Abstract
In Tetrahymena cells, phosphorylation of linker histone H1 regulates transcription of specific genes. Phosphorylation acts by creating a localized negative charge patch and phenocopies the loss of H1 from chromatin, suggesting that it affects transcription by regulating the dissociation of H1 from chromatin. To test this hypothesis, we used FRAP of GFP-tagged H1 to analyze the effects of mutations that either eliminate or mimic phosphorylation on the binding of H1 to chromatin both in vivo and in vitro. We demonstrate that phosphorylation can increase the rate of dissociation of H1 from chromatin, providing a mechanism by which it can affect H1 function in vivo. We also demonstrate a previously undescribed ATP-dependent process that has a global effect on the dynamic binding of linker histone to chromatin.
Keywords: H1; phosphorylation; ATP remodeling; FRAP; Tetrahymena
Introduction
Linker histones, also known as H1s, are associated with the DNA located between nucleosome core particles in eukaryotic chromatin. Although they have been intensely studied in diverse organisms, there is little agreement or understanding of the mechanisms by which linker histones affect chromatin structure and function. Until recently, most of the effects ascribed to linker histones were based largely on in vitro analyses or descriptive studies of their behavior in different cell types, cell fractions, or physiological states. In vitro studies suggest linker histones affect chromatin structure in at least four ways: they (a) stabilize DNA entering the nucleosome core (Simpson, 1978; Woodcock et al., 1993); (b) affect nucleosome positioning and spacing (Meersseman et al., 1991; Blank and Becker, 1995); (c) restrict nucleosome mobility (Pennings et al., 1994); and (d) stabilize higher order structure (Thoma et al., 1979; Carruthers et al., 1998). Interestingly, many of the effects of linker histone on chromatin in vitro can be partially or completely mimicked by increased salt concentrations, suggesting that most of the effects of H1 are due to its cationic nature (Widom, 1986; Blank and Becker, 1995; Bednar et al., 1998).
Early in vitro studies suggested that linker histones might be general inhibitors of transcription (Paranjape et al., 1994; for review see Owen-Hughes and Workman, 1994), and more recent studies have shown that the presence of H1 on chromatin globally inhibits the action of a number of ATP-dependent chromatin remodeling enzymes (Peterson, 2002). However, most recent evidence, from a number of organisms that contain structurally diverse linker histones, has shown that linker histones can have highly specific effects on transcription in vivo. For example, gene disruption experiments have shown that linker histone is both a positive and a negative regulator of transcription in Tetrahymena (Shen and Gorovsky, 1996). A global analysis of the effects of linker histone disruption in Saccharomyces cerevisiae showed that H1 is responsible for the repression of only a few genes, whereas most genes are indifferent to the presence of H1, and the expression of a sizable subset of genes actually decreases in its absence (Hellauer et al., 2001). Similar gene-specific effects of H1 depletion were also demonstrated during early embryonic development of Xenopus (Steinbach et al., 1997), and specific roles of some linker histone variants in germline development have been reported in Caenorhabditis elegans (Jedrusik and Schulze, 2001) and in tobacco (Prymakowska-Bosak et al., 1999).
Given that linker histones are found in all eukaryotes and have been shown to affect many features of chromatin structure and function, it is surprising that the effect of complete disruption of linker histone genes in unicellular eukaryotes has been small, resulting in little or no effect on growth or on chromatin structure (Shen et al., 1995; Ushinsky et al., 1997; Patterton et al., 1998; Barra et al., 2000; Ramon et al., 2000). One possible explanation for these results is that the linker histones of unicellular eukaryotes are diverse and many lack the typical tripartite structure (NH2-terminal tail, central globular domain, COOH-terminal tail) of linker histones in multicellular organisms (Wolffe, 1998). Thus, the Tetrahymena linker histone lacks a globular domain, and the yeast linker histone consists almost entirely of two closely linked globular domains. However, this explanation seems unlikely in light of the observation that disruption of the typical, tripartite linker histone of Aspergillus is also without significant effect (Ramon et al., 2000). In addition, whereas complete elimination of the multiple genes encoding linker histones in a multicellular eukaryote has not yet been reported, deletion of five of the six genes in chicken tissue culture cells does not effect their growth (Takami and Nakayama, 1997), and deletion of a testis-specific H1 in mice has no effect on spermiogenesis (Rabini et al., 2000).
Another feature of linker histones that has been intensely studied is phosphorylation which, in all cases studied to date, occurs on either or both of the terminal tails, but not on the globular domain. Based on temporal correlations between hyperphosphorylation of H1 and mitosis in mammalian cells and on similar studies in Physarum, it was originally suggested (Bradbury, 1992) that H1 phosphorylation played an active role in mitotic chromosome condensation. However, in subsequent studies, mitotic H1 hyperphosphorylation could be dissociated from chromosome condensation, and linker histone dephosphorylation was shown to be associated with chromatin condensation, leading to the hypothesis that linker histone phosphorylation causes decondensation rather than condensation of chromatin (Roth and Allis, 1992).
We have developed Tetrahymena thermophila as a system for studying the function of H1 phosphorylation in vivo. Tetrahymena H1 has many features of a typical linker histone (perchloric acid solubility, lysine richness, linker location, dissociation from chromatin at moderate salt concentration, growth-dependent phosphorylation by a Cdc2 kinase) but lacks the central globular domain. It can be viewed as a model for linker histone tails and their phosphorylation. In Tetrahymena, large changes in H1 phosphorylation levels are correlated with dramatic shifts in gene expression patterns in different physiological states (Glover et al., 1981; Roth et al., 1988). Five phosphorylation sites have been mapped and shown to be the only phosphorylation sites on the molecule (Mizzen et al., 1999).
We have performed extensive mutagenesis studies showing that the phosphorylation of H1 in Tetrahymena mimics the H1-null phenotype in its positive and negative effects on transcription (Dou et al., 1999). Additional studies showed that the effects of phosphorylation on gene expression probably function by modulation of the coulombic interactions between H1 and DNA (Dou et al., 1999; Dou and Gorovsky, 2000, 2002). In particular, the robust expression of the CyP1 gene in starved Tetrahymena cells was shown to require dephosphorylation of the macronuclear linker histone. Phosphorylation of H1 was shown to regulate CyP1 expression by altering the net charge of a 19-residue region (residues 35–54) of H1 containing the five phosphorylation sites. When the total number of charges in that region was mutagenized to be the same as the fully phosphorylated H1, CyP1 expression was inhibited. When the total charges of the region were the same as unphosphorylated H1, CyP1 expression was strongly induced. These effects were independent of the hydrophobicity of the region and did not require any residues that structurally resembled phosphorylation; only the charge of the region was important. Further studies showed that the charge altering mutations placed anywhere in the molecule had the same effect on CyP1 transcription, as long as they were clustered in a small region (Dou and Gorovsky, 2002). The same number of charge-altering mutations spread throughout the molecule failed to regulate CyP1 expression. These studies demonstrated that phosphorylation of H1 in Tetrahymena regulated the expression of specific genes by changing the overall charge of a small domain. This domain could be located anywhere in the H1 molecule and the only feature of phosphorylated H1 that regulates transcription was the creation of a localized region of high relative negative charge that we refer to as a charge patch. To explain these observations, we proposed that clustered charges produced by phosphorylation acted synergistically to facilitate the dissociation of H1 from the chromatin (Dou and Gorovsky, 2002).
The dynamics of H1 dissociation from chromatin can be addressed using FRAP. This method can be used to visualize protein dynamics and, in combination with kinetic modeling, it can also be used to determine the biophysical properties of nuclear protein in vivo including its interaction with chromatin. FRAP studies in cultured mammalian cells demonstrated that most GFP-tagged linker histone H1 in stably transfected human cell lines undergoes rapid association/dissociation with chromatin (Lever et al., 2000; Misteli et al., 2000). Rapid exchange occurs in both heterochromatin and euchromatin and does not require direct contact between chromatin fibers. Deletion of the H1 globular domain or the highly positively charged COOH-terminal domain dramatically increased the mobility of H1, suggesting that different regions of the H1 molecule can contribute to its interaction with chromatin. In addition, treatment with a deacetylase inhibitor or a kinase inhibitor also increased the rate of fluorescence recovery, suggesting that dynamic binding of H1 could be modulated by other processes.
