Abstract
The current advances in fluorescence microscopy, coupled with the development of new fluorescent probes, make fluorescence resonance energy transfer (FRET) a powerful technique for studying molecular interactions inside living cells with improved spatial (angstrom) and temporal (nanosecond) resolution, distance range, and sensitivity and a broader range of biological applications.
Keywords: FRET microscopy; data analysis; cameleons; dimerization; FRET assays
Fluorescence resonance energy transfer (FRET)* is a distance-dependent physical process by which energy is transferred nonradiatively from an excited molecular fluorophore (the donor) to another fluorophore (the acceptor) by means of intermolecular long-range dipole–dipole coupling. FRET can be an accurate measurement of molecular proximity at angstrom distances (10–100 Å) and highly efficient if the donor and acceptor are positioned within the Förster radius (the distance at which half the excitation energy of the donor is transferred to the acceptor, typically 3–6 nm). The efficiency of FRET is dependent on the inverse sixth power of intermolecular separation (Förster, 1965; Clegg, 1996; Lakowicz, 1999), making it a sensitive technique for investigating a variety of biological phenomena that produce changes in molecular proximity (dos Remedios et al., 1987). Technological advances in light microscopy imaging, combined with the availability of genetically encoded fluorescent proteins provide the tools necessary to obtain spatial and temporal distribution of protein associations inside living cells (Heim and Tsien, 1996; Day, 1998; Elangovan et al., 2002, 2003).
The widely used donor and acceptor fluorophores for FRET studies come from a class of autofluorescent proteins, called GFPs. The spectroscopic properties that are carefully considered in selecting GFPs as workable FRET pairs include sufficient separation in excitation spectra for selective stimulation of the donor GFP, an overlap (>30%) between the emission spectrum of the donor and the excitation spectrum of the acceptor to obtain efficient energy transfer, and reasonable separation in emission spectra between donor and acceptor GFPs to allow independent measurement of the fluorescence of each fluorophore (Pollok and Heim, 1999). GFP-based FRET imaging methods have been instrumental in determining the compartmentalization and functional organization of living cells and for tracing the movement of proteins inside cells (Hanson and Kohler, 2001).
In this paper, we give a brief description of FRET microscopic imaging techniques and data analysis. In addition, important biological applications of FRET microscopy, such as protein interaction studies, Ca2+ signaling, nucleic acid studies, characterization of gene expression, and real-time PCR assays, are discussed.
Supplemental material available
More technical details of various intensity-based FRET and fluorescence lifetime imaging techniques are available online at http://www.jcb.org/cgi/content/full/jcb.200210140/DC1. A discussion about the successes of other FRET microscopy techniques is also included.
FRET imaging microscopy
Whereas light microscopy initiated our understanding of cellular structure and the associated function, molecular biological studies over the past few decades have shown that cellular events, such as signal transduction and gene transcription, require the assembly of proteins into specific macromolecular complexes. Traditional biophysical or biochemical methods did not provide direct access to the interactions of these protein partners in their natural environment. Intensity-based imaging techniques applying the method of FRET microscopy (wide field, confocal, and multiphoton [MP]) were subsequently developed, facilitating the study of these interactions inside intact living cells (Periasamy, 2001). New imaging technologies, coupled with the development of new genetically encoded fluorescent labels and sensors and the increasing capability of computer software for image acquisition and analysis, have enabled more sophisticated studies of protein functions and processes ranging from gene expression to second-messenger cascades and intercellular signaling (Roessel and Brand, 2002).
FRET microscopy relies on the ability to capture fluorescent signals from the interactions of labeled molecules in single living or fixed cells. If FRET occurs, the donor channel signal will be quenched and the acceptor channel signal will be sensitized or increased (Herman, 1998). With FRET microscopic imaging, not only colocalization of the donor- and acceptor-labeled probes within ∼0.09 μm2 can be seen, but molecular associations at close distances can be verified.
