Abstract
Tim23p is imported via the TIM (translocase of inner membrane)22 pathway for mitochondrial inner membrane proteins. In contrast to precursors with an NH2-terminal targeting presequence that are imported in a linear NH2-terminal manner, we show that Tim23p crosses the outer membrane as a loop before inserting into the inner membrane. The Tim8p–Tim13p complex facilitates translocation across the intermembrane space by binding to the membrane spanning domains as shown by Tim23p peptide scans with the purified Tim8p–Tim13p complex and crosslinking studies with Tim23p fusion constructs. The interaction between Tim23p and the Tim8p–Tim13p complex is not dependent on zinc, and the purified Tim8p–Tim13p complex does not coordinate zinc in the conserved twin CX3C motif. Instead, the cysteine residues seemingly form intramolecular disulfide linkages. Given that proteins of the mitochondrial carrier family also pass through the TOM (translocase of outer membrane) complex as a loop, our study suggests that this translocation mechanism may be conserved. Thus, polytopic inner membrane proteins, which lack an NH2-terminal targeting sequence, pass through the TOM complex as a loop followed by binding of the small Tim proteins to the hydrophobic membrane spanning domains.
Keywords: protein import; TIM complex; Saccharomyces cerevisiae; mitochondria; protein translocation
Introduction
Most mitochondrial proteins are coded in the nucleus, synthesized in the cytosol as precursors with a cleavable NH2-terminal presequence, and imported into the mitochondria via the general import pathway (Schatz and Dobberstein, 1996; Paschen and Neupert, 2001; Pfanner et al., 2001). This pathway consists of chaperones in the cytosol and two translocons in the inner and outer membrane of mitochondria. The heterooligomeric translocase of outer membrane (TOM)* complex transports the precursor across the outer membrane. In the mitochondrial inner membrane, the translocase of inner membrane (TIM)23 complex facilitates import into the matrix. Tim23p and Tim17p form the pore, and the ATP-dependent import motor consisting of Tim44p, mHsp70, and GrpE provides the driving force for import. After translocation across both membranes, the presequence is generally cleaved and a number of chaperones mediate folding of the mature form of the precursor protein.
Proteins targeted to the mitochondrial inner membrane require an adapted second pathway (Káldi and Neupert, 1998; Pfanner, 1998; Koehler et al., 1999b). Substrates of this pathway are the mitochondrial carrier family including the ADP/ATP carrier (AAC) (Palmieri et al., 1996), Tim22p, and Tim23p (Koehler et al., 1998a; Sirrenberg et al., 1998; Leuenberger et al., 1999); these precursors are synthesized without a cleavable NH2-terminal presequence. Instead, the targeting information resides within the mature protein (Pfanner et al., 1987; Davis et al., 1998; Endres et al., 1999). The carrier protein sequence is a tripartite repeat, in which each repeat consists of a pair of membrane-spanning domains connected by a matrix-sided loop. Each module contains targeting information that cooperates in binding to the receptor Tom70 such that three Tom70 dimers are recruited per precursor (Ryan et al., 1999; Wiedemann et al., 2001). The carrier then passes through the TOM pore in a loop formation. After passing through the TOM pore, the hydrophobic precursors are escorted through the intermembrane space by two different soluble 70-kD protein complexes consisting of either Tim9p–Tim10p or Tim8p–Tim13p (Adam et al., 1999; Koehler et al., 1998b, 1999a; Leuenberger et al., 1999; Davis et al., 2000; Murphy et al., 2001). By binding to the hydrophobic transmembrane segments of AAC, the Tim9p–Tim10p complex potentially prevents the precursor from aggregating while passing through the aqueous intermembrane space (Curran et al., 2002). Insertion into the inner membrane is mediated by the 300-kD TIM22 complex, consisting of Tim12p, Tim18p, Tim22p, Tim54p, and a small fraction of Tim9p and Tim10p (Kerscher et al., 1997, 2000; Koehler et al., 1998a, 1998b, 2000; Sirrenberg et al., 1998; Adam et al., 1999).
The members of the family of small Tim proteins (Tim8p, Tim9p, Tim10p, Tim12p, and Tim13p) share ∼25% identity and 50% similarity and contain a conserved twin CX3C motif (Koehler et al., 1999a, 1999b). Previous studies suggested that the twin CX3C motif might coordinate zinc and form a zinc finger-like structure to mediate substrate binding (Sirrenberg et al., 1998; Adam et al., 1999; Paschen et al., 2000). Recently, we have shown that the cysteine residues in the Tim9p–Tim10p complex seemingly form disulfide bonds and do not coordinate zinc (Curran et al., 2002). Tim8p assembles with Tim13p and Tim9p assembles with Tim10p to form distinct 70-kD complexes with different substrate specificity: Tim8p–Tim13p binds to Tim23p, whereas Tim9p–Tim10p binds to members of the carrier family, Tim17p and Tim22p (Endres et al., 1999; Leuenberger et al., 1999; Davis et al., 2000; Paschen et al., 2000). The function of the 70-kD complexes is not essential for viability but facilitates import across the intermembrane space (Murphy et al., 2001).
To determine if the translocation mechanism via a loop formation for the carrier family is general for polytopic inner membrane proteins, we have extended our studies to characterize the Tim8p–Tim13p complex and its role in mediating the import of Tim23p. Tim23p does not consist of the tripartite repeat like the carrier proteins. Instead, the NH2-terminal half of Tim23p forms a soluble domain in the intermembrane space and potentially inserts in the outer membrane (Donzeau et al., 2000), and the COOH-terminal half consists of four membrane spanning domains, which contains the mitochondrial targeting and membrane insertion signals (Davis et al., 1998, 2000; Káldi et al., 1998). We report that Tim23p follows an import pathway similar to that of the carrier proteins and propose that this is a general translocation mechanism for polytopic inner membrane proteins, thus enabling the small Tim proteins to bind to the hydrophobic membrane domains in the aqueous intermembrane space to prevent subsequent aggregation.
Results
Tim23p passes through the TOM complex and inserts into the inner membrane as a loop
The insertion of Tim23p into the inner membrane is mediated by the TIM22 translocon in a membrane potential (Δψ)-dependent manner. Before reaching the TIM22 translocon, inner membrane precursors must first traverse the outer membrane and the intermembrane space. The carrier family, including the phosphate carrier and the AAC, passes through the TOM complex as a loop as shown with AAC–dihydrofolate reductase (DHFR) fusion constructs and crosslinking assays (Ryan et al., 1999; Wiedemann et al., 2001); however, it has not been shown whether the AAC–DHFR fusion constructs subsequently insert into the inner membrane. The Tim9p–Tim10p complex in the intermembrane space then binds to the membrane spanning domains of the substrate and escorts the precursor across the aqueous intermembrane space to the inner membrane (Curran et al., 2002). To investigate the mechanism by which Tim23p is translocated from the cytosol to the inner membrane, we performed in vitro import studies with a series of Tim23–DHFR fusion constructs and isolated mitochondria (Fig. 1 A). DHFR was fused in frame to Tim23p at the NH2 terminus, the COOH terminus, and at both termini. The radiolabeled fusion proteins were synthesized in vitro in the presence of methothrexate and incubated with uncoupled wild-type mitochondria followed by crosslinking and immunoprecipitation with antibodies monospecific for Tim8p (Fig. 1 B). Methotrexate binds tightly to DHFR and arrests translocation of the precursor at the TOM complex because the DHFR–methothrexate complex can not be unfolded at the outer membrane. With all fusion constructs, specific cross-linked products were obtained that were immunoprecipitated with antibodies against Tim8p. Antibodies against Tim13p yielded similar results for crosslinking and immunoprecipitation (unpublished data). To confirm that the DHFR fusion constructs arrested on the outer membrane, the import reactions were incubated with proteinase K prior to crosslinking. As expected, the crosslinked products were no longer detectable because the DHFR–Tim23p fusion constructs arrested on the outer membrane were accessible to protease. In the control reaction with Tim23p, crosslinks were obtained in both the presence and absence of protease because Tim23p completely crossed the outer membrane and arrested as a translocation intermediate in the intermembrane space. Therefore, Tim23p passes through the TOM complex, independently of the NH2 or COOH terminus, as a loop and binds to the Tim8p–Tim13p complex.
Figure 1.
