Abstract
The RanGTP gradient across the interphase nuclear envelope and on the condensed mitotic chromosomes is essential for many cellular processes, including nucleocytoplasmic transport and spindle assembly. Although the chromosome-associated enzyme RCC1 is responsible for RanGTP production, the mechanism of generating and maintaining the RanGTP gradient in vivo remains unknown. Here, we report that regulator of chromosome condensation (RCC1) rapidly associates and dissociates with both interphase and mitotic chromosomes in living cells, and that this mobility is regulated during the cell cycle. Our kinetic modeling suggests that RCC1 couples its catalytic activity to chromosome binding to generate a RanGTP gradient. Indeed, we have demonstrated experimentally that the interaction of RCC1 with the chromatin is coupled to the nucleotide exchange on Ran in vivo. The coupling is due to the stable binding of the binary complex of RCC1–Ran to chromatin. Successful nucleotide exchange dissociates the binary complex, permitting the release of RCC1 and RanGTP from the chromatin and the production of RanGTP on the chromatin surface.
Keywords: Ran; nuclear; chromatin; nucleotide exchange; fluorescence intensity
Introduction
The small GTPase Ran plays a key role in diverse cellular functions including nucleocytoplasmic transport (Mattaj and Englmeier, 1998), nuclear envelope formation, and spindle assembly (Dasso, 2002). Like many small GTPases, Ran exits as either a GDP- or GTP-bound state and functions as a molecular switch. Regulator of chromosome condensation (RCC1)* is the only known guanine nucleotide exchange factor (GEF) for Ran (Bischoff and Ponstingl, 1991; Seino et al., 1992; Seki et al., 1996; Nemergut et al., 2001; Renault et al., 2001). Previous studies showed that RCC1 is chromatin bound both in interphase and mitosis, whereas the proteins that stimulate RanGTPase activity, such as RanGAP1 and RanBP1, are cytoplasmic. This unique localization of the Ran regulators strongly suggests that there is a RanGTP concentration gradient across the interphase nuclear envelope and on the condensed mitotic chromosomes (Mattaj and Englmeier, 1998). Recent studies using Xenopus egg extract revealed that there is indeed a high RanGTP concentration in the interphase nuclei and on the condensed chromosomes in vitro, therefore lending support for the idea of a gradient (Kalab et al., 2002).
Although RanGTP gradient appears to exist in vitro, it is not clear whether the gradient also exists in living cells. Most importantly, the mechanism by which RCC1 catalyzes and maintains RanGTP in living cells remains unknown. As a result, little is known about how the cell regulates RanGTP production. Studies of other small GTPases have shown that a plethora of mechanisms are involved in regulating nucleotide exchange catalyzed by GEFs, and understanding these mechanisms have been crucial in deciphering the functions of small GTPases such as Ras, Rho, and Arf in vivo. Clearly, understanding the mechanisms regulating RanGTP production in living cells is essential to understand the Ran system.
Results and discussion
Establishing the cell line to study RCC1 function in vivo
To understand the mechanism by which RCC1 generates and maintains the RanGTP gradient on the dynamic chromosomes in vivo, we established a stable Swiss 3T3 cell line that expresses human RCC1 fused to the GFP at its COOH terminus (RCC1-GFP). We found that the localization of the RCC1-GFP was identical to endogenous RCC1 in interphase and mitosis (Fig. 1 A). Interestingly, in interphase, both endogenous RCC1 and RCC1-GFP are enriched in regions where there is strong DAPI staining. Live imaging of RCC1-GFP showed that it is localized on the chromosomes at all stages of mitosis (Fig. 1 B). Furthermore, both RCC1-GFP and endogenous RCC1 are extracted from the interphase nuclei at the same salt concentrations, suggesting that they bind to the chromatin with the same affinity (Fig. 1 C).
Figure 1.
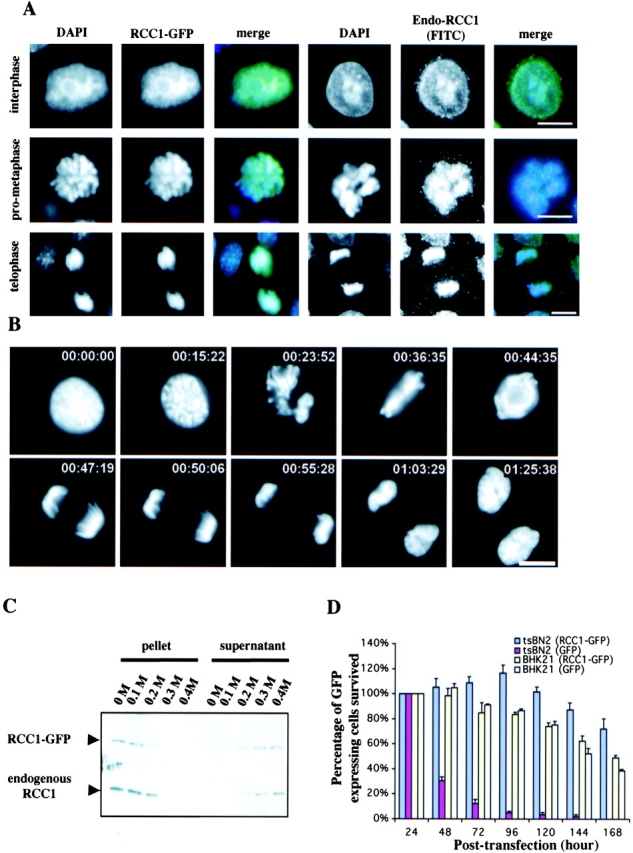
Characterization of RCC1-GFP. (A) RCC1-GFP–expressing or –nonexpressing Swiss 3T3 cells were fixed and stained with DAPI to visualize DNA. The nonexpressing cells were also stained with anti-RCC1 antibody to visualize endogenous RCC1. Both RCC1-GFP and endogenous RCC1 colocalized with DAPI in interphase and mitosis. Bar, 10 μm. (B) Time-lapse microscopy of RCC1-GFP–expressing cells during mitosis. Bar, 10 μm. (C) RCC1-GFP and endogenous RCC1 were extracted from isolated nuclei with increasing concentrations of salt and analyzed by Western blotting. (D) Plasmids expressing RCC1-GFP or GFP were transfected into tsBN2 cells, which harbor a temperature-sensitive mutation in RCC1, and the wild-type parental cells, BHK-21. The cells expressing RCC1-GFP or GFP were counted in five random fields with a 20× objective 24 h after transfection. Cells were then shifted to the nonpermissive temperature (39.5°C) to inactivate endogenous RCC1 in tsBN2 cells and were counted every 24 h. The tsBN2 cells transfected with GFP did not survive at the nonpermissive temperature. The same cells transfected with RCC1-GFP, and the control BHK-21 cells survived, leading to establishment of stable cell lines able to grow at 39.5°C.
