Abstract
Centrioles and basal bodies fascinate by their spectacular architecture, featuring an arrangement of nine microtubule triplets into an axial symmetry, whose biogenesis relies on yet elusive mechanisms. However, the recent discovery of new tubulins, such as δ-, ɛ-, or η-tubulin, could constitute a breakthrough for deciphering the assembly steps of this unconventional microtubule scaffold. Here, we report the functional analysis in vivo of ɛ-tubulin, based on gene silencing in Paramecium, which demonstrates that this protein, which localizes at the basal bodies, is essential for the assembly and anchorage of the centriolar microtubules.
Keywords: centriole duplication; basal body; tubulin; cytoskeleton; paramecium
Introduction
Centrioles and basal bodies have long fascinated cell biologists not only for their central role in cell division or motility, but also for their ability to duplicate at each cell cycle by an autoreplicative process that, by perpetuating structural, positional, and temporal information, results in two structurally identical, but functionally distinct, organelles. These cylindrical organelles are characterized by their symmetrical geometry, defined by nine microtubule triplets organized as blades around a central axis. The assembly process of this peculiar microtubule scaffold, whose mechanism remains enigmatic, has been well documented at the ultrastructural level in the ciliate Paramecium (Dippel, 1968; unpublished data). It involves discrete successive steps, beginning with the appearance of a generative disc, upon which is nucleated the inner ring of microtubules, or A-tubules, regularly positioned around a central structure called the cartwheel. Then, B- and C-tubules develop as short ribbons, which progressively elongate, curve, and eventually close back onto the next tubule circumference, thus forming incomplete microtubules, which share a quarter of their boundaries with their neighbors. During this process, the procentriole elongates, moves perpendicularly to its parent, and anchors to the cortex. The new structure is then likely stabilized (Bobinnec et al., 1998) and is necessarily matured to acquire its potential to nucleate microtubules (Fleury and Laurent, 1995) and appendages and to be able to duplicate further (Iftode et al., 1989; Lange et al., 2000).
On a molecular level, these assembly steps remain elusive due to an insufficient knowledge of the molecular components of the centriole. However, the development of genetic tools allowing a functional approach to understanding the basal body duplication in unicellular organisms, such as the green alga Chlamydomonas or the ciliate Paramecium, has recently proven to be very efficient in identifying structural proteins and, in particular, new tubulin types, shown to be involved in the assembly of centriolar microtubules (Dutcher and Trabuco, 1998; Ruiz et al., 1999, 2000; Garreau De Loubresse et al., 2001). Indeed, the ciliate Paramecium, with its 4,000 basal bodies regularly positioned on the cell cortex and whose duplication may be easily monitored by simple immunostaining, provides a favorable model for such a functional approach. Moreover, the development of functional genetics in this organism demonstrated that it is possible to obtain viable mutants affected in basal body duplication (Ruiz et al., 1987, 1999; Jerka-Dziadosz et al., 1998), thus giving the opportunity to genetically dissect the assembly process, which is difficult in the case of centriole duplication because of its intimate coupling with cell cycle progression.
Major contributions in the identification and functional characterization of centriolar tubulins have recently benefited from the use of this model organism. Thus, gene silencing experiments in Paramecium have demonstrated that γ-tubulin is essential for basal body duplication (Ruiz et al., 1999), thus assessing its role in the nucleation not only of the cytoplasmic microtubules but also of the microtubules that constitute the centriolar barrel. More recently, the same approach confirmed previous results obtained in Chlamydomonas, showing the role of δ-tubulin in patterning the C-tubule of the centriolar triplets (Garreau De Loubresse et al., 2001). Finally, the functional complementation of a thermosensitive mutant affected in basal body duplication led to the discovery of η-tubulin (Ruiz et al., 2000).
In the course of a PCR screening aiming to identify potential new tubulins in the Paramecium genome, we cloned the ɛ-tubulin–encoding gene. This new tubulin type, first identified via a homology-based search in the human genome, was immunolocalized at the centrosome, but its function remained to be elucidated (Chang and Stearns, 2000). Here, we analyze the localization of ɛ-tubulin at the ultrastructural level and describe its functional analysis by gene silencing, which demonstrates its essential role in basal body assembly, consistent with a function of stabilizing the microtubule triplets.
Results
Sequence analysis
Paramecium ɛ-tubulin was identified in the course of a PCR screening aiming to characterize putative new tubulins in the Paramecium genome. “Tubulin signatures” were thus defined from the alignment of α-, β-, γ-, and δ-tubulin by choosing the most conserved motifs among these proteins. Degenerated primers were designed from two sequences both located in the tubulin NH2-terminal nucleotide binding domain: GGTGSG, common to all tubulins, and GQCGNQ, highly conserved in all tubulin types except for α-tubulins. PCR experiments initiated from these primers led to the identification of a partial ɛ-tubulin sequence; this fragment being subsequently used to probe a Paramecium genomic library (Keller and Cohen, 2000), allowing the cloning and the sequencing of the respective entire gene. Its deduced amino acid sequence proved to correspond to the Paramecium ɛ-tubulin, based on its respectively remarkable 50% and 43% identity with the human (GenBank/EMBL/DDBJ accession no. NP057346) and Trypanosoma (AAF32302) counterparts, compared with an average of 30% identity with any other tubulin, including those from Paramecium. Moreover, the alignment of this protein with other tubulins reveals that ɛ-tubulins (from human, Trypanosoma, and Paramecium) share common distinctive sequence features, which one would expect to have important functional implications (Vaughan et al., 2000; Inclan and Nogales, 2001). The salient characteristic of ɛ-tubulin, when aligned with other Paramecium tubulins (Fig. 1), lies in its large insertion, common to all ɛ-tubulins, whose position, with respect to the three-dimensional structure of the α/β dimer, corresponds to the “+ surface” of the protein and is therefore expected to confer specific properties in terms of longitudinal contacts between ɛ-tubulin and other tubulins. In contrast to this peculiar + surface, the − surface of the ɛ-tubulin, as previously reported (Inclan and Nogales, 2001; McKean et al., 2001), shares structural features with α-tubulins, including a conserved E in a position analogous to the α-tubulin catalytic E254, involved in the hydrolysis of β-tubulin GTP, which suggests a possible interaction between the − end of ɛ-tubulin and the + end of β-tubulin.
Figure 1.
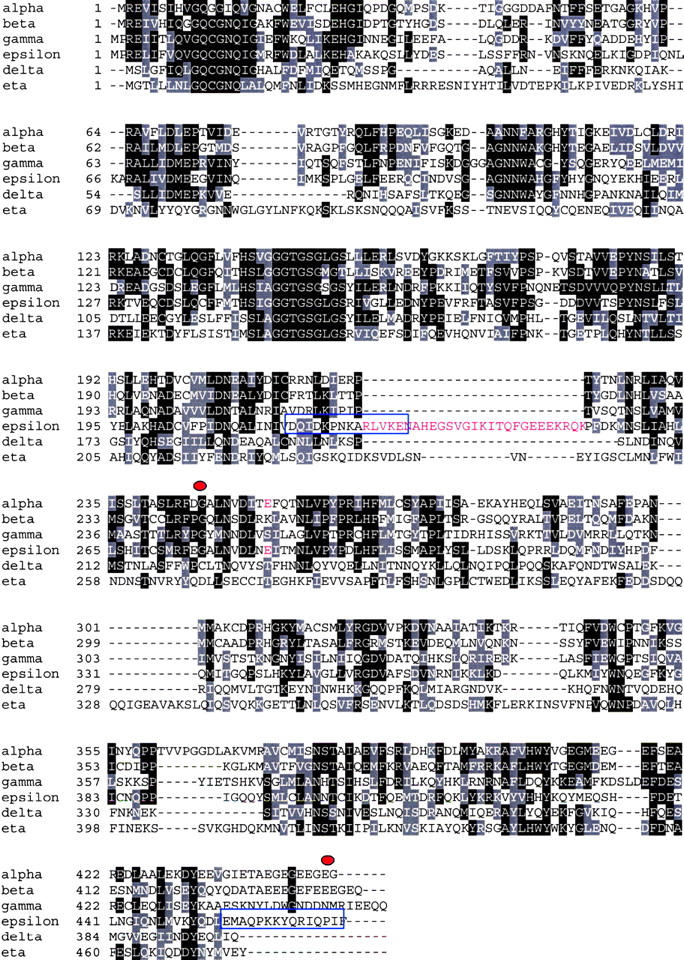
ɛ-Tubulin sequence features. The ɛ-tubulin was aligned along with the other Paramecium tubulins. Paramecium ɛ-tubulin shows the same characteristics as those from human and Trypanosoma, namely a large insertion (highlighted in red) whose position corresponds to the + end of the molecule with respect to the α/β-tubulin three-dimensional structure (Inclan and Nogales, 2001), as well as the presence of a glutamate (highlighted in red) in a position corresponding to the α-tubulin catalytic glutamate, involved in GTP hydrolysis at the E-site of β-tubulin. Blue boxes indicate the sequences of the peptides chosen to raise the polyclonal antibody. Red spots delimit the region used to trigger specific ɛ-tubulin RNAi in feeding experiments (see Results).
