Abstract
CHO1 is a kinesin-like protein of the mitotic kinesin-like protein (MKLP)1 subfamily present in central spindles and midbodies in mammalian cells. It is different from other subfamily members in that it contains an extra ∼300 bp in the COOH-terminal tail. Analysis of the chicken genomic sequence showed that heterogeneity is derived from alternative splicing, and exon 18 is expressed in only the CHO1 isoform. CHO1 and its truncated isoform MKLP1 are coexpressed in a single cell. Surprisingly, the sequence encoded by exon 18 possesses a capability to interact with F-actin, suggesting that CHO1 can associate with both microtubule and actin cytoskeletons. Microinjection of exon 18–specific antibodies did not result in any inhibitory effects on karyokinesis and early stages of cytokinesis. However, almost completely separated daughter cells became reunited to form a binulceate cell, suggesting that the exon 18 protein may not have a role in the formation and ingression of the contractile ring in the cortex. Rather, it might be involved directly or indirectly in the membrane events necessary for completion of the terminal phase of cytokinesis.
Keywords: actin; alternative splicing; cytokinesis; kinesin-like protein; midbody
Introduction
To ensure high fidelity of chromosome segregation, cells form a special device called mitotic spindles. The structural framework of the spindle is microtubules, and their assembly into the spindle and function in chromosome separation and cytokinesis are largely dependent on microtubule-based motor proteins associated with them. A number of kinesin-like proteins have been identified and further divided into distinct subclasses (Miki et al., 2001). One such subclass designated as a mitotic kinesin-like protein (MKLP)*1 subfamily contains plus-end–directed NH2-terminal motor proteins. Since the first member (CHO1) was identified in CHO cells (Sellitto and Kuriyama, 1988), other members of this subfamily have been shown to exist in various organisms, including human (MKLP1 [Nislow et al., 1992]), Caenorhabditis elegans (ZEN-4 [Raich et al., 1998]; CeMKLP1 [Powers et al., 1998]), Drosophila (PAV-KLP [Adams, 1998]), zebrafish (sequence data available from GenBank/EMBL/DDBJ under accession no. AF139990), and sea urchin (Chui et al., 2000). Although all of those proteins are composed of well-conserved three subdomains (NH2-terminal motor region and COOH-terminal globular tail connected with the α-helical coiled-coil central stalk), CHO1 has been noted to be unique in a sense that it contains extra ∼100 amino acids in the middle of the tail (Kuriyama et al., 1994). Since none of the homologs have the CHO1-specific tail sequence, it is not clear whether the proteins are heterogeneous among species or different forms of MKLP1/CHO1 coexist in a single cell type.
MKLP1/CHO1-related proteins show dynamic changes in their subcellular distribution during the cell cycle. In mammalian cells, CHO1 is present in interphase centrosomes and nuclei and becomes associated with the mitotic spindle. As chromosomes move toward poles, the protein shifts to the midzone and eventually concentrates into a bright spot in the middle of the intercellular bridge (Sellitto and Kuriyama, 1988). Since it is a plus-end–directed motor present in the interzonal region of the spindle, MKLP1/CHO1 was originally thought to function in chromosome separation and spindle elongation during karyokinesis (Nislow et al., 1992). In fact, microinjection of CHO1 antibodies caused mitotic arrest in mammalian cells (Nislow et al., 1990) and sea urchin embryos (Wright et al., 1993). However, genetic analysis has suggested involvement of zen-4 and pavarotti in cytokinesis rather than karyokinesis (Adams et al., 1998; Powers et al., 1998; Raich et al., 1998). Evidence has also been presented that overexpression of the mutant CHO1 and RNA-mediated interference specifically blocked completion of cytokinesis in mammalian cells (Matuliene and Kuriyama, 2002). The ability of the motor proteins to organize central spindles and the midbody appears to be essential for their function in cytokinesis, which could be achieved through their interaction with Aurora kinase AIR-2 (Schumacher et al., 1998; Severson et al., 2001) and RhoGAP Cyk-4 (Jantsch-Plunger et al., 2000).
To clarify the nature of molecular diversity among species and define the role of MKLP1/CHO1 during cell division, we examined the chicken genomic sequence. Here we report that heterogeneity of the COOH-terminal tail is derived from alternative splicing and that the sequence encoded by exon 18 is expressed in only the CHO1 isoform. Exon 18 includes a polypeptide capable of interaction with F-actin in vivo and in vitro, and microinjection of exon 18–specific antibodies blocked the terminal phase of cytokinesis. A possibility of CHO1 involvement in membrane events, rather than the actin-containing contractile ring in the cell cortex, is discussed.
