Abstract
EB1 is an evolutionarily conserved protein that localizes to the plus ends of growing microtubules. In yeast, the EB1 homologue (BIM1) has been shown to modulate microtubule dynamics and link microtubules to the cortex, but the functions of metazoan EB1 proteins remain unknown. Using a novel preparation of the Drosophila S2 cell line that promotes cell attachment and spreading, we visualized dynamics of single microtubules in real time and found that depletion of EB1 by RNA-mediated inhibition (RNAi) in interphase cells causes a dramatic increase in nondynamic microtubules (neither growing nor shrinking), but does not alter overall microtubule organization. In contrast, several defects in microtubule organization are observed in RNAi-treated mitotic cells, including a drastic reduction in astral microtubules, malformed mitotic spindles, defocused spindle poles, and mispositioning of spindles away from the cell center. Similar phenotypes were observed in mitotic spindles of Drosophila embryos that were microinjected with anti-EB1 antibodies. In addition, live cell imaging of mitosis in Drosophila embryos reveals defective spindle elongation and chromosomal segregation during anaphase after antibody injection. Our results reveal crucial roles for EB1 in mitosis, which we postulate involves its ability to promote the growth and interactions of microtubules within the central spindle and at the cell cortex.
Keywords: EB1; microtubule; mitosis; spindle; dynamics
Introduction
The microtubule cytoskeleton functions as an essential scaffold that helps to organize the cytoplasm of eukaryotic cells in interphase. Microtubules, which emanate in a radial pattern from the centrosome during interphase in most eukaryotic cells, provide tracks for microtubule motors carrying membrane, RNA, and protein complexes toward and away from the cell center. Microtubules also play an important role in establishing cell polarity, as they align in the direction of cell migration and toward the interface between immune and antigen-presenting cells (Drubin and Nelson, 1996; Hyman and Karsenti, 1996; Segal and Bloom, 2001; Wittman and Waterman-Storer, 2001). During mitosis, microtubules reorganize to create a mitotic spindle that, in conjunction with motor proteins, segregates chromosomes during cell division (Sharp et al., 2000b). Mitotic spindle orientation, which is a process important in development, tissue morphogenesis, and stem cell differentiation, also involves interactions between astral microtubules, motors, and other proteins at the cell cortex (Lu et al., 1998).
Microtubules are intrinsically dynamic, which allows the microtubule cytoskeleton to rapidly rearrange in response to internal or external cues. Within a population of microtubules at steady-state, individual microtubules undergo transitions between phases of prolonged polymerization and depolymerization. This behavior, known as “dynamic instability,” is enabled by the hydrolysis of GTP after monomeric tubulin becomes incorporated into the microtubule (Desai and Mitchison, 1997). Dynamic instability is modulated by various microtubule-associated proteins (MAPs)* and motor proteins, some of which act to promote microtubule assembly and stability, whereas others induce their depolymerization (Desai et al., 1999). Although many MAPs bind along the length of microtubules, two classes of MAPs localize selectively to the plus ends of growing microtubules: the Cap-Gly proteins (e.g., CLIP-170, p150glued subunit of dynactin) and the EB1 protein family (Schuyler and Pellman, 2001). The mechanism by which these proteins interact selectively with microtubule plus ends and their biological roles are poorly understood. Current work, however, suggests that microtubule plus end–binding proteins mediate interactions between microtubule ends and the cell cortex, kinetochores, endosomes, and dynein motor complexes (Tirnauer and Bierer, 2000; Schuyler and Pellman, 2001).
EB1 was first discovered in a yeast two-hybrid screen for proteins that interact with the human adenomatous polyposis coli (APC) tumor suppressor protein (Su et al., 1995). Homologous proteins have been identified subsequently in many organisms including budding and fission yeast, Drosophila, and Caenorhabditis elegans (Tirnauer and Bierer, 2000). The budding yeast EB1 homologue, BIM1, has received the most attention to date. In yeast, Bim1p is a nonessential gene product that performs at least three related functions: (1) it localizes to the plus ends of cytoplasmic microtubules, where it increases dynamic instability (Tirnauer et al., 1999); (2) Bim1p links microtubule ends to the cell cortex to facilitate orientation of the spindle toward the bud site by binding to a multiprotein complex containing Kar9 and myosin (Myo2p) (Korinek et al., 2000; Lee et al., 2000; Miller et al., 2000; Yin et al., 2000); and (3) through its participation in spindle orientation, Bim1p indirectly participates in a checkpoint that delays cytokinesis pending mitotic exit (Muhua et al., 1998). A mitotic function also has been assigned to the EB1 homologue Mal3 in Schizosaccharomyces pombe (Beinhauer et al., 1997).
In higher eukaryotes, the functions of EB1 proteins remain poorly understood. In epithelial cells of the early Drosophila embryo, EB1 is required to direct the axis of cell division (Lu et al., 2001), although the mechanism by which it performs this function was not resolved. In vertebrate cells, the only activity attributed to EB1 is its ability to bind the COOH terminus of the APC tumor suppressor protein and target it to the tips of growing microtubules (Mimori-Kiyosue et al., 2000a,b). The functional significance of these interactions has not been ascertained, although truncations of the COOH-terminal EB1 binding domain of APC are frequently associated with sporadic and familial colorectal cancers (Polakis, 1997).
Given the high degree of evolutionary conservation, EB1 proteins very likely perform important functions in higher eukaryotes. However, given that budding yeast and higher eukaryotes exhibit considerable differences both in their interphase microtubule organization and in their mechanisms of mitosis (Segal and Bloom, 2001), extrapolating results from yeast BIM1 to metazoan cells becomes precarious. In this study, we investigated the role of EB1 in Drosophila cells in culture by decreasing EB1 protein levels using RNA-mediated inhibition (RNAi) technology and in Drosophila embryos by injecting antibodies against EB1. These complementary techniques and preparations have allowed us to demonstrate that EB1 influences microtubule dynamics and plays a particularly critical role in the assembly, dynamics, and positioning of the mitotic spindle. Interference of EB1 function in these metazoan cells shows similar yet distinct phenotypes from those described in lower eukaryotes.
Results
Drosophila EB1 localizes to microtubule plus ends
To begin our analysis of EB1 function in Drosophila, we first examined the fly genome for genes that exhibited homology to human EB1 (MAPRE1) (Su and Qi, 2001). The Drosophila genome contains four predicted gene products that encode proteins with a high degree of sequence similarity (>40%) to human EB1: genes CG3265, CG18190, CG15306, and CG2955. One gene (CG3265, termed here Drosophila Dm EB1) exhibits a higher degree of sequence identity throughout its length to both human EB1 (52%) and Saccharomyces cerevisiae Bim1p (33%), making it the most likely orthologue. This gene encodes a predicted protein of 294 residues (32.5 kD) with a similar domain organization to human EB1 and Bim1p (see Fig. S1, available online at http://www.jcb.org/cgi/content/full/jcb.200202032/DC1, for comparative sequence alignment). Residues 1–134, which have the highest degree of sequence conservation among EB1 family members, constitutes the domain of the protein implicated in microtubule binding (Juwana et al., 1999). Residues 129–212 are enriched in serines and prolines and hence may be unstructured, and residues 213–273 are predicted to form a coiled coil. In Bim1p, the COOH-terminal coiled-coil domain binds to Kar9 (Miller et al., 2000), and it may mediate protein–protein interactions in other species as well.
As tools for immunolocalization and RNAi studies, we generated polyclonal antibodies against a Dm EB1–GST fusion protein. The affinity-purified antibodies recognized a protein with a molecular weight of 31 kD on immunoblots of extracts from Drosophila embryos and Schneider (S2) tissue culture cells (Fig. 1 a). To ensure that the 30-kD immunoreactive band was Drosophila EB1, S2 cells were treated with dsRNA corresponding to a 600-bp sequence of Dm EB1. Quantitative immunoblots showed that the band recognized by our anti–Dm EB1 antibodies decreased over time to 1% of controls after 6 d of dsRNA treatment. In contrast, this band was unaltered in cells treated for 6 d with dsRNA corresponding to either GFP (Fig. 1 b) or the most homologous member of the other three EB1 proteins, CG18190 (unpublished data). From these results, we conclude that our antibodies specifically recognize Drosophila Dm EB1 and that the RNAi treatment was effective in eliminating virtually all Dm EB1 protein from S2 cells.
Figure 1.
