Abstract
Aphysical and functional link between the nuclear pore complex (NPC) and the spindle checkpoint machinery has been established in the yeast Saccharomyces cerevisiae. We show that two proteins required for the execution of the spindle checkpoint, Mad1p and Mad2p, reside predominantly at the NPC throughout the cell cycle. There they are associated with a subcomplex of nucleoporins containing Nup53p, Nup170p, and Nup157p. The association of the Mad1p–Mad2p complex with the NPC requires Mad1p and is mediated in part by Nup53p. On activation of the spindle checkpoint, we detect changes in the interactions between these proteins, including the release of Mad2p (but not Mad1p) from the NPC and the accumulation of Mad2p at kinetochores. Accompanying these events is the Nup53p-dependent hyperphosphorylation of Mad1p. On the basis of these results and genetic analysis of double mutants, we propose a model in which Mad1p bound to a Nup53p-containing complex sequesters Mad2p at the NPC until its release by activation of the spindle checkpoint. Furthermore, we show that the association of Mad1p with the NPC is not passive and that it plays a role in nuclear transport.
Keywords: nucleoporins; cell cycle; kinetochore; mads; nuclear transport
Introduction
A defining feature of eukaryotic cells is the encapsulation of their genome by the nuclear envelope (NE)* membrane. Replication of the genome and the regulation of transcriptional activity require the exchange of massive amounts of macromolecules between the cytoplasm and the nucleoplasm across the NE. This transport occurs through channels formed by nuclear pore complexes (NPCs). These highly conserved structures are composed of multiple repetitive subunits that form an elaborate eightfold symmetrical structure (for review see Rout and Aitchison, 2001).
In budding yeast, the NPC is believed to be composed of ∼30 nucleoporins, or nups (Rout et al., 2000). 12 of these contain phenylalanine-glycine repeats. These “FG-nups” play a direct role in transport through the NPC by binding a family of proteins (termed karyopherins) that carry cargo molecules through the NPC (for reviews see Wozniak et al., 1998; Wente, 2000; Macara, 2001; Rout and Aitchison, 2001).
Several observations also point to a role for specific nucleoporins in chromosome segregation that may be independent of their functions in mediating transport through the NPC. Most notably in yeast, we have shown that strains containing mutations in the gene encoding Nup170p, an evolutionarily conserved nup, exhibit a chromosome loss phenotype. Moreover, in these mutants, transcription of a reporter gene through an assembled kinetochore was detected, suggesting a defect in kinetochore integrity (Kerscher et al., 2001). These defects were not seen in deletion mutants of NUP157, a paralogue of NUP170, suggesting they are specifically linked to NUP170 and not a general defect in nuclear transport.
Other links between chromosome segregation and nups have been uncovered by two localization studies conducted in vertebrate cells. In these papers, three nucleoporins, Nup107p, Nup133p (Belgareh et al., 2001), and Nup358 (RanBP2; Joseph et al., 2002) were found to be associated with kinetochores during mitosis. The functional relevance of recruiting nups to the kinetochores during mitosis is not clear. However, a clue may come from another work showing that two human proteins, hsMad1p and hsMad2p, are associated with the NPC during interphase (Campbell et al., 2001). These proteins are members of an evolutionarily conserved group of spindle checkpoint mediators that were first identified in Saccharomyces cerevisiae and include Mad1p, Mad2p, Mad3p, Bub1p, Bub2p, Bub3p, and Mps1p (for review see Millband et al., 2002). Subcellular localization studies in Xenopus laevis and humans have shown that Mad1p and Mad2p localize to kinetochores before chromosome alignment at the metaphase plate (Chen et al., 1996, 1999; Li and Benezra, 1996). These proteins and other checkpoint mediators transmit a signal that prevents anaphase entry and chromosome segregation by inhibiting the anaphase-promoting complex/cyclosome from targeting key proteins for ubiquitin-mediated proteolysis until all chromosomes have formed functional spindle attachments, thus preventing aneuploidy (for reviews see Hardwick, 1998; Shah and Cleveland, 2000; Hoyt, 2001). In vertebrates, a key player in this process is a complex containing Mad3p (BubR1), Bub3p, Cdc20p, and perhaps Mad2p, that inhibits the anaphase-promoting complex in early mitosis and during checkpoint activation (Sudakin et al., 2001; Tang et al., 2001; Fang, 2002). This complex in its active form could also be isolated from interphase cells (Sudakin et al., 2001), suggesting it is stored, perhaps at the NPC, in preparation for its role during mitosis.
In S. cerevisiae, little is known about the subcellular distribution of the checkpoint proteins or how their dynamic associations with kinetochores and possibly NPCs influence their function. In this paper, we begin to address this question by focusing on two checkpoint proteins, Mad1p and Mad2p. We show that both Mad1p and Mad2p are associated with NPCs through a subcomplex of nups, consisting of Nup53p, Nup59p, Nup157p, and Nup170p (termed the Nup53p-containing complex; Marelli et al., 1998). On activation of the spindle checkpoint, distinct changes are detected in the molecular interactions between components of the Nup53p-containing complex, Mad1p, and Mad2p that lead to the release of Mad2p, but not Mad1p, from the NPC and the subsequent recruitment of Mad2p to kinetochores. Biochemical and genetic data are presented that demonstrate the significance of these interactions both in checkpoint and NPC functions. These findings represent the first report that Mad1p and Mad2p may require specific nucleoporins as a scaffold for their function. Furthermore, we show that Mad1p plays a role in nuclear transport.
Results
Mad1p and Mad2p are associated with S. cerevisiae NPCs
Mad1p and Mad2p are members of a group of at least seven conserved yeast proteins that are critical for executing a mitotic checkpoint in response to kinetochore and spindle integrity defects. We have investigated the subcellular localization of these two proteins by attaching a GFP tag to the COOH terminus of the endogenous protein. Three observations suggest that the Mad1-GFP and Mad2-GFP proteins are functional in checkpoint control. First, cells expressing these proteins grew at similar rates to isogenic wild-type (WT) cells in the presence of the microtubule-depolymerizing drug benomyl (Fig. 1 A). This was in contrast to strains containing null mutations in MAD1 or MAD2 (Fig. 1 A; mad1Δ and mad2Δ), which exhibited a severe growth deficiency due to a defect in mitotic checkpoint arrest. Second, Mad1-GFP, but not Mad2-GFP, was hyperphosphorylated in a manner similar to the WT protein when the spindle checkpoint was activated by the microtubule-destabilizing drug nocodazole (Fig. 1 B; for review see Hardwick and Murray, 1995). Finally, chromosome segregation defects observed in mad1Δ and mad2Δ mutants were not detected in the MAD1-GFP– and MAD2-GFP–containing strains (unpublished data).