Results and discussion
Coexpression of CHO1 and MKLP1 isoforms generated by alternative splicing
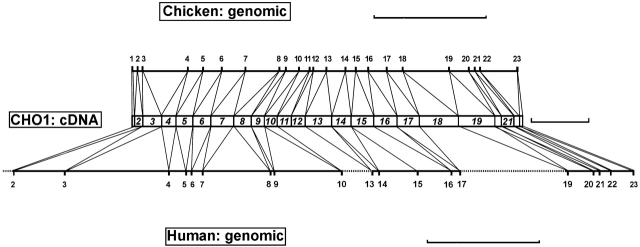
Although CHO1 is a member of the MKLP1 subfamily, it is significantly larger in size than other subfamily members. This suggested a possibility of alternative splicing, which was confirmed by cloning of chicken genomic sequences (Fig. 1) . The chicken CHO1/MKLP1 gene spans ∼17.1 kb in which 22 introns intervene between 23 exons flanked by GT-AG consensus splice sequences (unpublished data). The coding sequences of the motor (amino acids 1–441), stalk (amino acids 442–654), and tail (amino acids 655–954) domains are covered by exons 1–13, 13–17, and 17–23, respectively. By comparing with the human sequence available in the genome databanks, it was concluded that all exon-intron boundaries occur at the same position between two species (Fig. 1). Thus, the splicing pattern of CHO1/MKLP1 gene is well conserved in higher eukaryotes. The major diversity resides in the tail domain at the COOH terminus (Fig. 2) . Particularly prominent is the difference between CHO1 and MKLP1, and a nearly 100 amino acid sequence encoded by exon 18 is expressed in only CHO1 but not the MKLP1 isoform.
Figure 1.
MKLP1/CHO1 gene organization in chicken and human. The middle column represents the full-coding chicken cDNA. Numbers 1–23 indicate the position of exons intervening between 22 introns. Although human MKLP1/CHO1 gene (∼23 kb) is considerably larger than chicken (∼17 kb), almost all splicing sites occur at the same position between two species. A shaded column is a CHO1-specific exon 18, and dotted lines represent the gap detected in the human genomic sequence (sequence data available from EMBL/GenBank/DDBJ under accession no. NT_010222.1; mapped to chromosome 15). Bars: (top and bottom) 5 kb; (middle) 0.5 kb.
Figure 2.
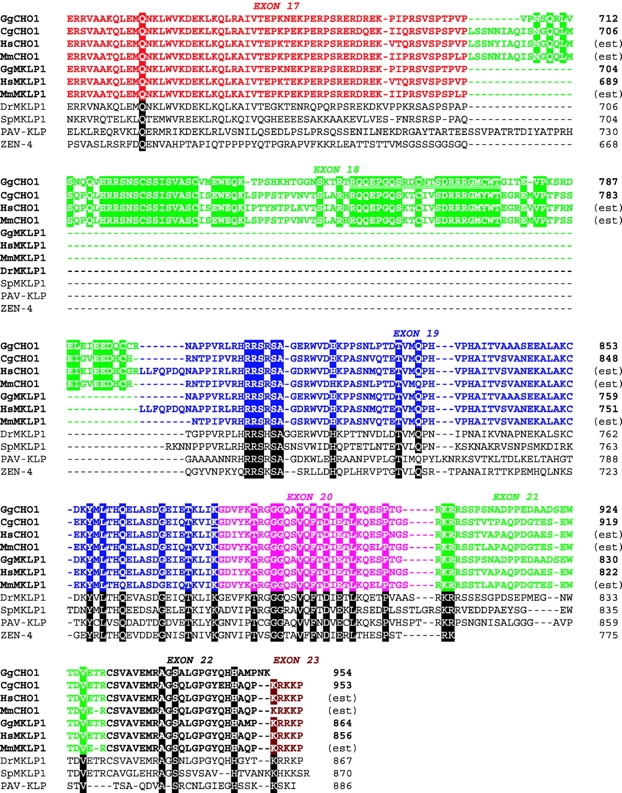
Sequence alignment of the tail among MKLP1 subfamily members. In chicken and human, the COOH-terminal tail is encoded by exons 17–23, which are differently colored. Sequences are derived from chicken (GgCHO1 and GgMKLP1; this paper), CHO cells (CgCH: sequence data available from EMBL/GenBank/DDBJ under accession no. X83575), human EST (HsCHO1: sequence data available from EMBL/GenBank/DDBJ under accession nos. AU123817 and BF897861), mouse EST (MmCHO1: sequence data available from EMBL/GenBank/DDBJ under accession nos. BE333860, AA856173, and BG068324; MmMKLP1: sequence data available from EMBL/GenBank/DDBJ under accession nos. AA798124 and AW907863), HeLa (HsMKLP1: sequence data available from EMBL/GenBank/DDBJ under accession nos. X67155 and S46300), zebrafish (DrMKLP1: sequence data available from EMBL/GenBank/DDBJ under accession no. AF139990), sea urchin (SpMKLP1: sequence data available from EMBL/GenBank/DDBJ under accession no. AAG18582), Drosophila (PAV-KLP: sequence data available from EMBL/GenBank/DDBJ under accession no. AJ224882), and C. elegans (ZEN-4: sequence data available from EMBL/GenBank/DDBJ under accession nos. AF057567 and AF057568). The E18 antibody was raised against the oligopeptide sequence encoded by exon 18 (double underlined).