We have used FRAP to test whether negative charges like those introduced by phosphorylation can directly affect H1 binding to chromatin in vivo. Because WT Tetrahymena H1 is phosphorylated to varying extents and phosphorylation is very sensitive to changes in physiological conditions, we compared the properties of two GFP-tagged mutant H1s. One (A5-GFP) mimics the unphosphorylated state by mutating all of the phosphorylation sites to alanine; the other (E5-GFP) mimics the fully phosphorylated state in charge by replacing all of the phosphorylation sites with glutamate residues. Our previous studies (Dou and Gorovsky, 2000) argue strongly that, with regard to their effects on gene expression, A5 and E5 H1 phosphorylation mutants mimic the function of unphosphorylated and phosphorylated H1, respectively. Therefore, we will refer to these mutations as phosphorylation mutations.
In this paper, we demonstrate that charge alterations that mimic phosphorylation can alter the interaction of H1 with chromatin in vivo, in detergent-extracted cells and in isolated nuclei, providing a mechanism by which phosphorylation can affect H1 function. We also demonstrate that the dynamic binding of linker histone can be markedly affected by a heretofore undescribed ATP-dependent remodeling process. These results are discussed in terms of the properties of H1.
Results
Gene replacement with two GFP-tagged H1 phosphorylation mutations
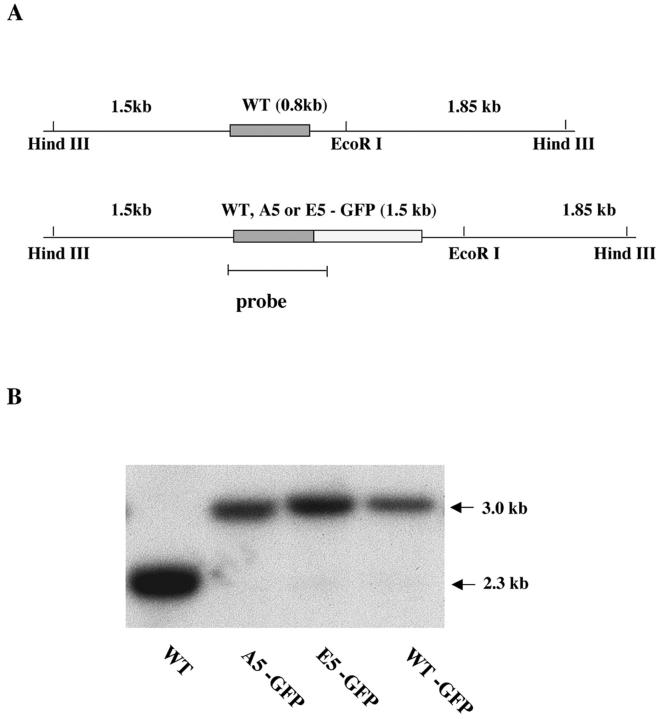
Two mutated versions of H1 were used in this study. In A5-GFP, all five phosphorylation sites in H1 were mutated to alanine (A), to eliminate phosphorylation. In E5-GFP, they were mutated to glutamic acid (E), to mimic the constitutively phosphorylated state (Dou et al., 1999). We also made GFP-tagged WT H1 as a control. The GFP coding sequence was introduced at the COOH terminus of these H1s (Fig. 1 A) and the tagged constructs containing a flanking selectable marker were biolistically transformed into the polyploid macronuclei of Tetrahymena thermophila. Transformed cell lines were selected in paromomycin until all of the WT copies of the gene were completely replaced by the mutated ones. On a Southern blot (Fig. 1 B), DNA from WT cells showed a single 2.3-kb band. Mutant cell DNA lacked this band but had a new 3-kb band, the size expected if the mutant genes had integrated by homologous recombination. This result indicates that GFP-tagged genes had completely replaced the endogenous HHO1 gene in the macronuclei of the transformed strains. The HHO1 locus of each strain was PCR amplified and sequenced, confirming that they lacked unwanted mutations and encoded H1 proteins that have GFP tags at their COOH termini. Fluorescence microscopy showed that H1-GFP chimeric proteins were localized uniformly in macronuclei and excluded from the cytoplasm and from micronuclei, which contain a different linker histone (Fig. 2 A).
Figure 1.
GFP-tagged H1s replace endogenous H1 by homologous recombination. (A) Maps of the macronuclear HHO1 locus before and after gene replacement. The WT HHO1 gene is shown as a box in a 4.15-kb Hind III fragment. In WT and A5 or E5, EcoRI will cleave the original 4.15-kb fragment into 2.3- and 1.85-kb fragments. The position of the probe used in the Southern blot is shown at the bottom. (B) Southern blot analysis shows complete gene replacement in WT-GFP, A5-GFP, and E5-GFP strains. Genomic DNAs were isolated from WT, WT-GFP, A5-GFP, and E5-GFP cells and digested with HindIII and EcoRI. The 2.3-kb band observed in WT cells has been completely replaced by a 3.0-kb band in the GFP-tagged cells.
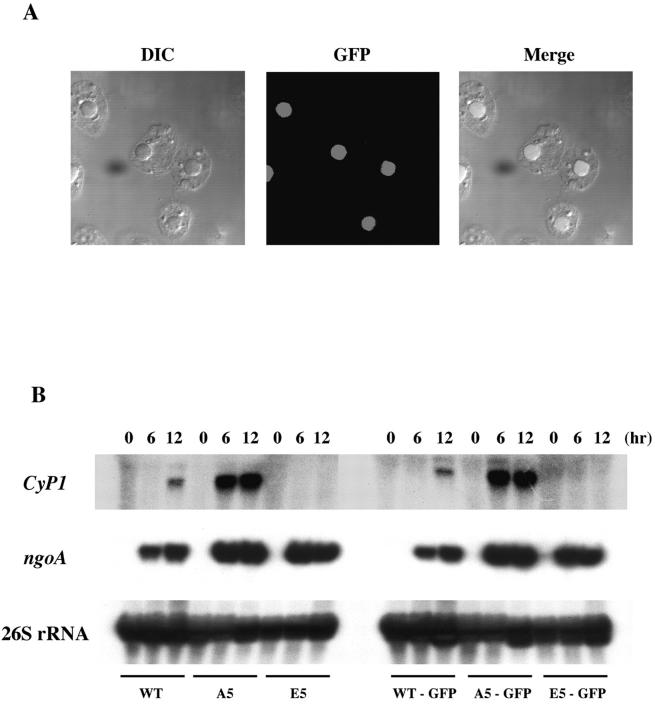
Figure 2.
GFP-tagged H1 is correctly targeted to macronuclei and does not affect the transcription regulation of CyP1 by H1 phosphorylation. (A) Nuclear localization of GFP-tagged H1 in A5 cells. A DIC image is shown on the left, GFP fluorescence in the middle and the merged image is shown on the right. WT and E5 cells gave the same results (not depicted). (B) The function of H1 in the regulation of CyP1 expression is not disturbed by GFP tagging. Whole-cell RNAs isolated from WT, A5, E5, WT-GFP, A5-GFP, and E5-GFP strains after 0, 6, and 12 h of starvation were analyzed on a Northern blot probed with a CyP1-specific probe and a ngoA-specific probe. A 26S rRNA probe was used as a loading control. The expression patterns of CyP1 and ngoA were similar to the results obtained from WT, A5, and E5 cells without GFP tag.
To test whether the GFP tag disrupted the physiological function of H1 in gene regulation, we analyzed the expression of a well-characterized reporter gene, CyP1, in WT-GFP, A5-GFP, and E5-GFP cells (Fig. 2 B). In all three GFP-tagged strains, CyP1 showed the same expression pattern as proteins without the tag. In WT cells, CyP1 was expressed only when H1 became dephosphorylated after long starvation (Dou et al., 1999). CyP1 was easily detectable shortly after starvation was begun in A5-GFP cells but was expressed at a much lower level even after long starvation in E5-GFP cells. These results are indistinguishable from those obtained with mutant H1s lacking the GFP tag (Dou et al., 1999; Dou and Gorovsky, 2000, 2002). This effect was gene specific, as the induction of the ngoA gene during starvation was not affected. We conclude that GFP-tagged H1s properly regulate the expression of CyP1. For convenience, we will refer to WT-GFP as WT, to A5-GFP as A5 and to E5-GFP as E5 in the following sections.