Tim23p passes through the TOM complex as a loop and is inserted into the inner membrane. (A) Diagram of Tim23p-DHFR fusions constructed to investigate the import pathway of Tim23p. DHFR–Tim23p, DHFR fused at NH2 terminus of Tim23p; Tim23p–DHFR, DHFR fused to COOH terminus of Tim23p; DHFR–Tim23p–DHFR, DHFR fused to both the NH2- and COOH-termini of Tim23p. (B) Radiolabeled Tim23p and fusion constructs listed in A were synthesized in vitro and incubated with wild-type mitochondria in the absence of a Δψ and in the presence of methotrexate. Half of the import reaction was treated with 5 μg/ml proteinase K (protease) to ensure that the precursor arrested across the outer membrane. Subsequently, the chemical crosslinker DSS was added at 1 mM. After quenching, the sample was denatured and immunoprecipitated with antibodies against Tim8p (IP). Bound proteins were eluted with SDS-containing sample buffer and analyzed by SDS-PAGE and fluorography. STD, 5% of the radioactive prescursor added to each assay. The arrow denotes the location of the monomeric precursor. (C) The radiolabeled precursors in A were imported into wild-type mitochondria at 25°C in the presence or absence of a Δψ. The mitochondria were lysed by osmotic shock at 4°C in the presence of 20 μg/ml proteinase K, followed by incubation with 1 mM PMSF, and extraction with 0.1 M Na2CO3 (pH 11.0). Tim23f, 14 kD COOH-terminal protease-resistant fragment that is inserted into the inner membrane.
We tested whether the Tim23p–DHFR fusion constructs inserted into the mitochondrial inner membrane. The fusion constructs were synthesized in the absence of methotrexate so that the precursor would not arrest at the outer membrane and imported into wild-type mitochondria in the presence or absence of a Δψ (Fig. 1 C). To confirm that the constructs were inserted into the inner membrane, the mitochondria from import reactions were treated by osmotic shock in the presence of protease to disrupt the outer membrane. Results from SDS-PAGE showed that the fusion constructs inserted into the inner membrane by the presence of a 14-kD protease-resistant fragment (Tim23f), consisting of the four membrane spanning domains. Taken together, these data suggest that import and insertion of Tim23p into the inner membrane is not dependent on the NH2 or COOH terminus and that Tim23p crosses the outer membrane as a loop.
Recombinant and native Tim8p–Tim13p complex have similar properties
To study the mechanism by which the Tim8p–Tim13p complex mediates the import of Tim23p, we developed a method to purify the recombinant Tim8p–Tim13p complex from an Escherichia coli strain. Specifically, TIM8 and TIM13 with their own E. coli ribosomal binding site were cloned in tandem in an expression plasmid and transformed into the E. coli host BL21(DE3). Affinity tags were not utilized because Tim8p and Tim13p have a molecular mass near 10 kD; the addition of tags to the monomers would significantly increase their molecular mass and could interfere with assembly of the complex. The Tim8p–Tim13p complex was purified to homogeneity from an E. coli lysate by subsequent chromatography steps using an anion exchange column, followed by a cation exchange column and a gel filtration column. The Tim8p–Tim13p complex eluted at 60–80 mM NaCl on ion exchange columns and eluted in the 60–70 kD mass range from the gel filtration column; the complex was purified to 95% homogeneity as shown by SDS-PAGE and Coomassie staining (Fig. 2 A). The Tim8p–Tim13p complex seemingly assembled efficiently in the E. coli cytoplasm, because Tim8p and Tim13p were only detectable in a 70-kD complex when separated by blue native gel electrophoresis (Fig. 2 B). In addition, the recombinant complex and the Tim8p–Tim13p complex from an intermembrane space fraction migrated identically in a native gel, indicating that the two complexes were identical with regard to shape and charge (Fig. 2 C).
Figure 2.
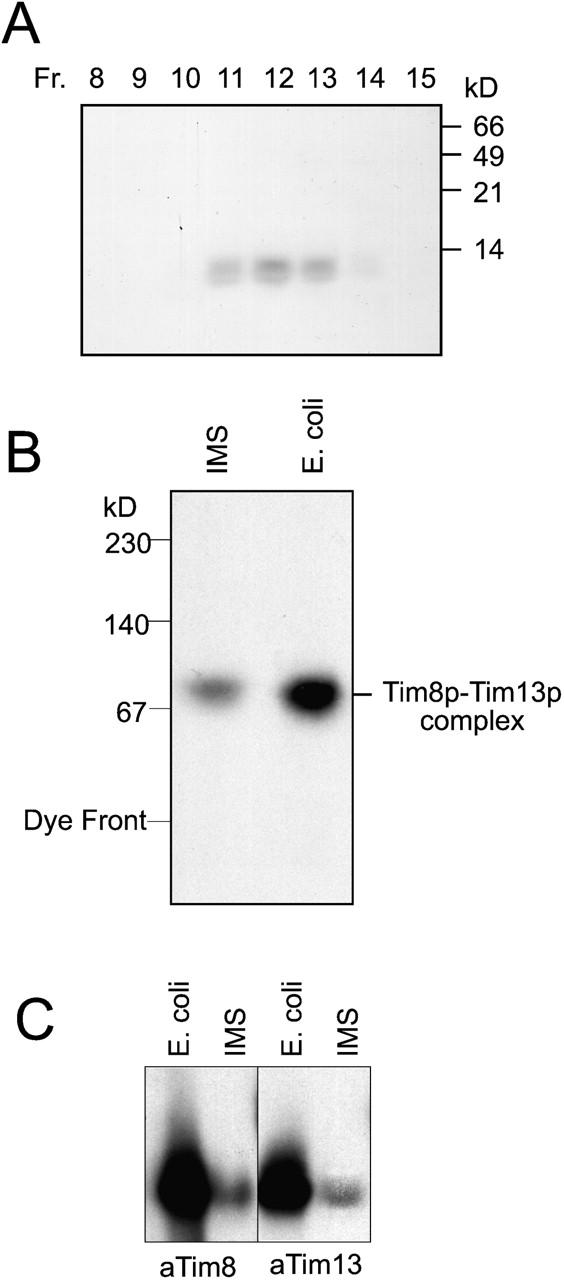
Tim8p and Tim13p assemble into a 70-kD complex when coexpressed in E. coli. (A) An expression plasmid was constructed in which TIM8 and TIM13 were each placed behind an E. coli ribosomal binding site. After transformation into E. coli host BL21(DE3) and induction, the Tim8p–Tim13p complex was purified using column chromatography (Materials and methods). The fractions from the final purification step (Superose 12 column) were analyzed for Tim8p and Tim13p by SDS-PAGE and Coomassie staining. Tim8p and Tim13p are denoted by the arrow. (B) Fraction 12 (as shown in A) and a mitochondrial intermembrane space sample (IMS) prepared by osmotic shock were analyzed on a 6–16% blue native gel followed by immunoblotting with antiserum for Tim8p. Proteins were identified by incubation with [125I]-protein A. (C) The recombinant Tim8p–Tim13p complex and a mitochondrial IMS were separated on a 6.5% native gel. Immunoblot analysis was performed with monospecific antiserum against Tim8p (αTim8) and Tim13p (αTim13) and detected with [125I]-protein A.
To characterize the oligomeric state of the Tim8p–Tim13p complex, the recombinant complex and an intermembrane space fraction were incubated with 0.1% glutaraldehyde on ice for 30 min (Fig. 3 A; van Dijl et al., 1998). The crosslinked intermediates were analyzed by SDS-PAGE and immunoblot analysis with antibodies against Tim8p. We observed six distinct species that were interpreted as monomer to hexamer because their electrophoretic mobility (M w/Mm) was linearly related to the logarithm of subunit number and the distance each oligomer had migrated from the origin (Fig. 3 A, graph). As the reaction proceeded, the primary crosslinked product was a hexamer. Also, an identical crosslinking pattern was observed with antibodies against Tim13p (unpublished data). Quantitative amino acid analysis confirmed that Tim8p and Tim13p were present in equal molar ratios in the recombinant Tim8p–Tim13p complex (unpublished data). Therefore, the hexameric Tim8p–Tim13p complex is composed of three Tim8p and three Tim13p.
Figure 3.