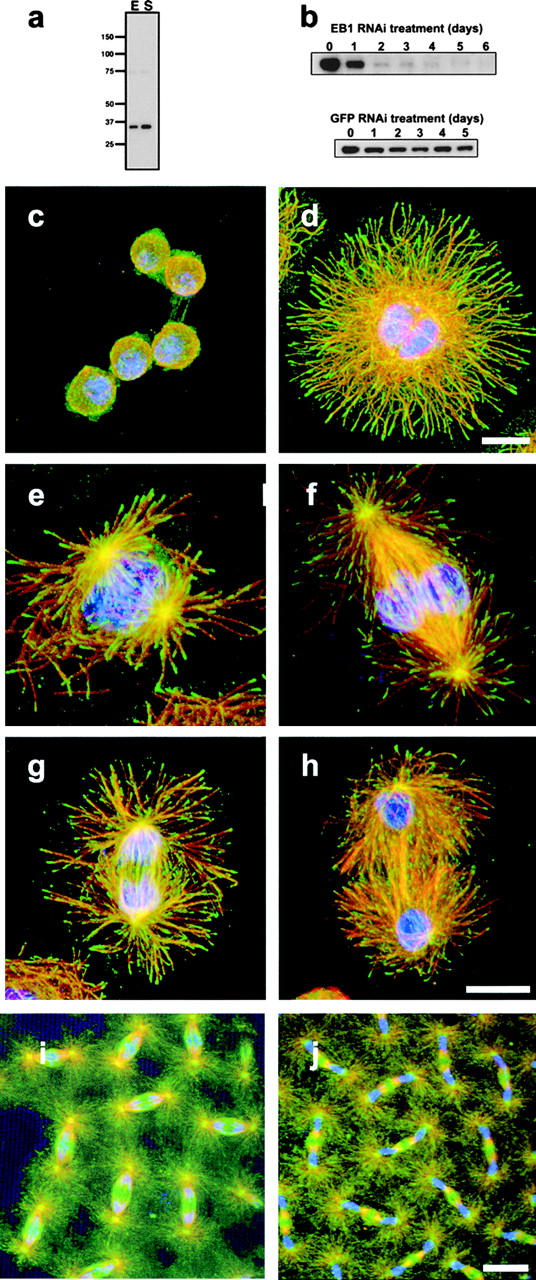
DmEB1 is localized to the plus ends of microtubules in S2 cells. (a) Immunoblot of Dm EB1 in extracts prepared from Drosophila embryos (E) (0–4 h old) and Schneider cells (S). Molecular masses (in kD) are provided to the left. (b) Quantitative immunoblotting for Dm EB1 protein in cultures subjected to RNAi for 0–6 d for EB1 (top) and GFP (bottom). All lanes contained an equal load of protein as determined by blotting for tubulin (not depicted). (c–h) Immunolocalization of Dm EB1 (green), microtubules (red), and DNA (blue) in Drosophila S2 cells. (c) Cells plated on poly-l-lysine before fixation and staining. (d) A cell plated on concanavalin A and allowed to spread for 2 h before fixation shows considerable improvement in imaging of the microtubule cytoskeleton. The magnification in panels c and d are the same. Bar, 5 μm. S2 cells in prophase (e), metaphase (f), anaphase (g), and telophase (h). Bar, 5 μm. Immunofluorescent localization of Dm EB1 (green), tubulin (red), and DNA (blue) in Drosophila synctitial blastoderm-stage embryos during anaphase (i) and telophase (j). Dm EB1 shows a marked accumulation at spindle poles and on the midbody. Bar, 10 μm.
We first sought to examine the intracellular localization of Dm EB1 in S2 cells. However, under routine growth conditions, these cells adopt a spherical morphology (10-μm diameter) and possess a thin rim of cytoplasm that encircles the nucleus. As a result, S2 cells have been considered relatively poor for cytological examination. When we examined these cells by immunofluorescence, the microtubule cytoskeleton appeared as a dense basket-like network without visible organization (Fig. 1 c); Dm EB1 also was difficult to visualize, but it clearly colocalized along short (1–2 μm) stretches of microtubules. The crowded packing of microtubules, however, made it difficult to discern unambiguously whether the colocalization corresponded to the microtubule plus ends.
To improve the cytology of the S2 cells, we tested various substrates for their ability to promote cell adhesion and spreading. One of the substrates tested, concanavalin A, promoted S2 cell attachment to coverslips and caused them to adopt a flattened, discoid morphology (∼20 μm in diameter) within 1–2 h. In these preparations, S2 cells elaborated a well-developed, radial interphase microtubule network with readily discernible tips extending toward the cell periphery (Fig. 1 d). Because of the considerable improvement in cytology, we employed this cell preparation for subsequent examination of Dm EB1 and microtubules.
In concanavalin A–treated cells, Dm EB1 staining clearly coincided with individual microtubules and exhibited a comet-like gradient of staining, with the greatest intensity at the most distal tip of the microtubule (Fig. 1 d). During all stages of mitosis, Dm EB1 also was localized at microtubule plus ends (Fig. 1, e–h). Additionally, puncta of Dm EB1 staining were found at the duplicated centrosomes of prophase cells as they began to separate from one another (Fig. 1 e). During metaphase, Dm EB1 localization to the tips of astral microtubules was particularly prominent (Fig. 1 f). In addition, as cells progressed to telophase, EB1 staining was enriched on the interpolar microtubule bundles that separated each chromosomal mass (Fig. 1 h). The distribution of Dm EB1 in S2 cells is, therefore, very similar to the localization that has been described in vertebrate cell lines (Morrison et al., 1998; Tirnauer et al., 1999; Mimori-Kiyosue et al., 2000b).
We also examined the distribution of Dm EB1 in synctitial blastoderm embryos. Consistent with observations in S2 cells, antibodies against Dm EB1 decorated the mitotic spindle and showed prominent staining of the spindle poles and astral microtubules (Fig. 1 i). Embryos in late anaphase and telophase also showed a dramatic accumulation of Dm EB1 staining on interpolar microtubule bundles and midbodies (Fig. 1 j).
Depletion of Dm EB1 affects microtubule dynamics but causes minimal perturbation of microtubule organization in interphase cells
To gain insight into the cellular functions of EB1, we investigated whether RNAi depletion of Dm EB1 affected microtubule organization by fluorescence microscopy. As discussed above, 6 d of dsRNA treatment was sufficient to reduce Dm EB1 protein to very low levels (Fig. 1 b). When plated on concanavalin A–coated coverslips, Dm EB1 dsRNA–treated cells attached and spread as well as control cells and displayed no obvious morphological abnormalities. Tubulin staining revealed that the interphase microtubule organization in these cells was indistinguishable from controls (see Fig. 3 g).
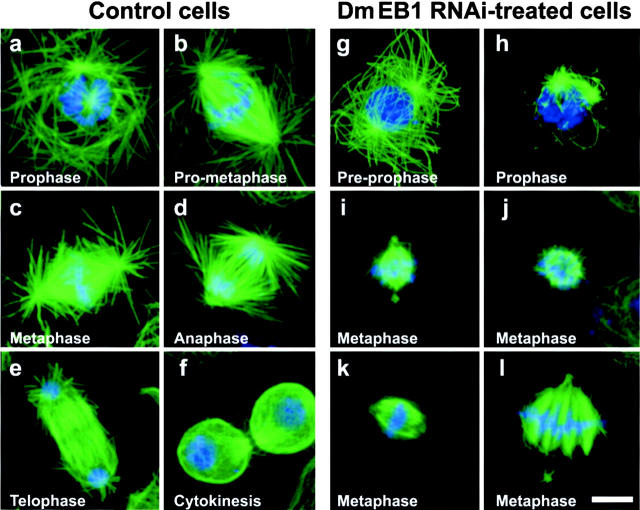
Figure 3.
Microtubule organization during mitosis in control and Dm EB1 RNAi–treated cells. Control cells stained for tubulin (green) and DNA (blue) show a normal progression through the cell cycle from prophase (a) to prometaphase (b), metaphase (c), anaphase (d), telophase (e), and cytokinesis (f). Note the well-developed arrays of microtubules nucleated from both centrosomes in preprophase (a) and the halo of astral microtubules present at both poles throughout mitosis. In Dm EB1 RNAi–treated cells in preprophase (g), both poles nucleate a typical interphase array of microtubules. Upon progression to prophase (h), however, microtubules at each pole are far shorter, and fragmented microtubules may often be seen in the cytoplasm (arrow). Panels i–l catalog the predominant defects observed in metaphase spindles. Note the absence of astral microtubules in all cases. Bar, 5 μm.