H1 phosphorylation mutations affect the dynamic binding of H1 to chromatin in vivo
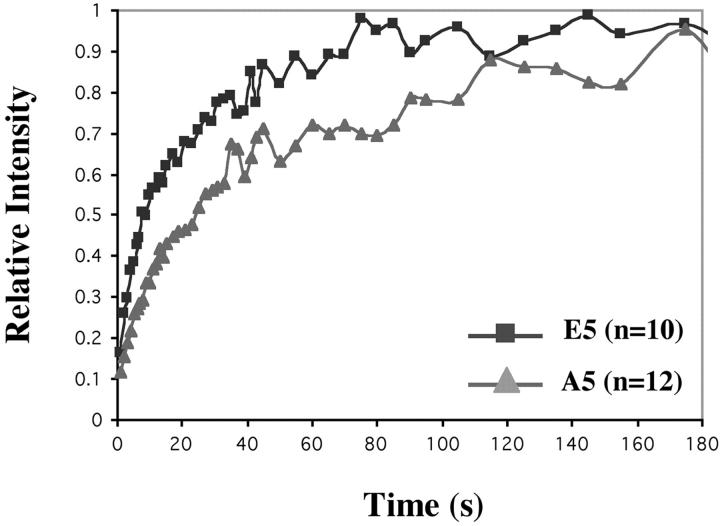
We used FRAP analysis (Lever et al., 2000; Misteli et al., 2000) to directly analyze the mobility of H1s with mutations mimicking the fully phosphorylated or dephosphorylated state in living cells. After photobleaching a small, randomly chosen area in the macronucleus of A5 or E5 cells, the recovery of fluorescence signal in the bleached area was recorded by time-lapse imaging (Fig. 3 A). Our results showed that the initial rate of recovery in both A5 and E5 strains was very fast. Fifty percent recovery was reached within seconds (Fig. 3 B). Tetrahymena H1 lacks a globular domain (Wu et al., 1986) and this rapid rate is similar to that observed with a mammalian H1 lacking the globular domain (Misteli et al., 2000).
Figure 3.
Phosphorylation affects the dynamics of H1 binding in vivo. (A) Time-lapse images were taken before and during recovery after bleaching macronuclei in Tetrahymena. An A5 cell expressing A5-GFP is used as an example. Images were taken immediately before photobleaching, immediately after photobleaching, and at indicated intervals during recovery. The bleached region is shown within the square. (B) Quantitative analysis of FRAP experiments after bleaching macronuclei in WT-GFP, A5-GFP, or E5-GFP cells. The number of cells analyzed for each curve is indicated.
The fluorescence recovery rate of A5 H1 (T1/2 = 2.64 ± 0.26 s) was slower than that of E5 H1 (T1/2 = 1.61 ± 0.23 s). These values were significantly different from each other (P < 0.0001 using Student's t test). We also tested the fluorescence recovery of WT H1 (Fig. 3 B). The rate for WT H1 was intermediate between E5 and A5 (T1/2 at 2.20 ± 0.45 s) as expected from WT H1 that is heterogeneous in terms of phosphorylation. We believe that this difference in fluorescence recovery rates reflects the difference in the binding affinity of H1 for chromatin caused by the mutations mimicking the different phosphorylation states of H1. About 80% of the H1 in the E5 strain and ∼70% in A5 recovered at this rapid rate. This rapid rate was followed by an extremely slow recovery to ∼90% in E5 and ∼80% in A5. Unfortunately, because Tetrahymena are motile cells that must be immobilized in an extremely thin drop of liquid covered by mineral oil, we were unable to maintain these preparations alive long enough (assayed by ciliary beating which still occurs in immobilized but still living cells) to determine whether there was a small amount of a completely immobile fraction of H1 (as in mammalian cells) or only a fraction that recovered extremely slowly. In mammalian cells, heterochromatin had a significantly higher percentage of immobile H1 than euchromatin, consistent with the interpretation that a faction of H1 in the transcriptionally inert heterochromatin was in a much less dynamic state. Even though there is no cytologically distinct heterochromatin in the Tetrahymena macronucleus, they do contain the heterochromatin protein HP1 (Huang et al., 1998) and the biphasic fluorescence recoveries clearly indicates the existence of two pools of H1 with very different mobilities. The difference in the size of the immobile/slow fraction in the two phosphorylation mutants suggests that dephosphorylation may drive more H1 into the low mobility fraction, which could have physiological implications. For example, global changes in H1 phosphorylation state, which are induced by processes like starvation, heat shock, or conjugation, could regulate the expansion and contraction of the H1 low-mobility pool, possibly contributing to shifts in gene expression patterns observed under these conditions.
ATP affects H1 binding to chromatin in vivo
In the course of our experiments, we observed that cells that had rounded up, or appeared damaged, had slower recovery rates than intact, pyriform cells. This suggested to us that an ATP-dependent process might affect the dynamic binding of H1 to chromatin. To test this hypothesis, we examined a number of inhibitors of ATP production to determine if they had reversible effects on Tetrahymena. We found that treating Tetrahymena cells with 180 μM rotenone (Bizzozero et al., 1999) resulted in reversible ATP depletion, as indicated by the cessation of ciliary beating (Hill, 1972) in the drug and resumption of movement and growth after its removal.
A dramatically reduced rate of H1 exchange was observed in rotenone-treated cells. Quantitative analyses of FRAP recovery for A5 and E5 cells are shown in Fig. 4. The t1/2 of recovery was 24.6 ± 6.62 s and 10.5 ± 2.99 s for A5 and E5 treated cells, respectively. These rates were significantly slower than the recovery time of untreated cells (Table I). Similar reduction in recovery rates was also seen in cells treated with potassium cyanide (unpublished data). The difference in fluorescence recovery rates in the presence of inhibitors of ATP production suggests the existence of an ATP-dependent remodeling process that affects binding of H1 to chromatin.
Figure 4.
The dynamic binding of H1 to chromatin is an ATP-dependent process. FRAP analysis on A5-GFP or E5-GFP in macronuclei of living cells after treatment with the ATP depleting drug, Rotenone. Cells were treated with 180 μM Rotenone until they stopped swimming, an indicator of ATP depletion.
Table I. Time for 50% recovery of fluorescence.
| t 1/2 (s) | A5 (no phosphorylation) | E5 (phosphorylation) |
|---|---|---|
| Cells | ||
| – | 2.64 ± 0.26 | 1.61 ± 0.23 |
| + Ra | 24.6 ± 6.62 | 10.5 ± 2.99 |
| Nuclei | ||
| – | 73.8 ± 13.3 | 31.0 ± 8.70 |
| + Eb | 25.6 ± 9.44 | 14.3 ± 4.70 |
| Cell ghosts | ||
| – | 9.71 ± 3.70 | 6.00 ± 2.48 |
| + Eb | 3.50 ± 0.74 | 2.55 ± 0.61 |
| + ATPc | 3.78 ± 0.56 | 2.57 ± 0.34 |
Rotenone treatment.
Energy mix is added.
ATP (100 uM) is added.
Phosphorylation and ATP regulate H1 binding to chromatin in vitro
Because inhibitors of ATP can have pleiotropic effects on living cells, we set up two different in vitro systems to confirm the effect of ATP on H1 binding to chromatin. In the first system, freshly isolated macronuclei were incubated in a buffer with or without an energy mix (which includes NTPs, an ATP-regeneration system, cAMP, and cGMP). This isolation procedure preserved the structural integrity of macronuclei and similar preparations have been used for in vitro transcription studies of gene expression (Stargell et al., 1990). Also, no leakage of H1 was detected during the observation period.