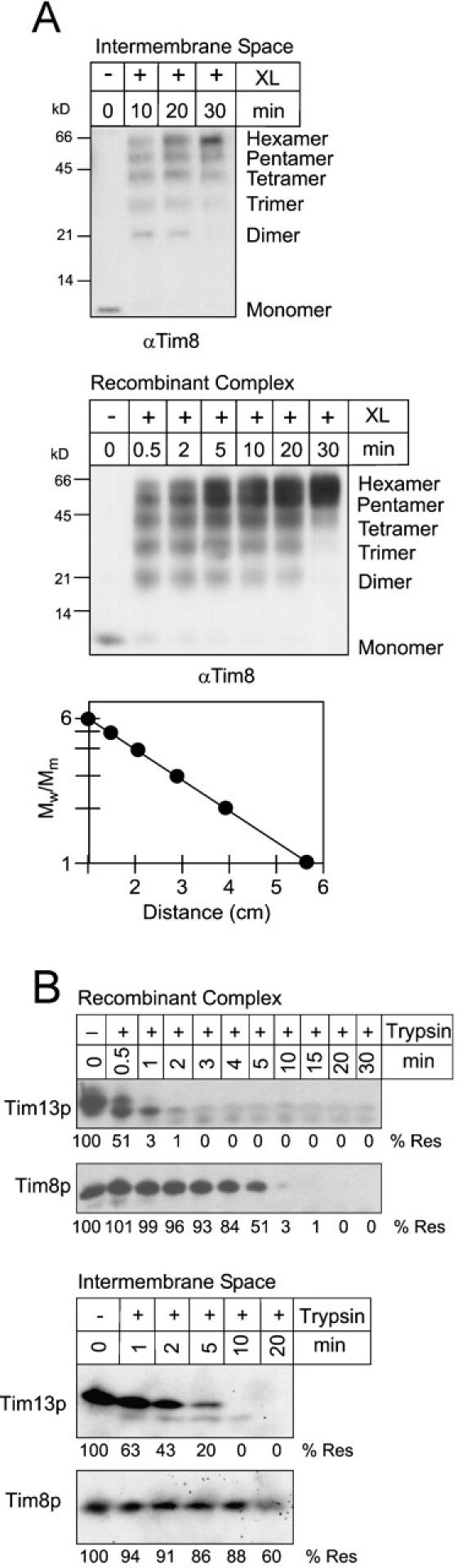
The recombinant and native Tim8p–Tim13p complexes have identical properties. (A) For intermolecular crosslinking assays, an intermembrane space sample and the recombinant Tim8p–Tim13p complex were incubated with 0.1% glutaraldehyde (XL) and aliquots were removed at the indicated time points. Crosslinked products were separated on 16% Tricine gels followed by immunoblot analysis with monospecific antisera against Tim8p (αTim8). The number of subunits is indicated at the right. Electrophoretic mobility (lowest panel) of the products from the 10-min time point of crosslinking was plotted against the logarithm of Mw/Mm, where Mw is the relative molecular mass of the crosslinked species and Mm is the average molecular mass of the Tim8p or Tim13p monomer (11 kD). (B) The recombinant Tim8p–Tim13p complex and a mitochondrial intermembrane space sample were incubated with 5 μg/ml trypsin at 37°C. At the indicated periods, aliquots were removed and soybean trypsin inhibitor was added. Samples were separated by SDS-PAGE followed by immunoblot analysis with antibodies for Tim8p or Tim13p. Protease resistance (% Res) was quantified by scanning laser densitometry and the zero time point was set at 100%.
To further compare assembly of the recombinant and native Tim8p–Tim13p complexes, we investigated the stability of Tim8p and Tim13p in limited proteolysis experiments. The recombinant complex and a mitochondrial intermembrane space fraction were treated with 5 μg/ml trypsin at 37°C (Fig. 3 B). Aliquots were removed as the reaction proceeded, stopped with the addition of soybean trypsin inhibitor, and analyzed by SDS-PAGE and immunodetection with polyclonal antisera specific to Tim8p and Tim13p. Tim13p was more sensitive to trypsin than Tim8p, although the stoichiometry and distribution of trypsin cleavage sites were approximately equal. The proteolysis pattern was similar when the complexes were treated with chymotrypsin or V8 protease (unpublished data). These findings confirmed additionally that the native and recombinant complexes have similar properties. Tim8p potentially might form a core complex because Tim8p was more protease-resistant than Tim13p.
Because the native and recombinant complexes were seemingly identical, the physical properties of the recombinant Tim8p–Tim13p complex were examined in further detail. The secondary structure of the recombinant Tim8p–Tim13p complex was analyzed by circular dichroism spectroscopy (Fig. 4, top). Computational analysis of the spectra (curves a and c) between 195 and 250 nm showed that the Tim8p–Tim13p complex was composed of 70.7% alpha helix, 5.2% beta sheet, 10.7% turn, and 13.4% undetermined (Fig. 4, bottom) (Perczel et al., 1992; Sreerama and Woody, 1993). Under thermal denaturation conditions of 95°C (curve b), the proteins adapted a random coil confirmation as expected. When cooled back to 4°C the subunits refolded and reassociated into the native complex with similar secondary structure to the undenatured sample (Fig. 4, curve c). In addition, analysis of the refolded complex on blue native gels showed that Tim8p and Tim13p were only detectable in a 70-kD complex (unpublished data) and thus confirmed that refolding and reassembly were efficient.
Figure 4.
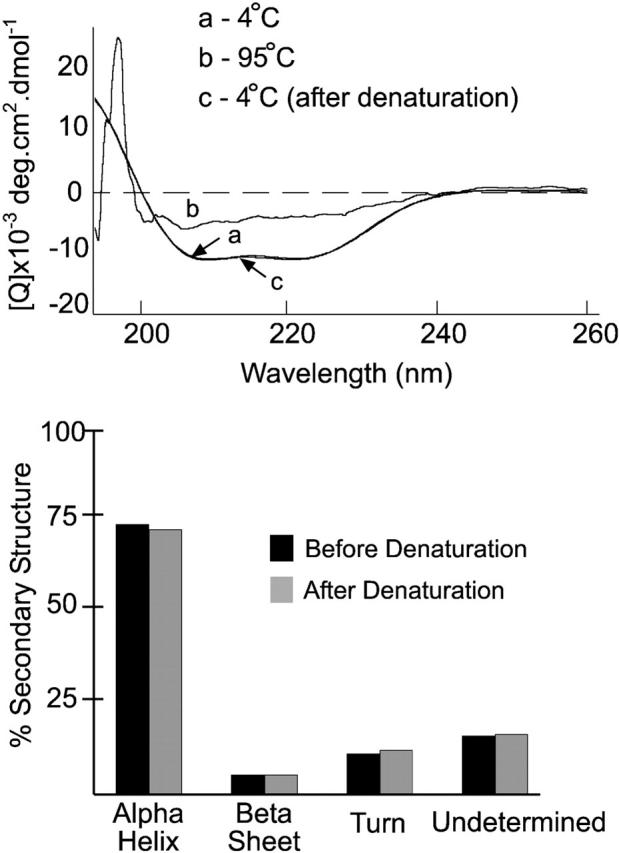
CD studies on the purified Tim8p–Tim13p complex. The CD spectra of the recombinant Tim8p–Tim13p complex at (a) 4°C, (b) 95°C, and (c) 4°C after heating to 95°C. Each spectrum was an average of eight scans. The secondary structure of the spectra at 4°C (curve a) before and after thermal denaturation (curve c) was predicted by a self-consistent method and the convex constraint algorithm (Perczel et al., 1992; Sreerama and Woody, 1993) (bottom).
The molecular mass and sedimentation coefficient of the recombinant Tim8p–Tim13p complex was investigated by analytical ultracentrifugation. From sedimentation velocity and equilibrium analysis experiments, the molecular mass of the hexameric Tim8p–Tim13p complex was calculated as 63 kD ± 3 kD with a sedimentation coefficient, S20,w = 3.83 (unpublished data); the actual molecular mass of the recombinant Tim8p–Tim13p complex is 62.4 kD. Sedimentation velocity ultracentrifugation analysis also showed that the sample was homogenous suggesting that both Tim8p and Tim13p were present in a hexameric complex and not complexes of other oligomeric states. Thus, these molecular mass studies confirm the results obtained by gel filtration and blue native gel electrophoresis.
The Tim8p–Tim13p complex does not coordinate zinc
The small Tim proteins contain the conserved twin CX3C motif, in which two cysteine residues are separated by three amino acids and the spacing between each CX3C is 11–16 amino acids. (Koehler et al., 1999b) Previous studies have yielded conflicting results: the small Tim proteins as monomers coordinated zinc in a 1:1 molar ratio (Sirrenberg et al., 1998; Paschen et al., 2000; Rothbauer et al., 2001), but the assembled Tim9p–Tim10p complex did not contain zinc (Curran et al., 2002). We investigated the state of the cysteine residues using several techniques, including thiol-trapping assays, reductant and metal chelator sensitivity assays, inductively coupled plasmon–atomic emission spectrometry (ICP-AE), and protein import assays.