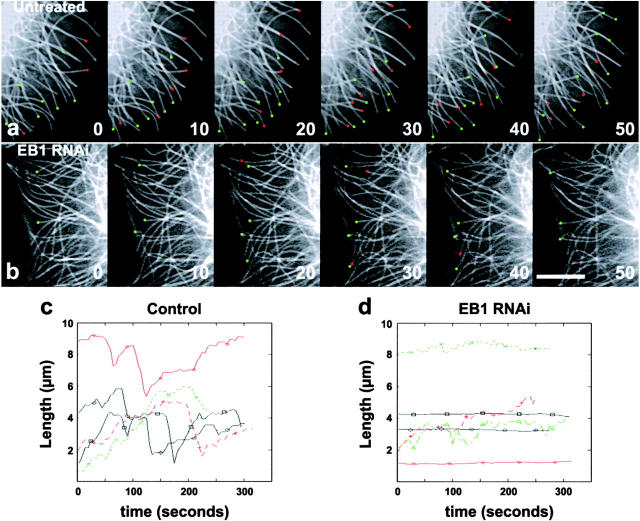
To probe the effects of Dm EB1 depletion more carefully, we used live cell fluorescence microscopy to observe microtubule behavior in control and Dm EB1 dsRNA–treated S2 cells transfected with GFP–tubulin. Microtubules in untreated control cells exhibited dynamic instability, asynchronously transiting between phases of elongation and shrinkage. The rates of microtubule growth and shrinkage were 3.8 ± 0.9 μm/min and 8.7 ± 2.8 μm/min, respectively (Table I). The rate of catastrophe was 0.021 transitions from growth (or pause) to shrinkage per second, whereas the rate of rescue was 0.029 transitions from shrinkage (or pause) to growth per second (Table I). The populations of microtubules in control cells spent, on average, 55.2% of the time in growth, 27.5% of the time in shrinkage, and 17.3% of the time in a paused state (neither growing nor shrinking) (Fig. 2, a and c; Table I; Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200202032/DC1). These parameters of dynamic instability measured for S2 cell microtubules are similar to those measured in other cell types using GFP-tagged tubulin, with velocities of growth and shrinkage intermediate to those measured in mammalian cells and yeast (Tirnauer et al., 1999; Yvon et al., 1999; Rusan et al., 2001). These results represent the first direct measurements of microtubule dynamic instability in Drosophila cells.
Table I. Kinetic parameters of microtubule polymerization dynamics in untreated cells and cells depleted of EB1 by RNAi.
| Untreated cells (n) | EB1-depleted cells (n) | |
|---|---|---|
| Shrinkage rate (μm/min) | 8.7 ± 2.8 (60) | 8.6 ± 3.1 (50) |
| Growth rate (μm/min) | 3.8 ± 0.9 (50) | 3.7 ± 1.0 (60) |
| Catastrophe frequency | 0.021 (101) | 0.007 (30) |
| Rescue frequency | 0.029 (120) | 0.011 (33) |
| Time in growth (%) | 55.2 | 30.0 |
| Time in shrinkage (%) | 27.5 | 9.3 |
| Time in pause (%) | 17.3 | 60.6 |
Velocities are presented as the mean ± SD. Lifetime percentage measurements were calculated from microtubule life history plots and represent the percentage of time spent in growth, shrinkage, or pause. The n refers to the number of microtubules observed for control and EB1-depleted cells.
Figure 2.
Time-lapse fluorescence imaging of microtubule dynamics in S2 cells. (a) This sequence of images shows microtubule behavior in a control cell at 10-s intervals. Green circles denote the ends of microtubules that are elongating, whereas the red circles mark the tips of shortening microtubules. (b) This image series illustrates microtubule dynamics in an S2 cell that has been treated with dsRNA for 7 d to eliminate Dm EB1. Note that microtubules in this cell are less dynamic than in the control cell, above. Bar, 5 μm. Dynamic life history plots that track the positions of the plus ends of microtubules over time in relation to a fiduciary point proximal to the tip. Plots were generated by plotting distance in microns versus time in seconds for five microtubules each in control (c) or Dm EB1–depleted cells (d).
Microtubule behavior was very different in cells depleted of Dm EB1 by dsRNA (Fig. 2, b and d; Table I; Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200202032/ DC1). Rates of microtubule growth (3.7 ± 1.0 μm/min) and shrinkage (8.6 ± 3.1 μm/min) were similar compared with untreated control cells. However, the frequencies of catastrophe in Dm EB1–depleted cells were approximately threefold lower compared with control cells (Fig. 2, b and d; Table I). The most notable effect of EB1 depletion was that microtubules spent the majority (55.2%) of their lifetimes in a paused state relative to growth (30%) or shrinkage (9.3%) (Table I). These results indicate that Dm EB1 promotes microtubule dynamics in Drosophila cells. The effects of EB1 RNAi on microtubule dynamics in S2 cells are qualitatively similar to interphase microtubule behavior observed in bim1Δ S. cerevisiae as reported by Tirnauer et al. (1999). In both cases, microtubule catastrophe and rescue frequencies were decreased in the absence of EB1/Bim1p and microtubules spent the majority of their lifetimes in a state of pause.
Loss of Dm EB1 function causes defects in mitotic spindle structure
Given the role of EB1 family members in mitosis in yeast, we also examined how RNAi inhibition of Dm EB1 expression affected mitosis in Drosophila cells. Mitosis in untreated or GFP dsRNA–treated cells progressed in a very reproducible manner. At prophase, the two spindle poles were in close proximity to condensing chromosomes and always nucleated asters of long, radial microtubules (Fig. 3 a). As the cells proceeded to prometaphase (Fig. 3 b), the spindles assumed a typical bipolar organization and chromosomes were positioned between each pole. At this stage, and for all successive stages, bipolar spindles nucleated highly developed radial arrays of astral microtubules, many of which extended to the cell cortex. The chromosomes congressed to the metaphase plate (Fig. 3 c), and subsequently migrated to the spindle poles during anaphase (Fig. 3 d) and telophase (Fig. 3 e). At cytokinesis (Fig. 3 f), the two incipient cells assumed a more rounded shape.
In cells lacking Dm EB1, defects in microtubule organization were readily apparent. During preprophase, Dm EB1–deficient cells duplicated centrosomes normally and the two centrosomes migrated to opposite sides of the nucleus as in control cells (Fig. 3 g). At this stage, each centrosome nucleated a normal radial array of long microtubules that extended toward the cell periphery. However, when dsRNA-treated cells progressed to prophase, the long cytoplasmic microtubules disappeared (Fig. 3, compare h with a), and instead, only very short (<1 μm) astral microtubules were observed clustered around the two poles. Short microtubule fragments unattached to the poles were often present in the cytoplasm of Dm EB1–deficient cells. These phenotypes were observed in 74% of the Dm EB1–deficient prophase cells examined (n = 100), but were never observed in untreated (n = 100) or GFP RNAi control cells (n = 100). From these observations, we conclude that Dm EB1 is required for stabilizing microtubules and creating astral arrays in mitosis.
The loss of Dm EB1 also produced aberrant spindle phenotypes in metaphase cells that could be classified into four general categories. The most common defect was a complete loss of astral microtubules (Fig. 3 i) (35% of cells, n = 264). These spindles maintained their bipolar symmetry, but commonly exhibited detachment of centrosomes from the spindle (Fig. 3 i, arrow). The second class of defects (observed in 33% of the cells) lacked astral microtubules and exhibited an overall compaction of the spindle into a basket-like meshwork of microtubules surrounding the chromosomes (Fig. 3 j). In these structures, the poles could not be clearly distinguished, but mitotic chromosomes maintained their position at the center of the spindle. The third type of defect (30% of the cells) was a detachment of a spindle pole from the bundles of microtubules that were connected to the kinetochores (Fig. 3 l). These spindles exhibited a “splayed” morphology. The fourth category of defect (2% of cells) was “barrel-shaped” spindles that maintained their symmetry, but failed to focus the microtubules at the poles and also lacked astral microtubules. These phenotypes did not appear to be due to gross centrosome defects, as immunofluorescent staining with antibodies against centrosomin protein revealed spindle poles to be present and intact (unpublished data). In all four classes of defective spindles, the distance from pole to pole was significantly smaller (5.4 ± 1.1 μm) than in GFP dsRNA–treated cells (7.7 ± 0.9 μm, P < 0.0001, t test). These results indicate that Dm EB1 plays a critical role during spindle assembly.