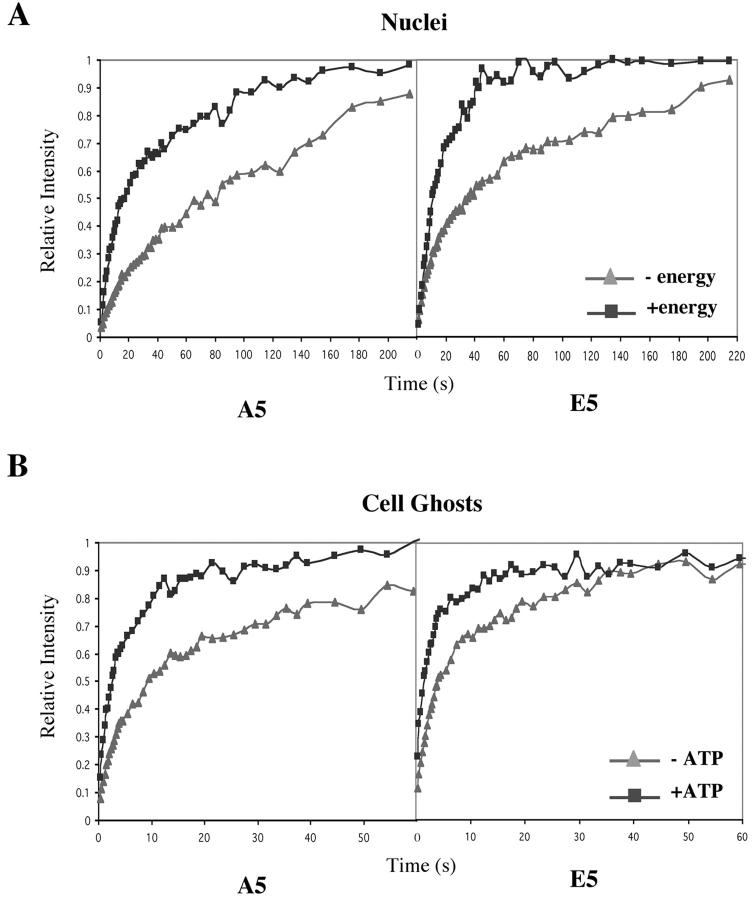
The fluorescence recovery rates for nuclei incubated with or without energy mix were compared for both A5 and E5 cells (Fig. 5 A; Table I). Although the exchange rates under all conditions were slower than those observed in intact cells for both strains, adding the energy mix dramatically increased the recovery rate. In addition, the recovery of nuclei isolated from A5 cells was slower than that of nuclei from E5 cells under all conditions.
Figure 5.
ATP-dependent H1 binding to chromatin in two in vitro systems. (A) ATP-dependent binding of H1 in isolated nuclei. Quantitative analysis of FRAP experiments after bleaching A5-GFP and E5-GFP nuclei incubated with or without energy mix. Nuclei were isolated immediately before the analysis, and images were taken within 2 h of isolation. (B) ATP-dependent binding of H1 in Tetrahymena cell ghosts. The fluorescent recovery of bleached A5-GFP cell ghosts and E5-GFP cell ghosts was determined in the presence or absence of ATP (100 μM) mix.
The second in vitro assay involved the use of Tetrahymena cell ghosts. To prepare cell ghosts, treatment with nonionic detergent, Triton X-100 was used to extract the soluble cytoplasm components of the cell leaving the cytoskeleton framework and nuclei intact (unpublished data). These preparations were designed originally for in vitro run off transcription analyses in Tetrahymena (Love et al., 1988).
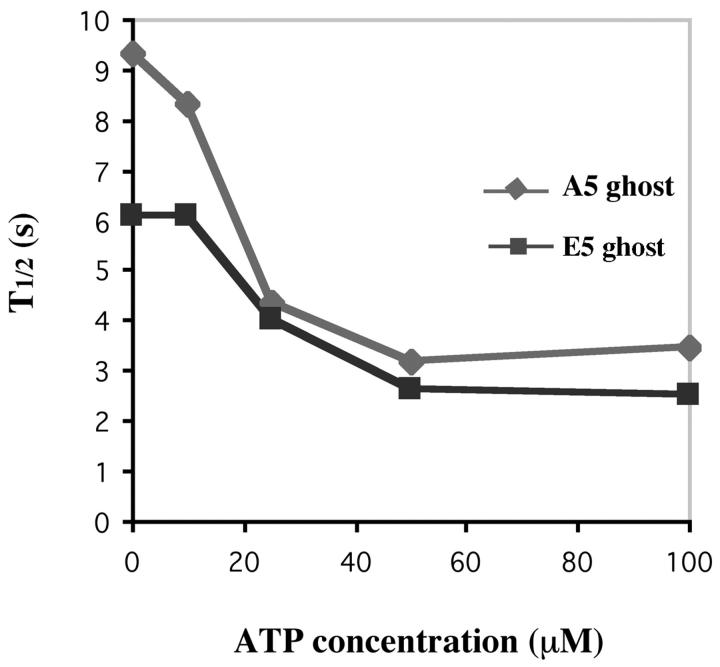
The cell ghosts of A5 and E5 strains were incubated with or without the energy mix. As for isolated nuclei, increased fluorescent recovery rates were observed for both strains in the presence of the energy mix (Table I). Because the energy mix contains components (in addition to ATP) that support transcription and could affect other nuclear processes, we also tested the recovery rates of cell ghosts incubated only with an ATP-regenerating system (Fig. 5 B). For ghosts of both A5 and E5 cells, the rate of recovery was highly stimulated by the ATP-regenerating system, and there was no statistically significant difference between adding the complete energy mix and adding the ATP-regeneration system alone (P > 0.1; Table I). The rates of recovery of A5 and E5 cell ghosts respond to ATP at physiological concentrations from 0 to ∼50 μM (Fig. 6).
Figure 6.
ATP titration of A5 and E5 cell ghosts. The 50% recovery times were calculated for A5 and E5 cell ghosts at 0, 10, 25, 50, and 100 μM ATP.
A number of conclusions can be made by combining and comparing the studies in living cells, isolated nuclei, and cell ghosts (Table I). First, in all three preparations, the dynamic binding of H1 to chromatin is greatly increased by an ATP-dependent process(es). Second, it is unlikely that transcription per se is the ATP-dependent process that affects H1 dynamics, as ATP alone has the same effect as a complete mixture that supports transcription. Third, in both the presence and absence of ATP, creation of a negative charge patch that mimics phosphorylation increases the rate of H1 exchange in chromatin.
Discussion
FRAP analysis of the dynamic binding of nuclear proteins to chromatin
FRAP experiments generally measure the mobility of a population of fluorescent molecules (Axelrod et al., 1976, Elson and Qian, 1989). Flow, or directional transport, could be an important factor in dissipating a macroscopic fluorescence intensity gradient. However, no case involving directional transport within the nucleus has been reported and we did not observe any significant asymmetry in the recovery of the bleached region. Therefore, in our system the mobility is probably determined only by the apparent diffusion rate of the protein being studied. Many factors may influence the apparent diffusion rate, causing it to differ from the rate of free diffusion, which is extremely rapid (Brown et al., 1999). These factors include the hydrodynamics of the microenvironment, steric hindrance by other macromolecules, formation of aggregates (Kucik et al., 1999), and reversible binding to large complexes. We have compared the behavior of two GFP-tagged H1s differing only in the charge state of a small region, making it unlikely that differences in the hydrodynamics or steric hindrance in the nuclei are responsible for our observations. Differences in the nuclear environment also seem unlikely to explain the effects we observed of small amounts of ATP on the rate of exchange in three different types of preparations. Likewise, changes in aggregation state are not likely to be responsible for any of our results, as H1 shows little tendency to aggregate, and the diffusion rate of proteins in FRAP experiments is relatively insensitive to the aggregation state of a protein until high levels of aggregation are reached (Kucik et al., 1999). Differences in the nuclear environment (e.g., viscosity of the nucleoplasm) could be responsible for the different rates of exchange seen between cells, isolated nuclei, and cell ghosts. Thus, the data obtained from each type of experiment (cells, nuclei, ghosts) are likely to be internally consistent, but absolute rates cannot be compared between the different experiments.