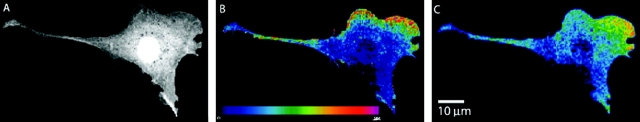
Several FRET microscopy techniques exist, each with its own advantages and disadvantages. They are used for various biological applications, including studies of organelle structure, conjugated antibodies, cytochemical identification, and oxidative metabolism (www.cyto.purdue.edu). Wide-field microscopy is the simplest and most widely used technique. It is used for quantitative comparisons of cellular compartments and time-lapse studies for cell motility, intracellular mechanics, and molecular movement (www.api.com). For example, new fluorescent indicators have allowed the measurement of Ca2+ signals in the cytosol and organelles that are often extremely localized (Miyawaki et al., 1997) and nondestructive imaging of dynamic protein tyrosine kinase activities in single living cells (Ting et al., 2001). Wide-field microscopy, however, suffers from a major drawback due to the generation of out of focus signals. Laser scanning confocal and MP-FRET microscopy provide the advantage of rejecting out of focus information. They also allow associations occurring inside the cell to be localized in three dimensions. A confocal FRET image (Fig. 1) , with improved lateral resolution, yields a wealth of spectral information with several advantages over a wide-field image, including controllable depth of field and the ability to collect serial optical sections from thick specimens. Owing to its nanometer depth resolution and nonintrusiveness, confocal FRET provides a new approach to measure viscoelasticity and biochemical responses of living cells and real-time monitoring of cell membrane motion in natural environments (unpublished data). Confocal microscopy, however, is limited to standard laser lines of defined wavelengths. MP-FRET microscopy overcomes this limitation by using a tunable laser (range 700–1000 nm), allowing excitation of a wide variety of fluorophores with higher axial resolution, greater sample penetration, reduced photobleaching of marker dyes, and increased cell viability. These advantages allow investigations on thick living tissue specimens that would not otherwise be possible with conventional techniques. However, all of these intensity-based FRET techniques require processing software to remove the unwanted bleedthrough components in the FRET image (Fig. 2) .
Figure 1.
Confocal FRET analysis demonstrates that integrins induce local Rac–effector coupling. NIH-3T3 cells were microinjected with cDNAs encoding the indicated GFP–V12-Rac fusion proteins and then with Alexa–PBD protein (Pozo et al., 2002). Donor (A), uncorrected FRET (B), and corrected FRET (C) images are shown. In the color scale, red represents a high FRET signal and blue represents a low signal.
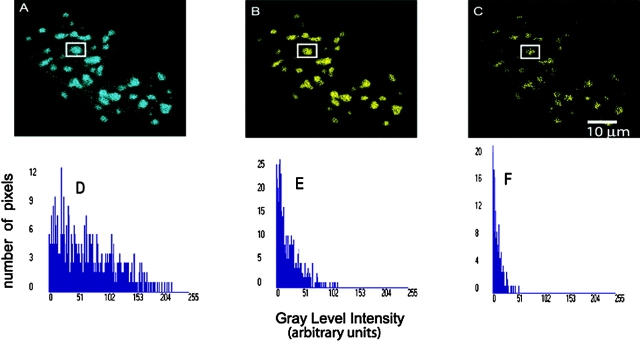
Figure 2.
Localization of CFP– and YFP–C/EBPα proteins expressed in live mouse pituitary GHFT1-5 cells studied using Bio-Rad Laboratories MP-FRET microscopy. The donor (A), the uncorrected FRET (B), and processed FRET (C) images and their respective histograms (D, E, and F) representing the signal strength of the selected protein (Elangovan et al., 2003).
Other FRET imaging techniques.
