–microtubule attachment (Janke et al., 2001; Nabetani et al., 2001). In contrast, we found for HeLa cells that depletion of hNuf2 not only prevents kinetochore microtubule formation, but it also blocks cells in mitosis, indicating that spindle checkpoint activity is not disrupted by depletion of hNuf2. To test this prediction, we examined spindle checkpoint proteins at kinetochores in hNuf2-depleted cells, and as shown in Fig. 4 , in hNuf2 siRNA transfected cells, both Mad2 and BubR1 were detectable at kinetochores. Mad2 and BubR1 appeared diminished compared to unattached kinetochores in prometaphase control cells (data not shown), however, both proteins showed strong localization to all kinetochores in nocodozole-treated hNuf2 siRNA transfected cells, indicating that hNuf2 depletion does not prevent their binding to kinetochores (data not shown).
We next tested if depletion of hNuf2 affected the localization to kinetochores of the motor proteins that help recruit spindle microtubules to the outer plate attachment sites. Immunofluorescence analysis revealed that kinetochores in hNuf2-depleted cells clearly exhibited high levels of the microtubule motor protein CENP-E (Fig. 4), and thus hNuf2 is not required for CENP-E localization to kinetochores. Previous studies have shown that after nocodazole-induced microtubule disassembly, CENP-E levels dramatically increase at kinetochores, forming “crescents” or “collars” that encircle the centromere (Thrower et al., 1996; Hoffman et al., 2001). As shown in Fig. 4 C, depletion of hNuf2 does not effect the formation of these structures, further suggesting that loss of hNuf2 does not result in a global loss of the kinetochore outer domain and its components. To test if loss of hNuf2 at kinetochores prevents dynein localization, we treated cells with nocodazole to depolymerize all microtubules, which increases dynein levels at kinetochores (Hoffman et al., 2001). Under these conditions, we could clearly detect dynein at kinetochores of control and hNuf2-transfected cells (Fig. 4 C); thus, kinetochores in Nuf2-depleted cells retain an ability to bind dynein.
Our results presented here highlight a specific role of hNuf2 for stable kinetochore–microtubule attachment and predict that hNuf2 is part of a molecular linker between the kinetochore attachment site and tubulin subunits within the lattice of attached plus ends. hNuf2 is an α-helical coiled-coil protein without an ATP binding or motor domain (Osborne et al., 1994; Nabetani et al., 2001). Whether it can bind microtubules directly or in concert with members of an Ndc80 complex or components of other complexes remain important unanswered questions. hNuf2 is a potentially attractive target for stopping the proliferation of tumor or cancer cells, because hNuf2 depletion appears selective for blocking cells in mitosis and producing subsequent cell death.
Materials and methods
Cell culture, cell lysates, and immunoblots
HeLa cells were maintained in DME (GIBCO BRL) containing 10% FCS, and the cells were maintained in a 37°C, 5% CO2 incubator. For the nocodazole experiments, HeLa cell growth media was exchanged for DME containing 20 μM nocodazole, and coverslips were incubated for 4 h at 37°C.
For lysates, cells were grown in 25-cm2 flasks at a confluency of ∼106 cells/ml. Cells were removed from the flasks with trypsin, pelleted in a tabletop clinical centrifuge at 5,000 rpm, washed with PBS (140 mM NaCl, 2.5 mM KCl, 10 mM Na2HPO4, 1.5 mM Na2HPO4, pH 7.8), and lysed by sonication. Samples were then clarified by centrifugation, and the protein concentration of the supernatants was determined. Samples were run on 10% SDS–polyacrylamide gels, transferred to nitrocellulose, and subjected to immunoblot analysis using hNuf2 antibodies (1:1,000; Wigge and Kilmartin, 2001) and actin antibodies (1:10,000; Sigma-Aldrich) for a loading control.