Immunolocalization of ɛ-tubulin
A polyclonal peptide antibody against ɛ-tubulin was generated by immunizing rabbits with two peptides designed from sequences highly specific to ɛ-tubulin (Fig. 1), namely the COOH terminus and the insertion, with the aim to minimize cross-reactions with other tubulins. The reactivity of the antibody, tested on blots of partially purified Paramecium cortices (see Materials and methods), revealed a single band of the expected size (theoretical molecular mass of 54 kD; unpublished data). The preincubation of the antibody with the two peptides resulted in the extinction of the signal, thus attesting the specificity of our anti–ɛ-tubulin antibody. Immunocytochemical experiments using the affinity-purified antibody displayed a labeling of the cortical and oral basal bodies (Fig. 2 a). Interestingly, no labeling could be detected in the nuclei, which are singular in that their division is driven by the development of intranuclear acentriolar spindles, thus suggesting that ɛ-tubulin is specifically associated with the centriolar structure. Similarly, the specificity of this immunolabeling of the basal bodies was verified by its extinction when the anti–ɛ-antibody was preincubated with the two peptides.
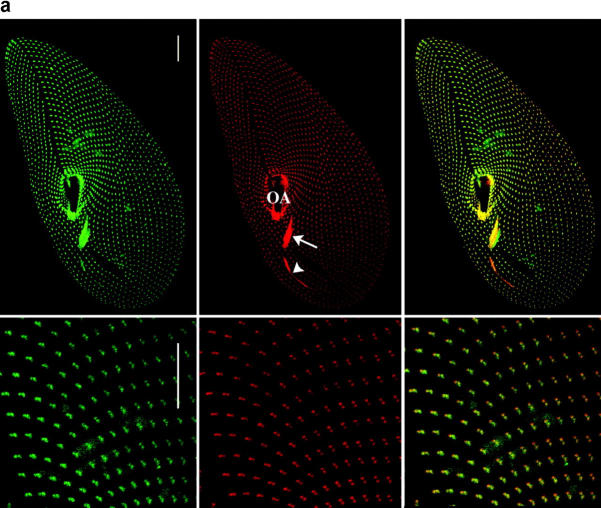
Figure 2.
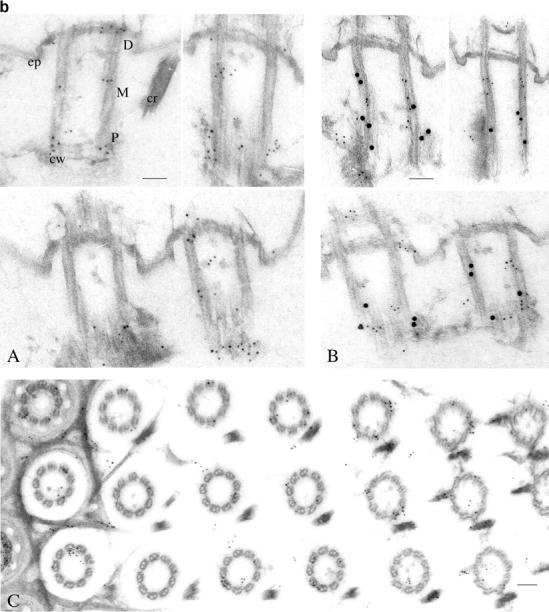
Cellular and ultrastructural localization of ɛ-tubulin. (a) The ɛ-tubulin is located at the basal bodies. The top panel displays projections of optical sections passing through the ventral side of a Paramecium cell, immunolabeled with the anti-ɛ (in green) and the ID5 (in red) antibodies as well as the merge of the two previous images (right panel). Bars, 10 μm. The anterior part of the cell is at the top of the picture. The cells are 120–140 μm long and 40–70 μm wide. The labeling with ID5 reveals the cortical single and paired basal bodies organized into parallel rows, those from the oral apparatus (OA; arrow) as well as the post-oral fiber (arrowhead), a massive microtubule bundle that emanates from the oral apparatus. The labeling with anti–ɛ-tubulin (in green) perfectly colocalizes with the basal body labeling (in red), as shown on the merge. In the bottom panel, this superimposition of the two labelings is confirmed on a higher magnification of the upper right part of the cell, where the paired basal bodies can be distinguished. Bars, 5 μm. (b) Ultrastructural localization of ɛ-tubulin. Immunogold electron microscopy of Paramecium cortices labeled with the anti–ɛ-tubulin/anti–rabbit 5-nm gold (A and C) or doubly labeled with the anti-ɛ/5-nm gold and ID5/10-nm gold (B). The distribution of ɛ-tubulin along the basal body length was analyzed by counting the 5-nm gold particles in the three respective regions shown in A: the proximal end (P), which contains the cartwheels (cw); the middle part (M); and the distal end (D), which protrudes through the epiplasm (ep). Statistical analysis demonstrated that although the ɛ-tubulin is distributed all along the basal body, it seems to accumulate at both ends of the structure (Table I). This distribution contrasts that of the 10-nm gold particles associated with antibody ID5, which are mostly localized along the P and M regions as shown in B and Table I. To analyze the localization of ɛ-tubulin further, cross sections (C) were examined in order to discriminate the 5-nm particles according to their position on versus in the vicinity of the microtubule sheath, and with respect to the section's level, namely through the regions P, M, and D. Panel C shows an oblique section through the oral apparatus where the regularly aligned basal bodies are crossed from their distal (left) to their proximal (right) end. This analysis showed that although ɛ-tubulin predominantly colocalizes with microtubules all along the basal body, it accumulates in the pericentriolar region, mostly associated with the fibrous matrix at their proximal end (see also A and B), but also at the edges of the epiplasm. Bars, 100 nm.
Double labeling experiments were performed using the anti–ɛ-tubulin antibody either with the ID5 anti–α-tubulin antibody, which specifically labels the basal bodies in Paramecium (Fig. 2 a), or with a monoclonal anti–acetylated α-tubulin (Callen et al., 1994), which reveals both basal bodies and cortical microtubule networks, such as the two short microtubule ribbons nucleated from each basal body (unpublished data). These experiments not only confirmed that the ɛ-tubulin labeling is restricted to the basal bodies but, in addition, the superimposition of the labelings with the anti–ɛ-tubulin and the ID5 suggested that ɛ-tubulin localizes at the core of the basal body.
To ascertain these results, the localization was studied at the ultrastructural level by postembedding immunostaining on isolated cortices (Fig. 2 b). The labeling with the anti–ɛ-tubulin antibody was unambiguously localized at the basal bodies and numerous gold particles (14 on average) were detected on longitudinal sections. To determine precisely the distribution of the ɛ-tubulin, the gold particles were counted on 52 longitudinal sections according to their position (proximal [P], middle [M], and distal [D]) along the cylindrical structure (Fig. 2 b, A). The counting, presented in Table I, revealed a heterogeneous distribution of the gold particles; respectively, 46, 17, and 37% in the proximal, middle, and distal parts of the basal body. When submitted to statistical analysis, these results demonstrated that the predominant localization of ɛ-tubulin at both ends (versus the middle part) of the centriolar structure is significant. Moreover, double labeling experiments demonstrated that this distribution contrasts with that of the antibody ID5 (expected to recognize the glutamylated α- and β-tubulins; Rudiger et al., 1999), which is homogeneously distributed along 2/3 of the proximal side of the basal bodies (Table I; Fig. 2 b, B).
Table I.