The mitotic defects we observed in the Dm EB1 RNAi–treated cells were severe enough that we suspected they might affect cell cycle progression by activating the spindle checkpoint. To test this possibility, fixed cells were stained for DNA and the number of cells with mitotic figures was scored as a percentage of the entire cell population. In Dm EB1 dsRNA–treated cultures, the mitotic index was 5.9% (n = 1,500 cells), approximately double that of control cultures at 2.7% (2,700 cells) (Table II). Although significant (P < 0.0001), this difference was not as dramatic as might be expected if mitotic progression were completely blocked. If the mitotic checkpoint were activated for prolonged periods of time, an increase in apoptotic cells might be expected. However, S2 cells exhibit macrophage-like properties (Ramet et al., 2002), and we observed that they consume their apoptotic neighbors, as judged by nuclear morphology (unpublished data). This property of S2 cells could give rise to artificially low mitotic index measurements. To determine at which stage of the cell cycle mitotic progression was interrupted, we next categorized all of the mitotic cells in these samples according to their stage of mitosis. In control-treated cultures, cells appeared to spend approximately the same amount of time in each stage of mitosis (Table II). In Dm EB1–depleted cells, however, there was an accumulation at metaphase (∼40% compared with 22% in controls) and in telophase (∼43% compared with 38% in controls). These data suggest that inhibition of Dm EB1 activated the spindle checkpoint. Further work will be required to understand potential checkpoint activation in response to loss of EB1 function, perhaps by live cell imaging.
Table II. Quantitation of the mitotic index and cell cycle stages of cells treated with control dsRNA or EB1 dsRNA after 7 d.
| Mitotic index | Prophase | Metaphase | Anaphase | Telophase | |
|---|---|---|---|---|---|
| % Control cells | 2.7a | 17.3 ± 1.6 | 22.3 ± 4.4 | 20.3 ± 5.1 | 38.3 ± 6.7 |
| % Dm EB1 RNAi | 5.9b | 8.2 ± 2.7 | 40.2 ± 6.1 | 7.5 ± 2.1 | 43.5 ± 2.1 |
Mitotic index is expressed as the percentage of the total population of cells in mitosis. Cell cycle stages were determined by scoring all mitotic cells by immunofluorescence for tubulin and chromosome staining. Values represent the mean ± SD.
n = 2,700.
n = 1,500.
Mitotic spindle positioning requires EB1 activity
During normal mitosis, the mitotic spindle positions itself at the geometric center of the cell (Fig. 4 a). In S2 cells lacking Dm EB1, however, the spindle was frequently mispositioned (Fig. 4 b). To quantitate this effect, we determined the spindle center by measuring the distance between the two poles in cells that had their mitotic spindle aligned parallel to the coverslip. We then calculated the centroid of the cell and determined the offset distance between the cell centroid and the spindle center for untreated cells and for Dm EB1– and GFP RNAi–treated cells (Fig. 4 c). In untreated and GFP dsRNA–treated cells, the average offset distances between the centroid and the spindle center were 0.42 μm ± 0.17 and 0.35 μm ± 0.15, respectively. In contrast, the average offset distance in Dm EB1–deficient cells was significantly greater, 1.93 μm ± 0.57 (P < 0.0001). From these data, we conclude that loss of Dm EB1 function causes mispositioning of the mitotic spindle.
Figure 4.
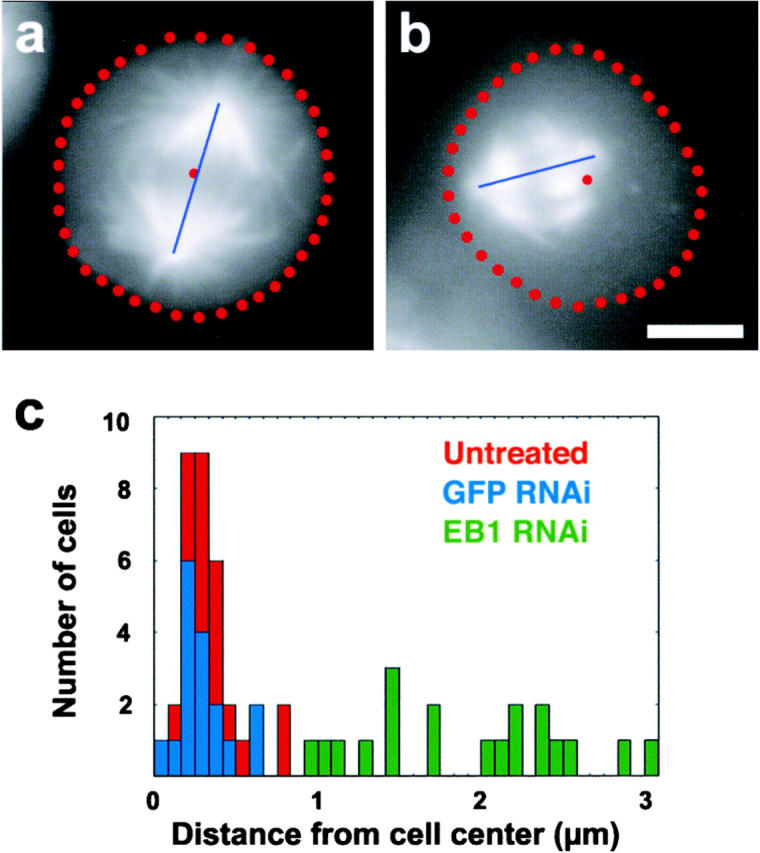
EB1 is necessary for proper spindle positioning. Positioning of the mitotic spindle in a control S2 cell (a) and in a cell treated with dsRNA against Dm EB1 (b). The cell borders are outlined in red circles, and the centroid of the cell is likewise marked. The blue line reflects the pole-to-pole distance. Self-centering of the metaphase spindle was evaluated by measuring the distances between the centroid of the cell and the midpoint of the spindle. (c) Histogram showing the measured distances in untreated (red), GFP RNAi (blue), and Dm EB1 (green) RNAi cells. Bar, 5 μm.
Dm EB1 is necessary for proper chromosomal segregation and spindle elongation during anaphase
Although the Schneider cell system provided a convenient method to generate loss-of-function phenotypes for Dm EB1, extended live cell imaging of the spindle proved technically difficult. To study the role of Dm EB1 in spindle dynamics, we used the Drosophila synctitial blastoderm as a model, because thousands of synchronous spindles that divide within a two-dimensional plane can be readily observed by confocal microscopy (Sullivan and Theurkauf, 1995). Furthermore, because preblastoderm embryos are not governed by the same mitotic checkpoint mechanisms as differentiated cells (Sullivan et al., 1993), the later stages of mitosis can be observed after treatments that might otherwise induce mitotic arrest by activation of the spindle checkpoint. To study the role of Dm EB1 in spindle dynamics in vivo, we microinjected anti–Dm EB1 antibodies into transgenic preblastoderm embryos expressing GFP–tubulin. Using time-lapse spinning disk confocal microscopy, we observed the effects of Dm EB1 inhibition on mitotic spindle formation, spindle elongation, and chromosomal segregation.
In control embryos, dynamics of the mitotic spindles followed a well-characterized, documented progression (Karr and Alberts, 1986; Kellogg et al., 1988; Sullivan and Theurkauf, 1995). During interphase of cycle 12, duplicated centrosomes moved to opposite sides of the nucleus to positions separated by ∼120°. Upon entry into prometaphase, the nuclear envelope broke down and the nuclear space was invaded by microtubules emanating from opposite poles (Fig. 5 c). These microtubules formed attachments either with chromosomes to form kinetochore fibers or intercalate with microtubules of opposite polarity to form interpolar bundles. A few minutes after chromosomes congressed to the metaphase plate, the spindles transited to anaphase and sister chromosomes segregated to opposite poles to complete mitosis. The pole-to-pole distances are highly reproducible in spindles throughout cycle 12 (Fig. 5). After nuclear envelope breakdown of cycle 12, the length of the spindle is ∼8 μm. As the cells progressed to metaphase, spindles elongated at a rate of ∼0.03 μm/s until reaching a separation of ∼12 μm. Upon anaphase onset (Fig. 5 c, asterisk), spindles further elongated at a rate ∼0.07 μm/s until reaching a maximal length of ∼16.5 μm. These measurements are in close agreement with a previous description of Drosophila embryo spindle dynamics (Sharp et al., 2000a).
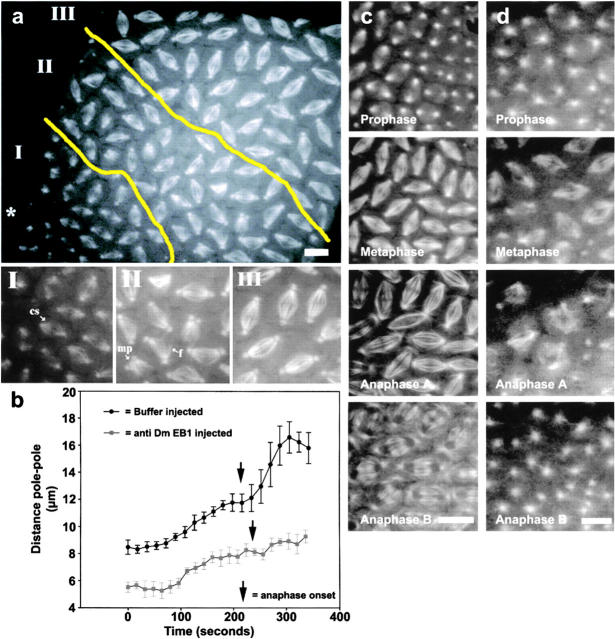
Figure 5.