Figure 1.
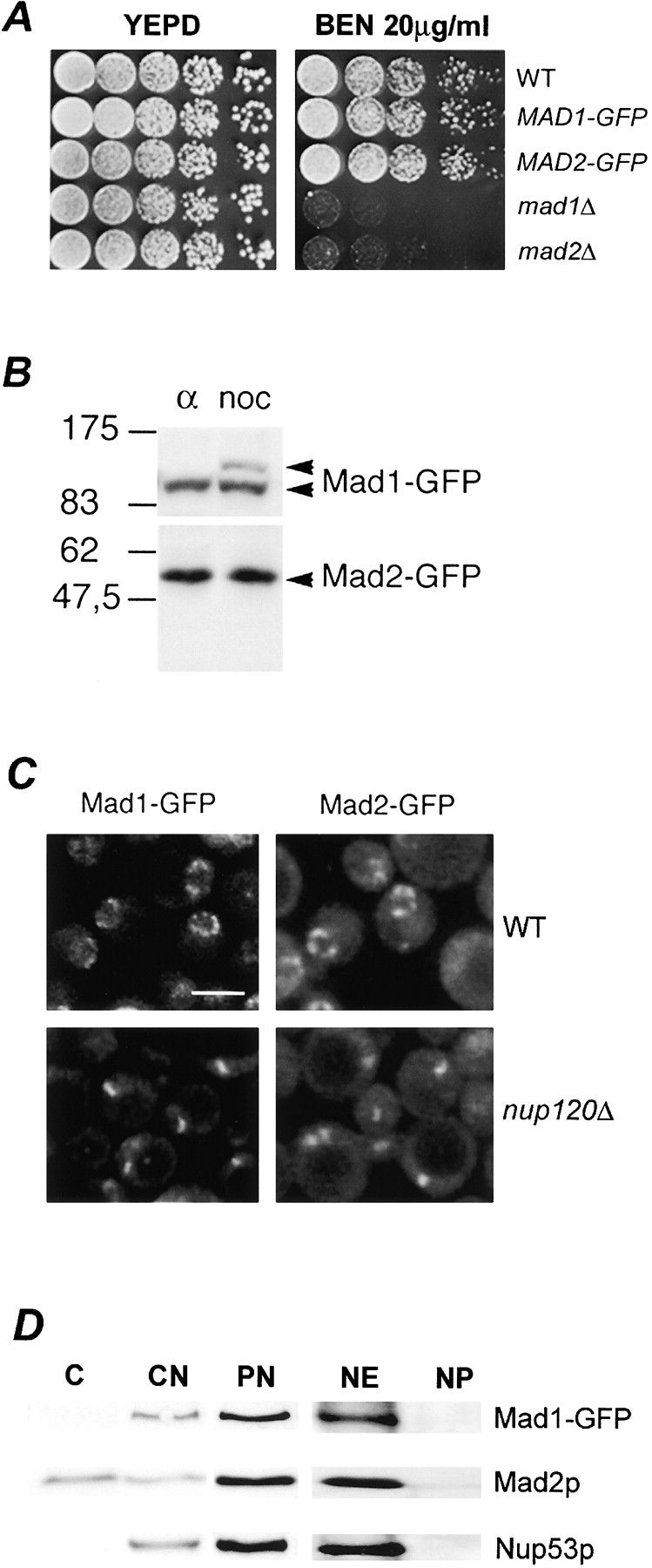
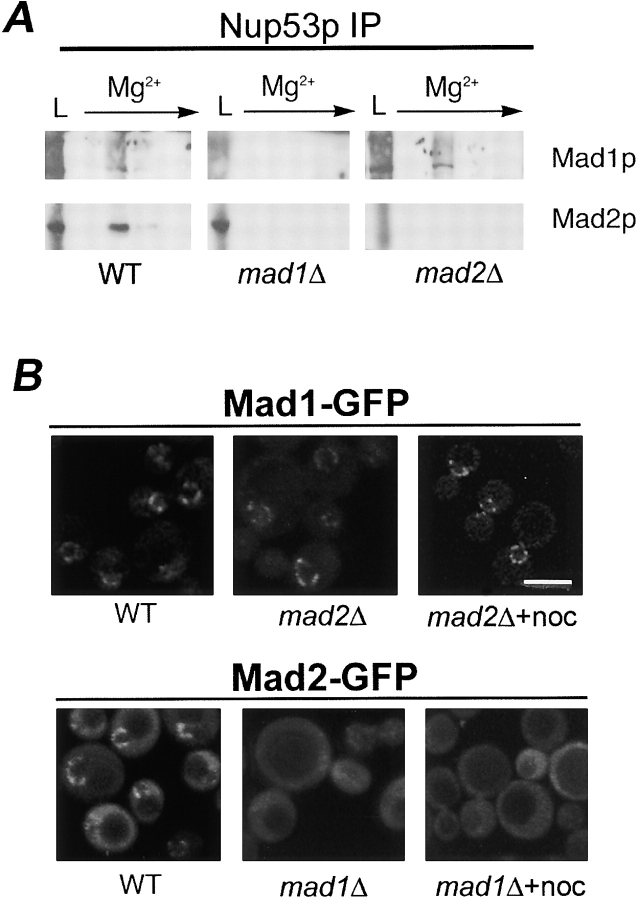
Association of Mad1-GFP and Mad2-GFP with NPCs. (A) Cells expressing Mad1-GFP and Mad2-GFP are not sensitive to the microtubule-destabilizing drug benomyl. The parental strain (WT), MAD1-GFP, MAD2-GFP, mad1Δ, and mad2Δ strains were grown to logarithmic phase in YEPD at 30°C, diluted, and spotted in 10-fold increments on YEPD and YEPD containing 20 μg/ml benomyl (BEN) and incubated for 4 d at 27°C. (B) Western blot analysis of Mad1-GFP– and Mad2-GFP–producing cells. Nuclear extracts were isolated from MAD1-GFP and MAD2-GFP stains arrested in G1 with α-factor (α) or G2/M with nocodazole (noc), and proteins were separated by double-inverted gradient PAGE. Western blots were performed using anti-GFP antibodies. The positions of molecular mass markers are indicated on the left in kD. (C) Mad1-GFP and Mad2-GFP are associated with NPCs. WT or nup120Δ stains synthesizing Mad1-GFP or Mad2-GFP were grown to logarithmic phase in YEPD at 30°C and examined by confocal fluorescence microscopy. Bar, 5 μm. (D) Mad1p and Mad2p are associated with isolated nuclear fractions. Western blot analysis was performed on subcellular fractions derived from the MAD1-GFP strain using anti-GFP, anti-Mad2p, and anti-Nup53p antibodies. Fractions are defined as follows: C, cytosol; CN, crude nuclei; PN, purified nuclei; NE, nuclear envelope pellet; NP, nucleoplasmic fraction. The materials loaded in the C and CN lanes were derived from equal cell equivalents and PN, NE, and NP from 10-fold higher cell equivalents.
