Abstract
There is increasing evidence that ubiquitination of receptors provides an important endosomal sorting signal. Here we report that mammalian class E vacuolar protein-sorting (vps) proteins recognize ubiquitin. Both tumor susceptibility gene 101 (TSG101)/human VPS (hVPS)28 and hepatocyte growth factor receptor substrate (Hrs) cytosolic complexes bind ubiquitin-agarose. TSG101 and hVPS28 are localized to endosomes that contain internalized EGF receptor and label strongly for ubiquitinated proteins. Microinjection of anti-hVPS28 specifically retards EGF degradation and leads to endosomal accumulation of ubiquitin–protein conjugates. Likewise, depletion of TSG101 impairs EGF trafficking and causes dramatic relocalization of ubiquitin to endocytic compartments. Similar defects are found in cells overexpressing Hrs, further emphasizing the links between class E protein function, receptor trafficking, and endosomal ubiquitination.
Keywords: endocytosis; EGF receptor; down-regulation; ubiquitin; multivesicular
Introduction
Ligand-induced down-regulation of mitogenic receptors involves their endocytosis via clathrin-coated vesicles. Mitogenic receptors are then sorted within the endosomal system to the lysosomal pathway. Sorting includes the relocalization of receptors to the internal vesicles of late endosomes or multivesicular bodies, which then fuse with the lysosome directly. Efficient endocytosis is regulated both by receptor tyrosine protein kinase activity and by ubiquitination (Waterman and Yarden, 2001). Consistent with this, the EGF receptor (EGFR)*–negative regulator c-Cbl was identified as an E3 ubiquitin ligase specific for autophosphorylated receptor tyrosine kinases (Joazeiro et al., 1999; Levkowitz et al., 1999). Receptor tyrosine kinase activity and ubiquitination have also been implicated in receptor sorting, since kinase-defective EGFR is recycled to the cell surface more efficiently than wild-type receptor (Felder et al., 1990) and ubiquitin ligase–defective Cbl mutants enhance EGFR recycling (Levkowitz et al., 1999).
Studies in yeast have identified a family of proteins important for endosomal sorting. Class E vacuolar protein-sorting (vps) mutants fail to transport newly synthesized hydrolases efficiently to the vacuole. Instead, hydrolases accumulate with endocytosed receptors in an exaggerated perivacuolar class E compartment (Raymond et al., 1992; Piper et al., 1995; Rieder et al., 1996). This compartment is unable to sort markers to internal vesicles, indicating that both receptor sorting and the inward invagination of membrane are directly or indirectly effected by class E vps proteins (Odorizzi et al., 1998). Mammalian homologues of class E vps proteins have been identified, including hepatocyte growth factor receptor substrate (Hrs; related to Vps27p) (Komada et al., 1997), tumor susceptibility gene 101 (TSG101; related to Vps23p) (Babst et al., 2000), and human vacuolar protein sorting (hVPS)28 (Bishop and Woodman, 2001). The precise roles that these proteins play in receptor sorting remain unclear.
Given the putative role for ubiquitination in mitogenic receptor sorting, we were intrigued by reports that the class E protein Vps23p (Babst et al., 2000) and its mammalian orthologue TSG101 (Li and Cohen, 1996) contain a domain resembling E2 ubiquitin-conjugating (Ubc) enzymes, albeit lacking the catalytic cysteine residue (Koonin and Abagyan, 1997; Ponting et al., 1997). Therefore, TSG101 might be a candidate for coupling the recognition of ubiquitinated moietie(s) to the sorting of receptors. This hypothesis formed the basis for the work presented in this article. Emr and colleagues have demonstrated independently that Vps23p assists the biosynthetic sorting of ubiquitinated vacuolar protease precursors (Katzmann et al., 2001).
Results
Two mammalian class E vps complexes bind independently to ubiquitin
Given the presence of a Ubc enzyme–like domain within TSG101 (Koonin and Abagyan, 1997; Ponting et al., 1997), we examined whether TSG101 might recognize ubiquitin. As a control, we measured the binding of the Ubc (E2) enzyme Ubc1 (Hershko et al., 1983). Ubc1 did not bind to ubiquitin-agarose when ATP was absent but bound ubiquitin-agarose (but not glutathione [GSH]-agarose) in the presence of ATP, which enables ubiquitin to bind covalently to E2 enzymes via the activity of cytosolic ubiquitin-activating (E1) enzymes (Hershko et al., 1983). Cytosolic TSG101 and hVPS28 form part of a high molecular weight complex (Babst et al., 2000; Bishop and Woodman, 2001). Both TSG101 and hVPS28 bound specifically to ubiquitin-agarose with comparable efficiency to Ubc1 (Fig. 1 A). However, binding was ATP independent, indicating that it did not require covalent modification of the components of the complex or other TSG101–hVPS28-interacting proteins. Binding was specific, since little binding of tubulin was detected (Fig. 1 A). In addition, binding of TSG101 and hVPS28 was substantially reduced by inclusion of soluble ubiquitin (Fig. 1 B).
Figure 1.
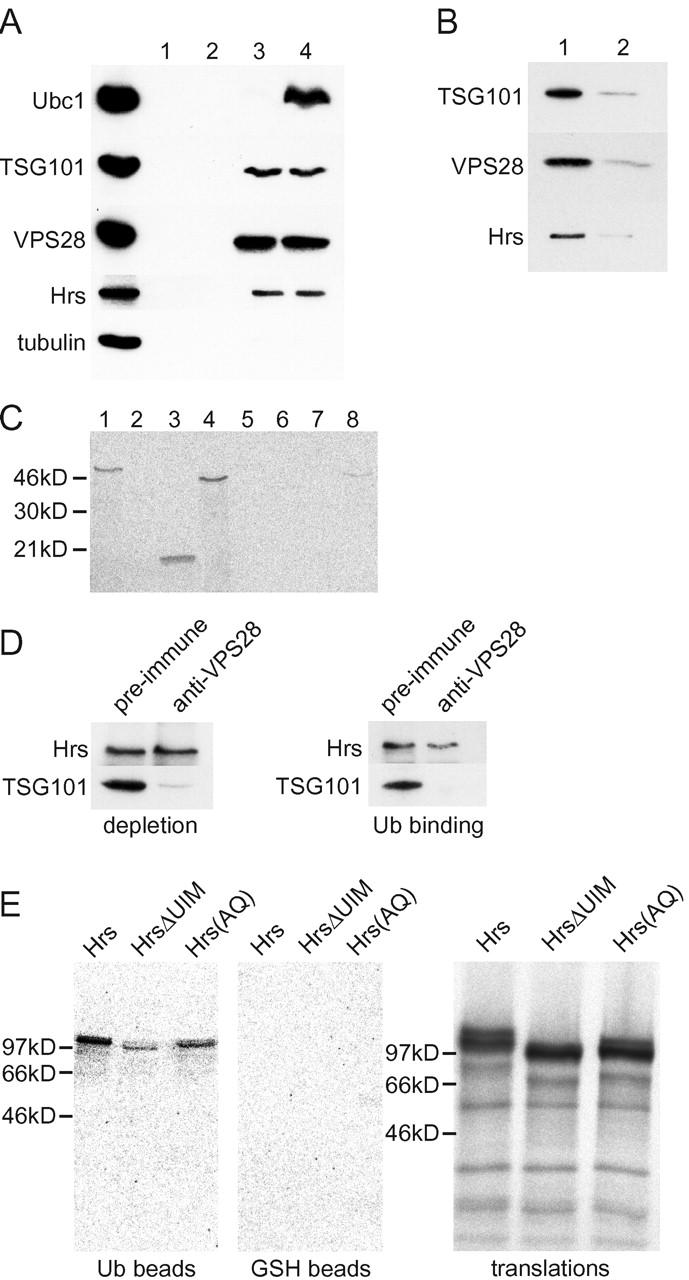
Mammalian class E vps proteins bind ubiquitin. (A) ATP-depleted HeLa cytosol was incubated with GSH- (lanes 1 and 2) or ubiquitin-agarose (lanes 3 and 4) with (lanes 1 and 3) or without (lanes 2 and 4) 1 mM Mg-ATP. Bound fractions were analyzed by Western blotting. 25% of the input was also blotted (T). (B) Cytosol was incubated with ubiquitin-agarose after preincubating for 15 min without (lane 1) or with (lane 2) 1 mg soluble ubiquitin. Bound fractions were analyzed as above. (C) Equal counts of in vitro–translated TSG101 (lanes 1 and 5), TSG101ΔUbc (lanes 2 and 6), the Ubc domain alone (lanes 3 and 7), or TSG101ΔHx (lanes 4 and 8) were incubated with ubiquitin- (lanes 1–4) or GSH-agarose (lanes 5–8). Bound fractions were analyzed by PAGE and phosphorimaging. (D) HeLa cytosol was immunodepleted using either preimmune or anti-hVPS28 IgG. The resulting cytosols were Western blotted for Hrs and TSG101 (left). Parallel samples were incubated with ubiquitin-agarose, and bound material was Western blotted (right). (E) Equal counts of in vitro–translated Hrs, HrsΔUIM, and Hrs(AQ) were incubated with GSH- or ubiquitin-agarose (left). Translation products, run on a separate gel, are also shown. Samples were analyzed as above.