Chicken cDNA encoding both CHO1- and MKLP1-type proteins was cloned from a library (Fig. 2, GgCHO1 and GgMKLP1), suggesting that two isoforms are likely to be expressed in a single organism. Indeed, cDNA fragments corresponding to each isoform were identified in EST databanks of not only human but also mouse. To confirm simultaneous expression of CHO1 and MKLP1, we double labeled CHO (Fig. 3 A) and HeLa (Fig. 3 B) cells with monoclonal anti-CHO1 antibody (mAb) and polyclonal antibody (E18) specific to exon 18. CHO1 probed by E18 was seen at the center of spindles and midbodies along with the antigen recognized by mAb, which is reactive to the COOH-terminal end of the stalk sequence (unpublished data). Midbodies isolated from CHO cells are also intensely stained with both types of antibodies (Fig. 3, C and D). Thus, CHO1 and MKLP1 are coexpressed and tightly attached to the midbody.
Figure 3.
Midbody staining with domain-specific antibodies. (A–D) Localization of the CHO1 isoform in CHO (A), HeLa cells (B), and isolated CHO midbodies (C and D) probed by E18 antibodies. The same cells/structures were also stained with either mAb (A, B, and D) or anti–α-tubulin antibody (C). Bars, 10 μm. (E) Immunoblot analysis of whole CHO (lanes 1 and 2), HeLa cells (lanes 3and 4), and isolated midbodies (lanes 5 and 6) stained with mAb (lanes 1, 3, and 5) and E18 (lanes 2, 4, and 6).
mAb recognizes polypeptides of 95 and 105 kD in CHO and HeLa cells (Fig. 3 E, lanes 1 and 3). In some occasions, the polypeptides are resolved as multiple bands, suggesting the possibility of protein phosphorylation (unpublished data). When the same protein fractions were probed with E18, only the band with the higher molecular mass was detected (Fig. 3 E, lanes 2 and 4). Both types of proteins are highly enriched in isolated midbodies (Fig. 3, lanes 5 and 6). It is notable that immunostaining (Fig. 3, A and B) and immunoblotting (Fig. 3 E, lanes 2 and 4) signals in whole cells obtained with E18 are far less intense than those with mAb. This may suggest that either CHO1 is minor or midbody staining with E18 is less effective than with mAb, or both. It was also noted that neither PAV-KLP nor ZEN-4/CeMKLP1 includes the sequence corresponding to exon 18 in their genomic sequences. However, immunostaining of Drosophila cultured cells revealed the presence of an E18-reactive molecule (∼200 kD) at the center of the intercellular bridge (unpublished data). Thus, a CHO1-related molecule with the exon 18 sequence might colocalize with PAV-KLP at the midzone and midbody in Drosophila cells.
CHO1-specific exon 18 encodes an actin-interacting domain
To characterize the CHO1-specific sequence, we expressed 91 amino acids encoded by exon 18 in CHO cells. The green fluorescent protein (GFP)-tagged exogenous polypeptide is located inside the cytoplasm in association with cytoskeletal fibers (Fig. 4 A). Double staining with Alexa-phalloidin (MFs) provided evidence that the fibers to which the exon 18 sequence attaches are actin filaments. When cells were treated with dihydrocytochalasin B, GFP was no longer detected along cytoskeletal fibers (Fig. 4 B, +DHCB). Instead, actin-containing dots formed by depolymerization of F-actin become visible by GFP fluorescence. In contrast, the exon 18 polypeptide does not reveal any affinity to the microtubule network (Fig. 4 C). This is in good agreement with our previous observation that CHO1 interacts with microtubules through microtubule-binding sites located at the NH2-terminal half of the protein (Matuliene and Kuriyama, 2002).
Figure 4.