Statistical analyses of ɛ-tubulin and ID5 labeling on longitudinal sections of basal bodies
| ID5a | anti–ɛ-tubulinb | |||
|---|---|---|---|---|
| M | SD | M | SD | |
| Basal body | 5.4 | 2.0 | 14.3 | 4.5 |
| Proximal part | 2.4 (44%) | 1.8 | 6.6 (46%) | 3.9 |
| Middle part | 2.5 (46%) | 1.8 | 2.4 (17%) | 2.5 |
| Distal part | 0.5 (10%) | 1.0 | 5.4 (37%) | 3.3 |
The gold particles associated, respectively, with ID5 and anti–ɛ-tubulin were counted by dividing the sections into three zones: proximal, middle, and distal. M, average number of gold particles.
n = 40 (n, number of sections examined).
n = 52.
The counting was also performed on the cross sections to assess the localization of the ɛ-tubulin with regard to the microtubule sheath. The cross sections (n = 94) were analyzed by counting the gold particles situated, respectively, on or in the vicinity of the microtubules, and with respect to their position along the cylinder (proximal, middle, and distal). The results (Table II; Fig. 2 b, C) showed that although the labeling situated on the microtubules does not vary over the length of the basal body (∼5 particles/cross sections on average), the labeling in their vicinity was significantly higher at the proximal and distal ends (on average 3.6 and 3.9 vs. 1.9 for the middle part). In conclusion, although the ɛ-tubulin is predominantly associated with the microtubules over the entire length of the centriolar structure, it accumulates at the pericentriolar material at both ends of the basal body, where it seems to be associated, respectively, with the fibrous material at the proximal end and the epiplasm at the distal end.
Functional analysis by gene silencing
The functional role of ɛ-tubulin in cellular processes was investigated by gene silencing. This phenomenon, conserved across phyla, can be induced by introducing DNA (transgene-induced gene silencing) or double-stranded RNA (RNA interference, first described in Caenorhabditis elegans; Fire et al., 1998) into a cell, which both lead to the sequence-specific degradation of endogenous homologous RNAs, and thus inactivation of the corresponding resident gene (Cogoni and Macino, 2000; Carthew, 2001). Such homology-dependent gene silencing can be induced by two different methods in Paramecium, microinjection and feeding, which both proved to efficiently trigger specific RNA degradation and subsequent gene silencing. The first method, which has been used routinely for functional analysis in Paramecium over the past 5 yr (Ruiz et al., 1999; Froissard et al., 2001; Garreau De Loubresse et al., 2001; Krzywicka et al., 2001; Vayssié et al., 2001), relies on the transformation of the somatic nucleus with supernumerary copies (typically 20 genome equivalents) of a nonfunctional version (depleted of its 3′untranslated region) of the gene to be silenced (Ruiz et al., 1998; Galvani and Sperling, 2001). These extragenic copies, replicated as minichromosomes throughout the successive somatic divisions, were shown to produce aberrant RNAs, which are subsequently processed into double-stranded RNAs (Galvani and Sperling, 2001). The second method, feeding, recently adapted from the one used for the nematode (Timmons and Fire, 1998), relies on the feeding of the paramecia with bacteria engineered to produce double-stranded RNAs from a plasmid containing the gene to be silenced (Galvani and Sperling, 2002). In both methods, the specificity of genetic interference was attested by the observation of an induced loss-of-function phenotype of the corresponding gene, provided that the homology between the introduced sequence and the resident gene was >85%.
In a first series of experiments, ɛ-tubulin silencing was thus induced by microinjecting into the macronucleus a highly concentrated solution of the cloned ɛ-tubulin restricted to its open reading frame (see Materials and methods). Six independent silencing experiments were performed, totaling ∼150 injected cells with an 80% average transformation efficiency. After microinjection, cells were individually collected and placed at 27°C in a fresh culture medium; their growth rate was recorded and the phenotype of their progeny compared with those of control cells (noninjected cells or injected with the cloning plasmid) grown in similar conditions. For each clone, the transformation was attested by PCR amplification designed specifically to detect the presence of the extragenic ɛ-tubulin in the progeny of the microinjected cells.
A second series of experiments took advantage of the feeding method to assess that the phenotypical effect was mediated by the sole genetic interference of the ɛ-tubulin gene. Two alternative recombinant plasmids were introduced in the bacteria: one carrying the entire ɛ-tubulin open reading frame and another that carried only its 3′ half whose nucleotide identity with other tubulins is <45% and that is devoid of any “tubulin” conserved motif (Fig. 1). The latter construct aimed to preclude any cross-interference with other tubulins. Feeding experiments corresponding to the two constructs were performed with, respectively, 110 and 21 Paramecium cells. In both cases, all the cells of the progeny turned out to be affected, when compared with control cells (see Materials and methods) with the same altered phenotype as that issued from microinjection experiments, as judged by morphological observation and immunocytochemical studies.
In each set of experiments, the phenotype induced by ɛ-tubulin silencing was highly reproducible, leading to the same progressive deterioration in the cell's morphology and corresponding basal body disorganization, described as follows. After three to four divisions, which is the time expected for the silencing to take place, cells from the transformed clones differed from the control cells by their less tapered shape and abnormal swimming. This phenotype then intensified along the next three to four divisions before death; the cells getting smaller and adopting a rounded shape and a swirling swimming. Because these characteristics were collectively suggestive of an impaired basal body duplication (Ruiz et al., 1987), an immunocytochemical analysis of the transformed cells was undertaken in order to reveal putative defects in the basal body patterning (Figs. 3–6).
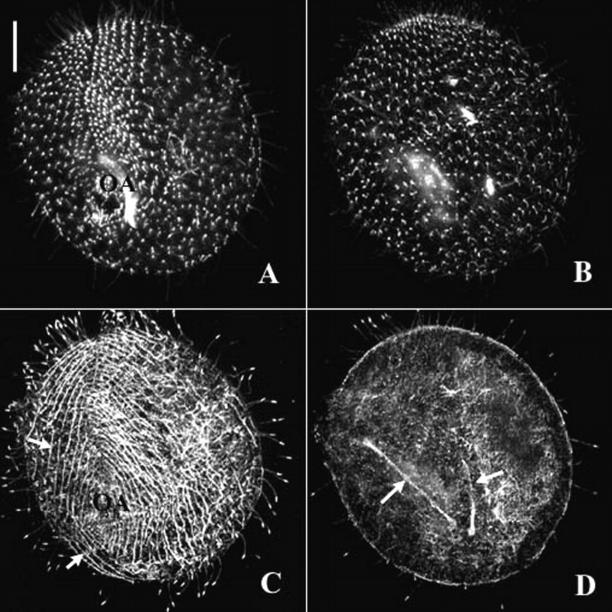
Figure 3.
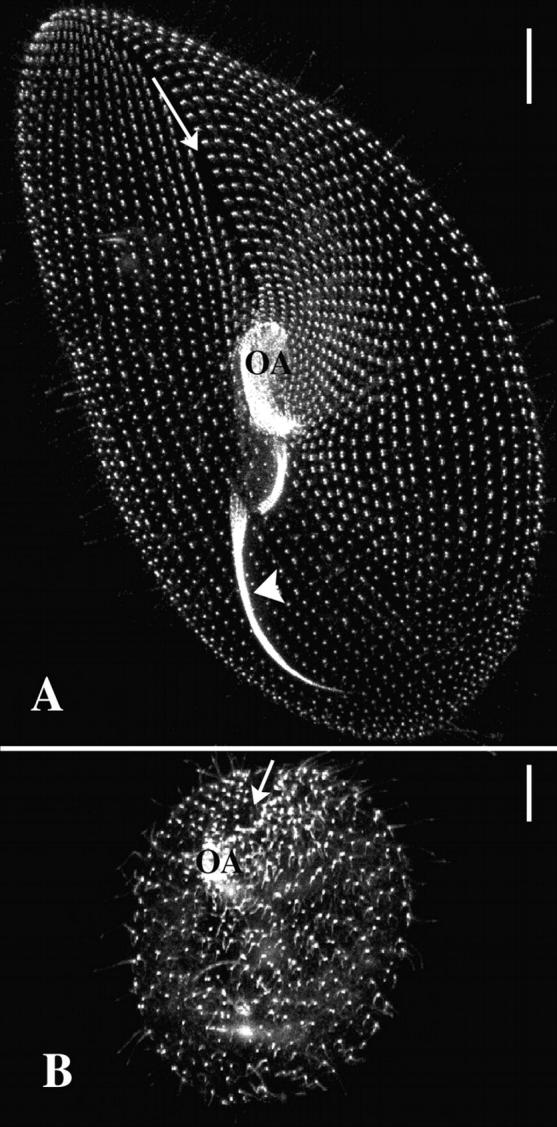
The silencing of the ɛ-tubulin gene blocks basal body duplication. Projections of optical sections passing through the ventral side of a control cell (A) and a silenced cell (B), immunolabeled with the ID5 antibody. Bars, 10 μm. In the control cell, the labeling with ID5 highlights the single and paired basal bodies patterned into anteroposterior curved rows that converge toward a meridian (arrow) where the oral apparatus (OA) is localized on the ventral side. Highly affected silenced cells (B) exhibit a round shape, a severely reduced number of basal bodies whose pattern is strongly affected. On the ventral side, single basal bodies are loosely patterned into rows, which do not converge toward the ventral meridian (arrow), in which a vestigial oral apparatus (OA), built up with few basal bodies, is detected.