In vitro–translated TSG101 also interacted with ubiquitin-agarose. Binding required the Ubc domain of TSG101, since TSG101ΔUbc failed to bind and the Ubc domain alone bound to the same extent as full-length protein. In contrast, deletion of the helical domain required for the interaction of TSG101 with hVPS28 (Bishop and Woodman, 2001) did not affect its binding to ubiquitin. In vitro–translated hVPS28 did not bind to ubiquitin-agarose (unpublished data). Together, these data are consistent with the complex binding to ubiquitin via the Ubc domain of TSG101, though we cannot rule out the possibility that this occurs through an intermediary factor rather than being direct.
The activities of class E vps proteins are likely to be linked, raising the possibility that other class E proteins might recognize ubiquitin. In particular, Hrs localizes to the sorting endosome (Komada et al., 1997; Raiborg et al., 2001) and interacts indirectly with a deubiquitinating enzyme (Kato et al., 2000). Indeed, Hrs binds ubiquitin (Fig. 1 A). As for TSG101, binding of cytosolic Hrs to ubiquitin was ATP independent and was reduced by inclusion of soluble ubiquitin (Fig. 1 B). Hrs binding was not mediated via the TSG101–hVPS28 complex. When cytosol was depleted of TSG101 with anti-hVPS28, there was little depletion of Hrs (Fig. 1 D, left). Therefore, Hrs does not appear to interact with the TSG101 complex in solution. Additionally, depletion of TSG101 had little impact on the association of cytosolic Hrs with ubiquitin (Fig. 1 D, right). Furthermore, in vitro–translated Hrs bound to ubiquitin-agarose specifically (Fig. 1 E). During the course of our studies, a putative ubiquitin-interacting motif (UIM) was identified within Hrs, with two such motifs in Vps27p (Hofmann and Falquet, 2001). Deletion of the UIM sharply reduced the efficiency with which Hrs bound ubiquitin (Fig. 1 E). Binding was also reduced by a double point mutation in which two charged amino acids within the UIM were substituted. Hence, the conserved UIM is mainly responsible for the interaction that we observe. A second UIM is present within Vps27p but is weakly conserved in Hrs. This may contribute to the residual ubiquitin binding we observed.
The TSG101 complex is localized to ubiquitin-enriched EGF-containing endosomes
Cells expressing reduced levels of TSG101 are impaired in mitogenic receptor down-regulation, consistent with the structural similarity between TSG101 and Vps23p (Babst et al., 2000). However, TSG101 has been implicated in several cellular processes, most likely involving its interaction with ubiquitinated substrate(s) (see Discussion). Hence, depletion of TSG101 may interfere with receptor sorting indirectly by allowing the accumulation of ubiquitinated intermediates in other cellular pathways and hence reducing the levels of available free ubiquitin and/or ubiquitin-interacting components. By analogy, it has been demonstrated by several laboratories that proteasomal inhibitors dramatically reduce cellular free ubiquitin levels, most likely by preventing the deubiquitination of proteasomal substrates (Patnaik et al., 2000; Schubert et al., 2000; Strack et al., 2000). Therefore, we looked for whether the TSG101–hVPS28 complex contributes directly to receptor trafficking. As a first step, we examined whether the complex was localized to EGFR-enriched endosomes.
A431 cells have a large complement of EGFR. At least some of these were ubiquitinated upon ligand binding, since EGFR immunoprecipitates isolated from cells shortly after EGF internalization but not from untreated cells were labeled with a monoclonal antibody against protein–ubiquitin conjugates, FK2 (Fig. 2 A). The intensity of ubiquitin staining fell sharply between 10 and 30 min after EGF internalization (though residual labeling was still detected). In addition, the appearance of antiubiquitin staining was not accompanied by any noticeable shift in mobility of the EGFR, suggesting that only a very small proportion of the receptor was ubiquitinated at any time. These data are consistent with previous reports that ligand-induced ubiquitination of the EGFR is rapid and transient (Levkowitz et al., 1998), though they might additionally indicate that receptor ubiquitination is substoichiometric. Immunofluorescence staining confirmed that EGF-rich endosomes contained ubiquitinated proteins (Fig. 2 B). Surprisingly, endosomes were strongly labeled for ubiquitinated proteins at later times when the amount of ubiquitinated EGFR had fallen. These data are consistent with ubiquitin functioning late on the endocytic pathway and with endosomally associated protein(s) distinct from the EGFR being ubiquitinated as a consequence of receptor internalization.
Figure 2.
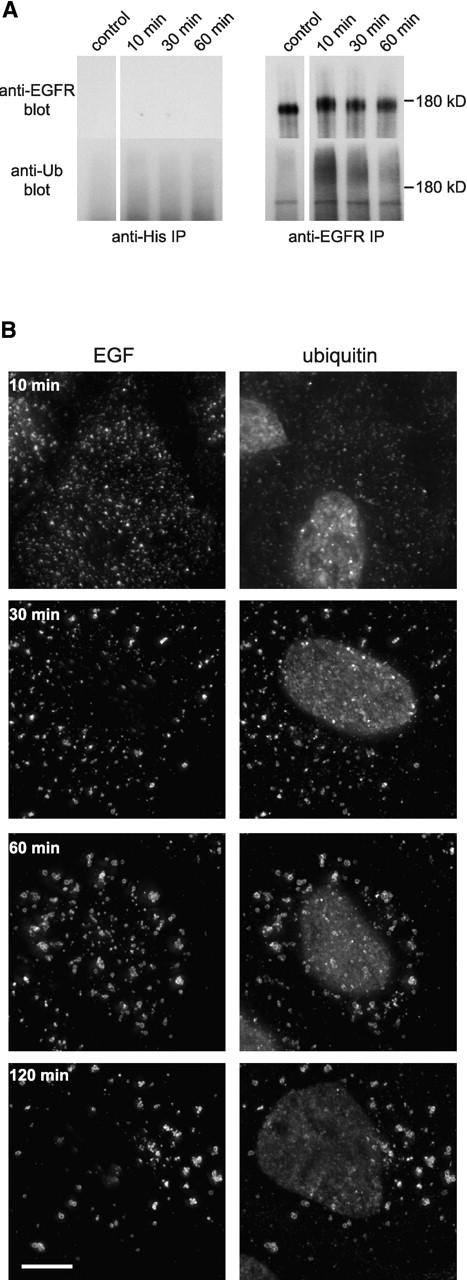
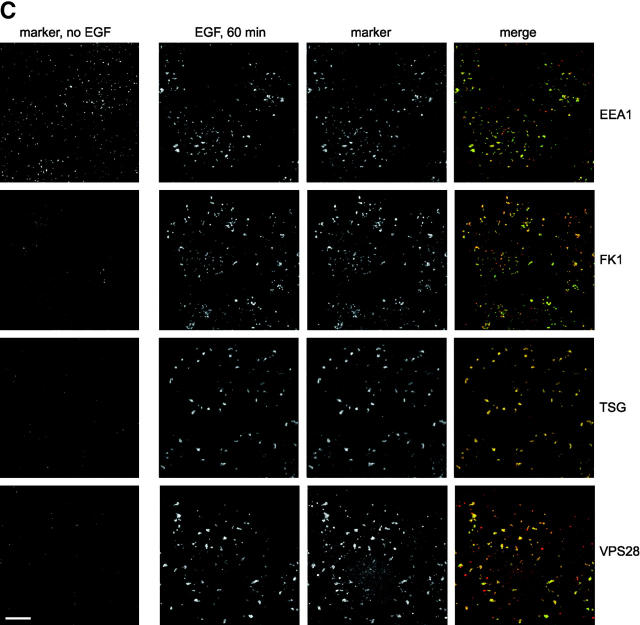
TSG101 and hVPS28 are localized to ubiquitin-rich EGF-positive endosomes. (A) A431 cells were starved overnight in serum-free medium and either left untreated (no EGF) or incubated with 0.4 μg/ml EGF at 4°C followed by the indicated time at 37°C. Cell lysates were immunoprecipitated with control (left) or anti-EGFR antibody (right). Bound fractions were Western blotted with anti-EGFR (top) or antiubiquitin (FK2; bottom). The band detected in the FK2 blot running just below the EGFR is an artifact, since it is found in EGFR immunoprecipitates from cells lacking EGFR (unpublished data). (B) Oregon green EGF was bound to A431 cells and internalized as indicated. Cells were chilled, incubated with saponin before fixation, and then analyzed for EGF content (left) and stained with FK2 antibody (right). (C) A431 cells were either left untreated (left) or had Oregon green EGF internalized for 60 min (right). Cells were chilled and incubated with saponin before fixation and then analyzed for EGF (green) or stained (red) for the following markers: EEA1, polyubiquitin–protein conjugates (FK1), TSG101, or hVPS28.