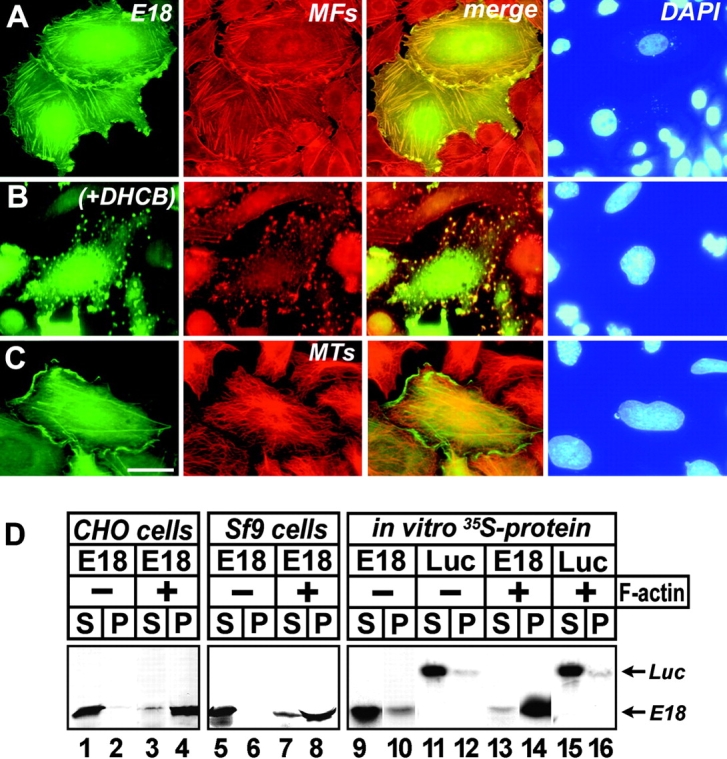
Interaction of GFP–exon 18 protein with F-actin. (A–C) CHO cells expressing the GFP-tagged exon 18 polypeptide were stained with Alexa-conjugated phalloidin (A and B) and α-tubulin antibodies (C). In B, the cells were treated with 5 μg/ml dihydrocytochalasin B (DHCB) for 30 min to depolymerize F-actin before fixation. Bar, 10 μm. (D) The proteins expressed in CHO (lanes 1–4), Sf9 cells (lanes 5–8), and reticulocyte lysates (lanes 9–16) were mixed with (+) or without (−) F-actin. Lanes 11, 12, 15, and 16 include 35S-labeled luciferase prepared as a control protein. GFP–exon 18 bands in supernatant (S) and pellet (P) fractions were visualized by either immunostaining with anti-GFP antibodies (lanes 1–8) or autoradiography (lanes 9–16). Note GFP–exon 18 prepared in the reticulocyte lysate (lane 10) was more easily sedimentable than that expressed in CHO (lane 2) and Sf9 cells (lane 6) without incubation with F-actin.
Affinity of the exon 18 sequence for F-actin was further confirmed by in vitro cosedimentation. Fig. 4 D shows CHO (lanes 1–4) and insect Sf9 cell lysates (lanes 5–8) containing the GFP-tagged exon 18 sequence mixed with F-actin prepared from rabbit skeletal muscles: the polypeptide was cosedimented with F-actin (lanes 4 and 8). Lanes 9–16 demonstrate interaction of F-actin with 35S-labeled GFP–exon 18 synthesized in vitro: the protein was recovered in the pellet along with F-actin (lane 14), whereas the control 35S-luciferase remained in the supernatant (lane 16). Association of GFP–exon 18 was also confirmed with platelet F-actin (unpublished data). These results clearly indicate that the sequence unique to the CHO1 isoform encodes a polypeptide capable of interaction with F-actin. Exon 18 does not appear to contain any known consensus motifs shared among actin-binding proteins. Thus, the question of whether CHO1 interacts with F-actin directly or indirectly still remains to be answered.
CHO1 is seen inside the nucleus, which is due to the presence of a nuclear localization signal at the COOH-terminal end of the tail (unpublished data). When the full-length CHO1 lacking nuclear localization signal was expressed in interphase cells, the tagged molecule was located in the cytoplasm in association with both microtubules and F-actin (unpublished data). However, in mitotic cells the protein was seen predominantly in the spindle, and virtually no fluorescent signal was detected in the cortex where the actin-containing contractile ring is assembled. Since the exon 18 sequence was targeted to the cortex, especially in the vicinity of the furrow during cytokinesis in mitotic cells (unpublished data), the interaction of CHO1 with actin-containing structures may be controlled in a cell cycle–dependent manner.