Microinjection of Dm EB1 antibodies into synctitial embryos inhibits elongation of the mitotic spindle. (a) A low magnification view of a GFP–tubulin-expressing embryo injected with polyclonal antibodies raised against Dm EB1. The injection site is indicated with an asterisk. The injection produces a gradient of effects in the embryo. Roman numerals indicate regions containing spindle defects depicted in the higher magnification images. Several spindle defects are annotated, including spindles lacking a central spindle (cs), monopolar spindles (ms), and frayed spindles (f). (b) Plot showing the pole-to-pole distances versus time in region I of two buffer- and five Dm EB1 antibody–injected embryos from prometaphase to telophase of cycle 12 mitosis. Errors bars represent the standard deviation of the average lengths. Time series of GFP–tubulin fluorescence showing mitotic spindle formation in buffer- (c) and Dm EB1 antibody–injected (d) embryos. Similar results were obtained for numerous spindles in >20 injected embryos. Bars, 10 μm.
To investigate whether Dm EB1 plays a role in mitosis in this system, we microinjected Dm EB1 antibodies into Drosophila embryos expressing GFP–tubulin. The injected antibodies produced a readily apparent gradient of effects on spindle structure and behavior, with the most severe defects centered around the injection site (Fig. 5 a, asterisk). As these embryos approached cycle 12 mitosis, it became readily apparent that the spindles closest to the injection site had fewer microtubules and had a shorter pole-to-pole distance than controls. (Fig. 5 a, I). This phenotype is similar to that produced by RNAi of EB1 in S2 cells. Given the general similarity in phenotypes observed with antibody microinjection and RNAi, we believe that the antibodies are manifesting their effects through EB1. However, we cannot exclude the possibility of effects manifest through other polypeptides (e.g., the weakly reactive 75-kD polypeptide observed after long exposure of immunoblots). We also do not know the exact mechanism of the antibody-induced defect, although the similarity to the RNAi phenotype makes us suspect that the antibody injection leads to a loss of EB1 function.
Spindles further from the injection site exhibited a pole-to-pole distance that more closely resembled controls, but also frequently displayed structural defects such as frayed (Fig. 5 a, II) and monopolar half spindles that had both centrosomes present at a single pole (Fig. 5 a, II). Observation of these defects over time revealed that spindle structure was dynamic and these frayed and monopolar spindles could sometimes correct themselves and complete mitosis (unpublished data). Regions of these embryos distal to the injection site supported formation of morphologically normal spindles that progressed through mitosis similar to controls.
We quantitated the effects of Dm EB1 inhibition on spindle elongation by measuring the pole-to-pole distances of spindles proximal to the injection site over time (Fig. 5 b). During the prophase-to-metaphase transition, spindles elongated twofold slower (∼0.015 μm/s) and achieved a shorter length (8.1 ± 0.5 μm) at metaphase. At anaphase, spindles elongated threefold slower (∼0.01 μm/s) and elongated to a maximal length 40% less than controls (9.2 ± 0.6 μm) (Fig. 5 b). In addition to reduced rates of elongation, spindles proximal to the injection site exhibited a striking overall reduction in associated microtubules and failed to form normal interpolar microtubule bundles or a midbody at the end of anaphase (Fig. 5 d).
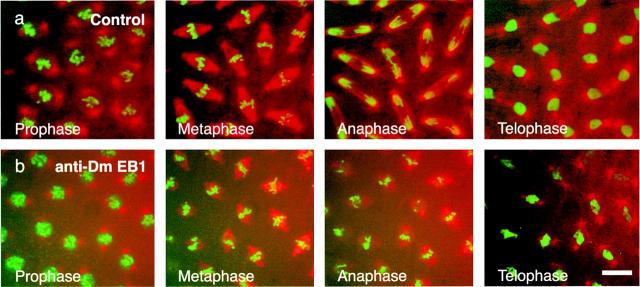
If interference with Dm EB1 activity disrupted normal spindle elongation at anaphase, we speculated that proper chromosome segregation could be affected as well. To test this hypothesis, we coinjected Dm EB1 antibodies and rhodamine-labeled histones into embryos expressing GFP–tubulin to simultaneously observe the behaviors of chromosomes and microtubules. In control embryos, fluorescent histones incorporated into chromatin and allowed observation of chromosome condensation at prometaphase, chromosome congression to the metaphase plate, and sister chromatid separation and segregation to each pole during anaphase (Fig. 6 a; Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200202032/DC1). Injection of Dm EB1 antibodies, however, disrupted chromosome segregation and produced a range of phenotypes (Fig. 6 b; Table III; Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200202032/DC1). The mildest defect caused by Dm EB1 antibody injection was the generation of lagging chromosomes during anaphase (30%). More deleterious effects were produced when the chromosomes began to segregate but failed during anaphase, producing bilobed (8.6%) or stretched (31%) chromosomal masses that failed to segregate and decondensed midway between the poles at the end of mitosis. The most extreme defect observed was complete inhibition of chromosomal segregation, leading to the formation of a tetraploid nucleus in between two spindle poles. Taken together, out results from microinjecting anti–Dm EB1 antibodies into preblastoderm embryos indicate that Dm EB1 plays a crucial role in mitotic spindle formation and elongation and is needed for the proper segregation of mitotic chromosomes during anaphase.
Figure 6.
Microinjection of antibodies against Dm EB1 prevents normal chromosome segregation during mitosis. (a) A series of still images illustrating proper chromosome segregation during cycle 13 mitosis in a control-injected embryo. The embryo is expressing GFP–tubulin to visualize microtubules (red) and was injected with rhodamine–histone (green) to image chromosome dynamics. (b) Panels of still images demonstrating failed chromosome-to-pole movements in an embryo prepared as above and injected with polyclonal antibodies raised against Dm EB1. Images were collected from living embryos using spinning disk confocal microscopy. Bar, 7.5 μm.
Table III. Quantitation of chromosome segregation defects induced by injection of Dm EB1 antibodies into cycle 13 Drosophila embryos.
| Nuclear phenotype | Control | Anti-EB1 | ||
|---|---|---|---|---|
| Proximal | Distal | Proximal | Distal | |
| % Normal (n) | 90.4 (57) | 98.3 (60) | 8.6 (14) | 79.0 (111) |
| % Lagging (n) | 9.5 (4) | 1.6 (1) | 30.0 (49) | 18.0 (26) |
| % Bilobed (n) | 0 | 0 | 8.6 (14) | 0.7 (1) |
| % Stretched (n) | 3.2 (2) | 0 | 31.0 (51) | 2.2 (3) |
| % No segregation (n) | 0 | 0 | 21.8 (34) | 0 |
The defects are expressed as percentages with the number of spindles observed provided in parentheses. Spindle defects were quantitated both proximal and distal to the site of microinjection in buffer- and antibody-injected embryos. Individual spindles were scored as “normal” if their mitosis occurred unperturbed by the injections, “lagging” if one or more of the chromosomes failed to segregate at anaphase, “bilobed” if the decondensed daughter nuclei were distinct at the end of mitosis but remained connected by a bridge of chromatin, or “stretched” if the sister chromosomes exhibited a partial segregation and decondensed to an elongated mass between the two poles.
Discussion
In this study, we investigated the role of EB1 in Drosophila cells using two different approaches for interfering with the function of this protein. Using RNAi, we depleted EB1 in S2 cells that were spread on concanavalin A–coated surfaces, which allowed the imaging of single microtubules. In addition, we injected anti-EB1 antibodies into early Drosophila embryos, which allowed live cell examination of many spindles simultaneously and without mitotic checkpoint arrest. Both of these techniques and preparations provided confirmatory results demonstrating that Dm EB1 plays an important role in the assembly and dynamics of the mitotic spindle. In both cases, we find that interfering with Dm EB1 has similar, albeit somewhat pleiotropic, effects on spindle structure, the most common being an overall decrease in the length of the spindle, a lack of astral microtubules, a defocusing of the spindle poles, and dissociation of centrosomes from the spindle. In the cell culture system, we also observe that mitotic spindles in Dm EB1–depleted cells lose the ability to self-center and are generally mispositioned within the cell. In living Drosophila embryos, where spindles proceed into anaphase without a checkpoint arrest, we also find that Dm EB1 plays an important role late in mitosis during anaphase spindle elongation and chromosome segregation.