We examined the localization of the TSG101 complex after EGF had been internalized for 1 h, after which time it had reached enlarged ubiquitin-rich endosomes and little degradation of the receptor-ligand complex had occurred. EGF was detected in early endosome-associated antigen 1 (EEA1)–positive compartments (Fig. 2 C). These also labeled with the polyubiquitinated protein-specific monoclonal antibody, FK1, and for TSG101 and hVPS28 (Fig. 2 C). The pattern of staining for ubiquitin, TSG101, and hVPS28 was significantly different from that in nonstimulated cells, indicating that EGFR internalization had affected the localization of both ubiquitin and the TSG101–hVPS28 protein complex.
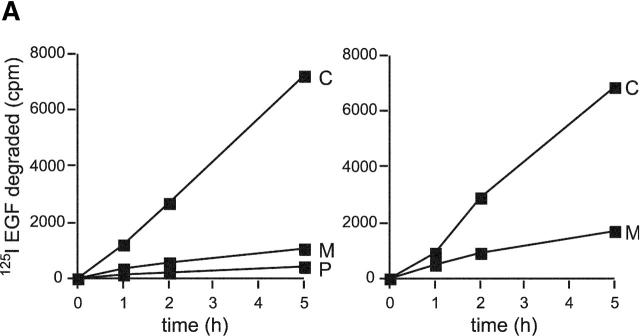
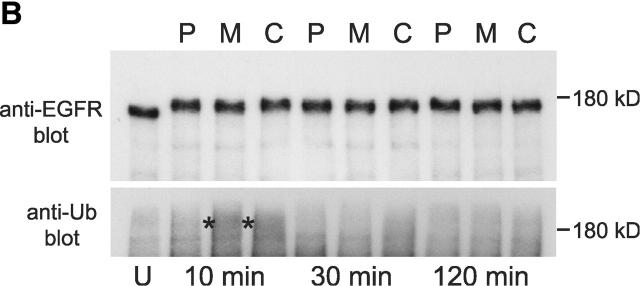
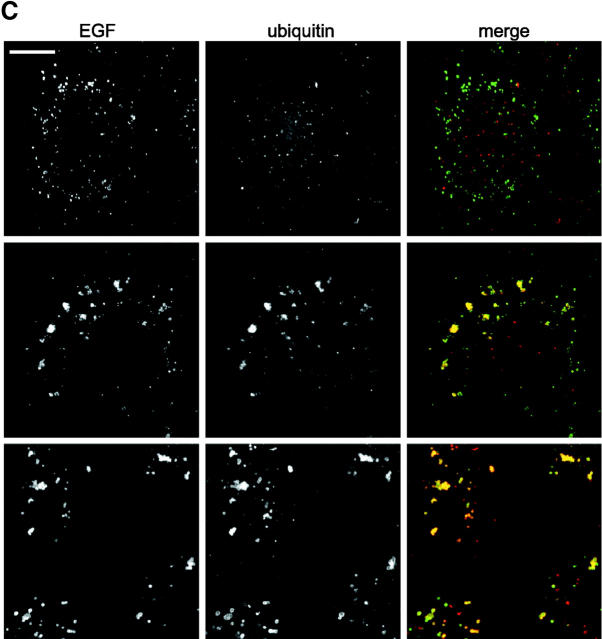
To further investigate links between ubiquitination of endosomal proteins and receptor degradation, we used the proteasome inhibitor, MG132, which has been demonstrated previously to block mitogenic receptor degradation (Rocca et al., 2001; van Kerkhof et al., 2001). Although it remains unclear whether proteasomes are directly involved in receptor degradation, it has been reported in the case of the interleukin 2 receptor β chain that inhibition of proteasome function is accompanied by defects in receptor sorting and the elevation of levels of ubiquitin–receptor conjugate (Rocca et al., 2001). Proteasome inhibition leads to a depletion of free ubiquitin (Patnaik et al., 2000; Schubert et al., 2000; Strack et al., 2000). This may arise from the inability of deubiquitinating enzymes to function in the context of stalled proteasomes, though other reasons are also possible. As expected, preincubation of A431 cells with MG132 for 3 h before EGF binding and during EGF internalization abolished EGF degradation (Fig. 3 A, left). EGF degradation was also substantially reduced upon incubating cells with another proteasome inhibitor, lactacystin (unpublished data). The small extent of EGF-dependent EGFR ubiquitination observed in control cells was abolished by prior incubation with MG132 so that ubiquitination of the receptor could not be detected over background levels found in untreated cells (Fig. 3 B, lane 1 compared with lane 2). Fluorescently labeled EGF accumulated in small endosomes that were only weakly labeled for ubiquitin (Fig. 3 C, top) and VPS28 (unpublished data). In contrast, when MG132 was added only after EGF binding internalized EGF (whose degradation was substantially impaired) (Fig. 3 A left) entered enlarged ubiquitin-enriched endosomes as in control cells (Fig. 3 C, middle). Much EGF remained associated with ubiquitin-enriched endosomes even after a 5-h internalization (unpublished data). However, interestingly, EGFR ubiquitination and deubiquitination was unaffected by MG132 (Fig. 3 B).
Figure 3.
Inhibition of proteasome function inhibits EGF degradation and prevents deubiquitination of endosomal proteins. (A, left) 125I EGF was bound and internalized into A431 cells in the absence (control; C) or presence (M) of 5 μM MG132 or with MG132 in cells which had also been preincubated for 3 h with 5 μM MG132 (P). After the indicated times of internalization, medium was removed and assayed for degraded 125I EGF. (A, right) 125I EGF was bound and internalized into untreated A431 cells (C) or into cells to which MG132 was added 30 min after internalization had begun (M). Samples were assayed for EGF degradation as above. All values are means of triplicate determinations ± SEM. (B) A431 Cells were incubated with 0.4 μg/ml EGF at 4°C followed by 10, 30, or 120 min at 37°C as indicated. Cells were incubated with 5 μM MG132 for 3 h before and during internalization (P), incubated with 5 μM MG132 during internalization only (M), or otherwise untreated (C). Cell lysates were immunoprecipitated with anti-EGFR antibody, and bound fractions were Western blotted with anti-EGFR (top) or antiubiquitin (FK2; bottom). The samples containing peak FK2 labeling are indicated by asterisks. An immunoprecipitate from untreated cells is also shown (U). (C) Oregon green EGF was bound to A431 cells and internalized. Cells were chilled, incubated with saponin before fixation, and then analyzed for EGF content (green) and stained with FK2 antibody (red). (Top) Cells preincubated for 3 h with MG132 and then EGF internalized for 1 h with MG132. (Middle) EGF internalized for 1 h with MG132. (Bottom) EGF internalized for 30 min followed by a further 4.5 h with MG132.