Previously, we employed a thiol-trapping method that allowed the separation and visualization of oxidized and reduced species on Tricine gels to show that the cysteines residues in the Tim9p–Tim10p were occupied (Fig 5 A, schematic) (Jakob et al., 1999; Curran et al., 2002). The modification of Tim8p was followed by immunoblot analysis (Fig. 5 A). First, all accessible thiol groups in the intermembrane space fraction were alkylated with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS). AMS is a thiol-reactive reagent that alkylates cysteines residues, thereby adding 0.5 kD to the molecular mass of each thiol group. As a control, completely reduced Tim8p (pretreated with DTT; Fig. 5 A, lane 2) showed a slower mobility on tricine gels due to the addition of four AMS molecules to the four cysteine residues in Tim8p; this translated into a 2-kD change in molecular mass that was easily observed in the 8-kD protein. In contrast, completely oxidized Tim8p (pretreated with hydrogen peroxide; Fig. 5 A, lane 3) migrated at the same molecular weight as the untreated complex (Fig. 5 A, lane 4) because the sulfhydryl residues were oxidized and unreactive to AMS. When the sample was alkylated with AMS, Tim8p migrated identically to the oxidized sample; AMS was not bound toTim8p. To further confirm that sulfhydryls might form disulfide linkages, the experiment was modified as follows. All accessible thiol groups were first blocked irreversibly with iodoacetamide (IAA). After removal of the excess IAA, disulfide bonds present in the protein were reduced with DTT, followed by AMS alkyation to trap any free sulfhydryl groups (Fig. 5 A, schematic). With this modification, Tim8p in the sample that was first pretreated with DTT was not modified by AMS (Fig. 5 A, lane 5), because the cysteines residues were already reduced by DTT and subsequently blocked by IAA. But Tim8p in the sample that was first oxidized with hydrogen peroxide was modified by AMS (Fig. 5 A, lane 6), because IAA was not able to block the thiols in the first step. As expected, when the sample was alkylated with IAA (avoiding both reduction and oxidation), Tim8p migrated at a molecular weight identical to that of the oxidized Tim8p (Fig. 5 A, lane 7); four AMS molecules covalently bound to Tim8p. This observation thus indicates that the cysteine residues are not reduced and accessible to IAA, but instead are occupied, potentially in disulfide bonds, because the cysteines residues were modified after reduction with DTT. Analysis of Tim13p by thiol-trapping yielded the same results; the cysteines residues in Tim13p are occupied (unpublished data).
Figure 5.
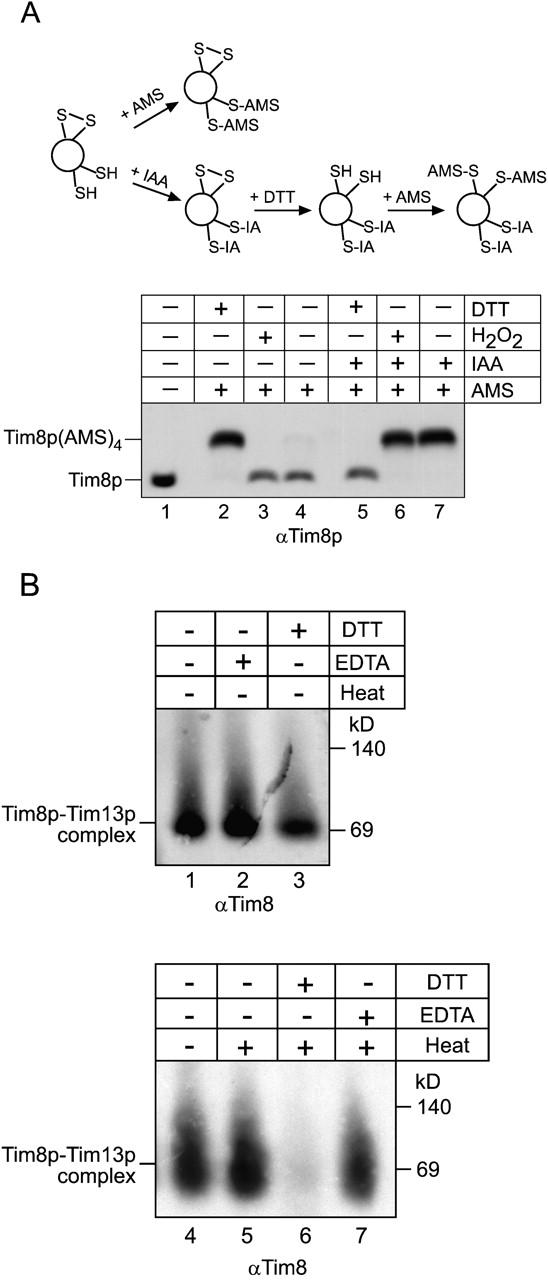
The Tim8p–Tim13p complex does not require Zn2 + for structural stability. (A) The sulfhydryl groups on the cysteines residues of the twin CX3C motif are occupied. A thiol-trapping method (Jakob et al., 1999) was used to assess the state of the cysteines residues (schematic). Mitochondrial intermembrane space fractions (200 μg/lane) were untreated (lanes 4 and 7), reduced with 10 mM DTT (lanes 2 and 5), or oxidized with 5% H2O2 (lane 3, 6). Half of each reaction was alkylated with AMS (lanes 2–4), which creates an increase in molecular mass of 0.5 kD. To confirm that AMS could access the cysteine residues, the other half of the reaction (lanes 5–7) was treated as follows: IAA was added to block free sulfhydryl groups, followed by treatment with DTT to reduce any disulfide bonds. The remaining free sulfhydryl groups were alkylated with AMS. As a control, an untreated sample was included (lane 1). Tim8p and Tim8p(AMS)4 are denoted. (B) The reductant DTT interferes with refolding of the Tim8p–Tim13p complex. The recombinant Tim8–Tim13p complex was incubated at 25°C in the presence of EDTA (lane 2), DTT (lane 3), or left untreated (lane 1) for 30 min. Additional samples were heated to 95°C in the presence of DTT (lane 6) or EDTA (lane 7) and quickly cooled by placing in an ice bath followed by blue-native gel electrophoresis. In control reactions, the Tim8p–Tim13p complex was mock-treated (heated and quickly cooled; lane 5) or incubated on ice (lane 4). Immunoblot analysis was performed with an antibody against Tim8p (αTim8).
To determine whether the Tim8p–Tim13p complex requires Zn2+ for structural stability we tested the ability of the complex to refold in the presence of the reductant DTT or metal chelator EDTA (Fig. 5 B). We first incubated the recombinant Tim8p–Tim13p complex in DTT or EDTA for 30 min at 25°C and then the samples were separated on blue native gels and visualized by immunodetection with antibodies specific for Tim8p (Fig. 5 B, lanes 1–3) and Tim13p (unpublished data). The samples incubated in EDTA and DTT were essentially stable at 25°C. Because the complex refolded efficiently after thermal denaturation (Fig. 4), we heated the Tim8p–Tim13p complex to 95°C for 10 min in the presence of DTT or EDTA and then chilled the complex in an ice bath. The complex refolded in the presence of the metal chelator EDTA (Fig. 5 B, lane 7), but not DTT (Fig. 5 A, lane 6). As controls, the stability of the complex was not affected when the complex alone was heated (Fig. 5 A, lane 5). Similar results were obtained with the Tim8p–Tim13p complex from an intermembrane space fraction (unpublished data). This suggests that the Tim8p–Tim13p complex does not coordinate Zn2+ and instead the cysteine residues may form disulfide linkages.
In addition, the Tim8p–Tim13p complex was separated on a denaturing nonreducing gel to determine if the thiol modifications were intra- or intermolecular. Tim8p and Tim13p migrated at a molecular mass near 10 kD that was almost indistinguishable from the separation profile on a reducing denaturing gel (unpublished data). This observation further suggests that the cysteine modifications are intramolecular.
Although Zn2+ is not essential for formation of the 70-kD complex, it is possible that the Tim8p–Tim13p complex coordinates zinc at sites other than the cysteine residues. To determine the total metal content of the Tim8p–Tim13p complex, we utilized ICP-AE spectrometry (Table I). Fractions containing the Tim8p–Tim13p complex from the final purification step on the size-exclusion column from three independent purifications were analyzed. As controls, fractions eluting before and after the Tim8p–Tim13p complex were included. The amount of protein in each sample was determined by quantitative amino acid analysis and added to the assay to provide a minimum detection of 30 ppb zinc, which is within the sensitivity of the instrument. This calculation was based upon the assumption that the Tim8p–Tim13p complex coordinates one Zn2+. It is possible that each Tim8p and Tim13p could bind a single zinc ion, which would result in detection of 180 ppb zinc. ICP-AE analysis of the recombinant Tim8p–Tim13p complex failed to detect significant levels of zinc above background (Table I). Manganese, iron, copper, and cobalt were also not detected at levels higher than that of the buffer blank (unpublished data). Because it is plausible that the availability of zinc was limiting during induction, the Tim8p–Tim13p complex was purified from cells that were induced and lysed in the presence of 1 mM zinc acetate. However, even under conditions of zinc loading, Zn2+ was not bound to the Tim8p–Tim13p complex (Table I).
Table I. Tim8p–Tim13 complex zinc complex.
| Mol zinc:mol complex | Actual (−Zn)a0:1 | Actual (+Zn)a0:1 | Predictedb1:1 | Predictedb6:1 |
|---|---|---|---|---|
| Zinc (sample/background, ppb)c | 0.792/0.352 | 0.821/0.443 | 7.34 | 44.06 |
| Zinc (μM)d | 0.007 | 0.006 | 1.12 | 6.72 |
| Protein (mg/ml)e | 0.071 | 0.074 | ||
| Protein (μM)d | 1.12 | 1.17 | ||
| Zinc per complex | .098 | .081 | 1 | 6 |
Values represent an average of three independent purifications from cells induced in the absence (−Zn) or presence of 1 mM zinc-acetate (+Zn).