Figure 6.
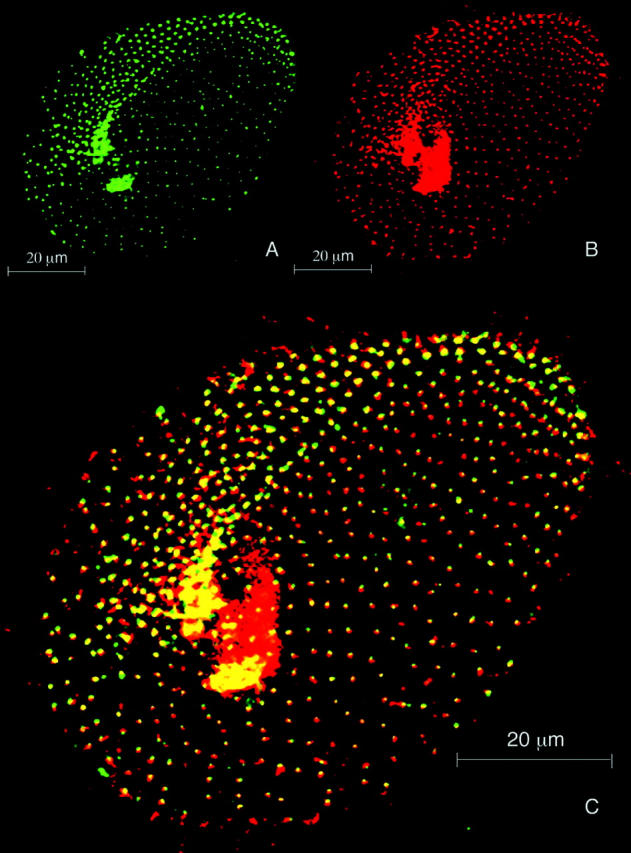
The amount of ɛ-tubulin in the basal bodies is reduced by gene silencing. Projections of optical sections passing through the ventral side of a silenced cell doubly labeled with the anti–ɛ-tubulin (A) and ID5 antibodies (B). (C) Merge of images A and B. The ID5 antibody reveals the altered cortical basal body pattern typical of the ɛ-tubulin–silenced cells, as well as the numerous basal bodies in the oral apparatus, which appear as a red patch. In this anterior division product, two regions can be distinguished: the invariant field, constituted with paired basal bodies, which is directly inherited from the mother cell and therefore unaffected by silencing, and the rest of the cortex, where the single basal bodies are issued from duplication events that occurred during gene silencing. The uniform labeling of the basal bodies by ID5 contrasts that obtained with the anti–ɛ-tubulin antibody, whose intensity is strongly reduced in the single basal bodies, attesting that ɛ-tubulin silencing resulted in the depletion of ɛ-tubulin in basal bodies.
In Paramecium, some 4,000 basal bodies are arranged according to a very precise pattern that has been extensively studied and mapped (Iftode et al., 1989). Their distribution is (Fig. 3 A) 3/4 on the cell cortex, where they are regularly spaced and strictly aligned into longitudinal rows, and 1/4 in the oral apparatus, where they are clustered into a precise pattern. This spatial organization of the basal bodies is essential to the coordinated beating of the associated cilia, and consequently for the cell's swimming, feeding, and thus survival. Its inheritance through cell division depends on the spatial and temporal control of the coordinated duplication of the basal bodies. During each individual duplication, the new basal body is strictly positioned along the ciliary row, anteriorly to the parental basal body, thus perpetuating the longitudinal organization. At the whole cell level, basal body duplication bears on cell elongation, morphogenesis and cell's swimming, and ensures the formation of the oral apparatus of one of the daughter cells (Iftode et al., 1989). Because of this very precise scenario, any failing in basal body duplication can be easily spotted by morphological observations and simple immunostaining experiments.
Basal bodies from control and transformed cells were immunorevealed with the ID5 anti–α-tubulin antibody (Rudiger et al., 1999), which specifically labels the basal bodies. When applied to the most severely affected cells (Fig. 3), this analysis confirmed that basal body duplication was affected, as attested by a strong reduction in cortical basal body number, the increased spacing between basal bodies, and the near absence of the oral apparatus in most of the cells. Moreover, in these cells, the remaining cortical basal bodies appear highly disorganized. Because the alignment of basal bodies into parallel rows rests on the duplication process itself, this suggests that the duplication has been inherently affected in the absence of ɛ-tubulin. These cells were also doubly labeled using ID5 and an anti–Paramecium α-tubulin, which binds to the most dynamic Paramecium microtubule networks (Dupuis-Williams et al., 1996), including those developed during division, such as the cortical cytospindle or the micronuclear division spindles (Fig. 4, C and D). These double-labeling experiments revealed an uncoupling of basal body duplication and cell cycle progression, as the complete inhibition of basal body duplication (as revealed by the ID5 immunolabeling; Fig. 4, A and B) does not interfere with an apparently appropriate setting up of the cortical and nuclear division microtubule networks. This also confirms that the failure in basal body duplication directly results from ɛ-tubulin depletion rather than being an indirect consequence of an impaired cell division and that ɛ-tubulin is not involved in the development of the nuclear division spindles, which are acentriolar in Paramecium.
Figure 4.
ɛ-Tubulin depletion does not affect cell cycle progression. Projection of optical sections passing through the ventral side (A and C), the dorsal side (B), and the interior (D) of a dividing silenced cell doubly immunodecorated with ID5 (A and B) and anti–Paramecium α-tubulin (C and D) antibodies. Bars, 10 μm. The ID5 labeling of the basal bodies shows that the ventral side of this silenced cell is similar to that described in Fig. 2 C, with no sign of basal body duplication on the ventral (A) or dorsal (B) cortices, as this would normally occur during the division morphogenesis. By contrast, transient microtubular networks specific for division morphogenesis are detected with the antitubulin labeling; a superficial set microtubular bundle on the ventral side (C, arrows) and the micronuclear spindles inside the cell (D, arrows).
To further characterize the primary effects of ɛ-tubulin silencing, “earlier phenotypes” were analyzed by collecting the cells to be immunostained at earlier steps with respect to the beginning of the silencing: starting from the first delayed division, before which no phenotypical defect could be detected on living cells and therefore considered as the silencing starting point, until the next second or third division. The immunocytochemical analysis of these cells revealed that the inhibition of basal body duplication seems to occur progressively along divisions, as attested by the only scarce defects detected in cells issued from the first division after the starting point, corresponding to localized missing or mispositioned basal bodies both on the cortex and in the new oral apparatus. These defects then amplify during the two next divisions, as seen in the cells shown in Fig. 5, whose immunostaining with ID5 reveals a clear reduction in basal body number and a disorganization of the basal body pattern, both on the cortex and in the newly formed oral apparatus. Once more, these cells are characterized by the progressive loss of basal body alignment, as attested by the existence of incomplete or unparallel ciliary rows or by the presence of basal bodies positioned outside of these rows.