Fluorescence microscopy of asynchronously growing cells revealed that Mad1-GFP and Mad2-GFP were predominately visible along the NE in a distinct punctate pattern reminiscent of proteins associated with NPCs (Fig. 1 C). Mad2-GFP also exhibited low levels of a diffuse signal throughout both the cytoplasm and the nucleoplasm. To confirm that the NE localization of Mad1-GFP and Mad2-GFP reflects their association with NPCs, we examined their distribution in a nup120Δ strain where NPCs cluster within a single region of the NE and, as a consequence, signals derived from NPC-associated proteins can be discriminated from other NE proteins (Aitchison et al., 1995a). In nup120Δ strains, both the Mad1-GFP and Mad2-GFP signals were clustered within a single patch of the NE, further supporting the conclusion that they are associated with the NPC (Fig. 1 C). Consistent with these data, subcellular fractionation experiments showed that Mad1-GFP was enriched in nuclear and NE fractions (Fig. 1 D). Both of these fractions also contained Mad2p; however, significant levels of Mad2p were also present in a cytosolic fraction (Fig. 1 D).
Spindle checkpoint activation induces the release of Mad2p, but not Mad1p, from NPCs and its accumulation at kinetochores
Numerous studies have shown that the mammalian orthologues of Mad1p and Mad2p are recruited to kinetochores during mitosis (Chen et al., 1996; Gorbsky et al., 1998). To explore whether Mad1p and Mad2p exhibit similar dynamics in yeast cells, we monitored the localization of Mad1-GFP and Mad2-GFP in an asynchronously growing cell population. We observed that the bulk of both Mad1-GFP and Mad2-GFP remained associated with the NPCs throughout the cell cycle (unpublished data), including mitosis (Fig. 2 A, representative images), with no detectable redistribution of either protein to kinetochore clusters (Fig. 2, B and C, Log) as judged by comparison to the CFP-tagged kinetochore protein Mtw1p (Goshima and Yanagida, 2000).
Figure 2.
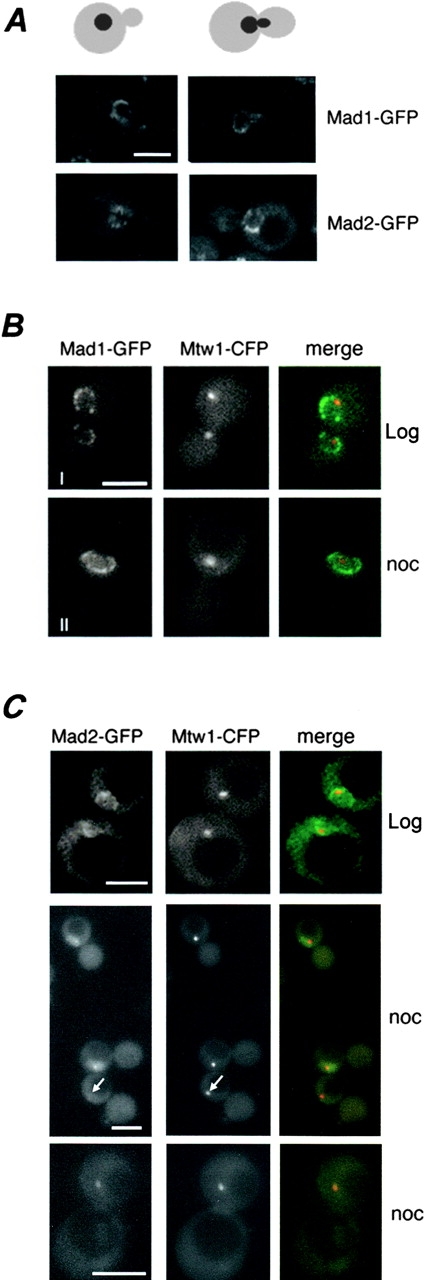
Spindle checkpoint activation induces the release of Mad2p-GFP, but not Mad1p, from the NPC and its recruitment to kinetochores. (A) Localization of Mad1-GFP and Mad2-GFP to NPCs throughout the cell cycle. Strains producing Mad1-GFP or Mad2-GFP were grown to logarithmic phase in YEPD at 30°C and examined by fluorescence microscopy. Representative images of cells in late S-phase (left) and M-phase (right) are shown. Corresponding shapes of cells (gray) and nuclei (black) are indicated. (B and C) Colocalization of Mad2-GFP and Mtw1-CFP to kinetochores on spindle checkpoint activation. Strains synthesizing the kinetochore marker Mtw1-CFP and either Mad1-GFP (B) or Mad2-GFP (C) were grown to early logarithmic phase of growth (control) or G2/M arrested with nocodazole (noc) and visualized by fluorescence microscopy. Subsequently, images were merged. G2/M arrest of these strains was confirmed by FACS® analysis (not depicted). Two separate magnifications of the Mad2-GFP/Mtw1-CFP cells are shown. In arrested cells, we observed Mtw1-CFP in 75% of the cells and Mad2-GFP foci in 50% of the cells. 94% of cells displaying both signals showed overlapping foci. Note, no bleed through of the Mtw1-CFP signal is observed in the GFP images (arrows). Bars, 5 μm.