siRNA
A 21-nucleotide siRNA duplex was synthesized by Dharmacon Research to target the hNuf2 sequence 5′-AAGCATGCCGTGAAACGTATA-3′. Transfections were performed following the protocol provided by Dharmacon Research using the oligofectamine transfection reagent (Invitrogen). For controls, cells were transfected with a Cy3-labeled luciferase GL2 duplex (Dharmacon Research), or cells were mock transfected with oligofectamine alone. Transfection efficiency was determined using the Cy3–luciferase GL2 siRNA; 48 h after transfection, cells were fluorescently imaged on an inverted Nikon TE30 using a 10× objective. Cells were counted from a total of four experiments, and, in each case, there were virtually no cells lacking the fluorescent label; thus the transfection efficiency was near 100%.
Immunofluorescence microscopy
Images were obtained using a Nikon 100×/NA 1.4 planapochromat oil immersion lens on a spinning disc confocal fluorescence microscope (Cimini et al., 2001). Mad2 antibodies were prepared as described by Waters et al. (1998) and used at a dilution of 1:100. Tubulin antibodies (DM1α; Sigma-Aldrich) were used at a dilution of 1:350. T.J. Yen (Fox Chase Cancer Center, Philadelphia, PA) provided BubR1 and CENP-E antibodies, both used at a dilution of 1:500. CREST serum, provided by B.R. Brinkley (Baylor College of Medicine, Houston, TX), was used at a dilution of 1:10,000.
For determination of cells with cold-stable microtubules, cell media was removed from the culture dishes 48 h after siRNA transfection and replaced with ice-cold media. Cells were then incubated on ice for 10 min to induce microtubule disassembly of all nonkinetochore microtubules and subsequently fixed and processed for immunofluorescence using hNuf2 and anti-tubulin antibodies and DAPI to stain cellular DNA. Cells were imaged as described by Howell et al. (2000).
Phase-contrast microscopy and time-lapse imaging
Cells were transfected with either hNuf2 siRNA or a control 21-nucleotide siRNA duplex and imaged 24, 48, and 72 h after transfection using a Nikon 10× apodized phase objective on a Nikon TE300 microscope with an inverted stand. For live-cell time-lapse imaging, coverslips were mounted in Rose chambers and the chambers were filled with L-15 medium (Sigma-Aldrich) supplemented with 7 mM Hepes, pH 7.2, and 10% FBS. Cells were time-lapse recorded using a Nikon 10× or 40× apodized phase objective on a Nikon inverted TE300 stand. Using a Ludl rotary-encoded scanning stage and a Hamamatsu Orca II camera with Metamorph digital imaging software (Universal Imaging Corp.), cells were imaged every 90 s for 8 h over 10 fields of view.
Flow cytometry and cell death analysis
For flow cytometry, cells were grown in 25-cm2 culture dishes and transfected with either control siRNA or hNuf2 siRNA. After transfection at various times, cells were trypsinized, pelleted, and washed in PBS. Cells were fixed with 1% paraformaldehyde for 20 min, pelleted, and washed in PBS. Cells were resuspended in ice-cold 70% ethanol and stored at 4°C. For analysis, cells were pelleted, washed with PBS, and resuspended in 500 μl propidium iodide (20 μg/ml)/RNase A (30 μg/ml). Cells were analyzed within 3 h on a Becton Dickinson FACScan® interfaced to a Cytomation data acquisition system.
For determination of cell death, mock-transfected and hNuf2 siRNA–transfected cells grown on coverslips were incubated for 10 min with 0.2% Trypan blue 72 h after transfection. Coverslips were mounted onto slides and observed by phase-contrast microscopy using a 20× objective and also by epifluorescence microscopy, as Trypan blue is a fluorescent dye that emits a red fluorescence upon excitation by blue light (Reno et al., 1997).
Acknowledgments
We thank the Salmon lab for helpful comments, advice, and support.
This work was supported by National Institutes of Health grants GM24364 to E.D. Salmon and GM66588 to J.G. DeLuca.
Footnotes
Abbreviation used in this paper: siRNA, small interfering RNA.