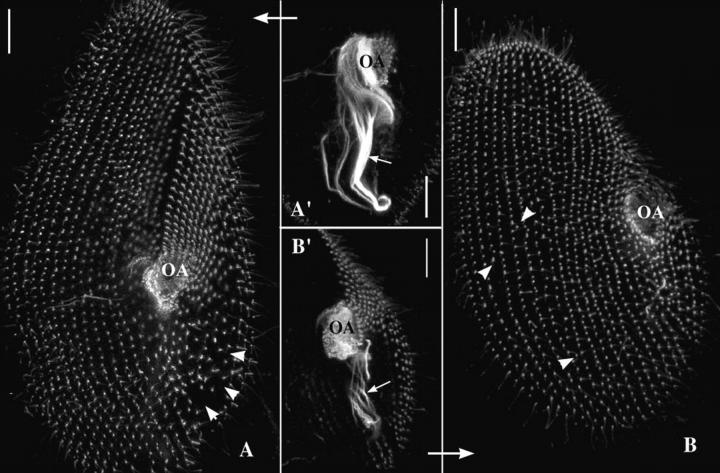
Figure 5.
Early phenotype of ɛ-tubulin gene silencing. To define better the early effects of ɛ-tubulin silencing on basal body pattern, the cells were collected as soon as the first cytological affects were visible and immunolabeled with ID5 to reveal the organization of cortical and oral cortices. A and B are two projections of optical sections passing through the ventral side of two division products: anterior (A) and posterior (B). A′ and B′ are projections of optical sections passing through the oral apparatuses of these two cells, respectively. Bars, 10 μm. During division, the anterior cell inherits the cortical anterior half as well as the oral apparatus from the parental cell, while its posterior half is transmitted to the posterior cell whose oral apparatus is newly assembled. Basal body proliferation not only replicates the characteristic distribution of cortical basal bodies in the two daughter cells but also drives the morphogenesis of the posterior third of the anterior cell and the anterior two thirds of the posterior cell. Consequently, the respective phenotypes of cells A and B may be precisely interpreted because the impairment of basal body duplication differently affects the two division products. Accordingly, the lower part of cell A and the upper part of cell B both appear less tapered. In A, the anterior part of the cell and the oral apparatus (A′) are nearly normal, whereas a strong depletion in basal bodies is noticeable in the lower right quarter of the cell (arrowheads). Conversely, cell B is characterized by a reduced anterior part and a strongly reduced oral apparatus (B′, OA). Arrow, post-oral fiber. Although reduced, the basal body duplication is not completely blocked at these stages after silencing. However, the defective alignment of the cortical basal bodies, particularly clear on cell B (arrowheads indicate basal bodies inserted between two rows and incomplete rows), attests that ɛ-tubulin silencing intimately affects the duplication process.
To check that these anomalies were consecutive to the ɛ-tubulin depletion, the silenced cells were immunostained with the anti–ɛ-tubulin antibody, showing that the signal was reduced in these cells but also that the progressive reduction of the labeling correlated with the number of basal body duplications (Fig. 6). In conclusion, if ɛ-tubulin silencing eventually leads basal body duplication to cease, the preceding steps likely correspond to a partial depletion of ɛ-tubulin, and the initial misorientation of the duplication process likely reveals structural anomalies of the neoformed basal bodies under these conditions. This contrasts with the drastic halt of the basal body duplication induced by γ-tubulin silencing (Ruiz et al., 1999), suggesting that ɛ-tubulin could be involved at a later stage of basal body biogenesis.
To analyze the consequences of ɛ-tubulin silencing further, the transformed cells were analyzed at the ultrastructural level by electron microscopy, aiming to detect putative defects in the organization of basal bodies or their associated appendages. In addition to the canonical centriolar structure shaped by the layout of nine microtubule triplets in an axial symmetry, the basal bodies in Paramecium and in all ciliates are characterized by three appendages (one ciliary rootlet and two microtubule bundles) that are nucleated on specific triplets of the basal body, with respect to the anteroposterior axis of the cell (Fig. 7 A; Beisson and Jerka-Dziadosz, 1999). Many structural anomalies in the basal bodies of the transformed cells could be detected that predominantly corresponded to erratic missing microtubules among the nine triplets; the anomalies ranged from one or two missing outer microtubules in one or several triplets to the lack of several microtubule sets (Fig. 7, A′, B′, and C′). The occurrence of these defects was appreciated by counting the respective defects on 58 cross sections. Globally, the occurrence and the intensity of these structural defects correlated with the severity of the corresponding cell's phenotype. These damages did not seem to interfere directly with the development of the appendages that mostly appear (for those whose nucleation point was visible on the pictures) to be nucleated in respective appropriate positions of the defective basal bodies. In some instances, however, some defects concerning the shape and point of nucleation of the ciliary rootlets or the length and orientation of the microtubule rootlets could be noted, which were interpreted as a consequence of the loss of the C-tubule (Garreau De Loubresse et al., 2001).
Figure 7.
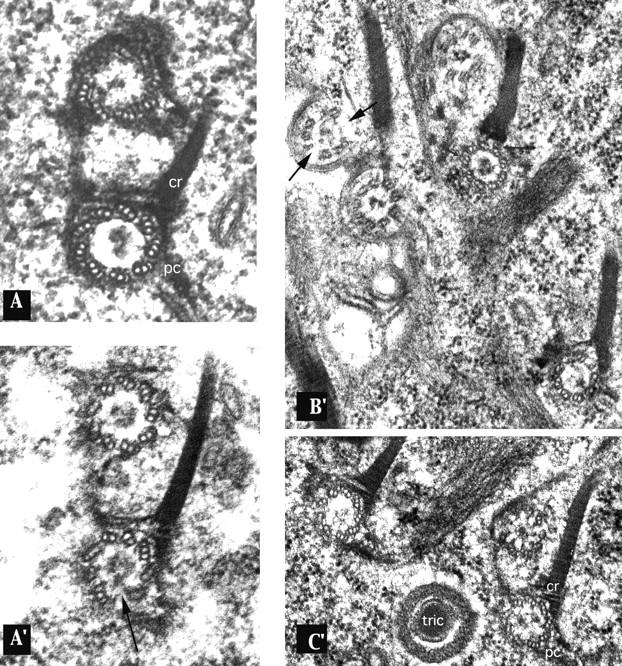
Basal bodies in the silenced cells display ultrastructural anomalies. Cross sections of basal bodies from a control cell (A) and silenced cells (A′, B′, and C′), whose phenotypes correspond to those reported in Fig. 3 or 5. (A) Control pair of basal bodies with the typical axial organization of its nine microtubule triplets. Two appendages of the parental basal body are visible: the ciliary rootlet (cr) (nucleated on triplets 5, 6, and 7) and the postciliary microtubule ribbon (pc) (nucleated on triplet 9). All of the basal bodies from the silenced cells (A′, B′, and C′) display ultrastructural anomalies in the organization of the microtubule triplets, ranging from a missing C-tubule in one or several triplets to the lack of entire triplets (black arrows). Another interesting observation, even less frequently noticed, is the presence of displaced microtubules, or alternatively of “ectopic” microtubules, which seem to be disconnected from the triplets (indicated by small arrows). Two facts may be pointed out: (1) the appendages, whose shape and length may be abnormal, are nevertheless nucleated on the respective correct triplets; and (2) the anomalies in the structural organization of the microtubule triplets are not inherited through duplication, as attested by A′, where the new basal body did not replicate the defects of its parent (identifiable by its appendages).
Among all the defects found on cross sections of basal bodies and cilia, a decreasing gradient of severity could be noted from the proximal side (cortex) to the distal side (cilium) of the basal body. Thus, although most of the proximal structures (36/37 cross sections) appeared highly affected, the distal part disclosed slighter damages (14 abnormal versus 7 normal cross sections), and the rare cilia's cross sections were essentially normal. This gradient was also noticeable in terms of the nature of the structural defects; whereas the most proximal sections (at the level of the cartwheels) had most of their B- and C-tubules missing, distal sections displayed a more or less extended lack of solely C-tubules, the erratic loss of entire triplets being noticed all along the basal body. This analysis suggests that the ɛ-tubulin depletion acts primarily on the assembly of the B- and C-tubules, which inhibits the elongation of the most affected microtubule structures. Thus, the rarer distal sections that could be examined are those that retain a stable configuration, namely those missing only the C-tubules or one or two triplets, which do not compromise the elongation or stabilization of the entire microtubule scaffold.