These data suggest that the kinetics of EGFR and endosomal protein deubiquitination may occur at different rates, with the receptor being deubiquinated much earlier than other endosomal proteins. This was further demonstrated by addition of MG132 30 min after EGF internalization when the population of ubiquitinated EGFR had already diminished. EGF degradation remained substantially reduced (Fig. 3 A, right), and much undegraded EGF was associated with clustered endosomes that labeled strikingly for ubiquitin (Fig. 3 C, bottom). Together, these data suggest that deubiquitination of the EGFR and loss of ubiquitin labeling from endosomes can be uncoupled experimentally and hence are consistent with EGFR sorting being accompanied by the ubiquitination of other endosomal proteins.
Impairing class E protein complexes retards EGF degradation and causes ubiquitinated proteins to accumulate on endosomes
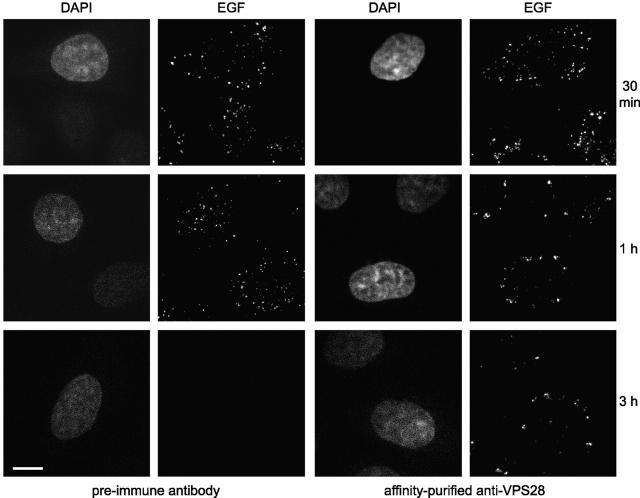
To provide functional evidence that the TSG101–hVPS28 complex acts directly in receptor trafficking, we examined the fate of EGF in cells microinjected with anti-hVPS28. Anti-hVPS28 did not impair EGF internalization, since EGF labeling was similar after a 30-min internalization in cells injected with either anti-hVPS28 or control antibody (Fig. 4). In HeLa cells, degradation of intracellular EGF is essentially complete after 3 h. However, cells that had been microinjected with anti-hVPS28 retained substantial EGF labeling at this time (Fig. 4). At 3 h, the maximal intensity of EGF staining in anti-hVPS28 injected cells (n = 35) was 467 ± 31% compared with that in control injected cells (n = 31).
Figure 4.
Anti-hVPS28 inhibits EGF degradation. HeLa cells were microinjected with preimmune IgG (left) or anti-hVPS28 (right). Texas red EGF was internalized as indicated, and cells were examined for EGF. Injected cells were detected by DAPI staining. Confocal settings were identical for all samples.
The effect of anti-hVPS28 on EGF degradation was specific, since it did not affect the kinetics of transferrin recycling. Compared with noninjected cells, cells injected with anti-hVPS28 took up comparable levels of Texas red–conjugated transferrin after 15 min (112 ± 9%; n = 30 compared with uninjected cells, n = 42). Likewise, loss of transferrin labeling after a 2-h chase was unaffected by microinjection of anti-hVPS28 (the residual signal being 102 ± 11% [n = 31] in injected cells relative to uninjected cells [n = 42]).
Given that the TSG101–hVPS28 complex binds to ubiquitin and contributes to endosomal sorting, we examined whether interfering with its function affected the release of ubiquitin from endosomally associated ubiquitin–protein conjugates that had formed as a consequence of receptor internalization. Indeed, microinjection of cells with anti-hVPS28 caused a dramatic increase in the level of FK2 labeling of EGF-positive endosomes (Fig. 5). The accumulation of ubiquitin–protein conjugates was largely dependent on EGFR internalization; the maximum intensity of FK2 labeling associated with cytoplasmic structures was 508 ± 23% in microinjected cells to which EGF had been bound and internalized (n = 21) compared with that observed in microinjected cells that had been mock treated (n = 22). In addition, little increase in endosomal FK2 labeling was observed in uninjected cells that had internalized EGF while being treated with the lysosomal protease inhibitor leupeptin or with primaquine, which blocks endosomal sorting by collapsing the pH gradient (Fig. 5). Hence, the accumulation of ubiquitinated moieties was not simply a consequence of preventing the degradation of receptor–ligand complexes.
Figure 5.
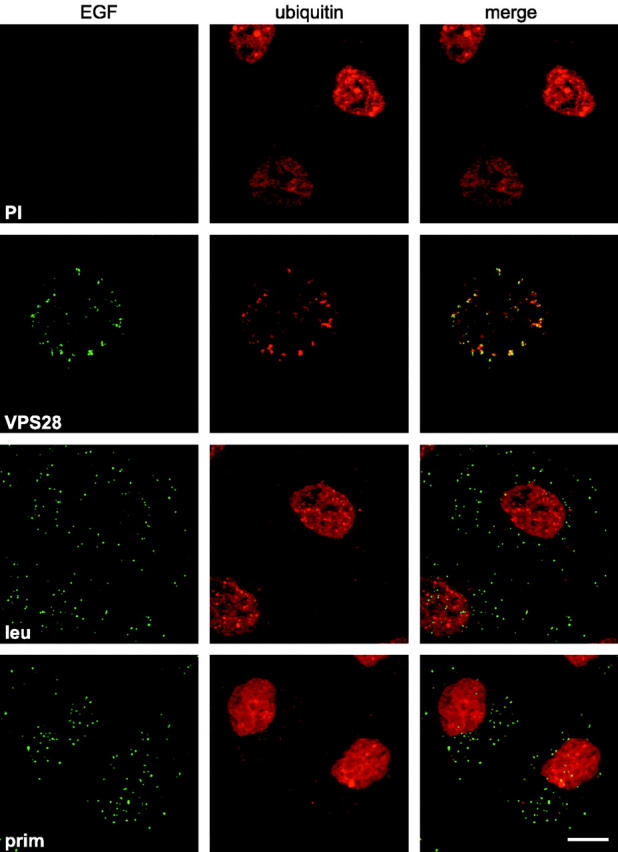
Anti-hVPS28 causes ubiquitinated proteins to accumulate on endosomes. HeLa cells were microinjected with preimmune IgG (PI) or anti-hVPS28 antibody (VPS28). Oregon green EGF was internalized for 3 h. Alternatively, Oregon green EGF was bound and internalized into cells treated with either 100 μM leupeptin (leu) or 0.3 mM primaquine (prim). Cells were permeabilized with saponin, fixed, visualized for EGF (EGF), and stained with FK2 monoclonal antibody. Microinjected cells were identified by DAPI staining (unpublished data). Confocal settings were identical for all EGF labeling but were altered for ubiquitin staining to prevent saturation.
The effects of disrupting the TSG101–VPS28 complex on receptor trafficking and endosomal ubiquitination were also investigated using small interfering RNA (siRNA) to deplete cellular TSG101. Transfection of HeLa cells with a siRNA duplex matching a nucleotide sequence within the coding region of Tsg101 caused a near total loss of cellular TSG101 protein after 48 h of culture (Fig. 6 A). The loss was specific, since levels of tubulin were unaffected (Fig. 6 A). Cell cultures transfected with the TSG101 siRNA were substantially impaired in their ability to degrade 125I-labeled EGF (unpublished data), consistent with data obtained using a mutant NIH 3T3 cell line that exhibits reduced expression levels of TSG101 (Babst et al., 2000).