We reasoned that if RCC1-GFP is fully active as the endogenous RCC1, it should be able to replace the function of the endogenous RCC1. We transfected RCC1-GFP or control vector into either wild-type (BHK-21) or RCC1 mutant (tsBN2) CHO cell lines. 1 d after transfection, the cells were shifted to restrictive temperature (39.5°C) to inactivate the endogenous RCC1 (through degradation) in the tsBN2 cells (Nishitani et al., 1991). The number of GFP-expressing cells was counted every 24 h for 7 d. We found that expression of RCC1-GFP in tsBN2 cells rescued the lethality caused by the temperature-sensitive mutation in RCC1 at 39.5°C, whereas GFP alone did not (Fig. 1 D). Stable RCC1-GFP expressing tsBN2 cells can survive at the restrictive temperature like the wild-type counterpart for as long as we have cultured them (months so far). Clearly, RCC1-GFP is fully functional in the hamster cells. Because mouse RCC1 is over 97% identical to the hamster RCC1, we conclude that RCC1-GFP expressed in Swiss 3T3 cells should also be fully functional. Therefore, RCC1-GFP can be used to faithfully report the endogenous RCC1 function in the mouse and hamster cells.
RCC1 is a highly mobile enzyme in interphase and mitosis
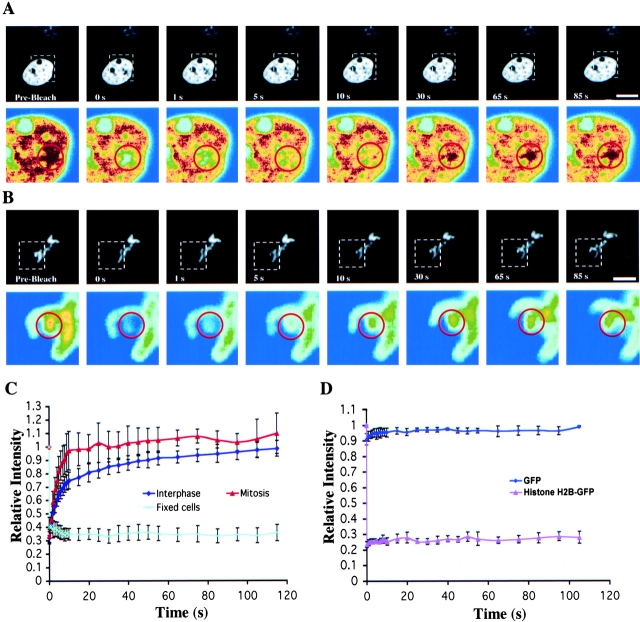
To decipher the mechanism of RanGTP gradient production in vivo, it is important to understand how RCC1 interacts with the chromatin in interphase and mitosis. Therefore, we first used FRAP to study the dynamics of RCC1-GFP. A defined region of the 3T3 cell nucleus or condensed mitotic chromosomes was bleached with a laser pulse of 500 ms. The kinetics of recovery, reflecting the mobility of the RCC1-GFP, was measured by sequential imaging (Fig. 2, A and B) . After photobleaching, ∼90% of the RCC1 fluorescence signal was recovered within 55 and 10 s, with a half time of ∼5 and ∼3 s in interphase and mitotic cells, respectively (Fig. 2 C). The recovery of RCC1-GFP was much faster than that of histone H2B-GFP, which was immobile during our observations (Kimura and Cook, 2001), but slower than the GFP molecule in the nucleus (Fig. 2 D), which freely diffused throughout the nucleus.
Figure 2.
FRAP. (A) Selected images of an interphase cell during FRAP of an area in the nucleus (red circle). Bottom panels show the enlargement of the indicated area (white dashed square) in pseudocolor. (B) Selected images of a mitotic cell during FRAP of an area on the chromosome. Bar, 10 μm. (C) RCC1-GFP FRAP kinetics in the interphase nucleus, on condensed chromosomes, and in the nuclei of methanol-fixed cells. (D) GFP and histone H2B-GFP FRAP kinetics in the interphase nucleus. Values in C and D represent means ± SD from at least five different cells.