Temporal resolution of the imaging modalities can be achieved by the technique of fluorescence lifetime imaging (FLIM). This technique monitors the localized changes in probe fluorescence lifetime and provides an enormous advantage for imaging dynamic events within the living cells. When combined with FRET, this approach provides direct evidence for the physical interactions between two or more proteins with very high spatial and temporal resolution (Bastiaens and Squire, 1999; Elangovan et al., 2002). Fluorescence correlation spectroscopy (FCS) (www.probes.com), in which spontaneous fluorescence intensity fluctuations are measured in a microscopic detection volume, has greatly increased the utility and scope of FRET. FRAP, based on the principle of observing the rate of recovery of fluorescence due to the movement of a fluorescent marker into an area of the membrane, is widely used to assess the structure of biological membranes and to measure the lateral diffusion of various membrane or cytoplasmic constituents. A related technique, fluorescence loss in photobleaching (FLIP), can reveal whether flow occurs between two compartments or not. FRAP and FLIP are valuable for studies of Golgi/ER trafficking in animal cells (Cole et al., 1996). With the development of new FRET modalities, like homotransfer or energy migration FRET, photochromic FRET (Giordano et al., 2002), bioluminescence resonance energy transfer (BRET) (www.lifesciences.perkinelmer.com), and a new class of luminophores, called long-wavelength long-lifetime high-quantum yield luminophores, the future of FRET is bright. More specifically, the future biological applications of FRET imaging microscopy may include cellular events coupled to specific molecular signaling processes and simultaneous observation of a series of reversible molecular processes in living cells (Truong and Ikura, 2001).
FRET data analysis
Intensity-based FRET imaging microscopy suffers from various drawbacks, including autofluorescence, detector noise, optical noise, and photobleaching. In addition, spectral bleedthrough (SBT) or contributions of donor and acceptor fluorescence emission into the FRET channel is a major problem. Due to these effects, the observed FRET signal is higher than the actual signal. To correct for these problems, various methods of FRET data analysis for wide-field microscopy have been developed (Gordon et al., 1998; Kraynov et al., 2000; Xia and Liu, 2001). These methods correct for SBT and for the dependence of FRET on donor and acceptor concentrations.
Recently, a new algorithm that removes both the donor and acceptor SBT problems and corrects the variation in fluorophore expression level for all intensity-based FRET techniques (wide field, confocal, and MP) has been developed to calculate FRET efficiency (in percent) and estimate the distance (in angstroms) between donor and acceptor molecules in a double-labeled cell. Both SBT and fluorophore expression level corrections are incorporated in mathematical calculations (Elangovan et al., 2003). From the data collected, the bleedthrough component is evaluated based on the individual donor and acceptor specimens and is then eliminated from the FRET data, pixel by pixel, to obtain the true (or precision) FRET signal (Fig. 2).
FRET applications in cell biology
FRET imaging using GFP spectral mutants provides the ability to localize and monitor ion binding and molecular protein–protein interactions in living cells. For example, FRET microscopy with a CFP/YFP FRET pair allows the detection of direct intermolecular integrin interactions in vivo. Integrins are important transmembrane receptor proteins involved in cell signaling and adhesion. FRET-based studies of Rac activation and localization revealed that integrins induce local Rac–effector coupling by directing Rac to membranes and dissociating it from Rho-GDI (guanine nucleotide dissociation inhibitors), and also that in spite of its homogeneous distribution in the cell, constitutively active Rac selectively interacts with effectors at specific regions of the cell edge (Pozo et al., 2002) (Fig. 1).
The onset and termination of Ca2+ signaling in specific cellular compartments like the cytoplasm, nucleus, or endoplasmic reticulum can be observed by measuring the change in the ratio of the fluorescence intensities of acceptor and donor molecules in live cells (Truong et al., 2001). Cameleons, a class of fluorescent indicators for Ca2+ based on GFPs and calmodulin (CAM), are useful tools in measuring the free Ca2+ concentrations in living cells. The traditional yellow cameleon consists of a fusion of CFP, CAM, the CAM-binding peptide of myosin light chain kinase (MLCKp), and a YFP. With the increase of free Ca2+ in solution, the CAM module of the cameleon binds Ca2+ and wraps around the fused MLCKp. This conformational change decreases the distance between CFP and YFP. Because FRET depends on both the proximity of the donor and acceptor fluorophores and the orientation of their relative dipoles, the interaction of cameleons with Ca2+ leads to changes in the degree of FRET between CFP and YFP. After calibration of the FRET response to known Ca2+ concentrations, the degree of FRET in vivo can theoretically reflect the absolute levels of Ca2+ present in cellular compartments.