Discussion
ɛ-Tubulin, which was initially identified in mammals (Chang and Stearns, 2000), appears, since its characterization in diverse protist lineages (Vaughan et al., 2000; Dutcher, 2001), to be an evolutionarily ancient protein, and therefore, like other tubulins, conceivably involved in conserved functions. The first clue about its functional role came from its localization in the human centrosome (Chang and Stearns, 2000). Interestingly, the systematic searching for homologues was noticeably unsuccessful in the genomes devoid of canonical centrioles, such as Arabidopsis, Caenorhabditis, or even yeast, whose duplication of the spindle pole body nevertheless display some similarity with that of the centriole, suggesting that this protein could be involved in properties peculiar to the centriolar structure, such as the remarkable organization of its microtubules into triplets or their arrangement in a ninefold symmetry. A second indication of its possible function came from a predictive analysis from Inclan and Nogales (2001) of the ɛ-tubulin conformation, based on the comparison of its sequence with those of α/β-tubulins. By virtue of the high similitude of its − surface with α-tubulin, as well as its very peculiar + end conformation, these authors postulated that ɛ-tubulin is susceptible to interact with the + end of the microtubules, but that its incorporation would further influence any longitudinal elongation with α/β dimers.
Our functional analysis in Paramecium demonstrates that ɛ-tubulin is necessary for the assembly or stabilization of the canonical centriolar structure. The ultrastructural defects of the basal bodies in the silenced cells consist in the more or less extended loss of microtubules among the triplets, mostly concerning the C- and B-tubules. The erratic nature of the microtubule loss, with incomplete triplets neighboring unaltered triplets, very likely reflects an incomplete depletion of the ɛ-tubulin at the time of basal body assembly. This is supported by the ɛ-tubulin immunolabeling of the basal bodies of the silenced cells (Fig. 6), which shows that its intensity is progressively reduced throughout divisions until the cells have reached their final syndrome when they are unable to duplicate their basal bodies any further (Fig. 3), thus signaling the exhaustion of the cellular pool of free ɛ-tubulin. This implies that basal body duplication requires ɛ-tubulin, which is likely to be involved specifically in the assembly of the B- and C–tubules; the rarer loss of the A-tubule being imputable to a subsequent destabilization, as previously observed after δ-tubulin silencing (Garreau De Loubresse et al., 2001). Interestingly, these structural defects are virtually identical to those observed in the Chlamydomonas BLD2RGN1 double mutant; their basal bodies have a mixture of no, singlet, doublet, and triplet microtubules, with more dramatic ultrastructural defects being localized at their proximal end (Preble et al., 2001). The comparison of the two systems is particularly interesting because it has been proposed recently that ɛ-tubulin could be the bld2 gene product (Dutcher et al., 2001). Thus, in Chlamydomonas, the likely partial rescue by rgn1 (whose function remains to be elucidated) of the ɛ-tubulin mutation bld2 mimics our incomplete depletion of ɛ-tubulin by gene silencing, and the apparent lethality of the null allele of bld2 confirms our observations that the complete exhaustion of ɛ-tubulin inhibits basal body duplication.
This essential role of ɛ-tubulin in the assembly of the B- and C-tubules, as well as its colocalization with microtubules all along the basal body length, suggests that ɛ-tubulin is structurally incorporated in the microtubule triplets. This hypothesis is consistent with the structural predictions of ɛ-tubulin (Inclan and Nogales, 2001) and its coimmunoprecipitation with α- and β-tubulins (Chang and Stearns, 2000), which both suggested a direct interaction of ɛ-tubulin with microtubules. Indeed, it is highly conceivable that the centriolar microtubules, with this peculiar arrangement in triplets, require specialized tubulins for their assembly. In particular, the unusual conformation of the hemitubules B and C suggests that either their nucleation or their “ribbon-shaped” elongation (which involves the branching and closing onto the next tubule circumference) requires specific tubulins with divergent structural motifs to allow these particular interactions between microtubules. By its regular distribution along the length of the microtubule triplets, ɛ-tubulin is expected to link, during their elongation, the hemitubules B and C with their neighbors. Moreover, both cosedimentation experiments (Chang and Stearns, 2000) and the normal γ-tubulin immunolabeling of the basal bodies obtained with the ɛ-tubulin–silenced cells (unpublished data) seem to rule out a direct interaction between ɛ- and γ-tubulin, which clearly distinguishes ɛ-tubulin from δ-tubulin or η-tubulin, whose putative interactions with γ-tubulin indicate that they are involved in microtubule nucleation (Ruiz et al., 2000; Smrzka et al., 2000). Finally, taking into account that the anchorage of the basal body in the cortex and the microtubule elongation are concurrent events, it is not surprising that the ɛ-tubulin silencing affects basal body positioning.
Lastly, our hypothesis about the presence of ɛ-tubulin in the pericentriolar material at both ends of the basal bodies is that this corresponds to the pool of free ɛ-tubulin, which is obviously needed at the site of its polymerization. Consistent with its putative role in the particular shaping of the microtubule triplets, the ɛ-tubulin is expected to be precisely directed (under an inactive state) to the site of the assembly of the basal bodies or centrioles. This is in agreement with our immunocytochemical data that display a very restricted localization of the ɛ-tubulin at the basal bodies, contrasting with that of the γ-tubulin, whose abundant soluble pool in the cytoplasm appeared as a high fluorescent background (Ruiz et al., 1999). Alternatively, the pericentriolar ɛ-tubulin could, via the interaction with other proteins, be involved in the anchorage of the triplets on the cortical structures, as suggested by the mispositioning of the basal bodies in the silenced cells. Consistent with both hypotheses, we recently identified several isoforms of the ɛ-tubulin, probably accounting for posttranslational modifications, which could modulate its interaction with different partners. The further identification of the posttranslational modifications of ɛ-tubulin and its molecular partners will hopefully help to decipher the assembly of this yet mysterious centriolar microtubule scaffold.
Materials and methods
Cells and culture conditions
The wild-type cells of stock d4-2 is a derivative of the wild-type stock 51 of P. tetraurelia (Sonneborn, 1974). The cells were grown at 27°C in a buffered infusion of wheat grass powder (Pines International Co.), supplemented with 0.4 μg/ml β-sitosterol and inoculated with Klebsiella pneumoniae.
DNA purification
Exponentially growing paramecia were pelleted and taken up to 50°C for at least 4 h in a lysis buffer (0.5 M EDTA, pH 9, 1% SDS, and 1 mg/ml proteinase K [Merck]). After three phenol extractions, the mixture was ethanol precipitated and resuspended in Tris 10 mM, EDTA 1 mM.
Molecular characterization of the tubulin genes
To identify tubulin sequences, PCR amplifications were initiated on genomic DNA using degenerated primers whose sequences (respectively, AAGKWGGWYAATGYGGWAAYYAA and CCWSWWCCWGTWCCWCCWSC) were designed from the most conserved regions among α-, β-, γ-, and δ-tubulins. The amplified fragments were then cloned in a pGEM-T plasmid and sequenced. The three newly identified fragments were then used as probes to screen an indexed genomic library (Keller and Cohen, 2000); the positive clones were then isolated, allowing the subsequent cloning and sequencing of the respective entire genes.
Gene silencing
DNA microinjection.
As previously demonstrated, a specific gene can be silenced by microinjecting large quantities of its coding sequence in the Paramecium macronucleus (Ruiz et al., 1998). In most of the experiments, gene silencing was induced by the microinjection into the macronucleus of the ɛ-tubulin coding sequence, cloned into a pGEM-T plasmid. Alternatively, PCR amplifications of the coding sequence were used, leading to the same phenotypes; the extent and the time of appearance of the phenotype being the two conditions mostly dependent on the quantity of DNA microinjected.
Feeding.
Sequences of interest were cloned into the L440 feeding vector between two T7 promoters (Timmons and Fire, 1998). The resulting constructs were used for transformation of an RNase III–deficient strain of Escherichia coli with an IPTG-inducible T7 polymerase, HT115 (DE3) (Timmons et al., 2001). From 1 to 10 cells were incubated in double-stranded RNA–expressing bacteria, as previously described (Galvani and Sperling, 2002). Phenotypes were screened after 48–72 h of feeding. Several control experiments were done by feeding the paramecia with three different bacteria: (1) HT115 carrying no plasmid; (2) HT115 carrying the L440 plasmid without insert; or (3) HT115 carrying the same construction as that of the experiment but without induction by IPTG. In each case, ∼30–50 cells were grown during >20 divisions without any alteration of their growth rate or morphology. The specificity of the silencing phenotype was attested by other feeding experiments performed on similar conditions with other tubulin genes (γ or θ), whose effects on cell growth and basal body duplication demonstrated that the phenotypical effects induced by the different tubulins were clearly distinguishable.