As the diffusion coefficient of a protein has only a weak dependence on molecular size and shape, all nuclear proteins should have a comparable value of free diffusion rate. Using the well-measured GFP diffusion coefficient of ∼100 μm2 s−1 (Brown et al., 1999), one can derive that free protein can recover a photobleached area with a diameter of 1 μm (the size of the bleached spot used in our experiments) in ∼1/10 of a second. All the chromatin binding proteins analyzed by FRAP so far (including Tetrahymena H1) have longer recovery times, with little or no indication that there exists a detectable population of freely diffusing molecules. This indicates that the size of their free pools is small and that something acts to retard the diffusion of the majority of molecules. Interaction with chromatin is likely to be the most important factor that affects the apparent diffusion rate of a chromatin binding protein. Being a huge complex, chromatin is generally assumed to be immobile. When a nuclear protein is bound to chromatin, it cannot contribute to diffusion. Therefore, for any nuclear protein with significant binding to chromatin, the apparent diffusion rate reflects the heavy influence of this interaction. Our FRAP experiments take advantage of this feature in trying to address the in vivo chromatin binding properties of the linker histone H1 in Tetrahymena.
Posttranslational modification affects the dynamic binding of histone H1 to chromatin
Charge-altering posttranslational modifications of chromatin binding proteins can directly affect the way that proteins interact with chromatin and change the binding equilibrium. Hyperacetylation of HMG-14 and HMG-17 results in their reduced affinity for nucleosomes (Herrera et al., 1999; Bergel et al., 2000), and FRAP analyses indicated that phosphorylation of these proteins prevents their binding to chromatin (Prymakowska-Bosak et al., 2001). Extensive acetylation greatly weakens, but does not abolish, the interaction of a histone H4 NH2-terminal peptide with DNA in vitro (Norton et al., 1989). Phosphorylated but not unphosphorylated H1 migrates rapidly from sea urchin sperm chromatin to rat liver chromatin at ionic strengths above 45 mM (Hill et al., 1991). In addition, phosphorylation of Ser10 results in reduced binding of the histone H3 tail to DNA in mitotic cells (Sauve et al., 1999).
In our experiments, we observed that a mutated H1 mimicking the fully phosphorylated state (E5) has an apparent diffusion rate that is greater than the one mimicking the unphosphorylated state. In the living cells and in two in vitro systems, comparable, approximately twofold differences between the apparent diffusion rates of the two H1s were obtained (Table I). These mutations are unlikely to alter the size or the shape of the GFP-tagged H1, and the expression level of the mutated H1s remained the same in both strains. Therefore, these results indicate that charge changes that mimic phosphorylation shifted the kinetic equilibrium constant in favor of dissociation and increased the concentration of free H1. These observations argue strongly that by adding negative charges to H1, phosphorylation probably weakens the electrostatic interactions between the negatively charged DNA and the overall positively charged H1 in vivo. This is consistent with in vitro salt elution studies showing that both E5 and A5 H1 remained bound to chromatin under physiological conditions, and that there was a small shift in the salt concentration at which half of the H1 eluted, from 0.24 (E5) to 0.26 M (A5) (unpublished data).
Our FRAP results provide the first evidence that introducing negative charges at the H1 phosphorylation sites, as would occur when H1 is phosphorylated, can influence H1 binding in vivo. Mutations mimicking the phosphorylation of H1 did not abolish H1 binding to chromatin, as was the case with HMG proteins (Prymakowska-Bosak et al., 2001) and mutations abolishing H1 phosphorylation did not abolish dissociation of H1 from chromatin. Instead, these mutations indicated that phosphorylation acts as a modulator that fine tunes the binding affinity of H1 to chromatin.
Phosphorylation of H1 is a highly conserved event that has long been suspected to play an important role in chromatin function. Our previous work established that changes in H1 phosphorylation states were critical for the activation of specific genes (Dou et al., 1999). The effect of phosphorylation on H1 binding described here provides a mechanistic link that connects this modification and gene regulation. We have shown that CyP1 expression is induced by starvation and requires that H1 be present on chromatin. Starvation induces dephosphorylation of H1. We hypothesize that, with enhanced affinity, dephosphorylated H1 can compete favorably with other factors (e.g., repressors) and gain access to key regulatory elements of the CyP1 gene, resulting in gene activation. The critical role of this regulation of H1 binding is supported by our findings that H1 phosphorylation mimics H1 removal. Thus, the E5 strain and a strain lacking H1 have the same phenotype—very low-level induced expression of CyP1 during starvation (Dou et al., 1999). This mechanism is likely to be a general one. Many analyses of chromatin crosslinked in vivo and immunoprecipitated by anti-H1 antibodies have indicated that H1 is either removed, reduced in amount, or somehow modified during transcription (Zlatanova and Van Holde, 1992).
A number of mechanisms might explain how the approximately twofold change in affinity could affect the expression of specific genes such as CyP1 or ngoA (Dou et al., 1999) either positively or negatively. It could tip the balance when H1 is in competition with gene-specific transcription factors whose recognition sites are in the linker regions. Alternatively, the increased time that phosphorylated H1 is dissociated from chromatin could enable conformational changes or complex formation necessary for these competing proteins to bind more stably to chromatin than H1, or could enable increased nucleosome mobility, exposing nucleosomal sites to transcription factors. Correlations between transcriptional potency and the increased koff for the binding of transcription factors have been reported for transcription factor STAT1. Mutations that reduced the residence time of STAT1 binding to DNA resulted in transcription inactivation (Yang et al., 2002). This supports the hypothesis that the rate of transcription factor binding to DNA relative to that of other chromatin proteins is important for transcription stimulation.
In theory, any changes that affect the interactions that tether the GFP-labeled protein to chromatin can affect its binding constant. These include modifications to the labeled protein as described here, or modifications to DNA, histones and other chromatin binding proteins participating in the formation of the complex. It has been shown that TSA-induced hyperacetylation of core histone tails, which results in the opening of chromatin and increased accessibility of DNA to remodeling factors (Struhl, 1998; Strahl and Allis, 2000), can increase the exchange rate of histone H1 in living cells (Misteli et al., 2000). The system we have described here, coupled with the ability to introduce specifically modified core histone tails (Ren and Gorovsky, 2001), should allow detailed analyses of the effects of specific modifications of core histone tails on the binding of H1.
The dynamic binding of histone H1 to chromatin is affected by an ATP-dependent process
In living cells, the active input of energy probably can shift the binding of chromatin proteins from the thermodynamic equilibrium dictated by the intrinsic properties of the factors involved. As most of the known ATP-dependent processes acting on chromatin tend to disrupt or interfere with the formation of chromatin complexes (Aalfs and Kingston, 2000), it is reasonable to assume that if such a process acts on H1 binding, it will drive the equilibrium to favor more free H1. Three independent approaches were used in our experiments to test the ATP dependency of H1 binding: energy depletion in living cells, energy addition to isolated nuclei, and energy or ATP addition to cell ghosts. The results in all three systems led to the conclusion that one or more ATP-dependent processes can significantly affect the binding of H1 to chromatin. This energy dependent process increased the apparent diffusion coefficient of H1. Based on the considerations described above, this likely occurred by facilitating the dissociation of H1 from chromatin. This effect is not likely to be related to DNA replication in the two in vitro systems, as dNTPs were not included. The observation that the effects of adding NTPs or of adding ATP alone to the cell ghosts are the same suggests that processes other than transcription elongation are involved. These results differ from previously published work showing that the binding of linker histone is ATP independent in mammalian culture cells (Lever et al., 2000). The basis for this difference is not known. Even though we can not yet exclude other explanations for the ATP effect on fluorescence recovery, our results suggest the exciting possibility that there exists a global ATP-dependent chromatin remodeling system that affects H1 dissociation from chromatin. The estimated Km for the ATP-dependent activity in our experiment is ∼25 μM, in the range of the Km for some known ATPase activities of chromatin remodeling complexes (Cairns et al., 1996; Aalfs et al., 2001). Also, it is worth noting that Tetrahymena has a high intracellular ATP concentration (∼100–200 μM; Rooney and Eller, 1969), at which level the putative ATP-dependent chromatin remodeling processes can be fully active. It remains to be determined whether this process is carried out by known ATP-dependent complexes that modify core nucleosomes (for review see Aalfs and Kingston, 2000; Wu and Grunstein, 2000) or is unique and has distinct structural effects on chromatin.