Vertebrate homologues of Mad1p and Mad2p are recruited to kinetochores during mitosis. We reasoned that in yeast their association with kinetochores might depend on activation of the spindle checkpoint. To test this, we examined the distribution of Mad1-GFP or Mad2-GFP in cells expressing MTW1-CFP after nocodazole treatment. As shown in Fig. 2 B, checkpoint arrest had no effect on the NPC localization of Mad1-GFP and little or no overlap was observed with the Mtw1-CFP signal. In contrast, checkpoint activation had a striking effect on Mad2-GFP. In nocodazole-arrested cells, Mad2-GFP was no longer visible at the NPC and instead colocalized with Mtw1-CFP at the kinetochores in >94% of cells that showed signals from both proteins (Fig. 2 C). Mad2-GFP was not detected at spindle pole bodies under these same conditions as judged by colocalization with the spindle pole body protein Bub2-CFP (unpublished data).
Mad1p and Mad2p are associated with a specific subset of nucleoporins
To further understand the functional significance of the associations of Mad1p and Mad2p with the NPC and the dynamics of Mad2p's localization to kinetochores, we focused on identifying nups that anchor Mad1p and Mad2p to the NPC. Clues as to the identity of these nups came from two previous observations. First, it was recently demonstrated that mutations that affect the function of Nup170p lead to defects in chromosome segregation and kinetochore integrity (Kerscher et al., 2001). Second, a paralogue of Nup170p (Nup157p) was shown to interact with Mad2p in a genome-wide two-hybrid screen (Uetz et al., 2000).
These observations led us to investigate whether Mad1p and Mad2p could be detected in association with Nup170p and Nup157p after their purification from disassembled NPCs. For these experiments, protein A (pA)–tagged versions of either nup (Nup157-pA or Nup170-pA) were purified from nuclear extracts derived from either logarithmically growing or α-factor–arrested cultures. Associated proteins were eluted with a step gradient of increasing MgCl2. Results from α-factor–arrested cultures are shown in Fig. 3, and are similar to those obtained with logarithmically growing cells (unpublished data). Consistent with our previous results, Nup170p was associated with Nup53p (Fig. 3 A; Nup170-pA, α-factor) (Marelli et al., 1998). Nup53p was also present in eluates from Nup157-pA (Fig. 3 A). Moreover, we detected Mad1p and Mad2p in association with both Nup170-pA and Nup157-pA. By comparison, neither protein was detected in experiments using strains lacking a pA tag or containing another tagged nup, Nup60-pA (Fig. 3 B), which interacts with Nup2p (Dilworth et al., 2001; Fig. 3 B).
Figure 3.
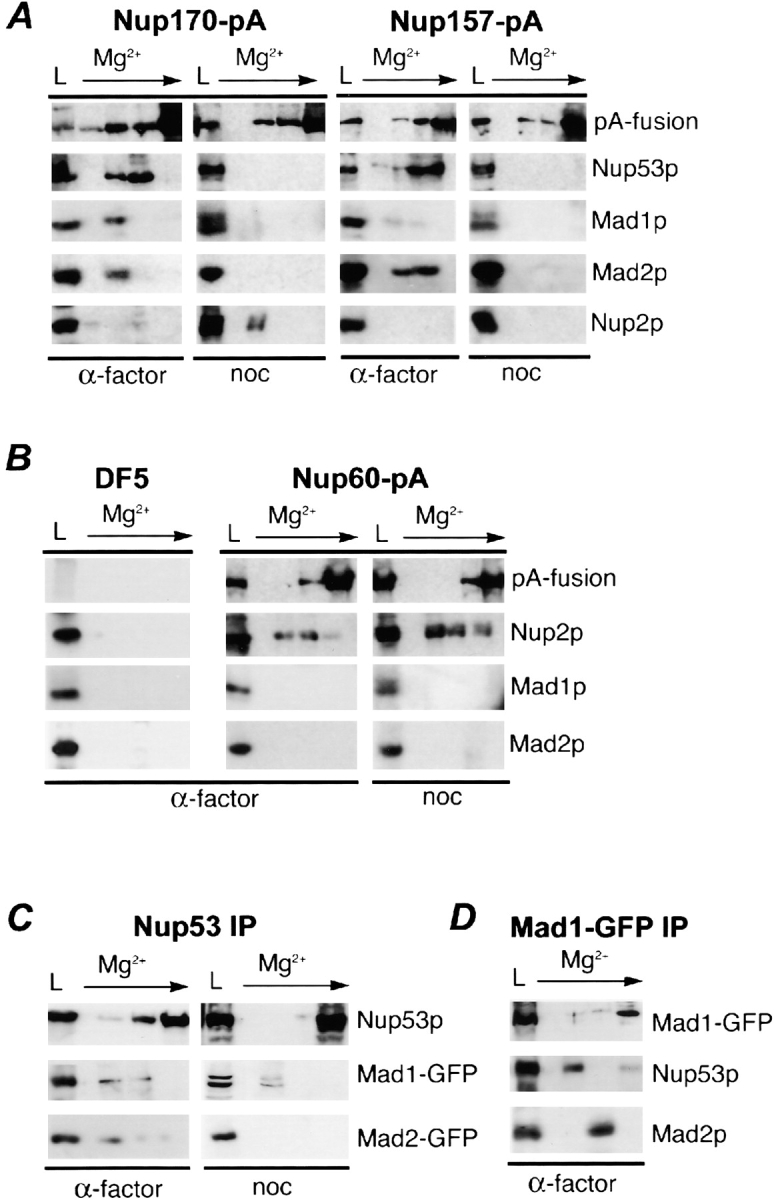
Mad1p and Mad2p associate with a specific subset of nucleoporins. (A) Nup170-pA or Nup157-pA was affinity-purified using IgG-Sepharose from nuclear extracts isolated from α-factor (α-factor)– and nocodazole (noc)-treated cells. Proteins were eluted with a MgCl2 step gradient and analyzed by Western blotting to detect Mad1p, Mad2p, Nup53p, Nup2p, and the pA fusion. The load fraction (L) is shown. (B) Similar experiments were performed with an untagged WT strain (DF5) and a strain synthesizing Nup60-pA. (C) Mad1p, but not Mad2p, is associated with Nup53p in nocodazole-treated cells. Nup53p was immunoprecipitated using Nup53p- specific antibodies out of nuclear extracts derived from α-factor (α-factor)– and nocodazole (noc)-treated cells synthesizing Mad1-GFP or Mad2-GFP. (D) Nup53p and Mad2p are associated with immunoprecipitated Mad1p. Mad1-GFP was immunoprecipitated from nuclear extracts of α-factor (α-factor)–treated cells synthesizing Mad1-GFP. In C and D, bound complexes were eluted and analyzed as described in A using antibodies direct against GFP (for all GFP fusions), Nup53p, and Mad2p.