Each twin CX3C motif (six per complex) in the Tim8p–Tim13p complex could potentially coordinate one Zn2+. The predicted levels of zinc are shown (in ppb) for hypothetical Tim8p–Tim13p complexes that bind one Zn2+ per complex or one Zn2+ per monomer (six zinc ions total). Predicted values are based on the same concentration of Tim8p–Tim13p complex (−Zn) used in actual measurements.
The average ICP-determined zinc concentration (ppb) is presented along with the average background level of zinc. Background levels of zinc were determined using buffer blanks from the same purification.
The micromolar concentrations of both protein and zinc were calculated assuming a molecular mass of 65.38 atomic mass units (amu) for zinc and 6.32 × 104 amu for the Tim8p–Tim13p complex.
The protein concentration of each sample was determined by quantitative amino acid analysis.
Measurement of Zn2+ content in the Tim8p–Tim13p complex
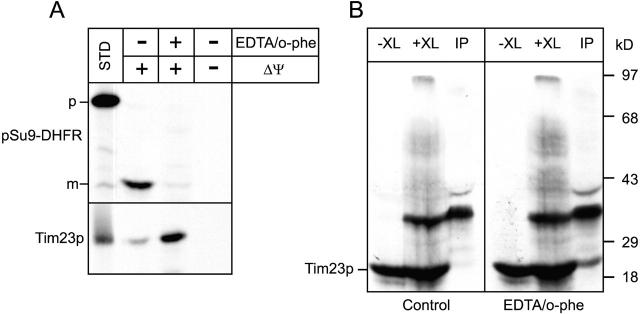
To assess the requirement of divalent metal ions in the native Tim8p–Tim13p complex in organello, we performed in vitro import and crosslinking assays with the Tim23p precursor in the presence of EDTA and 1,10-o-phenanthroline (o-phe) (Fig. 6); previous studies have shown that import of AAC and Tim23p is inhibited in the presence of these metal chelators (Sirrenberg et al., 1998; Paschen et al., 2000). Mitochondria were preincubated with the chelators for 10 min followed by import assays. To assess the effectiveness of chelator treatment, the fusion protein Su9–DHFR was imported as a control. Su9–DHFR is imported to the matrix and requires processing by the matrix processing peptidase. Because matrix processing peptidase requires a divalent metal ion for its activity, incubation with metal chelators results in subsequent inactivation (Bohni et al., 1983). As expected, Su9–DHFR import was inhibited in mitochondria that were pretreated with EDTA and o-phe. In contrast, Tim23p imported efficiently into wild-type mitochondria independent of chelator treatment; insertion into the inner membrane was confirmed by carbonate extraction (Fig 6 A, bottom). In crosslinking experiments, Tim23p was crosslinked to Tim8p, even in the presence of EDTA/o-phe (Fig. 6 B), in contrast to published studies (Sirrenberg et al., 1998; Paschen et al., 2000). The interaction of the Tim8p–Tim13p complex with Tim23p during translocation thus is not dependent upon divalent metal ions.
Figure 6.
The Tim8p–Tim13p complex does not require Zn2 + to facilitate import of Tim23p. (A) The radiolabeled fusion protein Su9-DHFR (top) and Tim23p (bottom) were synthesized in vitro and imported into wild-type mitochondria in the presence (+Δψ) or absence (−Δψ) of a Δψ. Where indicated, mitochondria were pretreated for 10 min with 10 mM EDTA and 2 mM o-phenanthroline (EDTA/o-phe). Samples were treated with proteinase K to remove nonimported precursor followed by addition of PMSF. Mitochondria in which Tim23p was imported were extracted with carbonate and the pellet after centrifugation was loaded. Import was analyzed by SDS-PAGE and fluorography. p, Su9-DHFR precursor; m, mature Su9-DHFR (processed by the matrix processing protease) (B) Radiolabeled Tim23p synthesized in vitro and imported into uncoupled wild-type mitochondria (−XL). Before import, mitochondria were pretreated for 10 min with 10 mM EDTA and 2 mM o-phenanthroline (EDTA/o-phe) or mock-treated (control). The reaction was crosslinked with 0.1 mM m-maleimidobenzoyl-N-hydroxysuccinimide ester for 30 min followed by quenching and immunoprecipitation with antibodies specific for Tim8p (IP).
The Tim8p–Tim13p complex binds preferentially to the membrane spanning domains of Tim23p
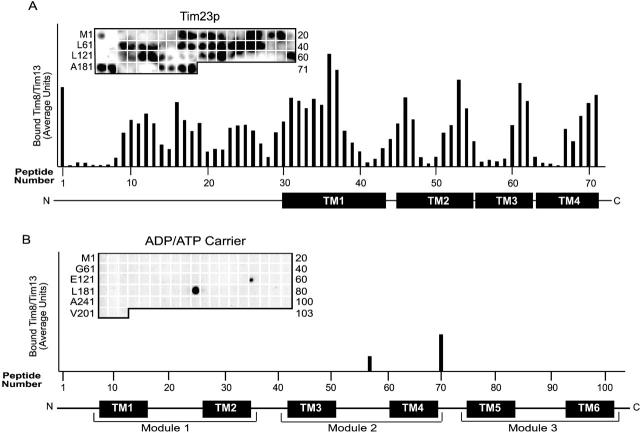
Previous studies to determine the domains in Tim23p that interact with the Tim8p–Tim13p complex have relied on cross-linking studies with truncated precursors and fusion constructs (Davis et al., 1998; Káldi et al., 1998; Davis et al., 2000). To identify specific sequences in Tim23p to which the Tim8p–Tim13p complex binds, we incubated a Tim23p peptide scan with the purified Tim8p–Tim13p complex (Fig. 7 A) (Brix et al., 1999, 2000). The peptide scan consisted of 13-mer peptides overlapping by 10 residues, which were attached covalently to a cellulose membrane. Thus, 71 peptides were synthesized for the 222 amino acids of Tim23p. The recombinant Tim8p–Tim13p complex was incubated to equilibrium with the cellulose-bound peptides, followed by electrotransfer and immunodetection with antibodies against Tim13p and [125I]-protein A (Fig. 7 A). Quantitative analysis was performed using laser scanning densitometry (Personal Densitometer SI; Molecular Dynamics) and the ImageQuaNT program, which corrects for the local background around each individual spot. The Tim8p–Tim13p complex bound preferentially to the membrane spanning domains of Tim23p, with strongest binding to transmembrane domain 1. Binding was also detectable within the soluble NH2-terminal domain of Tim23p, which confirms previous studies using a crosslinking approach (Davis et al., 2000; Paschen et al., 2000). Antibodies against Tim8p revealed a similar pattern of binding, confirming that the intact complex bound to the Tim23p peptides (unpublished data).
Figure 7.
Recombinant Tim8p–Tim13p complex binds to the transmembrane domains and intermembrane space domain of Tim23p but not AAC. (A) The recombinant Tim8p–Tim13p complex (200 nM) was incubated with a peptide scan consisting of 13-mers derived from Tim23p. The first peptide comprises amino acids 1–13 of Tim23p, the second peptide residues 4–16, and the third peptide residues 7–19, etc. The labeling on the left indicates the first amino acid of the left-most peptide of each row. The labeling on the right side indicates the number of the right-most peptide of each row. After washing the membrane, the bound Tim8p–Tim13p complex was transferred to a polyvinylidene difluoride membrane followed by immunoblot analysis with antiserum specific for Tim13p and [125I]-protein A. Binding was quantified by scanning laser densitometry from at least three independent experiments and plotted for each peptide. Transmembrane domains of Tim23p are plotted corresponding to the respective peptides. (B) As in A, except that the peptide scan consisted of 13-mers derived from AAC.
To confirm the specificity of the Tim23p-binding interactions, a peptide scan for AAC was synthesized and incubated with the Tim8p–Tim13p complex. The AAC peptide scan contained 103 peptides, again overlapping by 10 residues. As expected, after incubation with the Tim8p–Tim13p complex, significant binding to AAC was not detectable (Fig. 7 B). Although the Tim8p–Tim13p complex did bind to two peptides, the complex did not bind to the neighboring peptides with 10-residue overlaps, indicating that binding was most likely nonspecific. In previous studies, we also have shown that the Tim9p–Tim10p complex does not bind to the Tim23p peptide scan, confirming the specificity of the interaction of Tim8p–Tim13p with Tim23p (Curran et al., 2002). Taken together, the Tim8p–Tim13p complex binds the strongest to transmembrane domain 1 of Tim23p, suggesting that this region may cross the TOM complex first as a loop.