Beyond intracellular ion sensing, FRET constructs have been used to investigate the downstream events of second messenger signaling (Adams et al., 1991). Targeting the cAMP signaling pathway, a sensor construct has been designed in which the kinase-inducible domain of cAMP-responsive element–binding protein forms a linker between blue fluorescent protein and GFP. cAMP-induced PKA-dependent phosphorylation is known to cause a conformational change in the kinase-inducible domain, and FRET efficiency is altered in response to cAMP signaling.
FRET establishes the possibility of studying on a localized spatial scale the interactions between a receptor–ligand pair, dimerization of individual receptors, as well as transbilayer distribution of fluorescent lipid analogs and protein-mediated lipid transfer between vesicles. Changes in FRET efficiency induced by fatty acids, phospholipids, and cholesterol led to the identification of discrete binding sites for lipids on the membrane-bound nicotinic acetylcholine receptor protein (www.kenes.com/cholinergic). FRET is used to detect EGF receptor (EGFR) dimerization and its conformational state (Gadella and Jovin, 1995). Fluorescently labeled EGF molecules with fluorescein donor and rhodamine acceptor were allowed to bind EGFR present on cells. The extent of oligomerization of receptors was monitored by the spatially resolved FRET efficiency as a function of the donor/acceptor ratio and treatment conditions. The increase in average FRET efficiency indicated a minimal receptor dimerization for the subpopulation of high-affinity receptors. These results proved that binding of EGF leads to a fast and temperature-dependent microclustering of EGFR, which is present in a predimerized or oligomerized state.
FRET is also used to study the structure, conformation, hybridization, and automated sequencing of nucleic acids. Chromosome FISH, based on hybridization of a nucleic acid fragment to its complement, has become extremely important for gene mapping, identification of mutations, clinical diagnostics, and studies of chromosomal and nuclear architecture. A homogeneous DNA diagnostic assay based on template-directed primer extension detected by FRET, named the template-directed dye-terminator incorporation assay, has been developed for mutation detection and high throughput genome analysis (Chen et al., 1997).
A more recent approach to the characterization of gene expression involves the use of a “fluorescent timer,” a mutant of the dsRed fluorescent protein that shifts color from green to red over time. Green fluorescence indicates recently translated protein, which over the course of hours undergoes an oxygen-dependent autocatalytic reaction to generate a red fluorescence, denoting matured protein. As the timer protein switches fluorescence over time, it can be used as a timer for gene expression. A tissue thus indicates its fluorescent timer production history by its ratio of green to red fluorescence; tissues that have recently initiated gene expression appear green, those with continuous expression appear yellow to orange, and those that have ceased expression appear entirely red.
FRET also finds significant application in membrane fusion assays and real-time PCR assays. In the lipid-mixing assays based on NBD–rhodamine energy transfer (Struck et al., 1981), membranes labeled with a combination of FRET donor and acceptor lipid probes are mixed with unlabeled membranes. FRET decreases when the average spatial separation of the probes is increased upon fusion of labeled membranes with unlabeled membranes. In real-time PCR, the amount of fluorescence emission at each cycle is monitored as an indicator of amplicon production. Fluorescence monitoring of PCR for detection and quantification has become a standard method with many applications, including expression analysis and pathogen detection.