Because Nup53p is associated with both Nup157p and Nup170p, we also tested whether Mad1p and Mad2p could be detected in this complex by immunoprecipitating Nup53p from cells expressing MAD1-GFP or MAD2-GFP. As shown in Fig. 3 C (Nup53 IP, α-factor), both the Mad1-GFP and Mad2-GFP proteins were detected in association with Nup53p. Moreover, reciprocal immunoprecipitations performed with anti-GFP antibodies also detect Nup53p and Mad2p bound to Mad1-GFP (Fig. 3 D).
Changes in molecular interactions between the Mad1p–Mad2p complex and the NPC on spindle checkpoint activation
The activation of the spindle checkpoint leads to the release of Mad2-GFP from NPCs and its recruitment to kinetochores (Fig. 2 B), but does not affect the NPC association of Mad1p (Fig. 2 B) or Nup53p, Nup157p, and Nup170p (unpublished data). To investigate the biochemical basis for these events, we analyzed the effects of spindle checkpoint activation on interactions between these proteins. As predicted, after checkpoint activation induced by either nocodazole (Fig. 3 A, noc) or benomyl (unpublished data), Mad2p was no longer associated with affinity-purified Nup157pA, Nup170pA, or Nup53p (Fig. 3, A and C). In contrast, Mad1p remained associated with Nup53p; however, neither Mad1p nor Nup53p were detected in association with Nup170-pA or Nup157-pA (Fig. 3, A and C, noc). These results suggest that a complex containing Nup53p and Mad1p is no longer bound to Nup170p and Nup157p. We excluded the possibility that these were nonspecific effects caused by nocodazole or benomyl because the mere addition of benomyl to α-factor–arrested cells did not induce these changes (unpublished data). Moreover, checkpoint activation did not affect the association of Nup60p or Nup170p with Nup2p, even though Nup2p appears to be modified in these cells (Fig. 3, A and B), possibly by phosphorylation (Ficarro et al., 2002).
Because strains lacking individual members of the Nup53p–Nup170p–Nup157p complex are viable, we examined the localization of Mad1-GFP and Mad2-GFP in the deletion mutants. The localization of Mad1-GFP to the NPC in strains lacking NUP170 (nup170Δ), NUP157 (nup157Δ), or NUP59 (nup59Δ), a gene encoding a nup structurally similar to Nup53p, was not altered in asynchronous or nocodazole-treated cells (Fig. 4 A). In contrast, asynchronous cultures of cells lacking Nup53p (nup53Δ) exhibited reduced levels of NE-associated Mad1-GFP (Fig. 4 A). This effect was further exacerbated by nocodazole treatment where >80% of arrested cells exhibited weak or barely visible amounts of Mad1-GFP at the NPCs.
Figure 4.
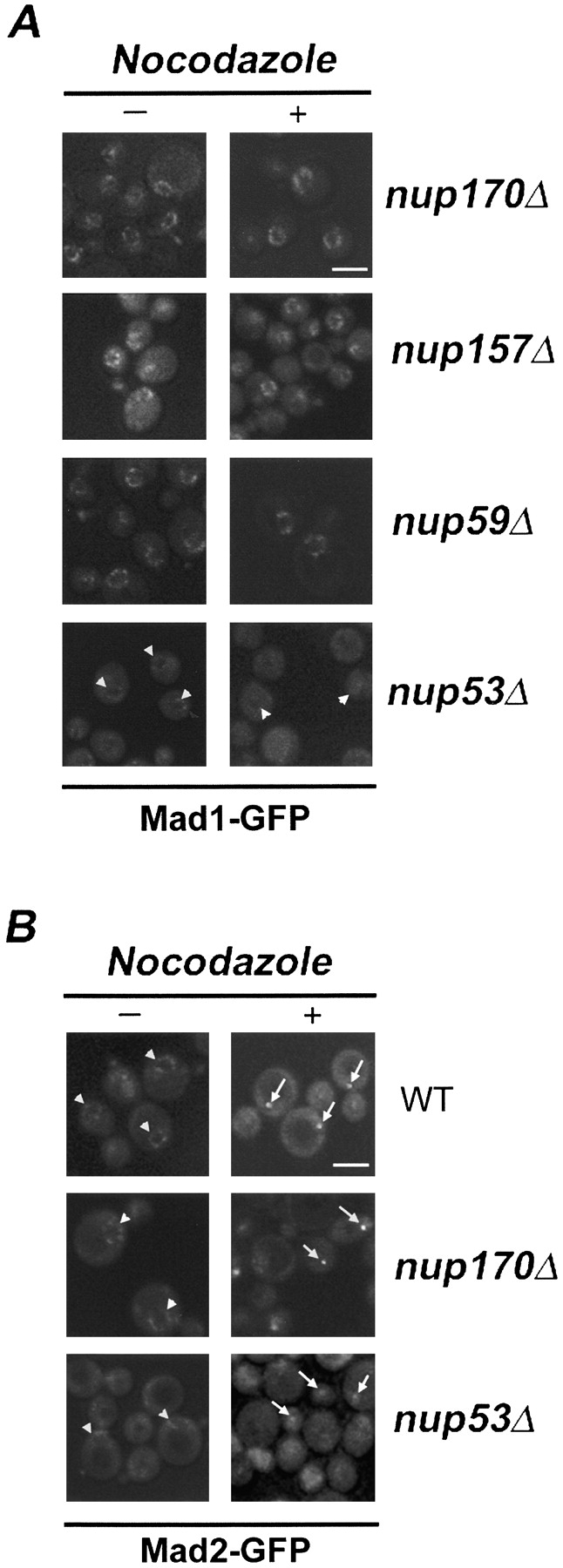
The effects of nup mutations on the subcellular distribution of the Mad1p–Mad2p complex. (A) The subcellular localization of Mad1-GFP in logarithmically growing (−) or nocodazole-treated (+) nup170Δ, nup157Δ, nup59Δ, and nup53Δ strains was examined using confocal fluorescence microscopy. (B) The localization of Mad2-GFP in logarithmically growing (−) or nocodazole-treated (+) WT, nup170Δ, and nup53Δ strains was examined using confocal fluorescence microscopy. Note, arrowheads point to the NE and arrows point to kinetochore signals. Bars, 5 μm.