Figure 6.
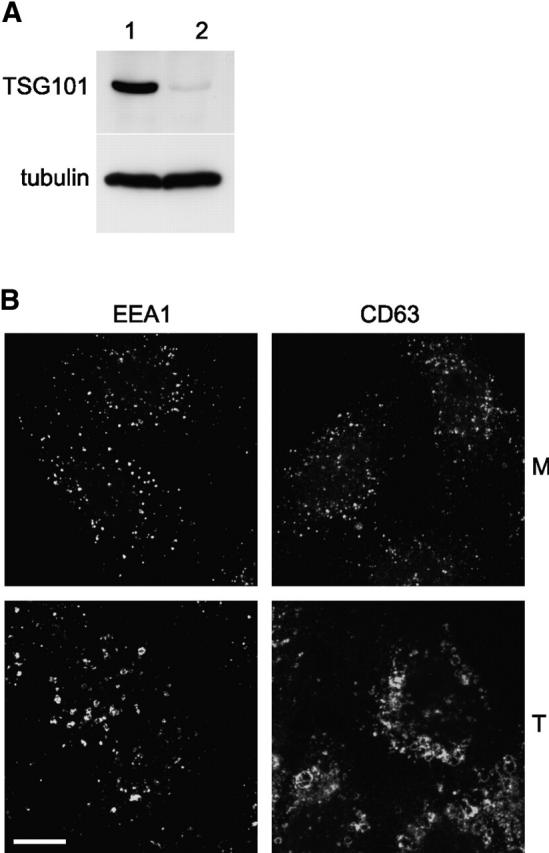
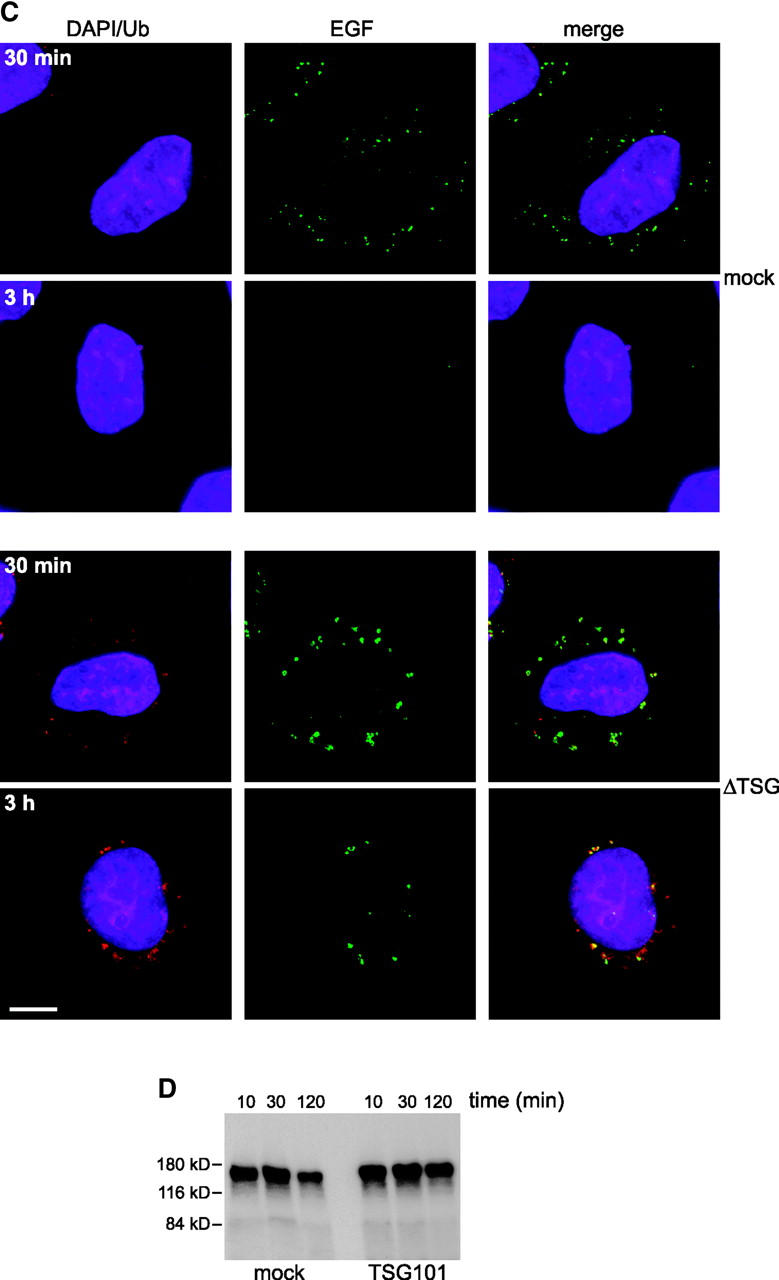
Depletion of TSG101 affects EGF trafficking and endosomal ubiquitination. (A) HeLa cells were mock transfected (lane 1) or transfected with siRNA against Tsg101 (lane 2). Cell extracts were Western blotted for TSG101 (top) or tubulin as a control (bottom). (B) HeLa cells were mock transfected (M; top) or transfected with siRNA against Tsg101 (T; bottom) and stained for EEA1 or CD63 as indicated. (C). Oregon green EGF was internalized for the indicated times into mock-transfected HeLa cells (top) or cells transfected with siRNA against Tsg101 (bottom). Cells were analyzed for EGF (green) and stained with DAPI (blue) and with FK2 monoclonal antibody followed by Texas red 2° antibody (red). (D). EGF was bound and internalized into mock-transfected HeLa cells (left) or cells transfected with siRNA against Tsg101 (right). Cell lysates were made after the indicated times of EGF internalization and Western blotted for EGFR.
Depletion of TSG101 had a striking effect on the morphology of endosomal compartments. Late endosomal compartments that stained for CD63 were highly vacuolated and clustered compared with mock-treated cells (Fig. 6 B). In addition, the early endocytic marker EEA1 was affected (Fig. 6 B). Although much EEA1 remained associated with small punctate structures, additional labeling was clustered in larger structures. A similar degree of redistribution was observed for the transferrin receptor (unpublished data). These alterations in membrane distribution were confined to endosomal compartments (unpublished data). The effect of TSG101 depletion on EGF trafficking was also observed by fluorescence microscopy (Fig. 6 C). Internalized EGF accumulated at foci, often toward the center of cells. Hence, although TSG101 depletion may enhance EGF recycling (Babst et al., 2000) a significant portion of EGF enters a compartment from which it can no longer be sorted efficiently. Significantly, label accumulated adjacent to vacuolated membranes, most likely endosomal compartments, which were highly enriched for ubiquitin–protein conjugates. Although knock-down of TSG101 caused a profound relocalization of cellular ubiquitin to endosomes in conjunction with impairment of EGF sorting, we found no evidence for alterations in the ubiquitination of the EGFR itself. In either mock-treated or TSG101 siRNA-treated cells, EGF binding and internalization was not accompanied by significant EGFR ubiquitination as judged by changes in molecular weight (Fig. 6 D). siRNA duplexes targeted against other coding sequences, including lamin A, affected neither TSG101 levels, EGF degradation, or ubiquitin accumulation. In addition, the TSG101 siRNA did not affect the rat cell line NRK (unpublished data).
