Abstract
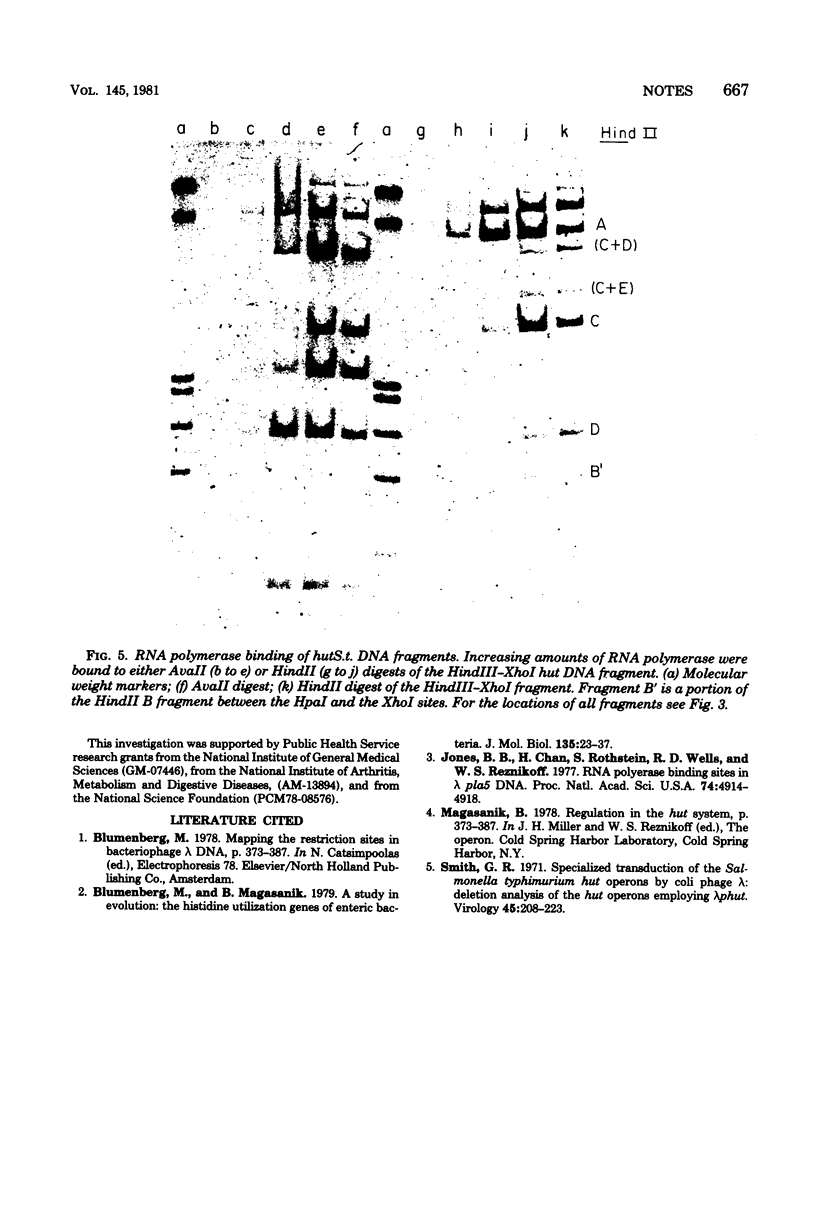
The recognition sites for several restriction endonucleases were mapped within deoxyribonucleic acid coding for histidine utilization (hut) genes of Salmonella typhimurium and Klebsiella aerogenes. Deoxyribonucleic acid fragments containing the two hut promoters were identified by ribonucleic acid polymerase binding.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blumenberg M., Magasanik B. A study in evolution: the histidine utilization genes of enteric bacteria. J Mol Biol. 1979 Nov 25;135(1):23–37. doi: 10.1016/0022-2836(79)90338-3. [DOI] [PubMed] [Google Scholar]
- Jones B. B., Chan H., Rothstein S., Wells R. D., Reznikoff W. S. RNA polymerase binding sites in lambdaplac5 DNA. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4914–4918. doi: 10.1073/pnas.74.11.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R. Specialized transduction of the Salmonella hut operons by coliphage lambda: deletion analysis of the hut operons employing lambda-phut. Virology. 1971 Jul;45(1):208–223. doi: 10.1016/0042-6822(71)90128-0. [DOI] [PubMed] [Google Scholar]