As noted above, in WT strains, Mad2-GFP is localized to the NPC in asynchronous cultures (Fig. 1 C and Fig. 2 A) and to kinetochores on checkpoint activation. A similar localization pattern for Mad2-GFP was observed in a nup170Δ strain (Fig. 4 B). However, in a nup53Δ strain, as was the case with Mad1-GFP, the levels of Mad2-GFP at the NPC were visibly reduced in asynchronous cultures (Fig. 4 B). Moreover, when this strain was treated with nocodazole, the accumulation of Mad2-GFP at kinetochores was attenuated, with many cells (∼63%) showing barely visible or nondetectable kinetochore signals. This effect did not appear to be a function of altered kinetochore localization as the signal intensity of Mtw1-CFP in a nup53Δ strain was the same as observed in WT cells (unpublished data).
The Mad1p–Mad2p complex associates with the NPC through Mad1p
We investigated whether Mad1p, Mad2p, or both proteins interacted with the Nup53p-containing complex. To do this, Nup53p was immunoprecipitated from WT, mad1Δ, and mad2Δ strains, and the eluates were probed with antibodies directed against Mad1p and Mad2p. As shown in Fig. 5 A, Mad1p was bound to Nup53p in the absence of Mad2p, but Mad2p was not detected in association with Nup53p in the absence of Mad1p. Consistent with these data, Mad1-GFP was localized to NPCs in vivo in the absence of Mad2p (Fig. 4 B). However, Mad2-GFP failed to concentrate at the nuclear periphery and was visible throughout the cell in a strain lacking Mad1p. Furthermore, Mad2-GFP was not recruited to kinetochores in nocodazole-treated mad1Δ cells (Fig. 4 B). Together, these data are consistent with a model in which the Mad1p–Mad2p complex binds the NPC through Mad1p (see Fig. 10).
Figure 5.
Mad1p links the Mad1p–Mad2p complex to the NPC. (A) The association of Nup53p with the Mad1p–Mad2p complex requires Mad1p. Nup53p was immunoprecipitated from nuclear extracts of WT, mad1Δ, and mad2Δ strains. Proteins were eluted with a MgCl2 gradient and analyzed by Western blotting with antibodies directed against Mad1p or Mad2p. The load fraction (L) is shown. (B) Mislocalization of Mad2-GFP in a mad1Δ strain. The localization of Mad1-GFP and Mad2-GFP was examined in WT, mad2Δ, or mad1Δ and nocodazole-treated mad2Δ or mad1Δ (+noc) strains using confocal fluorescence microscopy. Bar, 5 μm.
Figure 10.
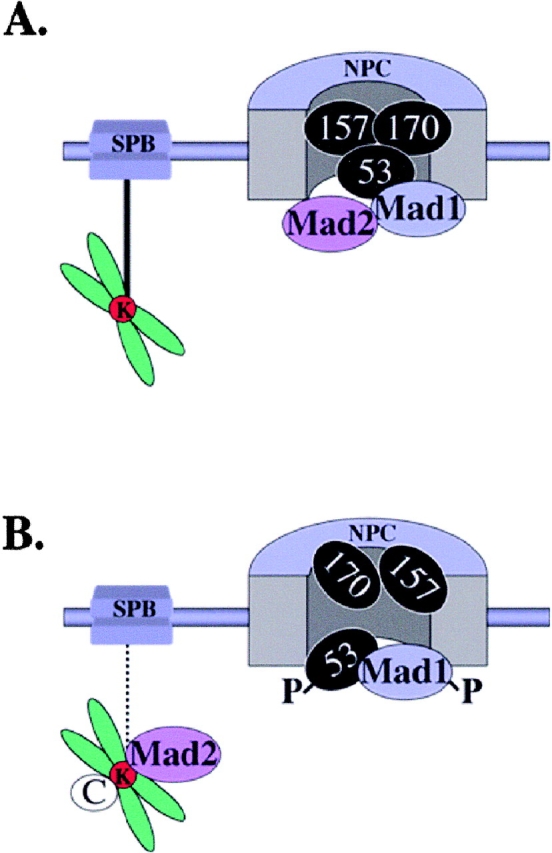
Model summarizing interactions between the Nup53p-containing complex, Mad1p and Mad2p. (A) Shown schematically is an NPC with Nup53p, Nup157p, and Nup170p (53, 157, and 170). The Mad1p–Mad2p complex is associated with the NPC through Mad1p. Additional interactions between Mad proteins and/or the NPC may also exist. Also depicted is one of the spindle pole bodies (SPB) connected to a chromosome's kinetochore (K) via a microtubule (black line). (B) Defects in microtubule interaction (interrupted line) of the SPB with the kinetochore lead to spindle checkpoint activation, hyperphosphorylation of Mad1p (P), dissociation of Mad2p from the NPC, and the recruitment of Mad2p to the kinetochores along with other checkpoint proteins (C). At the same time, Nup157p and Nup170p are no longer associated with phosphorylated Nup53p (P).
Genetic interactions between MAD1 and MAD2 and nucleoporin genes
We investigated genetic interactions between MAD1 and MAD2 and four nup genes (NUP170, NUP157, NUP53, and NUP59) to further assess the functional significance of the association of Mad1p and Mad2p with the NPC. Initially, we assayed the growth characteristics of each single deletion strain on rich media at 27 and 37°C. Our analysis revealed that all the strains showed growth characteristics similar to the WT strain with the exception of the nup170Δ strain, which grew slower at both temperatures (Fig. 6 A).
Figure 6.