EB1 influences microtubule dynamics in distinct ways in interphase and mitosis
During interphase, loss of Dm EB1 does not alter microtubule length or distribution and produces no obvious effect on cell morphology. However, by imaging GFP–tubulin, we find that loss of Dm EB1 causes the majority (60%) of microtubules to enter a “paused” state in which they are neither growing nor shrinking. Microtubules assembled from purified tubulin rarely exhibit such static behavior (Walker et al., 1988). Therefore, pausing most likely reflects the action of a cellular factor that suppresses microtubule dynamics, possibly by capping the microtubule end. Dm EB1 appears to promote dynamic behavior, at least in part, by antagonizing the actions of this yet unknown factor, either by directly competing for tubulin sites or by inducing a conformation at the microtubule end that prevents capping. Interestingly, our findings are similar to those obtained for Bim1p in S. cerevisiae, which show that microtubules in bim1-null cells are less dynamic in G1 of the cell cycle, spending >60% of their lifetimes in a paused state (Tirnauer et al., 1999). Although these effects of EB1 on interphase microtubule dynamics are not crucial to the formation of the microtubule network in S2 cells, we speculate that they may be important for dynamic rearrangements of the microtubule cytoskeletal network that occur during cell migration and other polarized cell shape changes.
Although Dm EB1 loss does not dramatically change the number of microtubules during interphase, it does decrease microtubule lengths and numbers in mitosis. In a study of microtubule dynamics at the G2/M transition in vertebrate cells, Zhai et al. (1996) observed that microtubule polymer levels dramatically decrease upon entry into prophase, but polymer levels increase as mitosis progresses and chromosomes become attached to microtubules. In EB1-depleted S2 cells, the extent of microtubule disassembly in prophase is more severe than in control cells, and may reflect an inability of the cell to reestablish microtubule polymer levels later in mitosis. This decrease in microtubule polymer was not observed with RNAi of another plus end–binding protein, CLIP-190 (Lantz and Miller, 1998; unpublished data). The basis for the mitotic-specific effect of Dm EB1 on microtubule stability may be due either to a change in how Dm EB1 interacts with microtubules or in the activities of other microtubule-associated proteins. We favor the latter possibility because it has been shown that assembly-promoting factors, such as XMAP215/TOG, are downregulated in mitosis, which allows depolymerization factors, such as the KIN-I kinesins or stathmin/OP18, to predominate (Andersen, 2000; Tournebize et al., 2000). Thus, EB1 may play a particularly important role in mitosis in counteracting microtubule depolymerization factors. The most direct way to test these ideas would be to observe microtubule behavior in Dm EB1–depleted mitotic cells in real time. However, due to the loss of astral microtubules and the bundling of microtubules in the interpolar regions, we were unable to resolve the behavior of individual microtubules in GFP–tubulin-transfected, Dm EB1–depleted cells.
This finding for DM EB1 differs from that obtained for Bim1p in S. cerevisiae, as bim1Δ cells do not exhibit significant defects in microtubule behavior in preanaphase or anaphase, even though there are subtle changes in microtubule dynamics and spindle positioning (Tirnauer et al., 1999). The role of Dm EB1 also differs from its orthologue Mal3 in fission yeast, as null mutants exhibit abnormally short cytoplasmic microtubules but no defects in their spindle morphology. These differences may not be due to different molecular mechanisms of EB1, but rather due to the distinct processes for creating the spindle and executing chromosome movements.
EB1 is needed for proper formation and positioning of mitotic spindles
The most frequent phenotype observed in Dm EB1–depleted mitotic spindles is the failure to form astral microtubules, which may underlie many of the aberrant spindle phenotypes produced by Dm EB1 RNAi– and antibody-injected embryos, although other unknown roles of EB1 (e.g., interactions with other proteins) may play a role as well. In the absence of Dm EB1, the central spindle still contains kinetochore fibers, often they are partially or fully detached from the centrosomes, which gives rise to defocused or “splayed apart” microtubules at the poles. In wild-type spindles, we speculate that astral microtubules nucleated from the spindle poles intercalate with microtubule bundles in the central spindle to focus them to the poles via microtubule cross-linking proteins or through motor proteins such as Ncd or cytoplasmic dynein (Sharp et al., 2000b).
Loss of astral microtubules is also likely to underlie the spindle positioning defects that we observe in Dm EB1–depleted cells. Spindle positioning has been speculated to involve a balancing of forces generated either by growing astral microtubules pushing against the cell cortex or by cortically bound motor complexes containing dynein and Lis1 pulling on astral microtubules (Faulkner et al., 2000; Segal and Bloom, 2001; Dujardin and Vallee, 2002). Similarly, yeast Bim1p has been shown to be important in orienting the mitotic spindle into the bud neck by linking microtubules to the cortically bound Kar9p complex and the actin cytoskeleton (for review see Bloom, 2000). Mammalian EB1 also has been shown to interact with dynein intermediate chain and with subunits of the dynactin complex, and so it may mediate motor microtubule linkages at the plasma membrane of higher eukaryotes as well (Berrueta et al., 1999).
Our results also shed light upon the recent observations of Lu et al. (2001) who demonstrated that Dm EB1 is required for spindle orientation in epidermoblasts of the Drosophila embryo. In this cell type, cell divisions are normally oriented within the plane of the tissue in response to lateral polarity cues established by adherens junctions formed between neighboring cells. When Dm EB1 was reduced by RNAi, epidermal cells instead divide randomly with respect to the plane of the tissue. It was generally assumed that this effect was due to impaired interactions of microtubules with adherens junction components that served as polarity cues. Although this may be true, our results also reveal a drastic reduction in the number and length of astral microtubules that also may underlie the defect observed in these asymmetric cell divisions.
Anaphase chromosome motion is impaired after inhibition of EB1 function
Inhibition of Dm EB1 in synctitial Drosophila embryos by injection of anti-EB1 antibodies also revealed important roles for this protein during the later stages of mitosis. In these cells, the most severe mitotic defects were observed closest to the injection site, and these included dramatically reduced rates of spindle elongation throughout mitosis and defective chromosome segregation. Spindles distal to the injection site exhibited less severe structural defects, but also exhibited lagging chromosomes during anaphase. These phenotypes were not directly observed in S2 cells depleted of Dm EB1, and we postulate that this is due to activation of the spindle checkpoint as the result of damage to the spindle.
Why do mitotic spindles fail to elongate during anaphase? The forces that drive spindle elongation during anaphase B are derived, at least in part, from the activities of cortical cytoplasmic dynein pulling on astral microtubules and from bipolar kinesins that push spindle poles apart by sliding antiparallel interpolar microtubule bundles. We demonstrated that inhibition of Dm EB1 suppresses the formation of both astral microtubules and interpolar microtubules and eliminates the formation of midbodies during late telophase. A role for Dm EB1 in the formation or stabilization of these subpopulations of spindle microtubules is supported by our immunolocalization data showing the protein enriched on astral microtubules and in interpolar bundles and midbodies in S2 cells and embryos (Fig. 1 h; unpublished data). The inhibition of anaphase after EB1 depletion may be a consequence of the failure to produce spindles that form the specialized microtubule structures required for elongation in anaphase. Another possible mechanism is suggested by the observation that anaphase B is accompanied by microtubule polymerization in the central spindle that may contribute to the forces that drive spindle poles apart (Shelden and Wadsworth, 1990). As Dm EB1 appears necessary to promote microtubule growth during mitosis, it may be that in the absence of this protein, anaphase microtubule polymerization is inhibited and spindle elongation fails. These two potential mechanisms are not mutually exclusive.
The question of why chromosome segregation fails when Dm EB1 is inhibited is also an important one. The simplest explanation is that, in the absence of EB1, spindle elongation during anaphase is crippled to such an extent that chromosome-to-pole movement is insufficient to drive their segregation, leading to an increased number of 4N nuclei. Alternatively, it is possible that Dm EB1 mediates interactions between kinetochores and microtubules and in the absence of this interaction, anaphase A is affected. This is an interesting possibility in light of recent work identifying APC as a kinetochore component (Fodde et al., 2001; Kaplan et al., 2001), although no evidence exists for a direct interaction between Drosophila APC/APC2 and Dm EB1 (Lu et al., 2001; unpublished data).