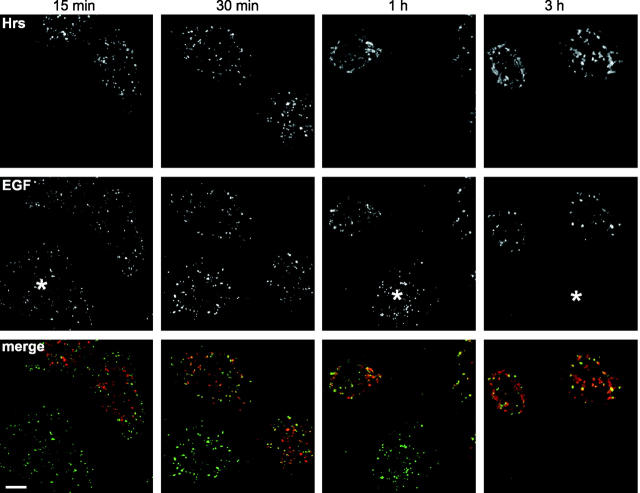
These data are consistent with impairment of TSG101–VPS28 preventing the release of ubiquitin from endosomal ubiquitin–protein conjugates. Likewise, impairing Hrs also blocked receptor sorting and release of ubiquitin. Consistent with previous reports (Komada et al., 1997), exogenously expressed Hrs localized to enlarged compartments (Fig. 7) that stained for early endosomal markers (unpublished data). These compartments partially colocalized with markers for the late endocytic pathway (unpublished data), implying that overexpression of Hrs causes defective endocytic sorting. To assess this in more detail, we examined the internalization and degradation of EGF. Hrs overexpression did not impair the internalization of EGF, since transfected cells were similarly labeled with EGF as neighboring untransfected cells after short periods of internalization (Fig. 7). At later times, internalized EGF reached compartments enriched in expressed Hrs (though not all Hrs-enriched compartments were labeled). A significant portion of EGF remained in these compartments even after 3 h internalization, in contrast to neighboring cells where the majority of EGF was degraded.
Figure 7.
Hrs expression inhibits EGF degradation. HeLa cells were transfected with Hrs. Oregon green EGF was bound and internalized for the indicated times. Cells were visualized for EGF (green) and stained with anti-Hrs IgG (red). Untransfected cells are indicated by an asterisk.
Cells expressing Hrs showed striking changes in ubiquitin distribution with dramatic localization of ubiquitin–protein conjugates to Hrs-enriched endosomes such that the endosomal ubiquitin staining was substantially more intense than over the nucleus (Fig. 8 A). No staining of Hrs-induced endosomes with antibodies against proteasome components was observed (Fig. 8 A), implying that the redistribution of ubiquitin was not simply a consequence of recruiting misfolded proteins to the Hrs endosomes. This apparent relocalization of cellular ubiquitin might be due merely to the accumulation of endosomal ubiquitinated protein(s) with unusual reactivity toward the FK monoclonal antibodies under the fixation conditions employed by us. However, this is unlikely since the localization of epitope-tagged ubiquitin was altered from a predominantly nuclear and cytosolic distribution to an endosomal distribution upon coexpression of Hrs (Fig. 8 B).
Figure 8.
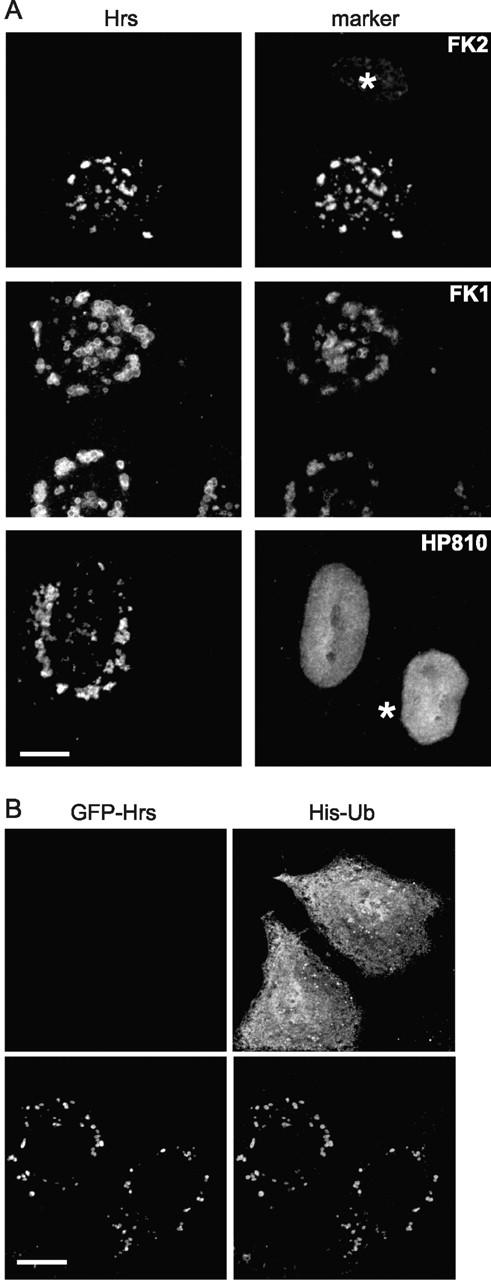
Hrs expression causes relocalization of ubiquitinated proteins. (A) HeLa cells were transfected with untagged Hrs, permeabilized in saponin, fixed, and stained for Hrs (left) and with FK2 or FK1 antiubiquitin–protein conjugates, or with HP810 antiproteasome as indicated (right). Untransfected cells are denoted by an asterisk. (B) HeLa cells were transfected with His6-tagged ubiquitin alone (top) or with His6-tagged ubiquitin and EGFP-Hrs (bottom). Cells were fixed directly in methanol without prior permeabilization and visualized for Hrs (left) and stained using anti-His antibody (right).
Discussion
The molecular machinery that directs receptors to the late endocytic pathway in mammalian cells is not known, though ubiquitination is thought to play an important role. We have focused our attention on mammalian orthologues of yeast class E vps proteins, since these are likely to act at this sorting step. Here we have shown that both the TSG101–VPS28- and Hrs-containing complexes bind ubiquitin. The affinity appears moderate as would be expected of interactions no doubt normally regulated by additional factors. The ATP-independent binding of cytosolic TSG101–hVPS28 to ubiquitin might be expected given that the TSG101 Ubc domain lacks the critical cysteine residue for formation of a ubiquitin thioester. However, this is in contrast to the case of the TSG101-related Mms2p, which interacts with ubiquitin in combination with Ubc13p (Hofmann and Pickart, 1999).
Cells with reduced levels of TSG101 are impaired in mitogenic receptor down-regulation and show enhanced EGF recycling (Babst et al., 2000). Our data using siRNA to deplete cellular TSG101 largely confirm these observations, though they show that a significant amount of internalized EGF is retained in a nondegradative endocytic compartment. In contrast to these earlier studies, we find that TSG101 depletion induces aberrant morphology in multiple endocytic compartments. The discrepancy between these findings may perhaps be accounted for by differences in the level to which TSG101 is depleted in each experimental system. Depletion experiments provide limited evidence that TSG101 acts directly to promote receptor trafficking, particularly since this event would be sensitive to changes in the levels of either ubiquitin or ubiquitin-interacting components that might be influenced by loss of TSG101. However, we have demonstrated that TSG101 and VPS28 are localized to ubiquitin-rich endosomes during ligand-induced EGFR internalization. Additionally, antibodies to VPS28 specifically impair EGF degradation without affecting other endosome-associated functions, such as delivery of receptors to the sorting endosome and transferrin receptor recycling. Importantly, though our data support a direct role for TSG101–VPS28 in receptor trafficking they do not preclude the complex from fulfilling other cellular roles involving ubiquitin recognition (Li and Cohen, 1996; Hittelman et al., 1999).