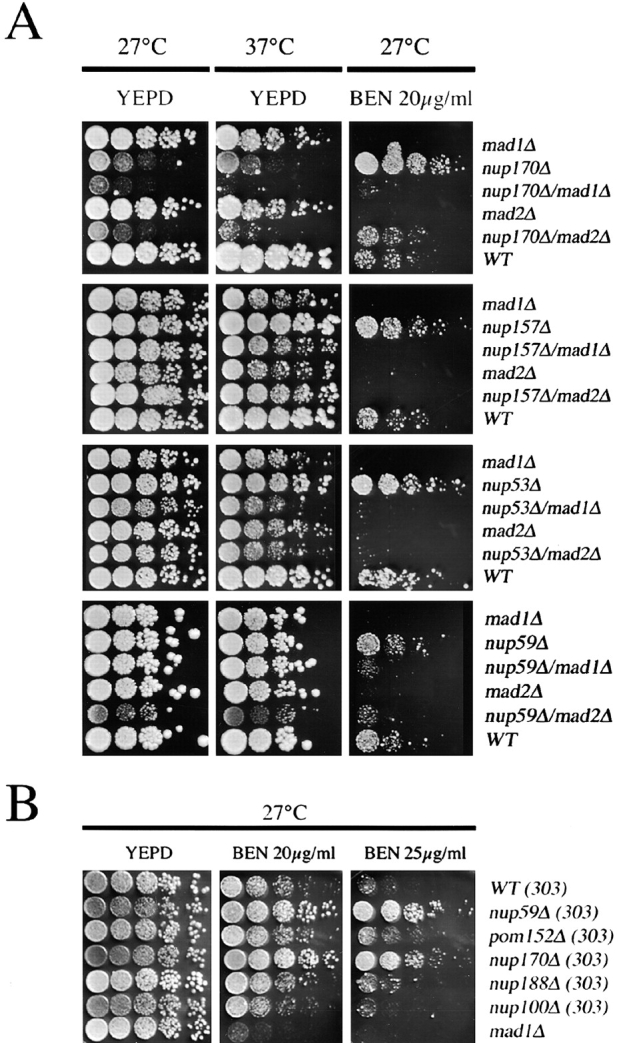
MAD1, MAD2, and nup-encoding genes of the Nup53p-containing complex interact genetically. (A) The growth of WT, mad1Δ, mad2Δ, nup170Δ, nup157Δ, nup53Δ, and nup59Δ strains and combinations of double mutants were assessed at 27, 37, and 30°C (not depicted). Strains were grown to early logarithmic phase in YEPD at 30°C, diluted, spotted on YEPD and YEPD containing 20 μg/ml benomyl (BEN), and incubated at the indicated temperatures (27 or 37°C). (B) Benomyl resistance of nup mutants. nup59Δ, pom152Δ, nup170Δ, nup188Δ, nup100Δ, and mad1Δ strains, using W303 as the parental WT strain (WT 303), were spotted on YEPD plates containing either 20 or 25 μg/ml of benomyl and incubated for 3 d at 27°C.
Each of the nup null haploids was crossed with the mad1Δ and mad2Δ deletion strains. We tested the growth characteristics of the haploid double deletion strains, and all of these strains showed similar growth characteristics (Fig. 6 A). Exceptions were the nup170Δ mad1Δ, nup170Δ mad2Δ, and nup59Δ mad2Δ deletion strains, which grew slower than the parents and exhibited impaired growth at 27°C and either slow (nup170Δ mad2Δ) or barely detectable (nup170Δ mad1Δ) growth at 37°C.
The benomyl sensitivity of various null mutants was also tested. As expected, benomyl inhibited growth of the mad1Δ and mad2Δ strains (Fig. 6, A and B). However, null mutants of NUP170, NUP157, NUP53, and NUP59 grew better than WT cells on benomyl-containing plates (Fig. 6, A and B). This was also the case for the viable double null strains nup59Δ nup53Δ, nup53Δ nup157Δ, and nup59Δ nup157Δ (unpublished data). Resistance to benomyl appears to be specific for these nups as several other nup null mutants, including two (nup188Δ and pom152Δ) that genetically interact with members of the Nup53p-containing complex (Aitchison et al., 1995b; Marelli et al., 1998), were not resistant (Fig. 6 B).
To determine whether the benomyl resistance of the nup null mutants required a functional checkpoint, we examined the growth of the mad/nup double mutants on benomyl-containing plates. As shown in Fig. 6 A, the mad1Δ and mad2Δ deletions eliminated the benomyl-resistant phenotype of the nup nulls. Most of the double null strains exhibited growth characteristics similar to the mad1Δ and mad2Δ mutants. The nup170Δ mad2Δ and nup59Δ mad2Δ strains were somewhat more resistant to benomyl than the mad2Δ mutant, with the nup170Δ mad2Δ strain growing similar to WT cells. From these analyses, we conclude that the increased benomyl resistance of the tested nup deletions is dependent on Mad1p and Mad2p. Thus, these genetic analyses suggest an important functional interplay between Mad1p, Mad2p and Nup53p, Nup59p, Nup170p, and Nup157p in spindle dynamics.
Nup53p is required for the hyperphosphorylation of Mad1p in nocodazole-treated cells
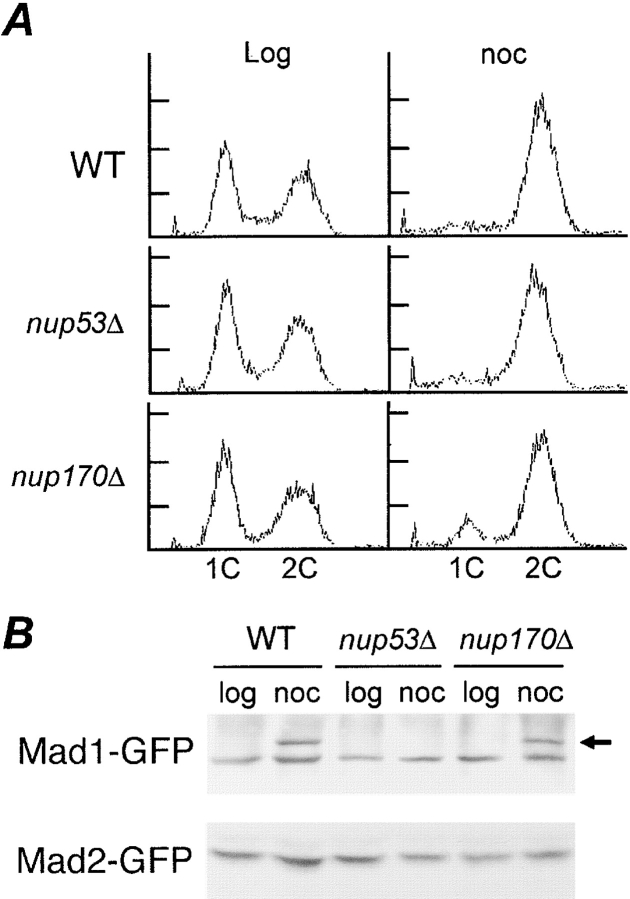
In response to checkpoint activation, Mad1p is hyperphosphorylated. Because Nup53p appears to play a role in anchoring Mad1p to the NPC during checkpoint activation, we examined the effects of removing Nup53p on the nocodazole-induced hyperphosphorylation of Mad1p. WT, nup53Δ, and nup170Δ strains containing Mad1-GFP or Mad2-GFP were treated with or without nocodazole. In the presence of nocodazole, each of these strains arrested with a 2C DNA content, suggesting that the spindle checkpoint was functional (Fig. 7 A). As shown in Fig. 7 B, a reduction in the mobility of Mad1-GFP, diagnostic of its phosphorylation, was visible in arrested samples from WT and nup170Δ strains. However, under the same conditions, no change in the mobility of Mad1-GFP was seen in the nup53Δ strain, suggesting that it was not hyperphosphorylated in nocodazole-arrested cells. These results suggest that the Nup53p-dependent association of Mad1p with the NPC is critical for its hyperphosphorylation.