Next, we used fluorescence loss in photobleaching (FLIP) to determine the dissociation kinetics of RCC1 from the chromatin. A spot (1-μm diam) in the 3T3 cell interphase nucleus, on the mitotic chromosome, or in the mitotic cytosol was bleached repeatedly with high laser power, and the cell was imaged after each round of bleaching (Fig. 3 A). The overall fluorescence loss was faster in mitotic cells than that of interphase cells, consistent with the FRAP analyses (Fig. 3 B). Then, we measured the decline of fluorescence signal at various distances from the bleach spot over time in both interphase and mitotic cells. In interphase nuclei, the rate of fluorescence loss decreased as the distance from the bleach spot increased (Fig. 3 C). This is presumably due to non-negligible diffusion transport limitations. However, the decline of fluorescence signal in mitosis was independent of the distance from the bleach spot (Fig. 3 D). This suggests that transport in mitosis is mainly regulated by the rate of RCC1 release from chromosomes. Based on these data, we developed kinetic models for the movement of RCC1 in interphase and mitosis (see supplemental material, available at http://www.jcb.org/cgi/content/full/jcb.200211004/DC1). We found that the effective diffusion coefficients of RCC1 in interphase and mitotic cells are 0.51 ± 0.11 and 2.86 ± 0.71 μm2/s, respectively. The smaller effective diffusion coefficient of RCC1 in mitosis reflects the faster dissociation of RCC1 from the chromosome, not that RCC1 diffuses five times faster in mitosis than in interphase. The mean residence times of RCC1 on chromatin, computed as the inverse of the dissociation rate of RCC1, are 50 ± 12 and 20 ± 5 s in interphase and mitosis, respectively. Also, we have performed the FRAP and FLIP experiments using tsBN2 cells expressing RCC1-GFP at 39.5°C and obtained similar results (unpublished data). Together, these works revealed that RCC1 associates with the chromatin transiently during the cell cycle, and that the association is regulated differently in interphase and mitosis in vivo.
Figure 3.
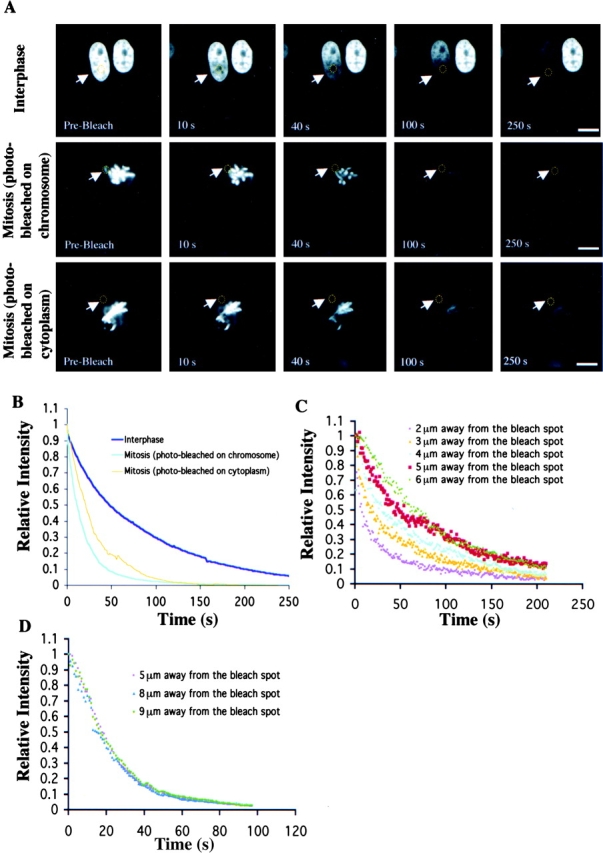
FLIP. (A) Selected images of interphase or mitotic cells during FLIP of the indicated area (yellow circles). Bars, 10 μm. (B) Kinetics of overall RCC1-GFP FLIP in interphase and mitotic cells. (C and D) Kinetics of RCC1-GFP FLIP in interphase (C) and mitotic (D) cells.
The binary complex of RCC1 and Ran binds stably to the chromatin in vivo
The finding that RCC1 transiently associates with the chromosome during the cell cycle raises an important question regarding whether and how RCC1 could generate a steep RanGTP concentration gradient in vivo. Previous studies showed that RCC1 in cell lysates or as a purified protein has a strong GEF activity toward Ran in vitro. Furthermore, histones H2A/H2B or isolated chromatin (RCC1 free) only stimulated the GEF activity of RCC1 by ∼20% as measured by the percentage of nucleotide released from Ran as a function of time (Nemergut et al., 2001). If the pool of free RCC1 can catalyze nucleotide exchange almost as efficiently as those bound to the chromatin in vivo, it could potentially prevent the formation of a steep RanGTP concentration gradient on the chromatin. Indeed, our kinetic modeling suggests that in order for the highly mobile RCC1 to generate a steep RanGTP gradient on the chromosomes, RCC1 may couple its binding to the chromosome to the nucleotide exchange on Ran (supplemental material and Fig. S3).
We hope to determine whether and how RCC1 may couple its binding to the chromosomes to the nucleotide exchange on Ran. Although little is known about how RCC1 catalyzes nucleotide exchange on Ran in living cells, the in vitro nucleotide exchange reaction is well documented. Studies using purified RCC1 and Ran showed that RCC1-catalyzed exchange is a multi-step process, involving the formation of ternary and binary complexes of RCC1, Ran, and guanine nucleotides (Klebe et al., 1995a, 1995b; Renault et al., 2001). Because RCC1 destabilizes the binding of nucleotide to Ran, the ternary complex of RCC1–Ran–nucleotide is of low affinity, which dissociates quickly (Klebe et al., 1995a, 1995b; Renault et al., 2001). When there is no free nucleotide, the ternary complex relaxes into the binary complex of RCC1–Ran. However, when the free nucleotide concentration is high, the ternary complex dissociates into RCC1– and Ran–nucleotide (Klebe et al., 1995a, 1995b; Renault et al., 2001). We reasoned that if nucleotide exchange on Ran occurred on the chromatin in vivo, we might find the binary complex on the chromatin.