In conclusion, our studies reveal that Dm EB1 is not essential for creating the microtubule network in interphase but is essential for microtubule organization in mitosis. Such cell cycle specificity, which is not common among MAPs, raises the possibility that EB1 might constitute an attractive target for small molecule inhibition of cell division in cancer chemotherapy. At least three different genes for EB1 family proteins exist in the human genome; one of which is ubiquitously expressed, and the other two are tissue specific. Selective inhibition of these mammalian genes will be required to evaluate the utility of EB1 inhibition as means of interfering with cancer growth.
Materials and methods
Cell culture
Schneider S2 cells were maintained in Schneider's Drosophila medium (GIBCO BRL) supplemented with 10% heat-inactivated FCS (GIBCO BRL) and penicillin/streptomycin. For microscopy, cells were plated on acid-washed No.1.5 coverslips (Corning) that had been treated with a solution of 0.5 mg/ml concanavalin A (Sigma-Aldrich) in water and allowed to air dry.
Antibodies
We obtained an EST, clone LD08743, from the Berkeley Drosophila Genome Project that contained the full reading frame for Drosophila EB1 (Research Genetics). Primers containing a 5′ BamHI site and a 3′ EcoRI site were used to amplify the EB1 coding sequence. This PCR product was inserted in frame into the expression vector pGEX-6-2P (Amersham Biosciences) to make a fusion protein with GST. The recombinant GST–Dm EB1 was expressed in Escherichia coli and purified by glutathione-Sepharose affinity chromatography per the manufacturer's instructions. Anti-Dm EB1–GST antisera were produced in rabbits by Covance, Inc. Polyclonal antibodies against GST were first removed by applying the serum to a GST–Sepharose column; the flowthrough was applied to a GST–Dm EB1–Sepharose column and the Dm EB1 polyclonal antibodies were eluted with low pH.
Immunofluorescence microscopy
For microtubule staining, S2 cells were rinsed in BRB80 (80 mM Pipes, pH 6.9, 1 mM MgCl2, 1 mM EGTA) and fixed in the same buffer containing 0.5% glutaraldehyde (EM Sciences), 3% formaldehyde (EM Sciences), and 1 mg/ml saponin for 10 min. The cells were then permeabilized in PBS containing 0.5% SDS, treated with sodium borohydride, and blocked with 5% normal goat serum in PBS/0.1% Triton X-100. In experiments examining the localization of Dm EB1, cells were fixed for 10 min by immersion in a solution of 90% methanol, 3% formaldehyde, 5 mM sodium carbonate (pH 9) chilled to −80°C. Samples were then rehydrated into PBS/0.1% Triton X-100 and blocked as above. All antibodies were diluted into 5% normal goat serum in PBS/Triton (DM1α, 1:500; rabbit anti-EB1, 1:1,000) and applied to the fixed cells for 1 h followed by extensive washing with PBS/Triton X-100. Fluorescent secondary antibodies (Cy2-conjugated anti–rabbit and rhodamine-X–conjugated anti–mouse; Jackson ImmunoResearch Laboratories) were used at a final dilution of 1:300. After antibody staining, cells were treated with DAPI (0.5 μg/ml in PBS) for 10 min, briefly rinsed with distilled water, and mounted in 90% glycerol, 10% 0.1 M borate, pH 9.0, plus 5% n-propyl gallate. Specimens were imaged by confocal microscopy (TCS; Leica) and presented as maximum intensity projections.
Double-stranded RNAi
RNAi was performed according to the methods of Clemens et al. (2000) using target sequences that exhibited minimal homology with other genes as determined by BLAST comparison. Templates for in vitro transcription were generated by using the primers 5′-GAATTAATACGACTCACTAT-AGGGAGAATGGCTGTAAACGTCTACTCCACAAATGTG-3′ and 5′-GAA-TTAATACGACTCACTATAGGGAGATGCCCGTGCTGTTGGCACAGGCG-TTTA-3′ to amplify the first 600 bp from the coding sequence of Dm EB1 from Drosophila EST clone LD08743 (Research Genetics), and the primers 5′-TAATACGACTCACTATAGGGAGAGATGTTAATGGGCACAAATTTTCT-3′ and 5′-TAATACGACTCACTATAGGGAGATTTGTATAGTTATCCATGCCATG-3′ to amplify a 650-bp segment of EGFP from the vector EGFP-C1 (CLONTECH Laboratories, Inc.). PCR products were used as templates for in vitro transcription using the Megascript T7 kit (Ambion) according to the manufacturer's instructions.
Microtubule dynamics in live S2 cells
In the experiments in which microtubule dynamics were observed, cells were treated with 15 μM dsRNA every 3 d for 6 d and transfected with a plasmid encoding GFP–α-tubulin (gift from Nicole Grieder, University of Basel, Basel, Switzerland) in the pAc5.1/His-V5B vector (Invitrogen) using the Cellfectin transfection reagent (Invitrogen) according to manufacturer's instructions. The cells were cultured for 2 d more in the presence of dsRNA, and then plated onto concanavalin A–coated coverslips 2 h before observation. Coverslips were fastened to microscope slides using warm VALAP (equal parts vaseline, lanolin, and paraffin) and fragments of broken coverslips as spacers. Microtubule dynamics were observed on a Nikon TE300 inverted microscope with a 100X/1.4 N.A. objective lens using an Orca II cooled CCD camera (Hamamatsu). Images were acquired for a period of 5 min at a frame capture rate of every 5 s using Simple PCI software (Compix, Inc.). Image sequences were converted to movies and the ends of microtubules were tracked over time using ImageJ (http://rsb.info.nih.gov/ij/). Microtubule dynamics were calculated as described in Tirnauer et al. (1999).
Embryo microinjection and live cell microscopy
Embryo microinjection was performed essentially as previously described (Sharp et al., 1999). Rabbit anti–Dm EB1 polyclonal antibodies were affinity purified on the day of injection and concentrated to ∼25 mg/ml using Ultrafree centrifugal concentrators (Millipore). Control embryos were injected with PBS alone. In some experiments, antibodies were coinjected with rhodamine-labeled histones prepared as previously described (Valdes-Perez and Minden, 1995). Time-lapse fluorescence microscopy was performed using an Ultraview spinning disk confocal microscope (PerkinElmer). Image series were manipulated and quantitated as previously described (Sharp et al., 1999).
Online supplemental material
Supplemental videos 1–4 are available online at http://www.jcb.org/cgi/content/full/jcb.200202032/DC1. Videos 1 and 2 show examples of microtubule behavior in cells expressing GFP–tubulin. Video 1 is a sequence acquired from a control cell, whereas Video 2 was acquired from a cell treated with RNAi to inhibit DmEB1. Videos 3 and 4 show mitotic spindle dynamics in embryos expressing GFP–tubulin and injected with rhodamine–histones to visualize chromosomes. Video 3 is a sequence of a control-injected embryo, whereas Video 4 is of an embryo injected with antibodies raised against DmEB1.
Supplemental Material
Acknowledgments
We are grateful to Nicole Greider for the gift of the pUASp-GFPS65C-α1-tub84B construct and to Thomas Kaufman (Indiana University, Bloomington, IN) for the gift of anti-centrosomin antibodies. We thank Dyche Mullins and Peter Walter (University of California, San Francisco [UCSF]) for the use of their microscopes. We are grateful to Kevin Slep (UCSF) for valuable input throughout this project.
This work was supported by grants from the National Institutes of Health (38499) and the Howard Hughes Medical Institute.
The online version of this article includes supplemental material.
Footnotes
Abbreviations used in this paper: APC, adenomatous polyposis coli; MAP, microtubule-associated protein; RNAi, RNA-mediated inhibition.