Cells treated with E18 antibodies failed to complete the terminal phase of cytokinesis
By mutating the mechanochemical motor domain of CHO1, we have shown previously that the protein is required for completion of cytoplasmic division in mammalian cells (Matuliene and Kuriyama, 2002). To examine whether inhibition of the actin-interacting domain results in the similar phenotypes, we microinjected the affinity purified E18 antibody into PtK1 cells. In Fig. 5 , the metaphase cell received E18 at time zero (Fig. 5 A) underwent normal chromosomes separation (Fig. 5 C) and cytoplasmic division (Fig. 5 D). Although two daughter cells appeared to be separated completely, the cell boundary became unclear (Fig. 5, E and F), and the two cells eventually merged together by ∼3 h after antibody injection (Fig. 5 G). mAb staining of the fixed cell clearly indicated the formation of a midbody between two separated nuclei (Fig. 5, arrows). Nonetheless, the furrow ultimately resumed to produce a binucleate cell (Fig. 5, DAPI), suggesting that E18 specifically inhibits the terminal phase of cytokinesis. The effect of E18 was specific because no major inhibition of cell division was observed in cells treated with injection buffer alone, commercially available nonspecific rabbit IgG, and protein A–purified E18 preimmune antibodies (Table I). These results contrast with those of Nislow et al. (1990) and Wright et al. (1993) who observed mitotic arrest in mAb-injected cells. The difference may be attributed to the specificity of the antibodies injected into cells (mouse monoclonal IgM recognizing the central stalk versus rabbit E18 IgG specific to the tail). The role of MKLP1/CHO1 in early stages of mitosis is left unsolved.
Figure 5.
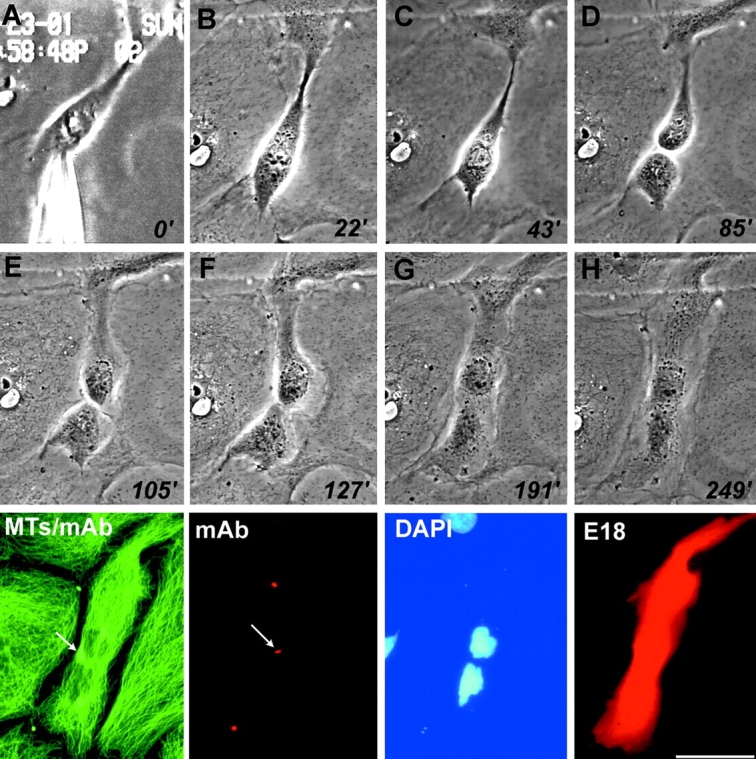
Microinjection of E18 antibodies into PtK 1 cells. Numbers at the corner are the time after antibody injection. After fixation with cold methanol, the cells were stained with mAb (probed by FITC-conjugated secondary antibodies, but its color was converted from green to red), DAPI, and anti-rabbit IgG secondary antibody (E18). The coverslip was further stained with anti–α-tubulin and FITC-conjugated secondary antibodies (MTs/mAb). Arrows indicate the position of a midbody formed between two separated nuclei. Bar, 50 μm.
Table I.
Effects of antibody injection into mitotic PtK 1 cells
| Cells counted | Normal cell division | Incomplete cell division a | Two cells appositioned b | Arrested/no changes c | |||||
|---|---|---|---|---|---|---|---|---|---|
| (%)
|
|||||||||
| E18d | 73 | 6(8) | 41(56) | 22(30) | 4(5) | ||||
| E18-pree | 29 | 26(90) | 0(0) | 1(3) | 2(7) | ||||
| Controlf | 41 | 35(85) | 5(12) | 0(0) | 1(2) | ||||
| Injection bufferg | 12 | 9(75) | 0(0) | 0(0) | 3(25) | ||||
Although the cells underwent normal chromosome segregation and cytokinesis, regression of the cleavage furrow and/or cell fusion resulted in the formation of multinucleate cells.