In further support of a direct role of class E vps proteins in ubiquitin-dependent receptor trafficking, we have shown that disruption of class E vps function by three independent means causes dramatic relocalization of cellular ubiquitin to endocytic compartments. The full cause of this relocalization remains unclear. Possibly, impairment of TSG101–VPS28 function prevents recruitment of further cytosolic components required for receptor sorting and that disassembly of the full complex is coupled to removal of ubiquitin chains. Consistent with this, expression of dominant negative mammalian VPS4, which may link the disassembly of such a complex to ATP hydrolysis (Babst et al., 1998; Bishop and Woodman, 2000), also causes dramatic endosomal accumulation of ubiquitinated proteins (unpublished data). Hrs might play a more direct role in releasing ubiquitin, since an Hrs-binding protein interacts with the deubiquitinating enzyme UBPY (Kato et al., 2000). Cytosolic Hrs is part of a 500-kD complex (unpublished data). We speculate that exogenously expressed Hrs, which binds to endosomes via its FYVE and coiled coil domains (Raiborg et al., 2001) (and which presumably interacts with ubiquitinated moieties on the endosome surface), would displace endogenous Hrs complexes and thus prevent recruitment of UBPY. We have been unable to analyze any further the functional significance of Hrs binding to ubiquitin since, as expected, the UIM is not solely necessary for endosomal localization of Hrs or the impairment of EGF trafficking by Hrs overexpression.
The cellular ubiquitinated substrates for TSG101 and Hrs remain to be identified, and we have been hampered in this respect by our inability to stabilize receptor-dependent ubiquitination events in cell extracts. Candidates include the mitogenic receptors themselves. By analogy, the yeast Vps23p-containing ESCRT-1 complex binds directly to ubiquitinated carboxypeptidase S precursor (Katzmann et al., 2001). Indeed, we have detected a weak coimmunoprecipitation of the EGFR with the TSG101 complex, though this is independent of receptor ubiquitination (unpublished data). However, we anticipate that recruitment of mammalian TSG101 may be more complex. Endocytic compartments within TSG101-depleted cells accumulate striking levels of ubiquitinated proteins, yet we find no evidence that the EGFR is hyperubiquitinated. In addition, we have shown using a proteasome inhibitor that accumulation of ubiquitinated proteins within EGFR-enriched endosomes can be experimentally uncoupled from ubiquitination of the EGFR itself. The failure to increase levels of ubiquitinated EGFR under conditions where deubiquitination of endosomal proteins is apparently so diminished raises the possibility that ubiquitinated proteins other than the receptor are important for endosomal sorting and hence might act as the principle substrates for Hrs and TSG101 binding. These data are also completely consistent with recent findings that endosomal sorting of a truncated growth hormone receptor is critically dependent on a ubiquitin-recruiting motif within the receptor but does not require receptor ubiquitination (van Kerkhof et al., 2001). Furthermore, TSG101 interacts via its Ubc-like domain with a highly conserved peptide motif within the Pr55Gag protein of HIV type 1 (Garrus et al., 2001; Martin-Serrano et al., 2001; VerPlank et al., 2001), which is absent from mitogenic receptors, including EGFR. Interestingly, artificial covalent attachment of ubiquitin to HIV Pr55Gag enhances TSG101 Ubc domain binding (Garrus et al., 2001).
Our data place mammalian class E vps proteins at the center of ubiquitination-dependent receptor sorting. They extend the finding that defects in yeast Doa4p, a deubiquitinating enzyme, are suppressed by class E vps mutants (Amerik et al., 2000). At present, we cannot discriminate between models that favor ubiquitin recruiting a specific endosomal sorting machinery and those that imply that proteasome function is important for endosomal sorting. However, our finding that the TSG101 complex contributes directly to mitogenic receptor sorting supports recent evidence that the yeast ESCRT-1 complex of Vps23/28/37p promotes the ubiquitin-dependent biosynthetic sorting of vacuolar precursors (Katzmann et al., 2001). In yeast, interfering with Vps23p blocks either directly or indirectly the localization of receptors to the internal vesicles of multivascular body. We now aim to address if this specific stage is blocked when mammalian class E vps protein function is impaired.
Materials and methods
DNA manipulations
Mouse Hrs (mHrs) cDNA was provided by Dr. Naomi Kitamura (Tokyo Institute of Technology, Tokyo, Japan) and cloned into pGEX-4T2 (Amersham Pharmacia Biotech) or into pcDNA3.1 lacking the epitope tag. HrsΔUIM-1 lacked amino acids 259–273, and Hrs(AQ) contained E260A/E261Q substitutions within the putative UIM. Alternatively, Hrs was cloned into the vector pEGFP (CLONTECH Laboratories, Inc.) to generate pEGFP-mHrs. TSG101 deletions were as described previously (Bishop and Woodman, 2001), with TSG101(Ubc) made by deleting amino acids 137–388. All mutagenesis (QuickChange [Stratagene] or cassette) was confirmed by sequencing. His-tagged ubiquitin was expressed using the pMT107 construct, a gift from Dirk Bohmann (European Molecular Biology Laboratory, Heidelberg, Germany).
Antibodies and reagents
Sheep were immunized with GST-mHrs purified from bacterial inclusion bodies. For fluorescence microscopy, cells were stained with an IgG fraction at a concentration that detected expressed but not endogenous Hrs. The following monoclonal antibodies were against ubiquitinated proteins (FK1, which recognizes only polyubiquitin–protein conjugates, and FK2, which recognizes both mono- and polyubiquitinated conjugates; Affiniti BioRegeants, Inc.), EEA1 (Transduction Laboratories), EGFR (clone 29.1.1; Sigma-Aldrich), 20S proteasome (HP810; Affinity BioReagents, Inc.), tubulin (TAT1; from K. Gull, University of Manchester, Manchester), TSG101 (GeneTex), CD63 (a gift from Fedor Berditchevski, University of Birmingham, Birmingham, UK). Also used were rabbit anti-Ubc1 (Affiniti BioRegeants, Inc.) and affinity purified anti-hVPS28 (Bishop and Woodman, 2001). HRP-conjugated antibodies (Dako) were used for ECL blotting. MG132 was purchased from Calbiochem and stored at –20°C in DMSO.
Cells
Cells were grown in DME (GIBCO BRL) containing 10% FCS. Cells were transfected with DNA constructs using FuGene (Roche) and analyzed after 16–24 h. For siRNA experiments, 21 nucleotide RNA duplexes corresponding to the Tsg101 coding nucleotides AACGATGGCAGTTCCAGGGAA with symmetric 2 nucleotide 3′ (2'-deoxy) thymidine overhangs (sense, 5′ CGAUGGCAGUUCCAGGGAAdTdT; antisense, 5′ UUCCCUGGAACUGCCAUCGdTdT) were synthesized and annealed (Dharmacon Research). HeLa cells were transfected with 6 μl 20 μM siRNA duplex per well of a 12-well dish using oligofectamine (Invitrogen) and standard procedures (Elbashir et al., 2001). Knock-down of cellular TSG101 was routinely observed after 48 h. For Western blot analysis of cell extracts, cells were solubilized in buffer A containing 1% Triton X-100, and proteins were precipitated immediately in chloroform/methanol before SDS PAGE. For immunoprecipitations, cells were lysed in 0.4 ml boiling buffer A (100 mM NaCl, 1 mM MgCl2, 20 mM Hepes-NaOH, pH 7.4) containing 50 mM NaF, 1 mM Na3VO4, and 1% SDS. Cleared samples were diluted into 5 vol of buffer A containing 2% Triton X-100, incubated at 4°C overnight with antibody and 10 μl protein A-sepharose, and then washed four times before preparation for SDS-PAGE. To measure degradation of 125I EGF, iodinated EGF (50 μCi/μg) was bound (50 ng/ml) to A431 cells for 1 h at 4°C with CO2-independent medium (GIBCO BRL) containing 2 mg/ml BSA (BM). Cells were washed and incubated in 1 ml BM. As indicated, medium was removed and TCA added to 10%. After 1–2 h on ice, samples were centrifuged, and the supernatant was counted for radioactivity.
Fluorescence microscopy
Cells were fixed in 3.5% (wt/vol) PFA with a 0.05% saponin pretreatment as indicated or with methanol. Secondary antibodies were from Jackson ImmunoResearch Laboratories. For EGF uptake, cells were washed in PBS and incubated in BM containing 0.4 μg/ml Oregon green or Texas red EGF (Molecular Probes), washed, and then incubated at 37°C. For transferrin uptake, 20 μg/ml Texas red transferrin (Molecular Probes) was bound and internalized in medium containing 0.1 mg/ml transferrin and 50 μM desferroxamine (Ciba). Cells were examined using a Leica NT confocal microscope or an inverted Olympus IX-70 linked to a Delta Vision system (Applied Precision, Inc.). Quantitation was performed using an Olympus BX60 microscope linked to a cooled slow-scan CCD camera (Roper Scientific) driven by Metamorph software (Universal Imaging Corp.). Images were processed using Adobe Photoshop® 5.0.