A recent in vitro study has demonstrated another possible mechanism by which linker histone phosphorylation might affect gene expression. Horn et al. (2002) found that whereas dephosphorylated linker histone could inhibit the chromatin remodeling by yeast SWI/SNF, linker histone phosphorylated by Cdc2/Cyclin B kinase could not. Because our studies demonstrate an effect of phosphorylation on H1 dynamics that is independent of ATP, and an effect of ATP that is independent of the phosphorylation state of H1, it is not likely that this mechanism contributes significantly to our results.
Mechanisms of transcription regulation in a dynamic chromatin environment
FRAP experiments provide direct visualization of the dynamic nature of chromatin. They have revealed that both structural chromatin-binding proteins and transcriptional activators are continuously and rapidly exchanging with chromatin (Berk, 1999; Phair and Misteli, 2000; Lever et al., 2000; Misteli et al., 2000; Boisvert et al., 2001). These observations are consistent with gene expression being regulated by the complex interactions of multiple stochastic processes (McAdams and Arkin, 1997; Misteli 2001). The probability for the assembly of transcription machinery (or machinery for other chromatin functions) at a specific locus depends on the accessibility of binding sites, the availability of each component at the assembly site, and the stability of the formed functional complex. Transcription regulation can be achieved by modulating any of these steps. The studies described here demonstrate that phosphorylation and an ATP-dependent effect on H1 binding are likely additions to the stochastic events that regulate gene activity.
Materials and methods
Cell and culture conditions
WT Tetrahymena thermophila strain CU428 mpr1-1/mpr1-1 (MPR1, mp-s, VII) was provided by P.J. Bruns (Cornell University, Ithaca, NY). Cells were grown in medium (Gorovsky et al., 1975) containing 1% proteose peptone (SPP). For FRAP analysis, log phase Tetrahymena cells were deposited under a drop of paraffin oil and excess media was aspirated away until the cells were immobilized as described (Chalker et al., 2000).
Construct information
HHO1/neo2, HHO1 A5, and HHO1 E5 constructs (Dou et al., 1999) were used to create the WT-GFP, A5-GFP, and E5-GFP constructs. The GFP construct was provided by M-C. Yao (Fred Hutchinson Cancer Research Center, Seattle, WA). GFP was inserted at the COOH terminus of the HHO1 coding region immediately 5′ of the terminator TGA by overlapping PCR (Sambrook and Russell, 2001).
The GFP coding sequence was amplified by primers GFP5′: 5′ CCTGCCAAGAAGAACAGTAAAGGAGAAGAAC 3′; and GFP3′: 5′ CTTTACTACAAAAAAATCATTTGTATAGTTCATCCATGCC 3′. The H1 5′ flanking and H1 coding sequence was amplified by primers HHO 5′ sense: 5′ CTTCCTCTTAGATATTTGATAAG 3′; and HHO 5′ antisense: 5′ CTCCTTTACTGTTCTTCTTGGCAGGC 3′. The H1 3′ flanking sequence (400 bp) was amplified using primers HHO 3′ sense: 5′ CTATACAAATGATTTTTTTGTAGTAAAGAAATTCCTC 3′; and HHO 3′ antisense: 5′ GGAATACACATTCATTGTATGC 3′. The three fragments were ligated together by overlapping PCR (94°C, 30 s; 42°C, 30 s; 72°C, 2 min) using the HHO 5′ sense and HHO 3′ antisense as primers. The final products were cleaved with SpeI and EcoRI and cloned into the HHO1/neo2 vector (Dou et al., 1999) digested by the same restriction enzymes.
Gene replacement by biolistic transformation
GFP fusion constructs containing a flanking neo2 gene conferring paromomycin (pm) resistance in Tetrahymena were digested with KpnI and SacII, biolistically transformed into CU428 cells as described (Dou et al., 1999), and selected in paromomycin sulfate to a final concentration of 2.5 mg/ml.
Southern blot analysis
Single cells were isolated and grown for 40–50 generations without pm. Under these nonselective conditions, cells containing any WT genes outgrow cells containing the neo2 gene (unpublished observations). However, if the endogenous WT gene had been completely eliminated from the genome of the selected cell, its progeny should contain only HHO1-GFP fusion genes flanked by the neo2 cassette. Total cellular DNA was isolated, digested with HindIII and EcoRI, separated by agarose gel electrophoresis, and blotted onto Magnagraph nylon membranes (Osmonics, Inc.) according to established protocols (Ausubel et al., 1988). To detect the introduced HHO1-GFP gene, the HHO1 coding sequence was labeled with (α-32P) dATP by random priming (Ausubel et al., 1988). Hybridization and washes were performed at 65°C.
Northern blot analysis
RNA was isolated from starved cells with Trizol (Invitrogen) electrophoresed in 2.2 M formaldehyde-1.2% agarose gels, blotted, and hybridized (Ausubel et al., 1988). (α-32P) dATP-labeled random primed probes were obtained as follows: the probe for rRNA was a 2.5-skb HindIII fragment from pBS26S encoding the Tetrahymena 26S rRNA (Engberg and Nielsen, 1990); the CyP1 probe was synthesized from two PstI fragments (0.5 kb, 0.7 kb) from pCyP1 (Karrer and Stein-Gavens, 1990); and the ngoA probe was a 1.1-kb PstI fragment from pC5.5 (Martindale and Bruns, 1983). Hybridizations were done at 42°C in 50% formamide, 5 × SSC, 1 × SPED, 1% SDS and 100 μg/ml salmon sperm DNA. Washes were in 2 × SSC-1% SDS at 42°C.
Rotenone and potassium cyanide treatment
Tetrahymena cells at 2 × 105 cells/ml were washed twice in 0.1 M tris-HCl buffer, pH 7.4, and then resuspended in Tris with KCN at final concentration of 1.25 mg/ml, or with Rotenone at final concentration of 180 μM. Experiments were carried out within 30 min of drug addition.
Preparation of Tetrahymena cell ghosts
A5 and E5 cell ghosts were prepared as described (Love et al., 1988). Growing cells were collected by pouring them into tubes packed with ice and pelleting at 250 g for 5 min. Pellets were suspended in ice-cold extract buffer (0.1 M sucrose, 0.1 M KCl, 2.5 mM MgCl2, 2.5 mM ethylene glyco-bis [β-aminoethyl ether]-N, N, N′, N′-tetra-acetic acid [EGTA], 10 mM Hepes [N-2-hydroxyethyl-piperazine-N'-2-ethanesulfonic acid], 1% Triton X-100, pH 6.8) and incubated on ice for 5 min. The pellet was then washed twice in extraction buffer without Triton X-100. The resulting cytoskeletal frameworks contain nuclei and can be used for in vitro nuclear runoff experiments. The final pellet was suspended in nuclear isolation buffer (10 mM Tris-HCl, pH 7.5, 50 mM KCl, 5 mM MgCl2, 1 mM CaCl2, 2 mM β-mercaptoethanol, 100 μg/ml BSA) with or without energy mix.
Nuclei isolation
Macronuclei of Tetrahymena thermophila were prepared as previously described (Stargell et al., 1990). The nuclei were then suspended in the nucleus isolation buffer as described above with or without energy mix. FRAP experiments were carried out within 1 h after nuclei isolation.
In vitro tests for energy and ATP dependence
Nuclei and cell ghosts were incubated in the nuclei isolation buffer described above with energy mix at final concentration of 100 μM each of ATP, CTP, GTP, TTP, cAMP, and cGMP; 30 mM phosphocreatine; and 1μg/ml creatine phosphokinase. The ATP mix used in the analysis of ghost cells contains 100 μM ATP, 30 mM phosphocreatine, and 1 μg/ml creatine phosphokinase. Experiments were carried out within 1 h of incubation. For ATP titration experiment, different ATP concentrations from 0 to 100 μM were used.