FRET immunoassays, comprising a Cy5 NH2-terminally labeled phosphopeptide, which is recognized by an antiphosphotyrosine primary mouse antibody, followed by a Cy3-labeled secondary antibody, are useful in measuring specific antibody–antigen interactions. FRET occurs when the components are sequentially bound together and excitation at Cy3 wavelengths produces emission at Cy5 wavelengths. Disruption of the interaction between the phosphopeptide and primary antibody will result in a reduction of the FRET signal observed. FRET is also used in the design and synthesis of FRET-based fluorogenic enzyme substrates, useful in monitoring the enzymatic activity.
Conclusion
Biology, imaging, and spectroscopy have been recently combined to provide powerful tools for research and clinical applications. FRET techniques and fluorescent reagents such as GFP, which are widely used to probe for the presence and activity of genes, cellular components, and metabolic or signaling pathways, can become more powerful and flexible when multispectral imaging tools are employed. Spectral imaging has started to improve chromosomal analysis and genotyping methods with easy and accurate detection of chromosomal rearrangements related to cancer and genetic abnormalities. Optical biopsy techniques use spectroscopic information present in intrinsic or exogenous chromophores and fluorophores to differentiate tumors and dysplasias from normal tissues. Potential applications for tissue FRET imaging are also burgeoning. With recent advances in fluorescent probes, instrumentation, and methodologies, FRET is sure to revolutionize scientific research in the near future.
Supplemental Material
Acknowledgments
We wish to acknowledge Drs. Richard Day and Martin Schwartz for their helpful discussions. We thank Colten Noakes and Ms. Ye Chen for their expert assistance.
This work was supported by the W.M. Keck Foundation.
The online version of this article includes supplemental material.
Footnotes
Abbreviations used in this paper: CAM, calmodulin; EGFR, EGF receptor; FRET, fluorescence resonance energy transfer; MP, multiphoton; SBT, spectral bleedthrough.
References
- Adams, S.R., A.T. Harootunian, Y.J. Buechler, S.S. Taylor, and R.Y. Tsien. 1991. Fluorescence ratio imaging of cyclic AMP in single cells. Nature. 349:694–697. [DOI] [PubMed] [Google Scholar]
- Bastiaens, P.I., and A. Squire. 1999. Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol. 9:48–52. [DOI] [PubMed] [Google Scholar]
- Chen, X., B. Zehnbauer, A. Gnirke, and P.Y. Kwok. 1997. Fluorescence energy transfer detection as a homogeneous DNA diagnostic method. Proc. Natl. Acad. Sci. USA. 94:10756–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg, R.M. 1996. Fluorescence resonance energy transfer. Fluorescence Imaging Spectroscopy and Microscopy. Vol. 137. X.F. Wang and B. Herman, editors. John Wiley & Sons Inc., New York. 179–251.
- Cole, N.B., C.L. Smith, N. Sciaky, M. Terasaki, M. Edidin, and J.L. Schwartz. 1996. Diffusional mobility of Golgi proteins in membranes of living cells. Science. 273:797–801. [DOI] [PubMed] [Google Scholar]
- Day, R.N. 1998. Visualization of Pit-1 transcription factor interactions in the living cell nucleus by fluorescence resonance energy transfer microscopy. Mol. Endocrinol. 12:1410–1419. [DOI] [PubMed] [Google Scholar]
- dos Remedios, C.G., M. Miki, and J.A. Barden. 1987. Fluorescence resonance energy transfer measurements of distances in actin and myosin: A critical evaluation. J. Muscle Res. Cell Motil. 8:97–117. [DOI] [PubMed] [Google Scholar]
- Elangovan, M., R.N. Day, and A. Periasamy. 2002. Nanosecond fluorescence resonance energy transfer-fluorescence lifetime imaging microscopy to localize the protein interactions in a single living cell. J. Microsc. 205:3–14. [DOI] [PubMed] [Google Scholar]
- Elangovan, M., H. Wallrabe, Y. Chen, R.N. Day, M. Barroso, and A. Periasamy. 2003. Characterization of one- and two-photon excitation fluorescence resonance energy transfer microscopy. Methods. 29:58–73. [DOI] [PubMed] [Google Scholar]
- Förster, T. 1965. Delocalized excitation and excitation transfer. Modern Quantum Chemistry. Vol. 3. O. Sinanoglu, editor. Academic Press Inc., New York. 93–137.