Microinjection
Antibodies were dialyzed against 48 mM K2HPO4, 4.5 mM KH2PO4, 14 mM NaH2PO4, pH 7.2, and concentrated. Antibody solutions (0.5 mg/ml) containing 25 μg/ml DAPI were injected using an Eppendorf 5171 micromanipulator and microinjector. Cells were normally incubated for ∼1 h at 37°C between microinjection and the start of internalization assays.
Ubiquitin binding
In vitro translation (Promega) products (5 μl) were diluted in 250 μl buffer A containing 1% Triton X-100 and incubated for 2 h at 4°C with 10 μl prewashed ubiquitin- (Affinity BioReagents, Inc.) or GSH-agarose beads, washed four times in buffer A, and boiled for SDS PAGE. Gels were analyzed by phosphorimaging. For native proteins, HeLa cytosols were incubated for 15 min with 50 IU/ml hexokinase and 5 mM glucose final to deplete ATP and desalted into buffer A. Samples (∼1 mg total protein) were incubated with 20 μl ubiquitin- or GSH-agarose beads as above and then analyzed by Western blotting.
Acknowledgments
We thank Martin Lowe and Sabine Hilfiker for helpful comments during preparation of the article.
N. Bishop is funded by a Wellcome Trust Career Development Fellowship (061045/Z/00/Z/CH/TH/lc), and A. Horman is funded by a Biotechnology and Biological Sciences Research Council studentship (99/A1/C/05291). This work is supported by the Medical Research Council (G117/153, G9722026, and G0001128).
N. Bishop and A. Horman contributed equally to this work.
Footnotes
Abbreviations used in this paper: EEA1, early endosome-associated antigen 1; EGFR, EGF receptor; GSH, glutathione; Hrs, hepatocyte growth factor receptor substrate; hVPS, human VPS; mHrs, mouse Hrs; siRNA, small interfering RNA; TSG101, tumor susceptibility gene 101; Ubc, ubiquitin-conjugating; UIM, ubiquitin-interacting motif; VPS, vacuolar protein sorting.
References
- Amerik, A.Y., J. Nowak, S. Swaminathan, and M. Hochstrasser. 2000. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell. 11:3365–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., B. Wendland, E.J. Estapa, and S.D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., G. Odorizzi, E.J. Estepa, and S.D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 1:248–258. [DOI] [PubMed] [Google Scholar]
- Bishop, N., and P.G. Woodman. 2000. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell. 11:227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, N., and P. Woodman. 2001. Tsg101/mammalian vps23 and mammalian vps28 interact directly and are recruited to vps4-induced endosomes. J. Biol. Chem. 276:11735–11742. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 411:494–498. [DOI] [PubMed] [Google Scholar]
- Felder, S., K. Miller, G. Moehren, A. Ullrich, J. Schlessinger, and C.R. Hopkins. 1990. Kinase activity controls the sorting of the epidermal growth factor receptor within the multivesicular body. Cell. 61:623–624. [DOI] [PubMed] [Google Scholar]
- Garrus, J.E., U.K. von Schwedler, O.W. Pornillos, S.G. Morham, K.H. Zavitz, H.E. Wang, D.A. Wettstein, K.M. Stray, M. Côté, R.L. Rich, et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 107:1–20. [DOI] [PubMed] [Google Scholar]
- Hershko, A., H. Heller, S. Elias, and A. Ciechanover. 1983. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 258:8206–8214. [PubMed] [Google Scholar]
- Hittelman, A.B., D. Burakov, J.A. Iniguez-Lluhí, L.P. Freedman, and M.J. Garabedian. 1999. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 18:5380–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, K., and L. Falquet. 2001. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci. 26:347–350. [DOI] [PubMed] [Google Scholar]
- Hofmann, R.M., and C.M. Pickart. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 96:645–653. [DOI] [PubMed] [Google Scholar]
- Joazeiro, C.A.P., S.S. Wing, H.-K. Huang, J.D. Leverson, T. Hunter, and Y.-C. Liu. 1999. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 286:309–312. [DOI] [PubMed] [Google Scholar]
- Kato, M., K. Miyazawa, and N. Kitamura. 2000. A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J. Biol. Chem. 275:37481–37487. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., M. Babst, and S.D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 106:145–155. [DOI] [PubMed] [Google Scholar]
- Komada, M., R. Masaki, A. Yamamoto, and N. Kitamura. 1997. Hrs, a tyrosine kinase substrate with a conserved double zinc finger domain, is localized to the cytoplasmic surface of early endosomes. J. Biol. Chem. 272:20538–20544. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V., and R.A. Abagyan. 1997. TSG101 may be the prototype of a class of dominant negative ubiquitin regulators. Nat. Genet. 16:330–331. [DOI] [PubMed] [Google Scholar]
- Levkowitz, G., H. Waterman, E. Zamir, Z. Kam, S. Oved, W.Y. Langdon, L. Beguinot, B. Geiger, and Y. Yarden. 1998. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 12:3663–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz, L., H. Waterman, S.A. Ettenberg, M. Katz, A.V. Tsygankov, I. Alroy, S. Lavi, K. Iwai, Y. Reiss, A. Ciechanover, et al. 1999. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell. 4:1029–1040. [DOI] [PubMed] [Google Scholar]
- Li, L., and S.N. Cohen. 1996. tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell. 85:319–329. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano, J., T. Zang, and P.D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313–1319. [DOI] [PubMed] [Google Scholar]
- Odorizzi, G., M. Babst, and S.D. Emr. 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 95:847–858. [DOI] [PubMed] [Google Scholar]
- Patnaik, A., V. Chau, and J.W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA. 97:13069–13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper, R.C., A.A. Cooper, H. Yang, and T.H. Stevens. 1995. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131:603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting, C.P., Y.-D. Cai, and P. Bork. 1997. The breast cancer gene product TSG101: a regulator of ubiquitination? J. Mol. Med. 75:467–469. [PubMed] [Google Scholar]
- Raiborg, C., B. Bremnes, A. Mehlum, D.J. Gillooly, A. D'Arrigo, E. Stang, and H. Stenmark. 2001. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J. Cell Sci. 114:2255–2263. [DOI] [PubMed] [Google Scholar]
- Raymond, C.K., I. Howald-Stevenson, C.A. Vater, and T.H. Stevens. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell. 3:1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, S.E., L.M. Banta, K. Köhrer, J.M. McCaffery, and S.D. Emr. 1996. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell. 7:985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca, A., C. Lamaze, A. Subtil, and A. Dautry-Varsat. 2001. Involvement of the ubiquitin/proteasome system in sorting of the interleukin 2 receptor β chain to late endocytic compartments. Mol. Biol. Cell. 12:1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, U., D.E. Ott, E.N. Chertova, R. Welker, U. Tessmer, M.F. Princiotta, J.R. Bennink, H.-G. Kräusslich, and J.W. Yewdell. 2000. Proteasome inhibition interferes with Gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA. 97:13057–13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack, B., A. Calistri, M.A. Accola, G. Palu, and H.G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA. 97:13063–13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kerkhof, P., C.M. Alves dos Santos, M. Sachse, J. Klumperman, G. Bu, and G.J. Strous. 2001. Proteasome inhibitors block a late step in lysosomal transport of selected membrane but not soluble proteins. Mol. Biol. Cell. 12:2556–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank, L., F. Bouamr, T.J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C.A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA. 98:7724–7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman, H., and Y. Yarden. 2001. Molecular mechanisms underlying endocytosis and sorting of ErbB receptor tyrosine kinases. FEBS Lett. 490:142–152. [DOI] [PubMed] [Google Scholar]