Two almost completely separated cells were closely aligned side-by-side. A part of the cell boundary was unclear, and the two cells appeared to be partially connected to each other.
The cells either arrested at mitosis or entered interphase without cell division.
5–13 mg/ml affinity purified E18 rabbit immunoglobulin molecules were prepared in injection buffer and introduced into PtK1 cells at prophase to anaphase. The cells were fixed at 2–6 h after antibody injection.
Protein A–purified preimmune E18 antibodies.
Two different batches of rabbit IgG fractions, which are commercially available.
Injection buffer alone.
Initiation and ingression of cleavage furrows proceeded normally in E18-injected cells, suggesting that the actin-interacting site of CHO1 may not be involved in the formation of contractile ring and the process of cell cleavage. Rather, the protein may function to link the microtubule-containing central spindle to the cortex/membrane at the cell equator during the late stage of cell division. Time-lapse analysis of dividing cells suggested that the midbody matrix could be a structure holding microtubules and the cell cortex/membrane together after the cessation of the furrowing and the disassembly of the contractile ring (Mullins and Biesele, 1977). Both the motor activity and microtubule-bundling capacity of CHO1 are required for completion of cytokinesis by organizing midzone microtubules and the electron-dense matrix in the center of the intercellular bridge (Matuliene and Kuriyama, 2002). Old and new observations clearly indicate that complete separation of two daughter cells is achieved through stretching and breaking the intercellular bridge at the narrowest region flanking the midbody center (Mullins and Biesele, 1977; Piel et al., 2001). Since microtubules derived from the remnant of the central spindle are still connected to each daughter cell, the microtubule bundle forming a core of intercellular bridge must be under tension between two cells. Therefore, for successful separation of daughter cells the microtubule rope must be tightly attached to the membrane/cortex during this tug-of-war. It might be CHO1 that is responsible for connection between the midbody matrix and cell membrane.
Alternatively, the motor protein could be involved in membrane events necessary for completion of cytokinesis. It is widely believed that targeted secretion, insertion, and fusion of membrane vesicles play a central role in progression and completion of cleavage furrows (for review see Straight and Field, 2000). Thus, cells lacking syntaxins and their associated proteins (Lukowitz et al., 1996; Heese et al., 2001), a phospholipid kinase (Brill et al., 2000), dynamin (Gu and Verma, 1996), or Golgi-associated proteins (Sisson et al., 2000), are defective in cytokinesis in a variety of cells. Rabkinesin-6/Rab6-KIFL, a Rab6-binding kinesin-like protein, is required for not only membrane traffic but also cytokinesis (Hill et al., 2000). Of particular interest is that this motor protein shares the highest degree of sequence identity to CHO1 (Echard et al., 1998). Skop et al. (2001) have reported recently that brefeldin, a potent inhibitor of vesicle secretion by targeting a small GTPase-binding protein Arf, specifically blocks the terminal phase of cytokinesis by regressing ingressed furrows in C. elegans. The phenotype detected in brefeldin-treated embryos is remarkably similar to what we saw in E18-injected mammalian cells. Importantly, CHO1/MKLP1 is capable of binding Arfs through the sequence encoded by exons 19–22, just downstream the actin-interacting domain of exon 18 (Boman et al., 1999). Since dividing cells contain dynamic membrane phospholipids tightly coupled with the actin cytoskeleton (Emoto and Umeda, 2000) and Arfs are believed to be involved in both membrane traffic and actin dynamics, it is likely that the actin- and Arf-interacting domains act in concert to achieve the CHO1 function during the late stage of cytokinesis. Further analysis of CHO1 would be of great benefit for our understanding of the mechanism and regulation of cell division.
Materials and methods
Cloning of cDNA and genomic sequences of chicken CHO1 and MKLP1
cDNA encoding chicken CHO1- and MKLP1-type of motor proteins were cloned by screening of a chicken cDNA library (Stratagene) with CHO1 probes derived from CHO cells (Kuriyama et al., 1994). To extend the 5′ sequence, mRNA was purified from cultured chicken lymphocytes (DT40 cells) and used for 5′-RACE with specific and degenerate primers matching the conserved ATP-binding consensus motif (amino acids 118–124). For cloning of genomic DNA, 10 primers (nucleotide positions 14 → 42, 3891 ← 3927, 3891 → 3927, 7031 ← 7073, 6294 → 6328, 9635 ← 9667, 9594 → 9633, 13863 ← 13902, 13851 → 13892, and 17097 ← 17138) were designed based on the nucleotide sequence of CHO1/MKLP1, and PCR was performed with DT40 genomic templates. Five fragments in a size between 3.2 and 4.3 kb were cloned and assembled after nucleotide sequence analysis.