Confocal microscopy and FRAP analysis
The confocal microscope (Leica TCS SP) uses the 488-nm argon laser (20 mW nominal output beam width at specimen 0.2 μm, detection 500–575 nm using a photon multiplier tube with pinhole setting at 1). All photobleaching experiments were performed using 100 × 1.4 N.A. objective. The experiments were done as described previously (Lever et al., 2000; Misteli et al., 2000). One imaging scan was acquired, followed by a single bleach pulse of 200–500 ms using a spot of 1 μm in diameter without scanning. Single-section images were then collected. FRAP recovery curves were generated and analyzed using Metamorph 4.0 (University Imaging) and Microsoft Excel as described (Misteli et al., 2000). Programs for collecting images varied for different samples, depending on their rates of recovery. For living cells, single-section images were collected at 0.3-s interval (15 images) followed by 2-s intervals (15 images). For cell ghosts, single-section images were collected at 0.3-s intervals (15 images), followed by 1-s intervals (15 images), 2-s intervals (10 images), and then by 5-s intervals (6 images). For Rotenone treated cells and nuclei, single-section images were first collected at 1-s intervals (15 images), then 2-s intervals (15 images), 5-s intervals (10 images), 10-s intervals (6 images), and 20-s intervals (3 images). The standard Student's t test was used to determine the statistical significance of the results. All quantitative values represent averages from at least 10 cells per experiment from two or more independent experiments. For each experiment, the average t1/2 and its standard deviation was calculated from the t1/2 from each individual recovery curve of each cell.
Acknowledgments
We are grateful to Hiram Lyon for help using the confocal microscope.
This work was supported by grant GM21793 from the National Institutes of Health.
References
- Aalfs, J.D., and K.E. Kingston. 2000. What does “chromatin remodeling” mean? TIBS. 25:548–555. [DOI] [PubMed] [Google Scholar]
- Aalfs, J.D., J.N. Geeta, and R.E. Kingston. 2001. Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J. Biol. Chem. 276:34270–34278. [DOI] [PubMed] [Google Scholar]
- Axelrod, D., D.E. Koppel, J. Schlessinger, E. Elson, and W.W. Webb. 1976. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16:1055–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1988. Current Protocols in Molecular Biology. L.M. Albright, D.M. Coen, A. Varki, and V.B. Chanda, editors. Wiley Interscience, New York.
- Barra, J.L., L. Rhounim, J.L. Rossignol, and G. Faugeron. 2000. Histone H1 is dispensable for methylation-associated gene silencing in Ascobolus immersus and essential for long life span. Mol. Cell. Biol. 20:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar, J., R.A. Horowitz, S.A. Grigoryev, L.M. Carruthers, J.C. Hansen, A.J. Koster, and C.L. Woodcock. 1998. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA. 95:14173–14178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergel, M., J.E. Herrera, B.J. Thatcher, M. Prymakowska-Bosak, A. Vassilev, Y. Nakatani, B. Martin, and M. Bustin. 2000. Acetylation of novel sites in the nucleosomal binding domain of chromosomal protein HMG-14 by p300 alters its interaction with nucleosomes. J. Biol. Chem. 275:11514–11520. [DOI] [PubMed] [Google Scholar]
- Berk, A.J. 1999. Activation of RNA polymerase II transcription. Curr. Opin. Cell Biol. 11:330–335 [DOI] [PubMed] [Google Scholar]
- Bizzozero, O.A., P. Sanchez, and S.U. Tetzloff. 1999. Effect of ATP depletion on the palmitoylation of myelin proteolipid protein in young and adult rats. J. Neurochem. 72:2610–2616. [DOI] [PubMed] [Google Scholar]
- Blank, T.A., and P.B. Becker. 1995. Electrostatic mechanism of nucleosome spacing. J. Mol. Biol. 252:305–313. [DOI] [PubMed] [Google Scholar]
- Boisvert, F.M., M.J. Kruhlak, A.K. Box, M.J. Hendzel, and D.P. Bazett-Jones. 2001. The transcription coactivator CBP is a dynamic component of the promyelocytic leukemia nuclear body. J. Cell Biol. 152:1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury, E.M. 1992. Reversible histone modifications and the chromosome cell cycle. Bioessays. 14:9–16. [DOI] [PubMed] [Google Scholar]
- Brown, E.B., E.S. Wu, W. Zipfel, and W.W. Webb. 1999. Measurement of molecular diffusion in solution by multiphoton fluorescence photobleaching recovery. Biophys. J. 77:2837–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns, B.R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R.D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell. 87:1249–1260. [DOI] [PubMed] [Google Scholar]
- Carruthers, L.M., J. Bednar, C.L. Woodcock, and J.C. Hansen. 1998. Linker histone stabilizes the intrinsic salt-dependent folding of nucleosome arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 37:14776–14787. [DOI] [PubMed] [Google Scholar]
- Chalker, D.L., J.G. Ward, C. Randolph, and M. Yao. 2000. Microinjection of Tetrahymena thermophila Tetrahymena thermophila. D.J. Asai, and J.D. Forney, editors. Academic Press, Inc., New York. 469–483. [DOI] [PubMed]
- Dou, Y., and M.A. Gorovsky. 2000. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol. Cell. 6:225–231. [DOI] [PubMed] [Google Scholar]
- Dou, Y., and M.A. Gorovsky. 2002. Regulation of transcription by H1 phosphorylation in Tetrahymena is position-independent and requires clustered sites. Proc. Natl. Acad. Sci. USA. 99:6142–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, Y., C.A. Mizzen, M. Abrams, C.D. Allis, and M.A. Gorovsky. 1999. Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Mol. Cell. 4:641–647. [DOI] [PubMed] [Google Scholar]
- Elson, E.L., and H. Qian. 1989. Interpretation of fluorescence correlation spectroscopy and photobleaching recovery in terms of molecular interactions. Methods Cell Biol. 30:308–332. [DOI] [PubMed] [Google Scholar]
- Engberg, J., and H. Nielsen. 1990. Complete sequence of the extrachromosomal rDNA molecule from the ciliate Tetrahymena thermophila strain B1868VII. Nucleic Acids Res. 18:6915–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover, C.V.C., K.J. Vavra, S.D. Guttman, and M.A. Gorovsky. 1981. Heat shock and deciliation induce phosphorylation of histone H1 in Tetrahymena pyriformis. Cell. 23:73–77. [DOI] [PubMed] [Google Scholar]
- Gorovsky, M.A., M.-C. Yao, J.B. Keevert, and G.L. Pleger. 1975. Isolation of micro- and macronuclei of Tetrahymena pyriformis Methods Cell Biol. IX:311–327. [DOI] [PubMed] [Google Scholar]
- Hellauer, K., E. Sirard, and B. Turcotte. 2001. Decreased expression of specific genes in yeast cells lacking histone H1. J. Biol. Chem. 276:13587–13592. [DOI] [PubMed] [Google Scholar]
- Herrera, J., K. Sakaguchi, M. Bergel, L. Trieschmann, Y. Nakatani, and M. Bustin. 1999. Specific acetylation of chromosomal protein HMG-17 by P/CAF alters its interaction with nucleosomes. Mol. Cell. Biol. 19:3466–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, C.S., J.M. Rimmer, B.N. Green, J.T. Finch, and J.O. Thomas. 1991. Histone-DNA interactions and their modulation by phosphorylation of -Ser-Pro-X-Lys/Arg- motifs. EMBO J. 10:1939–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, D.L. 1972. Cell Biology: Biochemistry and Physiology of Tetrahymena. Academy Press, Inc., New York. 83–84.