- Gadella, T.W.J., Jr., and T.M. Jovin. 1995. Oligomerization of epidermal growth factor receptors on A431 cells studied by time-resolved fluorescence imaging microscopy. A stereochemical model for tyrosine kinase receptor activation. J. Cell Biol. 129:1543–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano, L., T.M. Jovin, M. Irie, and E.A. Jares-Erijman. 2002. Diheteroarylethenes as thermally stable photoswitchable acceptors in photochromic fluorescence resonance energy transfer (pcFRET). J. Am. Chem. Soc. 124:7481–7489. [DOI] [PubMed] [Google Scholar]
- Gordon, G.W., G. Berry, X.H. Liang, B. Levine, and B. Herman. 1998. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 74:2702–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, M.R., and R.H. Kohler. 2001. GFP imaging: methodology and application to investigate cellular compartmentation in plants. J. Exp. Bot. 52:529–539. [PubMed] [Google Scholar]
- Heim, R., and R.Y. Tsien. 1996. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 6:178–182. [DOI] [PubMed] [Google Scholar]
- Herman, B. 1998. Fluorescence Microscopy. 2nd ed. Springer-Verlag New York Inc., New York. 170 pp.
- Kraynov, V.S., C. Chamberlain, G.M. Bokoch, M.A. Schwartz, S. Slabaugh, and K.M. Hahn. 2000. Localized Rac activation dynamics visualized in living cells. Science. 290:333–337. [DOI] [PubMed] [Google Scholar]
- Lakowicz, J.R. 1999. Principles of Fluorescence Spectroscopy. 2nd ed. Plenum Publishing Corp., New York. 692 pp.
- Miyawaki, A., J. Llopis, R. Heim, J.M. McCaffery, J.A. Adams, M. Ikura, and R.Y. Tsien. 1997. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 388:882–887. [DOI] [PubMed] [Google Scholar]
- Periasamy, A. 2001. Methods in Cellular Imaging. Oxford University Press, New York. 434 pp.
- Pollok, B.A., and R. Heim. 1999. Using GFP in FRET-based applications. Trends Cell Biol. 9:57–60. [DOI] [PubMed] [Google Scholar]
- Pozo, M.A.D., W.B. Kiosses, N.B. Alderson, N. Meller, K.M. Hahn, and M.A. Schwartz. 2002. Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat. Cell Biol. 4:232–239. [DOI] [PubMed] [Google Scholar]
- Roessel, P.V., and A.H. Brand. 2002. Imaging into the future: visualizing gene expression and protein interactions with fluorescent proteins. Nat. Cell Biol. 4:E15–E20. [DOI] [PubMed] [Google Scholar]
- Struck, D.K., D. Hoekstra, and R.E. Pagano. 1981. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 20:4093–4099. [DOI] [PubMed] [Google Scholar]
- Truong, K., and M. Ikura. 2001. The use of FRET imaging microscopy to detect protein-protein interactions and protein conformational changes in vivo. Curr. Opin. Struct. Biol. 11:573–578. [DOI] [PubMed] [Google Scholar]
- Truong, K., A. Sawano, H. Mizuno, H. Hama, K. Tong, T.K. Mal, A. Miyawaki, and M. Ikura. 2001. FRET-based in vivo Ca2+ imaging by a new calmodulin-GFP fusion molecule. Nat. Struct. Biol. 8:1069–1073. [DOI] [PubMed] [Google Scholar]
- Ting, A.Y., K.H. Kain, R.L. Klemke, and R.Y. Tsien. 2001. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc. Natl. Acad. Sci. USA. 98:15003–15008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Z., and Y. Liu. 2001. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys. J. 81:2395–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.