We used a mutant allele of Ran (RanT24N) to test whether the binary complex of Ran and RCC1 is chromatin bound in the cell. RanT24N has a greatly reduced affinity for nucleotide compared with wild-type Ran; therefore, it forms a stable binary complex with RCC1 due to lack of nucleotide exchange (Dasso et al., 1994; Kornbluth et al., 1994; Klebe et al., 1995a; Lounsbury et al., 1996). If the binary complex is chromosome bound, we should see a clear colocalization of RanT24N and RCC1-GFP on the chromatin in both interphase and mitosis. To test this possibility, we injected rhodamine-labeled (Rh) wild-type RanGDP, Rh-RanGTP, or Rh-RanT24N into interphase nuclei or mitotic cytoplasm of the 3T3 cells expressing RCC1-GFP. Using live imaging, we found that Rh-RanT24N clearly colocalized with RCC1-GFP in interphase nuclei and on mitotic chromosomes (Fig. 4 A). As expected, most Rh-RanGDP is found in the nucleus. Interestingly, in mitosis, Rh-RanGDP was found on condensed chromosomes and in the cytoplasm, resulting in a somewhat diffused distribution (Fig. 4 A). We believe that the localization of Rh-RanGDP to the condensed chromosomes is because RanGDP undergoes nucleotide exchange by forming the binary complex with RCC1, which binds to the condensed chromosomes. Consistent with this idea, we found that although Rh-RanGTP is localized in the interphase nucleus, in mitosis, Rh-RanGTP is diffusely distributed in the mitotic cytosol (Fig. 4 A). Based on these studies, we conclude that the binary complex is chromosome bound in vivo.
Figure 4.
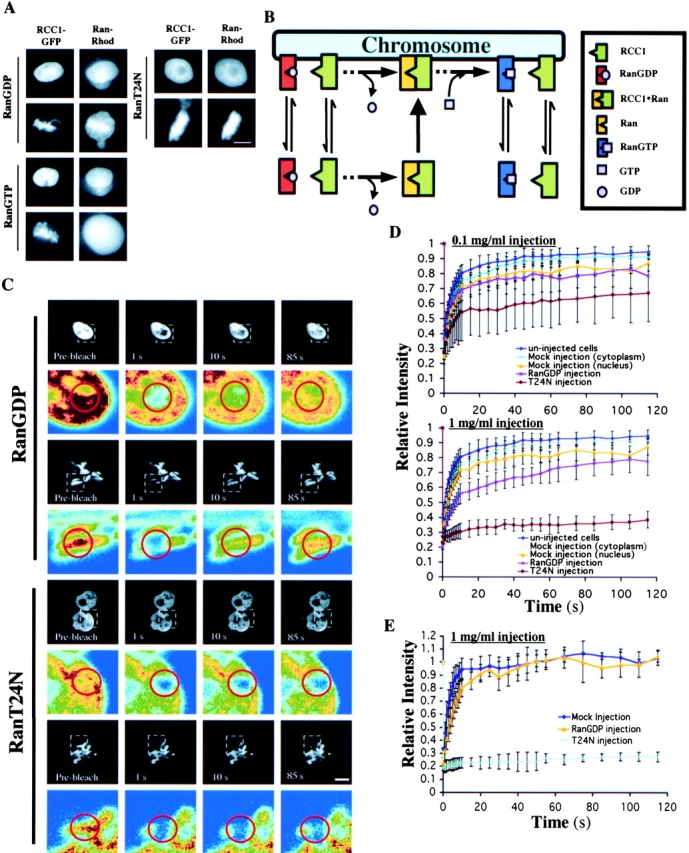
The binary complex of Ran–RCC1 binds stably to the chromatin in vivo. (A) Rh-RanGDP, Rh-RanT24N, or Rh-RanGTP was injected into interphase nuclei or mitotic cytoplasm of RCC1-GFP–expressing cells, followed by live imaging using fluorescence microscopy. Bar, 10 μm. (B) The model. RCC1, RanGDP, and RanGTP interact with the chromatin reversibly due to low affinity binding, whereas the RCC1–Ran binary complex binds to the chromatin stably. The ternary complexes of RCC1–Ran–GDP (or GTP) are omitted from the drawing for simplicity. (C) RanGDP or RanT24N was injected into the interphase nuclei or mitotic cytosol at 1 mg/ml followed by FRAP analysis of RCC1-GFP. Selected images before and after the bleach pulses (red circle denotes the bleached spot) were shown. Colored panels show the enlargement of the indicated area (white dashed square) in pseudocolor. Bar, 10 μm. (D) FRAP kinetics of injected interphase cells. Values represent means ± SD from at least five cells. (E) FRAP kinetics of injected mitotic cells. Values represent means ± SD from at least three cells and five independent FRAPs.
Recent studies showed that Ran and RCC1 interact with the chromatin via histones (Seino et al., 1992; Nemergut et al., 2001; Bilbao-Cortes et al., 2002). Although RCC1 binds to histones H2A and H2B, Ran binds to H3 and H4 (Seino et al., 1992; Nemergut et al., 2001; Bilbao-Cortes et al., 2002). These bindings are of low affinity in nature (Nemergut et al., 2001; Bilbao-Cortes et al., 2002). We reasoned that the binary complex of RCC1–Ran should have more of an increased affinity to the core histones on the chromatin than RCC1 and Ran alone, and that the stable binding of the binary complex could allow the coupling of nucleotide exchange to the chromatin (Fig. 4 B). We propose that successful nucleotide exchange, which dissociates RCC1 from RanGTP, is essential for the two proteins to return to the low affinity binding states. Consequently, RCC1 and RanGTP can dissociate from the chromatin, allowing the generation of RanGTP on the chromatin surface (Fig. 4 B).
If the above model is correct, excess RanT24N should immobilize RCC1 on the chromatin due to the formation of a stable binary complex that resists nucleotide exchange. On the other hand, excess wild-type RanGDP should only slow down the mobility of RCC1 because of the increased formation of the binary complex that allows nucleotide exchange. Therefore, we used FRAP to probe the mobility of RCC1-GFP in the 3T3 cells after microinjection of purified RanT24N or RanGDP (see Materials and methods). Perturbation by control microinjection into the nucleus had only a small effect on the mobility of RCC1-GFP when compared with cytoplasm-injected or uninjected cells (Fig. 4 D). Similarly, injecting RanGDP at 0.1 mg/ml into the interphase nuclei had no effect on FRAP of RCC1-GFP compared with controls. However, injecting the same amount of RanT24N significantly reduced the mobility of RCC1-GFP (Fig. 4 D).