Discussion
The protein import pathway for Tim23p
To determine the role of the Tim8p-Tim13p complex in the biogenesis of Tim23p, we have investigated the Tim23p import pathway in detail and purified the Tim8p–Tim13p complex from E. coli for structural characterization and binding studies. Published reports using fusion constructs and crosslinking approaches have shown that the Tim8p–Tim13p complex binds to the NH2-terminal region of Tim23p. Paschen et al. (2000) have shown that Tim8p and Tim13p crosslinked to residues 30–90, whereas Davis et al. (2000) have shown that Tim8p and Tim13p crosslinked to residues 25–75 and that Tim9p and Tim10p crosslinked, albeit less efficiently than Tim8p and Tim13p, to the COOH-terminal membrane spanning domains of Tim23p. With the purified Tim8p–Tim13p complex and a Tim23p peptide scan, we have shown that the purified Tim8p–Tim13p complex bound predominantly to the membrane spanning domains of Tim23p, with the highest binding affinity in transmembrane domain 1. The Tim8p–Tim13p complex also bound to residues 28–40, 46–55, and 64–75 in the NH2-terminal domain. However, the purified 70-kD Tim9p–Tim10p complex had no binding affinity for the Tim23p peptide scan (Curran et al., 2002) and we have failed to show a direct interaction between the Tim9p–Tim10p complex and Tim23p in our crosslinking studies in wild-type mitochondria (Leuenberger et al., 1999). Because Tim9p and Tim10p are both in a 70-kD complex and associated with the 300-kD Tim22p-containing membrane complex, the results reported by Davis et al. (2000) may reflect an interaction between Tim23p and Tim9p and Tim10p associated with the 300-kD complex.
Tim23p crossed the TOM complex as a loop and was inserted into the mitochondrial inner membrane. Fusion constructs containing DHFR on both the NH2- and COOH-termini were imported and inserted into the inner membrane. Specifically, the DHFR–Tim23p–DHFR precursor complexed with methotrextate was arrested at the outer membrane and was crosslinked to both Tim8p and Tim13p. When the DHFR–Tim23p–DHFR construct was synthesized in the absence of methotrexate (import competent because DHFR is no longer tightly folded), it was inserted into the inner membrane as shown by manipulations involving protease treatment, osmotic lysis, and resistance to carbonate extraction. Thus, these experiments confirm that the NH2 and COOH terminus are not required for import of Tim23p. Previous studies have shown that the polytopic carrier proteins, which also lack an NH2-terminal targeting sequence, cross the outer membrane as a loop. It has been demonstrated that AAC, when flanked by DHFR on both the NH2 and COOH termini, can span the TOM complex as a loop and be crosslinked to Tim9p and Tim10p in the intermembrane space (Ryan et al., 1999; Wiedemann et al., 2001); however, the ability of these AAC–DHFR fusion constructs to insert into the inner membrane was not tested. In addition, the central matrix loop of the carrier uncoupling protein-1 drives protein translocation across the outer membrane (Schleiff and McBride, 2000). Taken together, we propose that polytopic inner membrane proteins follow a conserved translocation mechanism to reach the inner membrane. The precursor crosses the TOM complex as a loop and then the small Tim proteins bind to the hydrophobic membrane spanning domains as they reach the aqueous intermembrane space. Thus, the small Tim proteins act as putative chaperones to potentially prevent aggregation and maintain the unfolded precursor in an import-competent state.
The Tim8p–Tim13p complex does not coordinate zinc
In addition to characterizing the interaction between the Tim8p–Tim13p complex and Tim23p, we characterized the biochemical and biophysical properties of the Tim8p–Tim13p complex. The Tim8p–Tim13p complex is a hexamer. Tim8p may form the core of the complex because Tim8p was more protease-resistant than Tim13p. From circular dichroism studies, the complex was mostly alpha-helical and could refold efficiently after thermal denaturation. The small Tim proteins contain the twin CX3C motif (Koehler et al., 1999b). This motif has been suggested to form a zinc finger-like structure despite its divergence from the canonical zinc finger in which two amino acids separate the cysteines (Mackay and Crossley, 1998; Laity et al., 2001). Mutations in the human homologue of Tim8p, DDP1/TIMM8a (deafness dystonia protein) result in Mohr-Tranebjaerg syndrome (Jin et al., 1996). The majority of patients with Mohr-Tranebjaerg syndrome have frameshift/nonsense mutations or deletions at the DDP1/TIMM8a locus resulting in a truncated or absent protein. Recently, a de novo mutation in DDP1/TIMM8a was identified in which the fourth cysteine in the twin CX3C motif was changed from cysteine-66 to tryptophan-66, C66W (Tranebjaerg et al., 2000). We made a similar mutation in yeast, designated Tim8pC68W, and showed that the mutant Tim8pC68W protein imported into mitochondria but failed to assemble into stable 70kDa complex (Roesch et al., 2001). These results further confirm that the cysteine residues are important for assembly of the Tim8p–Tim13p complex.
Conflicting results have been reported about whether the Tim9p–Tim10p and Tim8p–Tim13p complexes coordinate zinc. Monomeric Tim8p and Tim13p have been shown to bind zinc in a 1:1 molar ratio and binding of the Tim8p–Tim13p complex to Tim23p is abolished in mitochondria incubated with metal chelators (Paschen et al., 2000; Rothbauer et al., 2001). Recently, Millar and colleagues have developed an elegant assay in which they can import the carrier proteins into plant mitoplasts (mitochondria that lack the outer membrane); they show that the addition of Zn2+ or Cd2+ stimulates import of the carriers and treatment with metal chelators inhibits import (Lister et al., 2002). Because the mechanism by which the 70-kD complexes assemble in the intermembrane space is not known, a divalent metal ion may play a critical role in assembly of the complex; such a mechanism could explain why the monomers bind zinc. From our studies, the assembled Tim8p–Tim13p complex did not coordinate zinc or any other metal ion, even when expression of the Tim8p–Tim13p complex was induced in the presence of Zn2+. Furthermore, the chelators EDTA and o-phe did not affect the binding of the Tim8p–Tim13p complex to Tim23p in intact mitochondria. In similar studies, the Tim9p–Tim10p complex did not coordinate metal ions (Curran et al., 2002). While it is plausible that the metal ions were lost during purification, the Tim8p–Tim13p complex folded efficiently in the presence of the chelator EDTA. The cysteine residues most likely form disulfide bridges because the complex was unable to refold in the presence of DTT. In contrast, Harrison and colleagues have shown that Lck (a lymphoid-specific, Src family protein-tyrosine kinase) interacts with the T-cell co-receptor CD4 through a cocoordinated Zn2+ ion with two cysteines in Lck and two cysteines in CD4 (Huse et al., 1998). This interaction is disrupted with low concentrations of EDTA but DTT and β-mercaptoethanol have no effect. Taken together, these findings suggest that the cysteine residues in the twin CX3C do not coordinate zinc but instead may form disulfide bonds for structural stability. The elucidation of a high resolution structure of the Tim8p–Tim13p complex should provide helpful clues to the role of the cysteine residues.
Materials and methods
Plasmids and strains
Standard genetic techniques were used for growth, manipulation, and transformation of yeast strains (Sikorski and Hieter, 1989; Guthrie and Fink, 1991). For expression of the recombinant Tim8p–Tim13p complex in E. coli, the TIM8 was cloned into pET28a (Novagen) as an NcoI/SalI fragment and TIM13 was cloned into pET28a as an NcoI/NdeI fragment; only the open reading frames were cloned and no fusions or tags were constructed. TIM13 with the ribosomal binding site was then removed as a XbaI/NheI fragment and cloned into the XbaI site of the pET28a-TIM8 plasmid. As a result, a single transcript is synthesized in which both Tim8p and Tim13p are translated from their own ribosomal binding site. Expression was induced according to the manufacturer's protocols (Novagen).