Cell culture, protein expression, and immunostaining
CHO and HeLa cells were cultured in 10% FCS containing Ham's F-10 and DME medium, respectively. The exon 18 coding sequence (amino acid positions 696–786) was isolated by digestion of the full-length CHO-CHO1 with SspI and Eam1104 I and ligated into the eukaryotic expression vector, pEGFP-C1 (CLONTECH Laboratories, Inc.). CHO cells on a coverslip in a 35-mm dish were transfected by addition of 0.6–2 μg/ml purified plasmid DNA and cultured overnight (Matuliene and Kuriyama, 2002). To enrich mitotic cell populations, cells were partially synchronized by treatment with thymidine followed by accumulation at M phase by addition of nocodazole at a final concentration of 0.05 μg/ml (Sellitto and Kuriyama, 1988). After washing out the drug, cells were allowed to recover for 20–50 min before fixation with cold methanol.
For immunofluorescence staining, cells were rehydrated with 0.05% Tween-20 containing PBS and incubated with primary and secondary antibodies. Primary antibodies include monoclonal anti–α-tubulin antibodies (Sigma-Aldrich), monoclonal CHO1 (mAb [Sellitto and Kuriyama, 1988]), and rabbit polyclonal antibodies (E18) raised against 23 amino acids in exon 18 at amino acid positions 755–777 of chicken CHO1 (Fig. 2, double underlined). Immunoblotting analysis was done as described before (Kuriyama et al., 1994).
In vitro cosedimentation with F-actin
Actin was prepared from rabbit skeletal muscles (a gift from Drs. Albina Orlova and Ewa Prochniewicz, University of Minnesota, Minneapolis, MN) (Orlova et al., 1995), and platelet actin was purchased from Cytoskeleton, Inc. G-actin was polymerized and sedimented in a medium containing 5 mM Tris-HCl, pH 7.8, 0.2 mM ATP, 0.1 mM CaCl2, 150 mM KCl, and 1 mM MgCl2. For protein expression in insect cells, cDNA encoding GFP–exon 18 was subloned into pVL1392 (PharMingen) and used for infection of Sf9 cells as before (Kuriyama et al., 1994). 35S-labeled GFP–exon 18 was synthesized using a commercially available kit (TNT Coupled Reticulocyte Lysate System; Promega). CHO and Sf9 cells expressing exogenous proteins were washed once with PBS and lysed for 30–60 min at 0°C in a medium containing 10 mM Tris-HCl, pH 7.8, 0.5% Nonidet NP-40, and protease inhibitors. Cell extracts and reticulocyte lysates recovered after centrifugation at 200,000 g for 30 min were mixed with F-actin and further incubated for 30 min on ice. After layering on a 20% sucrose cushion, F-actin plus associated proteins were sedimented at 100,000 g for 30 min at 4°C.
Antibody injection
Affinity purified E18 was prepared in injection buffer (140 mM KCl, 100 mM glutamic acid, 40 mM citric acid, 1 mM MgCl2, 1 mM EGTA, pH 7.4) and injected into PtK1 cells cultured on a photoetched coverslip (Bellco). Microinjection was performed using a Narishige microinjector attached to a Nikon Diaphot inverted microscope, and time-lapse images were recorded with a Nikon Eclipse TE200 microscope using ImagePro software packages.
Acknowledgments
This work was supported by National Institutes of Health grant GM55735 to R. Kuriyama.
Footnotes
Abbreviations used in this paper: GFP, green fluorescent protein; MKLP, mitotic kinesin-like protein.