Antibodies and immunofluorescence
The following antibodies were used. The monoclonal antibody ID5 (dilution 1/10) raised against the 14 COOH-terminal amino acids of the detyrosinated pig brain tubulin (Wehland et al., 1984), which allows a precise observation of the pattern of basal bodies both on the cell cortex and in the oral apparatus. The polyclonal anti–Paramecium α-tubulin R200 directed against the 12 COOH terminus amino acids of the αPT1 gene product, previously shown to label the most dynamic microtubule networks in Paramecium, including those developed during cell division (Dupuis-Williams et al., 1996). To generate the polyclonal anti–ɛ-tubulin antibody 2942, two peptides, EP305 and EP306, whose respective amino acid sequences are DQIDKPNKARLVKENC and CEMAQPKKYQRIQPIF, were conjugated with KLH and injected simultaneously into rabbits following the immunization protocol “double X” from Eurogentec. The anti–ɛ-tubulin serum was immunopurified against the two peptides that had been transferred on nitrocellulose (Harlow and Lane, 1988) or on microchromatographic columns (Eurogentec). The secondary antibodies were from Molecular Probes.
Paramecium cortices were prepared according to Stelly et al. (1991). The crude pellets were loaded on 12.5% SDS-PAGE and subsequently transferred on nitrocellulose filters (Millipore) on a semidried system (Bio-Rad Laboratories). Immunological labeling of blots were made in PBS/Tween 0.1% with the immunopurified anti–ɛ-tubulin antibody diluted 1/20. The secondary anti–rabbit antibody was diluted 1/2,500. The immunoreaction was revealed with the ECL kit from Amersham Biosciences. The specificity of the reaction was ascertained by extinction experiments where the antibody was primarily incubated with 20 μM of both peptides.
For immunofluorescence, cells were first permeabilized for 3 min in 1% Triton X-100 in PHEM buffer (Schliwa and Van Blerkom, 1981) and rinsed twice in PBS, 3% BSA before being processed through primary and secondary antibodies. The ID5 antibody was diluted 1/10, the R200 1/400, the anti–ɛ-antibody 1/200, and the corresponding affinity-purified fraction 1/10. The specificity of the reaction with the antibody 2942 was attested by its extinction when the antibody was incubated with 1 μM of both peptides. After two washes in PBS/BSA, the cells were mounted in Citifluor (Citifluor, Ltd.) and analyzed alternatively with a Bio-Rad Laboratories MRC1024ES at the Service d'Imagerie Cellulaire in Orsay or with a Leica TCS-NT confocal microscope.
Ultrastructural analysis
The silenced cells were individually collected and fixed in 2% glutaraldehyde in 0.05 M cacodylate buffer, pH 7.2, for 90 min at 4°C. After washing in the same buffer, the cells were postfixed in 1% osmium tetroxide in 0.05 M cacodylate buffer for 60 min at 4°C. Postfixed cells were dehydrated by passage through a series of ethanol and propylene oxide baths before embedding in Epon. Thin sections were contrasted with ethanolic uranyl acetate and lead citrate, and finally examined using a Philips EM410. For postembedding immunolocalization, cortices (Klotz et al., 1997) were fixed in 2% paraformaldehyde, 0.15% glutaraldehyde in 0.05 M cacodylate buffer, pH 7.35, at room temperature for 2 h. After washing in the same buffer, cells were dehydrated by passage through a series of ethanol baths before embedding in LRW (London Resin Ltd.). Thin sections were collected on nickel grids and saturated and processed with 3% BSA in PBS. The affinity-purified anti–ɛ-tubulin and the ID5 antibodies were used at respective dilutions of 1/15 or 1/10. After washing, the sections were incubated with 5-nm colloidal gold–conjugated anti–rabbit immunoglobulins (GAR G5; Amersham Biosciences) at a 1/100 dilution. For double labeling experiments, 10-nm gold-conjugated anti–mouse immunoglobulins (GAM G10; Amersham Biosciences) diluted 1/25 were used to reveal the ID5 labeling. After extensive washing, the sections were contrasted with ethanolic uranyl acetate and examined in an EM Philips 410. It has to be noted that in these experiments, the fixation procedure was optimized for antibody recognition, at the expense of ultrastructure recognition.
The specificity of the anti–ɛ-tubulin antibody labeling was attested to by the absence of any signal in control experiments where the sections were incubated in similar conditions alternatively (a) without any primary antibody (in a mixture of GARG5 at 1/40 and GAMG10 at 1/20) and (b) with the preimmune serum diluted 1/200. Moreover, preincubation of the anti–ɛ-tubulin antibody with the corresponding peptide was shown to reduce the signal detected on the basal bodies, proportionally with the concentration of the peptide. Thus, the signal was reduced to the level of background signal when the antibody, used in the same conditions, had been preliminarily incubated with 1 mM peptide. For the immunolocalization of ɛ-tubulin on the basal body, gold particles were counted on individual basal bodies both on simple labeling with the anti–ɛ-tubulin antibody, or on double labeling with anti-ɛ and ID5, after having verified that the distribution was equivalent in the two conditions. The counting was done on selected images of complete longitudinal sections divided into three parts (proximal, middle, and distal) or on cross sections by distinguishing the gold particles in relation to their position on versus in the vicinity of the microtubule wall. Systematic statistical analysis (t test) was applied to the gold particle count to check the significance (99% confidence) of the respective labeling distributions.
GenBank/EMBL/DDBJ accession nos.
The GenBank/EMBL/DDBJ accession no. for Paramecium ɛ-tubulin is AJ427411.
Table II.
Statistical analyses of ɛ-tubulin labeling on cross sections of basal bodies
|
|
Anti–ɛ-tubulin
|
|||||
|---|---|---|---|---|---|---|
| Proximala | Middleb | Distalc | ||||
| M | SD | M | SD | M | SD | |
| Cross section | 9.0 | 4.3 | 7.7 | 5.0 | 9.6 | 4.6 |
| On microtubules | 5.4 (60%) | 3.5 | 5.8 (75%) | 3.4 | 5.7 (60%) | 4.2 |
| Around microtubules | 3.6 (40%) | 3.6 | 1.9 (25%) | 2.7 | 3.9 (40%) | 2.6 |
Gold particles associated with anti–ɛ-tubulin were counted by distinguishing their position on versus on the side of the microtubules in the three zones: proximal, middle, and distal. M, average number of gold particles.
n = 39 (n, number of sections examined).
n = 34.
n = 21.
Acknowledgments
We dedicate this article to the memory of André Adoutte (Centre de Génétique Moléculaire), his outstanding contributions to ciliate morphogenesis, and his contagious enthusiasm for research.
We acknowledge Dr. J. Wehland (German Research Center for Biotechnology, Braunschweig, Germany) for the gift of the monoclonal ID5. We are grateful to Dr. J. Beisson (Centre de Génétique Moléculaire) for helpful advice in immunocytochemistry and P. Neveu, G. Fryd, and A. Le Berre for excellent technical assistance. Thanks are due to Dr. J. Cohen (Centre de Génétique Moléculaire) and A.M. Keller for sharing of the genomic library and Dr. V. Redeker (Laboratoire de Neurobiologie) for the gift of Paramecium cortices. We thank S. Kuhlman, V. Gavric, and A. Bertin for their participation as undergraduate students in sequencing and immunolocalization and Y. Curé for his invaluable contribution in confocal imaging.
This work was partially supported by a Centre National de la Recherche Scientifique grant “Programme protéomique et génie des protéines.”
L. Vayssié's present address is Unité de Pathogénie Microbienne Moléculaire, INSERM U389, Institut Pasteur, 25-28 rue du Dr Roux, 75724 Paris cedex 15, France.