Purification of the Tim8p–Tim13p complex from E. coli
Frozen E. coli cell pellets containing the recombinant Tim8p–Tim13p complex were thawed and lysed by sonication on ice. Cell debris was removed by centrifugation (30 min, 100,000 g, 4°C) and proteins were precipitated by addition of polyethylene glycol (ave. MW 3350; J.T. Baker, Inc.) to 25% and NaCl to 250 mM. After centrifugation, the pellet was solubilized in Buffer A (20 mM Hepes-KOH, pH 7.4) and loaded onto a Q-Sepharose Fast Flow column (5 × 30 cm; Amersham Biosciences). The column was washed with Buffer A at a flow rate of 5 ml/min, and bound proteins were eluted with 500 ml of a linear gradient (0–200 mM NaCl) in Buffer A. 15-ml fractions were collected and analyzed by SDS-PAGE followed by immunoblotting, Coomassie staining, and blue native gel electrophoresis. After desalting and concentration in buffer A, the fractions containing the Tim8p–Tim13p complex were loaded onto a Source S column (1.6 × 10 cm; Amersham Biosciences). The column was washed with Buffer A at a flow rate of 5 ml/min and bound proteins were eluted with 50 ml of a linear gradient (0–100 mM NaCl) in buffer A. 1-ml fractions were collected and analyzed as described above. These fractions were concentrated to 200 μl in buffer A containing 100 mM NaCl and loaded onto a Superose-12 gel filtration column (1.0 × 30 cm; Amersham Biosciences). The column was developed with buffer A containing 100 mM NaCl at a flow rate of 0.2 ml/min. 1-ml fractions were collected and analyzed as described above. The molecular mass standards for the Superose 12 column were ribonuclease A (13.7 kD); chymotrypsinogen A (25 kD); ovalbumin (43 kD); bovine serum albumin (67 kD); and aldolase (158 kD).
Import of radiolabeled proteins into isolated mitochondria
Mitochondria were purified from lactate-grown yeast cells (Glick and Pon, 1995) and assayed for in vitro protein import as described (Rospert and Schatz, 1998). Proteins were synthesized in a rabbit reticulocyte lysate in the presence of [35S]-methionine after in vitro transcription of the corresponding gene by SP6 polymerase. The reticulocyte lysate containing the radiolabeled precursor was incubated with isolated mitochondria at the indicated temperatures in import buffer (1 mg/ml bovine serum albumin, 0.6 M sorbitol, 150 mM KCl, 10 mM MgCl2, 2.5 mM EDTA, 2 mM ATP, 2 mM NADH, 20 mM K+-Hepes, pH 7.4). Where indicated, the potential across the mitochondrial inner membrane was dissipated with 1 μM valinomycin. Nonimported radiolabeled proteins were removed by treatment with 100 μg/ml trypsin or 50 μg/ml proteinase K for 15–30 min on ice; trypsin was inhibited with 200 μg/ml soybean trypsin inhibitor and proteinase K with 1 mM PMSF, respectively. Cross-linking studies were performed as previously described (Leuenberger et al., 1999).
ICP-AE studies
Protein samples were diluted to 7 ml in metal-free water, and these samples were analyzed using a Thermo-Jarrel Ash Iris 1000 ICP-AE spectrometer.
Circular dichroism analysis
Circular dichroism (CD) analysis was performed on a JASCO J-600 spectropolarimeter by using a scan speed of 10 nm/min, a time constant of 8 s, and a bandwidth of 0.5 nm. Eight scans were averaged for each spectrum. The baseline correction option was used to subtract a buffer baseline. Spectra were recorded from 260 to 190 nm in 1-mm pathlength cells with protein concentrations of 0.1–0.2 mg/ml. Spectra were analyzed for secondary structure by using a self consistent method and/or the convex constraint algorithm for secondary structure prediction (Perczel et al., 1992; Sreerama and Woody, 1993).
Analytical ultracentrifugation
Sedimentation equilibrium measurements were performed at 20°C in an analytical ultracentrifuge (Beckman XLA) equipped with an optical absorption system. Sedimentation equilibrium was measured with 100 μl samples at 19,000 rpm; absorbance was recorded at 280 nm. The molecular mass was determined using a linear regression computer program that adjusts the baseline absorption to obtain the best linear fit of lnA versus r 2 (A = absorption, r 2 = radial distance from the rotor center). Measurements were in the presence of 20 mM potassium phospahte-KOH pH 7.4, 100 mM NaCl.
Screening of peptide scans with the Tim8p–Tim13p complex
The cellulose-bound peptide scans were prepared by automated spot synthesis (Jerini). Multiple 13-mer peptides with a 10 amino acid overlap were synthesized according to the sequence of Tim23p. The membranes were incubated with 200 nM of recombinant Tim8p–Tim13p complex or Tim9p–Tim10p complex in binding buffer (100 mM KCl, 5% sucrose, 1% bovine serum albumin, 30 mM Tris-HCl, pH 7.4) at 25°C for 2 h as described by Brix et al. (1999) After extensive washing, the bound Tim8p–Tim13p complex was transferred to a polyvinylidene difluoride membrane, followed by detection with antibodies against Tim8p or Tim13p and [125I]-protein A. Binding data was acquired by scanning laser densitometry (Personal Densitometer SI; Molecular Dynamics) and quantitated utilizing ImageQuaNT (version 4.2a; Molecular Dynamics). The mean of at least three independent experiments for each peptide spot was used and the local background of each peptide spot was subtracted. Binding data were identical for antibodies against Tim8p and Tim13p.
Blue native gel electrophoresis
Mitochondria (2.5 mg/ml) were solubilized in 20 mM K+-Hepes, pH 7.4, 50 mM NaCl, 10% glycerol, 2.5 mM MgCl2. 1 mM EDTA, 0.16% n-dodecylmaltoside (Boehringer Mannheim) for 30 min on ice. Insoluble material was removed by centrifugation at 100,000 g for 10 min, and the solubilized proteins were analyzed by blue native gel electrophoresis on a 6–16% linear polyacrylamide gradient (Schägger et al., 1994).
Miscellaneous
Mitochondrial proteins were analyzed by SDS-PAGE using a 10 or 16% polyacrylamide gel and a Tricine-based running buffer (Schägger and von Jagow, 1987). Proteins were detected by immunoblotting using nitrocellulose- or PVDF membranes and visualization of immune complexes with [125I]-protein A. Protein concentration was assayed by the bicinchoninic acid method (Pierce Chemical Co.) using bovine serum albumin as the standard. Thiol-trapping studies were previously described (Curran et al., 2002).
Acknowledgments
We would like to thank David Wong and Edward Legaspi for technical assistance, Amir Liba and Dr. Joan Valentine for ICP-AE analysis, and Dr. Martin Phillips for technical assistance with circular dichroism and analytical centrifugation.
C.M. Koehler is a Damon Runyon-Walter Winchell Scholar. This work was supported by the Damon Runyon-Walter Winchell Cancer Research Foundation (DRS18), the American Heart Association (0030147N), Burroughs Wellcome Fund New Investigator Award in the Toxicological Sciences (1001120), Research Corporation (RI0459), and the National Institutes of Health (1R01GM61721-01). S.P. Curran is funded by the United States Public Health Service National Research Service Award (GM07185). E. Schmidt is funded by the Dystonia Research Foundation.
Footnotes
Abbreviations used in this paper: Δψ, membrane potential; AAC, ATP/ADP carrier; AMS, 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid; CD, circular dichroism; DHFR, dihydrofolate reductase; ICP-AE, inductively coupled plasmon–atomic emission; o-phe, 1,10-orthophenanthraline; TIM, translocase of inner membrane; TOM, translocase of outer membrane.
References
- Adam, A., M. Endres, C. Sirrenberg, F. Lottspeich, W. Neupert, and M. Brunner. 1999. Tim9, a new component of the TIM22.54 translocase in mitochondria. EMBO J. 18:313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohni, P.C., G. Daum, and G. Schatz. 1983. Import of proteins into mitochondria. Partial purification of a matrix-located protease involved in cleavage of mitochondrial precursor polypeptides. J. Biol. Chem. 258:4937–4943. [PubMed] [Google Scholar]
- Brix, J., S. Rudiger, B. Bukau, J. Schneider-Mergener, and N. Pfanner. 1999. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J. Biol. Chem. 274:16522–16530. [DOI] [PubMed] [Google Scholar]
- Brix, J., G.A. Ziegler, K. Dietmeier, J. Schneider-Mergener, G.E. Schulz, and N. Pfanner. 2000. The mitochondrial import receptor Tom70: identification of a 25 kDa core domain with a specific binding site for preproteins. J. Mol. Biol. 303:479–488. [DOI] [PubMed] [Google Scholar]
- Curran, S.P., D. Leuenberger, W. Oppliger, and C.M. Koehler. 2002. The Tim9p-Tim10p complex binds to the transmembrane domains of the ADP/ATP carrier. EMBO J. 21:942–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, A.J., K.R. Ryan, and R.E. Jensen. 1998. Tim23p contains separate and distinct signals for targeting to mitochondria and insertion into the inner membrane. Mol. Biol. Cell. 9:2577–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, A.J., N.B. Sepuri, J. Holder, A.E. Johnson, and R.E. Jensen. 2000. Two intermembrane space TIM complexes interact with different domains of Tim23p during its import into mitochondria. J. Cell Biol. 150:1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donzeau, M., K. Káldi, A. Adam, S. Paschen, G. Wanner, B. Guiard, M.F. Bauer, W. Neupert, and M. Brunner. 2000. Tim23 links the inner and outer mitochondrial membranes. Cell. 101:401–412. [DOI] [PubMed] [Google Scholar]
- Endres, M., W. Neupert, and M. Brunner. 1999. Transport of the ADP/ATP carrier of mitochondria from the TOM complex to the TIM22.54 complex. EMBO J. 18:3214–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick, B.S., and L. Pon. 1995. Isolation of highly purified mitochondria from S. cerevisiae. Methods Enzymol. 260:213–233. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and G.R. Fink. 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego, CA. 933 pp.