References
- Chen, Y., D.J. Riley, P. Chen, and W. Lee. 1997. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol. Cell. Biol. 17:6049–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini, D., B. Howell, P. Maddox, A. Khodjakov, F. DeGrassi, and E.D. Salmon. 2001. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 153:517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 411:494–498. [DOI] [PubMed] [Google Scholar]
- He, X., D.R. Rines, C.W. Espelin, and P.K. Sorger. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 106:195–206. [DOI] [PubMed] [Google Scholar]
- Hoffman, D.B., C.G. Pearson, T.J. Yen, B.J. Howell, and E.D. Salmon. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, M., K.L. McDonald, D.G. Albertson, and B.J. Meyer. 2001. HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J. Cell Biol. 153:1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B.J., D.B. Hoffman, G. Fang, A.W. Murray, and E.D. Salmon. 2000. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 150:1233–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B.J., B.F. McEwen, J.C. Canman, D.B. Hoffman, E.M. Farrar, and E.D. Salmon. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., J. Ortiz, J. Lechner, A. Shevchenko, A. Shevchenko, M.M. Magiera, C. Schramm, and E. Schiebal. 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20:777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen, B.F., G.K.T. Chan, B. Zubrowski, M.S. Savoian, M.T. Sauer, and T.J. Yen. 2001. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell. 12:2776–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milliband, D.N., L. Campbell, and K.G. Hardwick. 2002. The awesome power of multiple model systems: interpreting the complex nature of spindle checkpoint signaling. Trends Cell Biol. 12:205–209. [DOI] [PubMed] [Google Scholar]
- Mills, J.C., N.L. Stone, and R.N. Pittman. 1999. Extranuclear apoptosis. The role of the cytoplasm in the execution phase. J. Cell Biol. 146:703–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, T.J., and E.D. Salmon. 1992. Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J. Cell Biol. 119:569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani, A., T. Koujin, C. Tsutsumi, T. Haraguchi, and Y. Hiraoka. 2001. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma. 110:322–334. [DOI] [PubMed] [Google Scholar]
- Nabha, S.M., R.M. Mohammad, M.H. Dandashi, B.C. Gerard, A. Aboukameel, G.R. Pettit, and A.M. Al-Katib. 2002. Combretastatin-A4 prodrug induces mitotic catastrophe in chronic lymphocytic leukemia cell line independent of caspase activation and poly(ADP-ribose) polymerase cleavage. Clin. Cancer Res. 8:2735–2741. [PubMed] [Google Scholar]
- Osborne, M.A., G. Schlenstedt, T. Jinks, and P.A. Silver. 1994. Nuf2, a spindle pole body–associated protein required for nuclear division in yeast. J. Cell Biol. 125:853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reno, F., E. Falcieri, F. Luchetti, S. Burattini, and S. Papa. 1997. Discrimination of apoptotic cells in flow cytometry using trypan blue and FDA. Eur. J. Histochem. 41:115–116. [PubMed] [Google Scholar]
- Rieder, C.L. 1981. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 84:145–158. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L. 1982. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 79:1–58. [DOI] [PubMed] [Google Scholar]
- Salmon, E.D., and C.L. Rieder. 1998. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8:310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower, D.A., M.A. Jordan, and L. Wilson. 1996. Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell Motil. Cytoskeleton. 35:121–133. [DOI] [PubMed] [Google Scholar]
- Waters, J.C., R. Chen, A.W. Murray, and E.D. Salmon. 1998. Localization of Mad2 to kinetochores depends upon microtubule attachment, not tension. J. Cell Biol. 141:1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge, P.A., and J.V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X., A. Abrieu, Y. Zheng, K.F. Sullivan, and D.W. Cleveland. 2000. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2:484–491. [DOI] [PubMed] [Google Scholar]
- Zhang, J., and M. Xu. 2002. Apoptotic DNA fragmentation and tissue homeostasis. Trends Cell Biol. 12:84–89. [DOI] [PubMed] [Google Scholar]