Next, RanT24N or RanGDP was injected at 10-fold higher concentration (1 mg/ml) into the interphase nuclei. We estimated that injecting at this concentration of Ran could deliver ∼4 × 106 molecules of Ran into the cells, which is similar to the estimated number of RCC1 molecules in the cell (see Materials and methods). We found that RanGDP injection allowed the recovery of 55% of the RCC1-GFP in the first 10 s, followed by a slow recovery of the fluorescence signal to the level of control injections in the next 110 s (Fig. 4 D). However, RanT24N injection completely blocked the recovery of RCC1-GFP in the bleached spot (Fig. 4, C and D). When the same amount of RanT24N or RanGDP was injected into the mitotic cells, we found that RanT24N completely immobilized RCC1-GFP on the condensed chromosomes, whereas RanGDP only reduced the mobility of RCC1 (Fig. 4, C and E). We also performed FRAP of fluorescently labeled RanGDP or RanT24N that were injected into cells, and found that although RanGDP is mobile, RanT24N is not (unpublished data). These results showed that nucleotide exchange on Ran is required for both RCC1 and Ran to dissociate from the chromatin in both interphase and mitosis. They further suggest that the binary complex of RCC1–Ran associates stably with the chromatin in vivo. Successful nucleotide exchange is required for the dissociation of RCC1 from RanGTP, which in turn allows RCC1 and RanGTP to dissociate from the chromatin.
The dissociation of the binary complex of Ran–RCC1 from the chromatin requires nucleotide exchange
The above in vivo studies revealed that the binary complex of Ran–RCC1 binds to chromosomes tightly and that nucleotide exchange is required for their dissociation from chromatin. We hope to biochemically confirm these findings by using Xenopus egg extracts that support the formation of chromatin structures from Xenopus sperm in vitro. Sperm were added to the egg extracts supplemented with or without RCC1-GFP in the presence of either RanT24N or RanGDP. We found that the addition of exogenous RanT24N and RanGDP strongly stimulated the binding of both the endogenous RCC1 (Fig. 5 A) and the exogenous RCC1-GFP (Fig. 5 B) to the chromatin assembled from the sperm, which is consistent with the idea that the formation of the binary complex of RCC1–Ran enhances the binding of RCC1 to the chromosomes. Next, we used competition assays to determine whether Ran and RCC1 bind to the chromatin tightly in the form of binary complex in the egg extracts. We added Xenopus sperm to the extracts supplemented with Rh-RanT24N, Rh-RanGD, or RCC1-GFP. Excess unlabeled RanT24N, RanGDP, or RCC1 was used as competitors. We found that unlabeled RanGDP and RCC1 readily competed for Rh-RanGDP and RCC1-GFP, respectively. However, unlabeled RanT24N and RCC1 only showed ∼40 and ∼20% competition of Rh-RanT24N and RCC1-GFP, even when the competitor concentrations reached 20- and 10-fold excess of the labeled proteins, respectively (Fig. 5, C and D). This suggests that RCC1 and Ran bind stably to the chromatin when they are locked into the RCC1–Ran binary complex in the form of RCC1–RanT24N.
Figure 5.
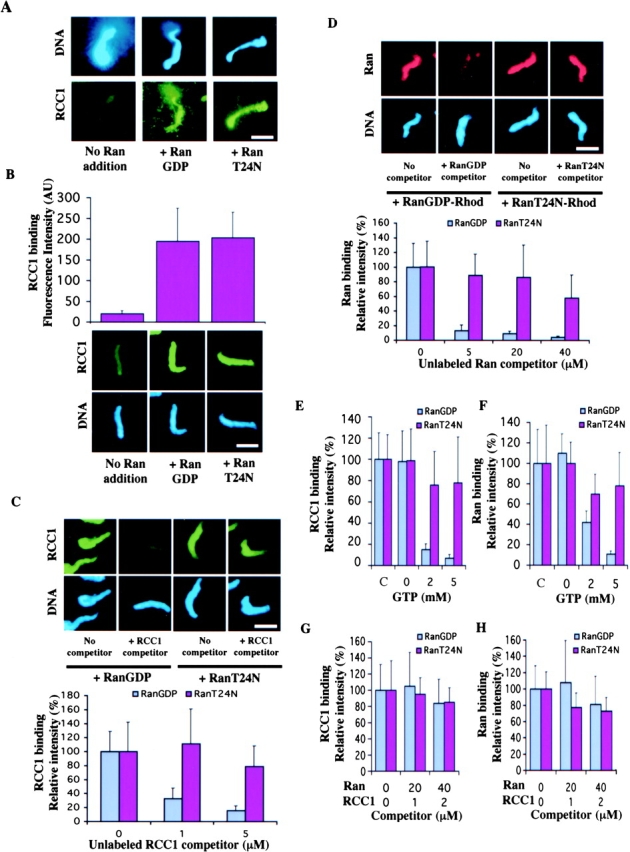
Biochemical characterization of the binding of Ran and RCC1 to the chromosomes. (A) Both RanGDP and RanT24N strongly stimulate the binding of the endogenous RCC1 in the egg extract to the mitotic chromosomes assembled from the sperm chromatin. The binding of the endogenous RCC1 to the chromatin is detected by immunofluorescence using an anti-RCC1 antibody. (B) RCC1-GFP and sperm was added to the egg extract supplemented with purified RanGDP or RanT24N. The fluorescence intensity of RCC1-GFP on the sperm chromatin was quantified as arbitrary unit (AU). (C) RCC1 competition. The sperm was incubated with egg extracts supplemented with RCC1-GFP and either RanGDP or RanT24N in the presence of unlabeled RCC1 at the indicated concentrations. The amount of RCC1-GFP bound to the chromatin was quantified. (D) Ran competition. Sperm was incubated with egg extracts containing Rh-RanGDP or Rh-RanT24N and unlabeled RanGDP or RanT24N at the indicated concentrations, respectively. The amount of labeled Ran bound to the chromatin was quantified. (E and F) Buffer containing 0–5 mM GTP was incubated with sperm chromatin with bound Rh-Ran or RCC1-GFP on coverslips for 20 min. The amount of labeled proteins on the sperm chromatin was quantified either before (c, control) or after incubation. (G and H) The same sperm chromatin as in E and F were incubated with buffer containing indicated concentrations of unlabeled Ran and RCC1 as competitors in the absence of free GTP. The amount of chromatin-bound RCC1-GFP (G) or Rh-Ran (H) was quantified after incubation. Error bars represent SD.