References
- Andersen, S.S. 2000. Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol. 10:261–267. [DOI] [PubMed] [Google Scholar]
- Beinhauer, J.D., I.M. Hagan, J.H. Hegemann, and U. Fleig. 1997. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J. Cell Biol. 139:717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrueta, L., J.S. Tirnauer, S.C. Schuyler, D. Pellman, and B.E. Bierer. 1999. The APC-associated protein EB1 associates with components of the dynactin complex and cytoplasmic dynein intermediate chain. Curr. Biol. 9:425–428. [DOI] [PubMed] [Google Scholar]
- Bloom, K. 2000. It's a kar9ochore to capture microtubules. Nat. Cell Biol. 2:E96–E98. [DOI] [PubMed] [Google Scholar]
- Clemens, J.C., C.A. Worby, N. Simonson-Leff, M. Muda, T. Maehama, B.A. Hemmings, and J.E. Dixon. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA. 97:6499–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., and T.J. Mitchison. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83–117. [DOI] [PubMed] [Google Scholar]
- Desai, A., S. Verma, T.J. Mitchison, and C.E. Walczak. 1999. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 96:69–78. [DOI] [PubMed] [Google Scholar]
- Drubin, D.G., and W.J. Nelson. 1996. Origins of cell polarity. Cell. 84:335–344. [DOI] [PubMed] [Google Scholar]
- Dujardin, D.L., and R.B. Vallee. 2002. Dynein at the cortex. Curr. Opin. Cell Biol. 14:44–49. [DOI] [PubMed] [Google Scholar]
- Faulkner, N.E., D.L. Dujardin, C.Y. Tai, K.T. Vaughan, C.B. O'Connell, Y. Wang, and R.B. Vallee. 2000. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2:784–791. [DOI] [PubMed] [Google Scholar]
- Fodde, R., J. Kuipers, C. Rosenberg, R. Smits, M. Kielman, C. Gaspar, J.H. van Es, C. Breukel, J. Wiegant, R.H. Giles, and H. Clevers. 2001. Mutations in the APC tumor suppressor gene cause chromosomal instability. Nat. Cell Biol. 3:433–438. [DOI] [PubMed] [Google Scholar]
- Hyman, A.A., and E. Karsenti. 1996. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell. 84:401–410. [DOI] [PubMed] [Google Scholar]
- Juwana, J.P., P. Henderikx, A. Mischo, A. Wadle, N. Fadle, K. Gerlach, J.W. Arends, H. Hoogenboom, M. Pfreundschuh, and C. Renner. 1999. EB/RP gene family encodes tubulin binding proteins. Int. J. Cancer. 81:275–284. [DOI] [PubMed] [Google Scholar]
- Kaplan, K.B., A.A. Burds, J.R. Swedlow, S.S. Bekir, P.K. Sorger, and I.S. Nathke. 2001. A role for the adenomatous polyposis coli protein in chromosome segregation. Nat. Cell Biol. 3:429–432. [DOI] [PubMed] [Google Scholar]
- Karr, T.L., and B.M. Alberts. 1986. Organization of the cytoskeleton in early Drosophila embryos. J. Cell Biol. 102:1494–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, D.R., T.J. Mitchison, and B.M. Alberts. 1988. Behavior of microtubules and actin filaments in living Drosophila embryos. Development. 103:675–686. [DOI] [PubMed] [Google Scholar]
- Korinek, W.S., M.J. Copeland, A. Chaudhuri, and J. Chant. 2000. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 287:2257–2259. [DOI] [PubMed] [Google Scholar]
- Lantz, V.A., and K.G. Miller. 1998. A class VI unconventional myosin is associated with a homologue of a microtubule-binding protein, cytoplasmic linker protein-170, in neurons and at the posterior pole of Drosophila embryos. J. Cell Biol. 140:897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, L., J.S. Tirnauer, J. Li, S.C. Schuyler, J.Y. Liu, and D. Pellman. 2000. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 287:2260–2262. [DOI] [PubMed] [Google Scholar]
- Lu, B., L.Y. Jan, and Y.N. Jan. 1998. Asymmetric cell division: lessons from flies and worms. Curr. Opin. Genet. Dev. 8:392–399. [DOI] [PubMed] [Google Scholar]
- Lu, B., F. Roegiers, L. Jan, and Y. Jan. 2001. Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature. 409:522–525. [DOI] [PubMed] [Google Scholar]
- Miller, R.K., S.C. Cheng, and M.D. Rose. 2000. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell. 11:2949–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., N. Shiina, and S. Tsukita. 2000. a. Adenomatous polyposis coli (APC) protein moves along microtubules and concentrates at their growing ends in epithelial cells. J. Cell Biol. 148:505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., N. Shiina, and S. Tsukita. 2000. b. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 10:865–868. [DOI] [PubMed] [Google Scholar]
- Morrison, E.E., B.N. Wardleworth, J.M. Askham, A.F. Markham, and D.M. Meredith. 1998. EB1, a protein which interacts with the APC tumor suppressor, is associated with the microtubule cytoskeleton throughout the cell cycle. Oncogene. 17:3471–3477. [DOI] [PubMed] [Google Scholar]
- Muhua, L., N.R. Adames, M.D. Murphy, C.R. Shields, and J.A. Cooper. 1998. A cytokinesis checkpoint requiring the yeast homologue of an APC- binding protein. Nature. 393:487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis, P. 1997. The adenomatous polyposis coli (APC) tumor suppressor. Biochim. Biophys. Acta. 1332:F127–F147. [DOI] [PubMed] [Google Scholar]
- Ramet, M., P. Manfruelli, A. Pearson, B. Mathey-Prevot, and R.A. Ezekowitz. 2002. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 416:644–648. [DOI] [PubMed] [Google Scholar]
- Rusan, N.M., C.J. Fagerstrom, A.M. Yvon, and P. Wadsworth. 2001. Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-α tubulin. Mol. Biol. Cell. 12:971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler, S.C., and D. Pellman. 2001. Microtubule “plus-end-tracking proteins”: the end is just the beginning. Cell. 105:421–424. [DOI] [PubMed] [Google Scholar]
- Segal, M., and K. Bloom. 2001. Control of spindle polarity and orientation in Saccharomyces cerevisiae. Trends Cell Biol. 11:160–166. [DOI] [PubMed] [Google Scholar]
- Sharp, D.J., K.L. McDonald, H.M. Brown, H.J. Matthies, C. Walczak, R.D. Vale, T.J. Mitchison, and J.M. Scholey. 1999. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 144:125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D.J., H.M. Brown, M. Kwon, G.C. Rogers, G. Holland, and J.M. Scholey. 2000. a. Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell. 11:241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D.J., G.C. Rogers, and J.M. Scholey. 2000. b. Microtubule motors in mitosis. Nature. 407:41–47. [DOI] [PubMed] [Google Scholar]
- Shelden, E., and P. Wadsworth. 1990. Interzonal microtubules are dynamic during spindle elongation. J. Cell Sci. 97:273–281. [DOI] [PubMed] [Google Scholar]
- Su, L.K., and Y. Qi. 2001. Characterization of human MAPRE genes and their proteins. Genomics. 71:142–149. [DOI] [PubMed] [Google Scholar]
- Su, L.K., M. Burrell, D.E. Hill, J. Gyuris, R. Brent, R. Wiltshire, J. Trent, B. Vogelstein, and K.W. Kinzler. 1995. APC binds to the novel protein EB1. Cancer Res. 55:2972–2977. [PubMed] [Google Scholar]
- Sullivan, W., and W.E. Theurkauf. 1995. The cytoskeleton and morphogenesis of the early Drosophila embryo. Curr. Opin. Cell Biol. 7:18–22. [DOI] [PubMed] [Google Scholar]
- Sullivan, W., P. Fogarty, and W. Theurkauf. 1993. Mutations affecting the cytoskeletal organization of syncytial Drosophila embryos. Development. 118:1245–1254. [DOI] [PubMed] [Google Scholar]
- Tirnauer, J.S., and B.E. Bierer. 2000. EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J. Cell Biol. 149:761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer, J.S., E. O'Toole, L. Berrueta, B.E. Bierer, and D. Pellman. 1999. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 145:993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize, R., A. Popov, K. Kinoshita, A.J. Ashford, S. Rybina, A. Pozniakovsky, T.U. Mayer, C.E. Walczak, E. Karsenti, and A.A. Hyman. 2000. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2: 13–19. [DOI] [PubMed] [Google Scholar]
- Valdes-Perez, R.E., and J.S. Minden. 1995. Drosophila melanogaster synctitial nuclear divisions are patterned: time-lapse images, hypothesis and computational evidence. J. Theor. Biol. 175:525–532. [DOI] [PubMed] [Google Scholar]
- Walker, R.A., E.T. O'Brien, N.K. Pryer, M.F. Soboeiro, and W.A. Voter. 1988. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 107:1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittman, T., and C.M. Waterman-Storer. 2001. Cell motility: can Rho GTPases and microtubules lead the way? J. Cell Sci. 114:3795–3803. [DOI] [PubMed] [Google Scholar]
- Yin, H., D. Pruyne, T.C. Huffaker, and A. Bretscher. 2000. Myosin V orientates the mitotic spindle in yeast. Nature. 406:1013–1015. [DOI] [PubMed] [Google Scholar]
- Yvon, A.C., P. Wadsworth, and M.A. Jordan. 1999. Taxol suppresses dynamics of individual microtubules in living human tumor cells. Mol. Biol. Cell. 10:947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, Y., P.J. Kronebusch, P.M. Simon, and G.G. Borisy. 1996. Microtubule dynamics at the G2/M transition: abrupt breakdown of cytoplasmic microtubules at nuclear envelope breakdown and implications for spindle morphogenesis. J. Cell Biol. 135:201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.