- Huse, M., M.J. Eck, and S.C. Harrison. 1998. A Zn2+ ion links the cytoplasmic tail of CD4 and the N-terminal region of Lck. J. Biol. Chem. 273:18729–18733. [DOI] [PubMed] [Google Scholar]
- Jakob, U., W. Muse, M. Eser, and J.C. Bardwell. 1999. Chaperone activity with a redox switch. Cell. 96:341–352. [DOI] [PubMed] [Google Scholar]
- Jin, H., M. May, L. Tranebjaerg, E. Kendall, G. Fontan, J. Jackson, S.H. Subramony, F. Arena, H. Lubs, S. Smith, et al. 1996. A novel X-linked gene, DDP, shows mutations in families with deafness (DFN-1), dystonia, mental deficiency and blindness. Nat. Genet. 14:177–180. [DOI] [PubMed] [Google Scholar]
- Káldi, K., and W. Neupert. 1998. Protein translocation into mitochondria. Biofactors. 8:221–224. [DOI] [PubMed] [Google Scholar]
- Káldi, K., M.F. Bauer, C. Sirrenberg, W. Neupert, and M. Brunner. 1998. Biogenesis of Tim23 and Tim17, integral components of the TIM machinery for matrix-targeted preproteins. EMBO J. 17:1569–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher, O., J. Holder, M. Srinivasan, R.S. Leung, and R.E. Jensen. 1997. The Tim54p-Tim22p complex mediates insertion of proteins into the mitochondrial inner membrane. J. Cell Biol. 139:1663–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher, O., N.B. Sepuri, and R.E. Jensen. 2000. Tim18p is a new component of the Tim54p-Tim22p translocon in the mitochondrial inner membrane. Mol. Biol. Cell. 11:103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, C.M., E. Jarosch, K. Tokatlidis, K. Schmid, R.J. Schweyen, and G. Schatz. 1998. a. Import of mitochondrial carriers mediated by essential proteins of the intermembrane space. Science. 279:369–373. [DOI] [PubMed] [Google Scholar]
- Koehler, C.M., S. Merchant, W. Oppliger, K. Schmid, E. Jarosch, L. Dolfini, T. Junne, G. Schatz, and K. Tokatlidis. 1998. b. Tim9p, an essential partner subunit of Tim10p for the import of mitochondrial carrier proteins. EMBO J. 17:6477–6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, C.M., D. Leuenberger, S. Merchant, A. Renold, T. Junne, and G. Schatz. 1999. a. Human deafness dystonia syndrome is a mitochondrial disease. Proc. Natl. Acad. Sci. USA. 96:2141–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler, C.M., S. Merchant, and G. Schatz. 1999. b. How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem. Sci. 24:428–432. [DOI] [PubMed] [Google Scholar]
- Koehler, C.M., M.P. Murphy, N. Bally, D. Leuenberger, W. Oppliger, L. Dolfini, T. Junne, G. Schatz, and E. Or. 2000. Tim18p, a novel subunit of the inner membrane complex that mediates protein import into the yeast mitochondrial inner membrane. Mol. Cell. Biol. 20:1187–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity, J.H., B.M. Lee, and P.E. Wright. 2001. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11:39–46. [DOI] [PubMed] [Google Scholar]
- Leuenberger, D., N.A. Bally, G. Schatz, and C.M. Koehler. 1999. Different import pathways through the mitochondrial intermembrane space for inner membrane proteins. EMBO J. 17:4816–4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister, R., B. Mowday, J. Whelan, and A.H. Millar. 2002. Zinc-dependent intermembrane space proteins stimulate import of carrier proteins into plant mitochondria. Plant J. 30:555–566. [DOI] [PubMed] [Google Scholar]
- Mackay, J.P., and M. Crossley. 1998. Zinc fingers are sticking together. Trends Biochem. Sci. 23:1–4. [DOI] [PubMed] [Google Scholar]
- Murphy, M.P., D. Leuenberger, S.P. Curran, W. Oppliger, and C.M. Koehler. 2001. The essential function of the small Tim proteins in the TIM22 import pathway does not depend on formation of the soluble 70-kilodalton complex. Mol. Cell. Biol. 21:6132–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri, F., F. Bisaccia, L. Capobianco, V. Dolce, G. Fiermonte, V. Iacobazzi, C. Indiveri, and L. Palmieri. 1996. Mitochondrial metabolite transporters. Biochim. Biophys. Acta. 1275:127–132. [DOI] [PubMed] [Google Scholar]
- Paschen, S.A., and W. Neupert. 2001. Protein import into mitochondria. IUBMB Life. 52:101–112. [DOI] [PubMed] [Google Scholar]
- Paschen, S.A., U. Rothbauer, K. Káldi, M.F. Bauer, W. Neupert, and M. Brunner. 2000. The role of the TIM8-13 complex in the import of Tim23 into mitochondria. EMBO J. 19:6392–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perczel, A., K. Park, and G.D. Fasman. 1992. Analysis of the circular dichroism spectrum of proteins using the convex constraint algorithm: a practical guide. Anal. Biochem. 203:83–93. [DOI] [PubMed] [Google Scholar]
- Pfanner, N. 1998. Mitochondrial import: crossing the aqueous intermembrane space. Curr. Biol. 8:R262–R265. [DOI] [PubMed] [Google Scholar]
- Pfanner, N., P. Hoeben, M. Tropschug, and W. Neupert. 1987. The carboxyl-terminal two-thirds of the ADP/ATP carrier polypeptide contains sufficient information to direct translocation into mitochondria. J. Biol. Chem. 262:14851–14854. [PubMed] [Google Scholar]
- Pfanner, N., A. Geissler, E. Schleiff, and H. McBride. 2001. Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2:339–349. [DOI] [PubMed] [Google Scholar]
- Roesch, K., S.P. Curran, L. Tranebjaerg, and C.M. Koehler. 2001. Human deafness dystonia syndrome is caused by a defect in assembly of the DDP1/TIMM8a-TIMM13 complex. Hum. Mol. Genet. 11:477–486. [DOI] [PubMed] [Google Scholar]
- Rospert, S., and G. Schatz. 1998. Protein translocation into mitochondria. In Cell Biology: A Laboratory Handbook. Vol. 2. J.E. Celis, editor. Academic Press, San Diego. 277–285.
- Rothbauer, U., S. Hofmann, N. Muhlenbein, S.A. Paschen, K.D. Gerbitz, W. Neupert, M. Brunner, and M.F. Bauer. 2001. Role of the deafness dystonia peptide 1 (DDP1) in import of human Tim23 into the inner membrane of mitochondria. J. Biol. Chem. 276:37327–37334. [DOI] [PubMed] [Google Scholar]
- Ryan, M.T., H. Muller, and N. Pfanner. 1999. Functional staging of ADP/ATP carrier translocation across the outer mitochondrial membrane. J. Biol. Chem. 274:20619–20627. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368–379. [DOI] [PubMed] [Google Scholar]
- Schägger, H., W.A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220–230. [DOI] [PubMed] [Google Scholar]
- Schatz, G., and B. Dobberstein. 1996. Common principles of protein translocation across membranes. Science. 271:1519–1526. [DOI] [PubMed] [Google Scholar]
- Schleiff, E., and H. McBride. 2000. The central matrix loop drives import of uncoupling protein 1 into mitochondria. J. Cell Sci. 113:2267–2272. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg, C., M. Endres, H. Folsch, R.A. Stuart, W. Neupert, and M. Brunner. 1998. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11 and Tim12/Mrs5. Nature. 391:912–915. [DOI] [PubMed] [Google Scholar]
- Sreerama, N., and R.W. Woody. 1993. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 209:32–44. [DOI] [PubMed] [Google Scholar]
- Tranebjaerg, L., B.C. Hamel, F.J. Gabreels, W.O. Renier, and M. Van Ghelue. 2000. A de novo missense mutation in a critical domain of the X-linked DDP gene causes the typical deafness-dystonia-optic atrophy syndrome. Eur. J. Hum. Genet. 8:464–467. [DOI] [PubMed] [Google Scholar]
- van Dijl, J.M., E. Kutejova, K. Suda, D. Perecko, G. Schatz, and C.K. Suzuki. 1998. The ATPase and protease domains of yeast mitochondrial Lon: roles in proteolysis and respiration-dependent growth. Proc. Natl. Acad. Sci. USA. 95:10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann, N., N. Pfanner, and M.T. Ryan. 2001. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 20:951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]