Next, we examined whether guanine nucleotide is required for the dissociation of Ran–RCC1 binary complex from the chromatin. We assembled sperm chromatin in the egg extracts supplemented with Rh-RanGDP, Rh-RanT24N, or RCC1-GFP. The sperm were then spun onto coverslips through a glycerol cushion containing no GTP or GDP. Under these conditions, a fraction of the chromatin-bound RCC1-GFP and wild-type Rh-Ran is in the form of the binary complex due to nucleotide depletion, and therefore, should bind stably to the chromatin (Klebe et al., 1995b; Renault et al., 2001). Indeed, when the sperm was extracted with buffer, Rh-Ran and RCC1-GFP dissociated from the chromatin only in the presence, but not in the absence, of free GTP or GDP. As expected, Rh-RanT24N and RCC1-GFP did not dissociate from the chromatin efficiently, even in the presence of excess GTP or GDP (Fig. 5, E and F). Next, we assessed the stability of the binding of the binary complex to the chromatin in the absence of free GTP or GDP. We repeated the above experiments using unlabeled Ran and RCC1 as competitors. Quantification revealed that, in the absence of free GTP or GDP, over 80% of wild-type Rh-Ran and RCC1-GFP remained bound to the chromatin, even when the concentrations of unlabeled Ran (40 μM) and RCC1 (2 μM) were higher than those found in the egg extract (Fig. 5, G and H; see also Materials and methods). This shows that the binary complex binds stably to the chromatin and that nucleotide exchange is required for its dissociation.
We have shown that RCC1 is a highly mobile enzyme, and the binding of RCC1 to the chromatin appears to be subjected to cell cycle regulation. More importantly, as a mobile enzyme, RCC1 couples nucleotide exchange on Ran with chromosome docking to generate RanGTP in vivo. The coupling is established through the stable binding of the RCC1–Ran binary complex to chromosomes. Successful nucleotide exchange on the chromatin-bound binary complex dissociates the complex, liberates RCC1, and generates RanGTP on the chromatin. Ran and RCC1 bind to the chromatin via the core histones, which are present in ∼100-fold molar excess of RCC1 in the cell. Therefore, the chromatin has sufficient capacity to bind to all RCC1–Ran binary complexes to support the chromosome-coupling exchange mechanism. Eukaryotic chromosomes are highly dynamic structures that undergo remodeling throughout the cell cycle. A mobile RCC1 and a chromosome-coupled exchange mechanism may be necessary to coordinate the dynamic chromatin reorganization with the production of RanGTP gradient during the cell cycle.
Although the chromosome-coupled nucleotide exchange by RCC1 described in this paper can significantly influence the production of RanGTP gradient, it does not exclude other mechanisms that may also influence RanGTP gradient. For example, the chromosomes were shown to stimulate the GEF activity of RCC1 modestly (Nemergut et al., 2001). This stimulation, in combination with the chromosome-coupled exchange, should further enhance RanGTP production on the chromosomes. In addition, RanGAP1 and RanBP1 present in the cytosol hydrolyze RanGTP into RanGDP, and therefore, should further sharpen the RanGTP gradient across the nuclear envelope in interphase and on the condensed chromosomes in mitosis. Finally, although free RCC1 in cell lysates is active in vitro, the free RCC1 in vivo may be negatively regulated. It will be important to understand the relative contributions of the different mechanisms toward the production of RanGTP gradient.
Materials and methods
Plasmids and cell transfection
Human RCC1 was cloned into pEGFP-N3 (CLONTECH Laboratories, Inc.) and transfected into Swiss 3T3, tsBN2, or BHK21 cells using FuGENE™ 6 (Roche). Stable cell lines expressing RCC1-GFP were selected and maintained in MEM (GIBCO BRL) with 10% FBS, 1% penicillin/streptomycin, and 0.6 mg/ml Geneticin, all purchased from GIBCO BRL. 6His-tagged wild-type Ran, RanT24N, RCC1, and RCC1-GFP were subcloned into pET-30a (for Ran and RanT24N) or pET-15a (for RCC1; Novagen).
Fluorescence microscopy and analysis of salt-extracted RCC1
Immunofluorescence microscopy on cells was performed as described previously (Zhang et al., 2000) using a commercial mAb against RCC1 (MBL International Corporation). Images were obtained with a CCD camera equipped with the MetaMorph® Imaging System (Universal Imaging Corp.). To extract RCC1, nuclei were isolated from cells expressing RCC1-GFP and extracted with increasing salt concentrations as described previously (Ohtsubo et al., 1989). The supernatant and pellet fractions were subjected to SDS-PAGE followed by Western blotting and probing with the anti-RCC1 antibody.