References
- Beisson, J., and M. Jerka-Dziadosz. 1999. Polarities of the centriolar structure: morphogenetic consequences. Biol. Cell. 91:367–378. [PubMed] [Google Scholar]
- Bobinnec, Y., M. Moudjou, J.P. Fouquet, E. Desbruyeres, B. Edde, and M. Bornens. 1998. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil. Cytoskeleton. 39:223–232. [DOI] [PubMed] [Google Scholar]
- Callen, A.-M., A. Adoutte, J.M. Andrew, A. Baroin-Tourancheau, M.-H. Bré, P.C. Ruiz, J.-C. Clérot, P. Delgado, A. Fleury, R. Jeanmaire-Wolf, et al. 1994. Isolation and characterization of libraries of monoclonal antibodies directed against various forms of tubulin in Paramecium. Biol. Cell. 81:95–119. [DOI] [PubMed] [Google Scholar]
- Carthew, R.W. 2001. Gene silencing by double-stranded RNA. Curr. Opin. Cell Biol. 13:244–248. [DOI] [PubMed] [Google Scholar]
- Chang, P., and T. Stearns. 2000. δ-Tubulin and ɛ-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat. Cell Biol. 2:30–35. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and G. Macino. 2000. Post-transcriptional gene silencing across kingdoms. Curr. Opin. Genet. Dev. 10:638–643. [DOI] [PubMed] [Google Scholar]
- Dippel, R.V. 1968. The development of basal bodies in Paramecium. Proc. Natl. Acad. Sci. USA. 61:461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis-Williams, P., C. Klotz, H. Mazarguil, and J. Beisson. 1996. The tubulin gene family of Paramecium: characterization and expression of the α PT1 and α PT2 genes, which code for α-tubulins with unusual C-terminal amino acids GLY and ALA. Biol. Cell. 87:83–93. [DOI] [PubMed] [Google Scholar]
- Dutcher, S.K. 2001. The tubulin fraternity: α to η. Curr. Opin. Cell Biol. 13:49–54. [DOI] [PubMed] [Google Scholar]
- Dutcher, S.K., and E.C. Trabuco. 1998. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes δ-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell. 9:1293–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher, S.K., J. Stanga, C. Rackley, and A.M. Preble. 2001. ɛ-Tubulin is required for basal body/centriole assembly. Mol. Biol. Cell. 12:440a. [Google Scholar]
- Fire, A., S. Xu, M.K. Montgomery, S.A. Kostas, S.E. Driver, and C.C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811. [DOI] [PubMed] [Google Scholar]
- Fleury, A., and M. Laurent. 1995. Microtubule dynamics and morphogenesis in Paramecium Europ. J. Protistol. 31:190–200. [Google Scholar]
- Froissard, M., A.M. Keller, and J. Cohen. 2001. ND9P, a novel protein with armadillo-like repeats involved in exocytosis. Physiological studies using allelic mutants in paramecium. Genetics. 157:611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani, A., and L. Sperling. 2001. Transgene-mediated post-transcriptional gene silencing is inhibited by 3′ non-coding sequences in Paramecium. Nucleic Acids Res. 29:4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani, A., and L. Sperling. 2002. RNA interference by feeding in Paramecium. Trends Genet. 18:11–12. [DOI] [PubMed] [Google Scholar]
- Garreau De Loubresse, N., F. Ruiz, J. Beisson, and C. Klotz. 2001. Role of δ-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol. 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: a Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 726 pp.
- Iftode, F., J. Cohen, F. Ruiz, A. Torres Rueda, L. Chen-Shan, A. Adoutte, and J. Beisson. 1989. Development of surface pattern during division in Paramecium I. Mapping of duplication and reorganization of cortical cytoskeletal structures in the wild type. Development. 105:191–211. [Google Scholar]
- Inclan, Y.F., and E. Nogales. 2001. Structural models for the self-assembly and microtubule interactions of γ-, δ- and ɛ-tubulin. J. Cell Sci. 114:413–422. [DOI] [PubMed] [Google Scholar]
- Jerka-Dziadosz, M., F. Ruiz, and F. Beisson. 1998. Uncoupling of basal body duplication and cell division in crochu, a mutant of Paramecium hypersensitive to nocodazole. Development. 125:1305–1314. [DOI] [PubMed] [Google Scholar]
- Keller, A.M., and J. Cohen. 2000. An indexed genomic library for Paramecium complementation cloning. J. Eukaryot. Microbiol. 47:1–6. [DOI] [PubMed] [Google Scholar]
- Klotz, C., N. Garreau de Loubresse, F. Ruiz, and J. Beisson. 1997. Genetic evidence for a role of centrin-associated proteins in the organization and dynamics of the infraciliary lattice in Paramecium. Cell Motil. Cytoskeleton. 38:172–186. [DOI] [PubMed] [Google Scholar]
- Krzywicka, A., J. Beisson, A.M. Keller, J. Cohen, M. Jerka-Dziadosz, and C. Klotz. 2001. KIN241: a gene involved in cell morphogenesis in Paramecium tetraurelia reveals a novel protein family of cyclophilin-RNA interacting proteins (CRIPs) conserved from fission yeast to man. Mol. Microbiol. 42:257–267. [DOI] [PubMed] [Google Scholar]
- Lange, B.M., A.J. Faragher, P. March, and K. Gull. 2000. Centriole duplication and maturation in animal cells. Curr. Top. Dev. Biol. 49:235–249. [DOI] [PubMed] [Google Scholar]
- McKean, P.G., S. Vaughan, and K. Gull. 2001. The extended tubulin superfamily. J. Cell Sci. 114:2723–2733. [DOI] [PubMed] [Google Scholar]
- Preble, A.M., T.H. Giddings, Jr., and S.K. Dutcher. 2001. Extragenic bypass suppressors of mutations in the essential gene BLD2 promote assembly of basal bodies with abnormal microtubules in Chlamydomonas reinhardtii. Genetics. 157:163–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger, A.H., M. Rudiger, J. Wehland, and K. Weber. 1999. Monoclonal antibody ID5: epitope characterization and minimal requirements for the recognition of polyglutamylated α- and β-tubulin. Eur. J. Cell Biol. 78:15–20. [DOI] [PubMed] [Google Scholar]
- Ruiz, F., N. Garreau de Loubresse, and J. Beisson. 1987. A mutation affecting basal body duplication and cell shape in Paramecium. J. Cell Biol. 104:417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz, F., L. Vayssié, C. Klotz, L. Sperling, and L. Madeddu. 1998. Homology-dependent gene silencing in Paramecium. Mol. Biol. Cell. 9:931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz, F., J. Beisson, J. Rossier, and P. Dupuis-Williams. 1999. Basal body duplication in Paramecium requires γ-tubulin. Curr. Biol. 9:43–46. [DOI] [PubMed] [Google Scholar]
- Ruiz, F., A. Krzywicka, C. Klotz, A. Keller, J. Cohen, F. Koll, G. Balavoine, and J. Beisson. 2000. The SM19 gene, required for duplication of basal bodies in Paramecium, encodes a novel tubulin, η-tubulin. Curr. Biol. 10:1451–1454. [DOI] [PubMed] [Google Scholar]
- Schliwa, M., and J. Van Blerkom. 1981. Structural interaction of cytoskeletal components. J. Cell Biol. 90:222–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrzka, O.W., N. Delgehyr, and M. Bornens. 2000. Tissue-specific expression and subcellular localization of mammalian δ-tubulin. Curr. Biol. 10:413–416. [DOI] [PubMed] [Google Scholar]
- Sonneborn, T.M. 1974. Paramecium aurelia. Handbook of Genetics. R. King, editor. Plenum Publishing Corp., New York. 469–594.
- Stelly, N., J. Mauger, M. Claret, and A. Adoutte. 1991. Cortical alveoli of Paramecium: a vast submembranous calcium storage compartment. J. Cell Biol. 113:103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L., D.L. Court, and A. Fire. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 263:103–112. [DOI] [PubMed] [Google Scholar]
- Timmons, L., and A. Fire. 1998. Specific interference by ingested dsRNA. Nature. 395:854. [DOI] [PubMed] [Google Scholar]
- Vaughan, S., T. Attwood, M. Navarro, V. Scott, P. McKean, and K. Gull. 2000. New tubulins in protozoal parasites. Curr. Biol. 10:R258–R259. [DOI] [PubMed] [Google Scholar]
- Vayssié, L., N. Garreau De Loubresse, and L. Sperling. 2001. Growth and form of secretory granules involves stepwise assembly but not differential sorting of a family of secretory proteins in Paramecium. J. Cell Sci. 114:875–886. [DOI] [PubMed] [Google Scholar]
- Wehland, J., H.C. Schröder, and K. Weber. 1984. Amino acid sequence requirements in the epitope recognized by the α-tubulin-specific rat monoclonal antibody YL 1/2. EMBO J. 3:1295–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]