References
- Adams, R.R., A.A. Tavares, A. Salzberg, H.J. Bellen, and D.M. Glover. 1998. pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 12:1483–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman, A.L., J. Kuai, X. Zhu, J. Chen, R. Kuriyama, and R.A. Kahn. 1999. Arf proteins bind to mitotic kinesin-like protein 1 (MKLP1) in a GTP-dependent fashion. Cell Motil. Cytoskeleton. 44:119–132. [DOI] [PubMed] [Google Scholar]
- Brill, J.A., G.R. Hime, M. Scharer-Schuksz, and M.T. Fuller. 2000. A phospholipid kinase regulates actin organization and intercellular bridge formation during germline cytokinesis. Development. 127:3855–3864. [DOI] [PubMed] [Google Scholar]
- Chui, K.K., G.C. Rogers, A.M. Kashina, K.P. Wedaman, D.J. Sharp, D.T. Nguyen, F. Wilt, and J.M. Scholey. 2000. Roles of two homotetrameric kinesins in sea urchin embryonic cell division. J. Biol. Chem. 275:38005–38011. [DOI] [PubMed] [Google Scholar]
- Echard, A., F. Jollivet, O. Martinez, J.-J. Lacapere, A. Rousselet, I. Janoueix-Lerosey, and B. Gould. 1998. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science. 279:580–585. [DOI] [PubMed] [Google Scholar]
- Emoto, K., and M. Umeda. 2000. An essential role for a membrane lipid in cytokinesis: regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J. Cell Biol. 149:1215–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X., and D.P. Verma. 1996. Phragmoplastin, a dynamin-like protein associated with cell plate formation in plants. EMBO J. 15:695–704. [PMC free article] [PubMed] [Google Scholar]
- Heese, M., X. Gansel, L. Sticher, P. Wick, M. Grebe, F. Granier, and G. Jurgens. 2001. Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J. Cell Biol. 155:239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, E., M. Clarke, and F.A. Barr. 2000. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J. 19:5711–5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger, V., P. Gonczy, A. Romano, H. Schnabel, D. Hamill, R. Schnabel, A.A. Hyman, and M. Glotzer. 2000. CYK-4: a Rho family GTPase activating protein (GAP) required for central spindle formation and cytokinesis. J. Cell Biol. 149:1391–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama, R., S. Dragas-Granoic, T. Maekawa, A. Vassilev, A. Khodjakov, and H. Kobayashi. 1994. Heterogeneity and microtubule interaction of the CHO1 antigen, a mitosis-specific kinesin-like protein. Analysis of subdomains expressed in insect Sf9 cells. J. Cell Sci. 107:3485–3499. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., U. Mayer, and G. Jurgens. 1996. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 84:61–71. [DOI] [PubMed] [Google Scholar]
- Matuliene, J., and R. Kuriyama. 2002. Kinesin-like motor protein CHO1 is required for the formation of midbody matrix and the completion of cytokinesis in mammalian cells. Mol. Biol. Cell. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, H., M. Setou, K. Kaneshiro, and N. Hirokawa. 2001. All kinesin superfamily protein, KIF, genes in mouse and human. Proc. Natl. Acad. Sci. USA. 98:7004–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, J.M., and J.J. Biesele. 1977. Terminal phase of cytokinesis in D-98s cells. J. Cell Biol. 73:672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow, C., C. Sellitto, R. Kuriyama, and J.R. McIntosh. 1990. A monoclonal antibody to a mitotic microtubule-associated protein blocks mitotic progression. J. Cell Biol. 111:511–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow, C., V.A. Lombillo, R. Kuriyama, and J.R. McIntosh. 1992. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 359:543–547. [DOI] [PubMed] [Google Scholar]
- Orlova, A., E. Prochniewicz, and E.H. Egelman. 1995. Structural dynamics of F-actin: II. Cooperativity in structural transitions. J. Mol. Biol. 245:598–607. [DOI] [PubMed] [Google Scholar]
- Piel, M., J. Nordberg, U. Euteneuer, and M. Bornens. 2001. Centrosome-dependent exit of cytokinesis in animal cells. Science. 291:1550–1553. [DOI] [PubMed] [Google Scholar]
- Powers, J., O. Bossinger, D. Rose, S. Strome, and W. Saxton. 1998. A nematode kinesin required for cleavage furrow advancement. Curr. Biol. 8:1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich, W.B., A.N. Moran, J.H. Rothman, and J. Hardin. 1998. Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol. Biol. Cell. 9:2037–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellitto, C., and R. Kuriyama. 1988. Distribution of a matrix component of the midbody during the cell cycle in Chinese hamster ovary cells. J. Cell Biol. 106:431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, J.M., A. Golden, and P.J. Donovan. 1998. AIR-2: an aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 143:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson, A.F., D.R. Hamill, J.C. Carter, J. Schumacher, and B. Bowerman. 2001. The Aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr. Biol. 10:1162–1171. [DOI] [PubMed] [Google Scholar]
- Sisson, J.C., C. Field, R. Ventura, A. Royou, and W. Sullivan. 2000. Lava Lamp, a novel peripheral Golgi protein, is required for Drosophila melanogaster cellularization. J. Cell Biol. 151:905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop, A.R., D. Bergmann, W.A. Mohler, and J.G. White. 2001. Completion of cytokinesis in C. elegans requires a brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr. Biol. 11:735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight, A.F., and C.M. Field. 2000. Microtubules, membranes and cytokinesis. Curr. Biol. 10:R760–R770. [DOI] [PubMed] [Google Scholar]
- Wright, B.D., M. Terasaki, and J.M. Scholey. 1993. Roles of kinesin and kinesin-like proteins in sea urchin embryonic cell division: evaluation using antibody microinjection. J. Cell Biol. 123:681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]