Microscopy of FRAP and FLIP
Cells were plated on a coverslip and mounted onto a glass slide with a depression containing culture medium (DME, 10% FBS, 10 U/ml penicillin/streptomycin, and 25 mM Hepes without phenol red). FRAP and FLIP were performed on a confocal microscope (TCS-SP2; Leica) using the 488-nm laser line of an argon laser at 37 and 23°C with similar results. In FRAP experiments, cells were scanned twice, followed by a single bleach pulse of 500 ms using a spot 1 μm in diameter. Single section images were then collected at 1-s intervals for the first 10 images, followed by 5-s intervals for the next 10 images, and 10-s intervals for the final 10 images with the laser power attenuated to ∼9% of the bleach intensity. In FLIP experiments, cells were repeatedly imaged and bleached at intervals of 1 s with each bleaching for 250 ms and with imaging identical to those used in FRAP. To determine the relative fluorescence intensity in a region of interest, the fluorescence intensity in the region at each time point was normalized to the change in total fluorescence caused by bleaching and imaging as described previously (Misteli et al., 2000; Phair and Misteli, 2000).
Recombinant protein and microinjection
6his-Ran and 6his-RanT24N were expressed in bacteria and purified using Ni-agarose adopting the methods as described previously (Dasso et al., 1994; Wilde and Zheng, 1999). 6his-RCC1 and 6his-RCC1-GFP were purified using Ni-agarose. Ran and RCC1 were labeled using tetramethyl-rhodamine and fluorescein, respectively. Unlabeled RanGDP or RanT24N in PBS was coinjected with tetramethyl-rhodamine succinididyl ester (Molecular Probes, Inc.) into the nuclei of interphase cells or the cytoplasm of mitotic cells with an InjectMan® microinjection system (Eppendorf) followed by FRAP. Control injections were performed with tetramethyl-rhodamine alone. For localization studies, both proteins were labeled with tetramethyl-rhodamine succinimidyl ester (Molecular Probes, Inc.) and desalted into PBS before injecting into the interphase nucleus or mitotic cytoplasm followed by live imaging and microscopy.
According to our Western blotting analysis (Fig. 1 C), RCC1-GFP is expressed at about the same level as the endogenous RCC1. According to Bischoff and Ponstingl (1995), the number of both endogenous RCC1 and RCC1-GFP expressed in the 3T3 cells should be ∼106 per cell. Based on our injection condition and the manufacturer's calibration (Eppendorf), we estimated that ∼0.2 pl of Ran was delivered into the cell per injection. Therefore, at 1 mg/ml of Ran, we delivered ∼4 × 106 Ran into the cell, whereas at 0.1 mg/ml of Ran, only ∼0.4 × 106 Ran was injected. If one RanT24N can immobilize one RCC1, injecting Ran at 0.1 mg/ml should lead to partial RCC1 immobilization, whereas injecting Ran at 1 mg/ml should lead to a complete immobilization. Consistent with this prediction, we observed that injecting RanT24N at 0.1 mg/ml lead to partial immobilization of RCC1-GFP, whereas injecting at 1 mg/ml lead to a complete immobilization.
In vitro assays using Xenopus egg extracts
Xenopus egg extracts and sperm were prepared as described previously (Murray and Kirschner, 1989). For Ran competition, sperm was incubated with egg extracts supplemented with 2 μM Rh-RanGDP or Rh-RanT24N in the presence of 0–40 μM unlabeled RanGDP or RanT24N, respectively, for 40 min at RT. After incubation, the sperm was spun onto coverslips through a glycerol cushion (80 mM Pipes, pH 6.8, 1 mM EGTA, 1 mM MgCl2, and 30% glycerol), fixed with methanol, and stained with DAPI (Wilde and Zheng, 1999). The amount of labeled Ran that remained bound to the sperm chromatin was quantified by taking images using a cooled CCD camera at the same exposure time that is below the saturation limit of the camera. The fluorescence intensity of each sperm was measured, and the background was subtracted from the area next to each sperm using the MetaMorph® software. At least 20 sperm were quantified for each experiment. For RCC1 competition, sperm was incubated with egg extracts supplemented with 0.5 μM RCC1-GFP and 40 μM unlabeled RanGDP or RanT24N in the presence of 0–5 μM unlabeled RCC1 as competitors.
The following experiments were used to determine whether the binding of RCC1 and Ran to the chromatin is sensitive to free GTP. For assaying RCC1-GFP binding, sperm was added to extracts supplemented with 0.5 μM RCC1-GFP and 40 μM of RanGDP or RanT24N. For assaying Rh-Ran binding, sperm was added to extracts supplemented with 2 μM Rh-RanGDP or Rh-RanT24N. After a 40-min incubation, the sperm was spun onto coverslips through the glycerol cushion without GTP. 250 μl of buffer (10 mM Hepes, pH 7.7, 100 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, 50 mM sucrose, and 5 mM EGTA) containing 0–5 mM GTP was added onto the coverslips and incubated for 20 min at RT followed by fixation and DAPI staining. The amount of RCC1-GFP or Rh-Ran that remained on the sperm chromatin were quantified as above. Similar amounts of Rh-Ran and RCC1-GFP remained bound to the chromatin before and after extraction with buffer containing no GTP. This observation suggests that most of Rh-Ran and RCC-GFP remain bound to the chromatin after centrifugation formed the binary complex. To test the stability of the binding of the binary complex to the sperm chromatin in the absence of free GTP, the same experiments as above were performed. After spinning the sperm labeled with RCC1-GFP or Rh-Ran onto coverslips, the sperm was incubated with XB buffer containing 0–2 μM unlabeled RCC1 and 0–40 μM unlabeled Ran as competitors for 20 min followed by fixation and DAPI staining. Rh-Ran and RCC1-GFP that remained bound to the sperm chromatin were quantified. The unlabeled RCC1 and Ran competitors were used at the similar ratio as that present in the egg extracts. The concentrations of Ran (20 μM) and RCC1 (1.3 μM) in the egg extract were estimated using quantitative Western blotting with known amounts of purified Ran and RCC1, respectively.