- Horn, P.J., L.M. Carruthers, C. Logie, D.A. Hill, M.J. Solomon, P.A. Wade, A.N. Imbalzano, J.C. Hansen, and C.L. Peterson. 2002. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat. Struct. Biol. 9:263–267. [DOI] [PubMed] [Google Scholar]
- Huang, H., E.A. Wiley, C.R. Lending, and C.D. Allis. 1998. An HP1-like protein is missing from transcriptionally silent micronuclei of. Tetrahymena. Proc. Natl. Acad. Sci. USA. 95:13624–13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik, M.A., and E. Schulze. 2001. A single histone H1 isoform (H1.1) is essential for chromatin silencing and germline development in Caenorhabditis elegans. Development. 128:1069–1080. [DOI] [PubMed] [Google Scholar]
- Karrer, K.M., and S. Stein-Gavens. 1990. Constancy of adenine methylation in Tetrahymena macronuclear DNA. J. Protozool. 37:409–414. [DOI] [PubMed] [Google Scholar]
- Kucik, D.F., E.L. Elson, and M.P. Sheetz. 1999. Weak dependence of mobility of membrane protein aggregates on aggregate size supports a viscous model of retardation of diffusion. Biophys. J. 76:314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever, M.A., J.P.H. Th'ng, X. Sun, and M.J. Hendzel. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 408:873–877. [DOI] [PubMed] [Google Scholar]
- Love, H.D., A. Allen-Nash, Q. Zhao, and G.A. Bannon. 1988. mRNA stability plays a major role in regulating the temperature-specific expression of a Tetrahymena thermophila surface protein. Mol. Cell. Biol. 8:427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale, D.W., and P.J. Bruns. 1983. Cloning of abundant mRNA species present during conjugation of Tetrahymena thermophila: Identification of mRNA species present exclusively during meiosis. Mol. Cell. Biol. 3:1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams, H.H., and A. Arkin. 1997. Stochastic mechanisms in gene regulation. Proc. Natl. Acad. Sci. USA. 94:814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meersseman, G., S. Pennings, and E.M. Bradbury. 1991. Chromatosome positioning on assembled long chromatin. Linker histones affect nucleosome placement on 5S rDNA. J. Mol. Biol. 220:89–100. [DOI] [PubMed] [Google Scholar]
- Misteli, T. 2001. Protein dynamics: Implications for nuclear architecture and gene expression. Science. 291:843–847. [DOI] [PubMed] [Google Scholar]
- Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D.T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature. 408:877–881. [DOI] [PubMed] [Google Scholar]
- Mizzen, C.A., Y. Dou, Y. Liu, R.G. Cook, M.A. Gorovsky, and C.D. Allis. 1999. Identification and mutation of phosphorylation sites in a linker histone. J. Biol. Chem. 274:14533–14536. [DOI] [PubMed] [Google Scholar]
- Norton, V.G., B.S. Imai, P. Yau, and E.M. Bradbury. 1989. Histone acetylation reduces nucleosome core particle linking number change. Cell. 57:449–457. [DOI] [PubMed] [Google Scholar]
- Owen-Hughes, T., and J.T. Workman. 1994. Experimental analysis of chromatin function in transcriptional control. Crit. Rev. Eukaryot. Gene Expr. 4:403–441. [PubMed] [Google Scholar]
- Paranjape, S.M., R.T. Kamakaka, and T.T. Kadonaga. 1994. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu. Rev. Biochem. 63:265–297. [DOI] [PubMed] [Google Scholar]
- Patterton, H.G., C.C. Landel, D. Landsman, C.L. Peterson, and R.T. Simpson. 1998. The biochemical and phenotypic characterization of Hho 1p, the putative linker histone H1 of Sacccharomyces cerevisiae. J. Biol. Chem. 273:7268–7276. [DOI] [PubMed] [Google Scholar]
- Pennings, S., G. Meersseman, and E.M. Bradbury. 1994. Linker histone H1 and H5 prevent the mobility of positioned nucleosomes. Proc. Natl. Acad. Sci. USA. 91:10275–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, C.L. 2002. Chromatin remodeling enzymes: taming the machines. EMBO Rep. 3:319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair, R.D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature. 404:604–609. [DOI] [PubMed] [Google Scholar]
- Prymakowska-Bosak, M., M.R. Przewloka, J. Slusarczyk, M. Kuras, J. Lichota, B. Kilianczyk, and A. Jerzmanowski. 1999. Linker histones play a role in mail meiosis and the development of pollen grains in tobacco. Plant Cell. 11:2317–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prymakowska-Bosak, M., T. Misteli, J.E. Herrera, H. Shirakawa, Y. Birger, S. Garfield, and M. Bustin. 2001. Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol. Cell. Biol. 21:5169–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon, A., M.I. Muro-Pastor, C. Scazzocchio, and R. Gonzale. 2000. Deletion of the unique gene encoding a typical histone H1 has no apparent phenotype in aspergillus nidulans. Mol. Microbiol. 35:223–233. [DOI] [PubMed] [Google Scholar]
- Rabini, S., K. Franke, P. Saftig, C. Bode, D. Doenecke, and B. Drabent. 2000. Spermatogenesis in mice is not affected by histone H1.1 deficiency. Exp. Cell Res. 255:114–124. [DOI] [PubMed] [Google Scholar]
- Ren, Q., and M.A. Gorovsky. 2001. Histone H2A.Z acetylation modulates an essential charge patch. Mol. Cell. 7:1329–1335. [DOI] [PubMed] [Google Scholar]
- Rooney, D.W., and J.J. Eller. 1969. Effects of division-synchronizing hypoxic and hyperthermic shocks upon Tetrahymena, a respiration and intracellular ATP concentration. J. Cell Biol. 41:145–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, S.Y., and C.D. Allis. 1992. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem. Sci. 17:93–98. [DOI] [PubMed] [Google Scholar]
- Roth, S.Y., I.G. Schulman, R. Richman, R.G. Cook, and C.D. Allis. 1988. Characterization of phosphorylation sites in histone H1 in the amitotic macronucleus of Tetrahymena during different physiological states. J. Cell Biol. 107:2473–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and D.W. Russell. 2001. Molecular Cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sauve, D.M., H.J. Anderson, J.M. Ray, W.M. James, and M. Roberge. 1999. Phosphorylation-induced rearrangement of the histone H3 NH2-terminal domain during mitotic chromosome condensation. J. Cell Biol. 145:225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X.T., and M.A. Gorovsky. 1996. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 86:475–483. [DOI] [PubMed] [Google Scholar]
- Shen, X.T., L. Yu, J.W. Weir, and M.A. Gorovsky. 1995. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 82: 47–56. [DOI] [PubMed] [Google Scholar]
- Simpson, R.T. 1978. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 13:691–699. [DOI] [PubMed] [Google Scholar]
- Stargell, L.A., K.M. Karrer, and M.A. Gorovsky. 1990. Transcription regulation of gene expression in Tetrahymena thermophila. Nucleic Acids Res. 18:6637–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach, O.C., A.P. Wolffe, and R.A.W. Rupp. 1997. Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature. 389:395–399. [DOI] [PubMed] [Google Scholar]
- Strahl, B.D., and C.D. Allis. 2000. The language of covalent histone modifications. Nature. 403:41–45. [DOI] [PubMed] [Google Scholar]
- Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599–606. [DOI] [PubMed] [Google Scholar]
- Takami, Y., and T. Nakayama. 1997. A single copy of linker H1 gene is enough for proliferation of the DT40 chicken B cell line, and linker H1 variants participate in regulation of gene expression. Genes Cells. 2:711–723. [DOI] [PubMed] [Google Scholar]
- Thoma, F., T. Koller, and A. Klug. 1979. Involvement of histone H1 in the organization of the nucleosome and the salt-dependent superstructures of chromatin. J. Cell Biol. 83:407–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushinsky, S.C., H. Bussey, A.A. Ahmed, Y. Wang, J. Friesen, B.A. Williams, and R.K. Storms. 1997. Histone H1 in Saccharomyces cerevisiae. Yeast. 13:151–161. [DOI] [PubMed] [Google Scholar]
- Widom, J. 1986. Physiochemical studies of the folding of the 100A nucleosome filament into the 300 A filament. J. Mol. Biol. 190:411–424. [DOI] [PubMed] [Google Scholar]
- Wolffe, A. 1998. Chromatin. London Academic Press, London. 447 pp.
- Woodcock, C.L., S.A. Grigoryev, R.A. Horowitz, and N. Whitaker. 1993. A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc. Natl. Acad. Sci. USA. 90:9021–9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J., and M. Grunstein. 2000. 25 years after the nucleosome model: chromatin modifications. TIBS. 25:619–623. [DOI] [PubMed] [Google Scholar]
- Wu, M., C.D. Allis, R. Richman, R.G. Cook, and M.A. Gorovsky. 1986. An intervening sequence in an unusual histone H1 gene of Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA. 83:8674–8678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, E., M.A. Henriksen, O. Schaefer, N. Zakharova, and J.E. Darnell. 2002. Dissociation time from DNA determines transcriptional function in a STAT1 linker mutant. J. Biol. Chem. 277:13455–13462. [DOI] [PubMed] [Google Scholar]
- Zlatanova, J., and K. Van Holde. 1992. Histone H1 and transcription: still an enigma? J. Cell Sci. 103:889–895. [DOI] [PubMed] [Google Scholar]