Online supplemental material
Online supplemental material includes kinetic modeling of RCC1 mobility and RanGTP gradient. Figs. S1 and S2 show curve fits of FRAP and FLIP analyses. Fig. S3 shows the simulation of RanGTP concentration profile on the chromatin. Online supplemental material available at http://www.jcb.org/cgi/content/full/jcb.200211004/DC1.
Supplemental Material
Acknowledgments
We thank Bob Phair for advice on FRAP and FLIP analyses and for critical reading of the manuscript, Ona Martin for technical assistance, Hiroshi Kimura and Peter Cook for H2B-GFP, Tom Misteli, Jim McNally, and Tatinana Karpova for advice on FRAP microscopy, and Joe Gall, Doug Koshland, Judith Yanowitz, Max Guo, and members of the Zheng lab for critical comments on the manuscript.
This work was supported by the Howard Hughes Medical Institute (to Y. Zheng) and by the National Science Foundation (NSE/NIRT 0210718 to D. Wirtz).
The online version of this article includes supplemental material.
Footnotes
Abbreviations used in this paper: FLIP, fluorescence loss in photobleaching; GEF, guanine nucleotide exchange factor; RCC1, regulator of chromosome condensation.
References
- Bilbao-Cortes, D., M. Hetzer, G. Langst, P.B. Becker, and I.W. Mattaj. 2002. Ran binds to chromatin by two distinct mechanisms. Curr. Biol. 12:1151–1156. [DOI] [PubMed] [Google Scholar]
- Bischoff, F.R., and H. Ponstingl. 1991. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 354:80–82. [DOI] [PubMed] [Google Scholar]
- Bischoff, F.R., and H. Ponstingl. 1995. Catalysis of guanine nucleotide exchange of Ran by RCC1 and stimulation of hydrolysis of Ran-bound GTP by Ran-GAP1. Methods Enzymol. 257:135–144. [DOI] [PubMed] [Google Scholar]
- Dasso, M. 2002. The Ran GTPase: theme and variations. Curr. Biol. 12:R502–R508. [DOI] [PubMed] [Google Scholar]
- Dasso, M., S.T. Azuma, T. Ohba, and T. Nishimoto. 1994. A mutant form of the Ran/TC4 protein disrupts nuclear function in Xenopus laevis egg extracts by inhibiting the RCC1 protein, a regulator of chromosome condensation. EMBO J. 13:5732–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab, P., K. Weis, and R. Heald. 2002. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 295:2452–2456. [DOI] [PubMed] [Google Scholar]
- Kimura, H., and P.R. Cook. 2001. Kinetics of core histones in living cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe, C., F.R. Bischoff, H. Ponstingl, and A. Wittinghofer. 1995. a. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 34:639–647. [DOI] [PubMed] [Google Scholar]
- Klebe, C., H. Prinz, A. Wittinghofer, and R.S. Goody. 1995. b. The kinetic mechanism of Ran-nucleotide exchange catalyzed by RCC1. Biochemistry. 34:12543–12552. [DOI] [PubMed] [Google Scholar]
- Kornbluth, S., M. Dasso, and J. Newport. 1994. Evidence for a dual role for TC4 protein in regulating nuclear structure and cell cycle progression. J. Cell Biol. 125:705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounsbury, K.M., S.A. Richards, K.L. Carey, and I.G. Macara. 1996. Mutations with the Ran/TC4 GTPase. J. Biol. Chem. 271:32834–32841. [DOI] [PubMed] [Google Scholar]
- Mattaj, I., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265–306. [DOI] [PubMed] [Google Scholar]
- Misteli, T., A. Gunjan, R. Hock, B. Michael, and D. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature. 408:877–881. [DOI] [PubMed] [Google Scholar]
- Murray, A.W., and M.W. Kirschner. 1989. Cyclin synthesis drives the early embryonic cell cycle. Nature. 339:275–280. [DOI] [PubMed] [Google Scholar]
- Nemergut, M., C.A. Mizzen, T. Stukenberg, C.D. Allis, and I.G. Macara. 2001. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science. 292:1540–1543. [DOI] [PubMed] [Google Scholar]
- Nishitani, H., M. Ohtsubo, K. Yamashita, H. Iida, J. Pines, H. Yasudo, Y. Shibata, T. Hunter, and T. Nishimoto. 1991. Loss of RCC1, a nuclear DNA-binding protein, uncouples the completion of DNA replication from the activation of cdc2 protein kinase and mitosis. EMBO J. 10:1555–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo, M., H. Okazaki, and T. Nishimoto. 1989. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J. Cell Biol. 109:1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair, R., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature. 404:604–609. [DOI] [PubMed] [Google Scholar]
- Renault, L., J. Kuhlmann, A. Henkel, and A. Wittinghofer. 2001. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1). Cell. 105:245–255. [DOI] [PubMed] [Google Scholar]
- Seino, H., S. Hisamoto, T. Uzawa, T. Sekiguchi, and T. Nishimoto. 1992. DNA-binding domain of RCC1 protein is not essential for coupling mitosis with DNA replication. J. Cell Sci. 102:393–400. [DOI] [PubMed] [Google Scholar]
- Seki, T., N. Hayashi, and T. Nishimoto. 1996. RCC1 in the Ran pathway. J. Biochem (Tokyo). 120:207–214. [DOI] [PubMed] [Google Scholar]
- Wilde, A., and Y. Zheng. 1999. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 284:1359–1362 (see comment). [DOI] [PubMed] [Google Scholar]
- Zhang, L., T.J. Keating, A. Wilde, G.G. Borisy, and Y. Zheng. 2000. The role of Xgrip210 in gamma tubulin ring complex assembly and centrosome recruitment. J. Cell Biol. 151:1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.