Figure 7.
Requirement of Nup53p for hyperphosphorylation of Mad1p. (A) nup170Δ and nup53Δ strains arrest in G2/M after treatment with nocodazole. FACS® analysis was performed on logarithmically growing (log) and nocodazole (noc)-treated (1.5 h) WT, nup53Δ, and nup170Δ strains. The positions of 1C and 2C DNA peaks are indicated. (B) Mad1p is not hyperphosphorylated in a nup53Δ strain. Logarithmically growing WT, nup53Δ, and nup170Δ strains expressing either Mad1-GFP or Mad2-GFP were treated with (noc) or without (log) nocodazole for 1.5 h. Total cell lysates were analyzed by immunoblotting using an anti-GFP antibody. The position of hyperphosphorylation Mad1-GFP is indicated by an arrow.
Mad1p plays a role in NPC function
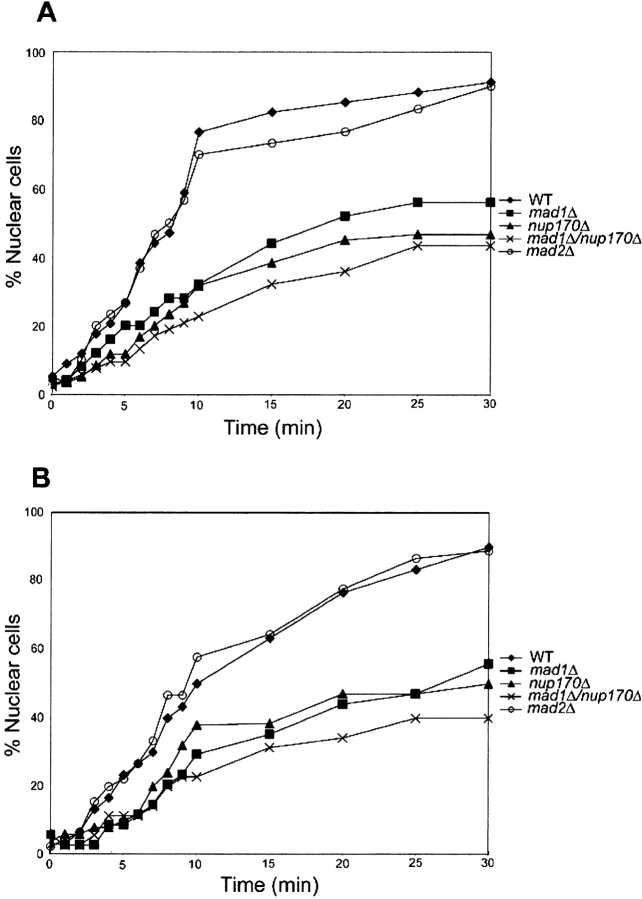
The association of Mad1p with the NPC raises the question of whether it plays a role in nuclear transport. To address this, we used an in vivo nuclear import assay developed by Shulga et al. (1996) to evaluate the effects of MAD1 deletion on transport. In this assay, a nuclear reporter is allowed to equilibrate across the NE by treatment of cells with inhibitors of glycolysis and mitochondrial respiration. After their removal and reinitiation of transport, the relative import rates can be assessed by counting the number of cells showing nuclear accumulation of the reporter at various time points. We examined the import of a reporter protein, Pho4-GFP that is imported by karyopherin Kap121p. Kap121p has been shown to specifically interact with the Nup53p-containing complex. As shown in Fig. 8, WT and mad2Δ strains exhibited similar rates of import, with ∼40% of the cells showing nuclear accumulation of the Pho4-GFP reporter after ∼6 min. Surprisingly, however, the mad1Δ and nup170Δ strains required 13 and 16 min, respectively. This reporter accumulation defect was further exacerbated in the mad1Δ nup170Δ strain, where 23 min were required for 40% of the cells to show a nuclear accumulation of the reporter. Similar results were also obtained when cells were first incubated at 37°C for 3 h before performing the assay (Fig. 8 B).
Figure 8.
Nuclear accumulation of Pho4-GFP is inhibited in strains lacking MAD1 and NUP170. Each of the indicated strains expressing a PHO4-GFP reporter were grown at 30°C (A) or at 30°C and then shifted to 37°C for 3 h (B). After treatment with and removal of metabolic poisons (2-deoxyglucose and sodium azide), relative rates of import were determined by counting the number of cells showing a nuclear accumulation of the reporter and plotting this versus time. The results shown are representative of those obtained in multiple experiments.
The nup170Δ and mad1Δ deletions also appear to affect the stability of the Nup53p-containing complex. When we examined the localization of Nup53-GFP in the mad1Δ, nup170Δ, and nup170Δ mad1Δ strains at 27°C, Nup53-GFP was clearly associated with the nuclear periphery (Fig. 9). However, after shifting to 37°C for 3 h, the levels of Nup53-GFP associated with the NE were decreased to barely detectable levels in each strain. This effect was specific, as temperature shift had no effect on the localization of two other nups, Nup49-GFP and Nup188-GFP. Moreover, shifting WT or mad2Δ cells to 37°C had no effect on the localization of Nup53p-GFP.
Figure 9.
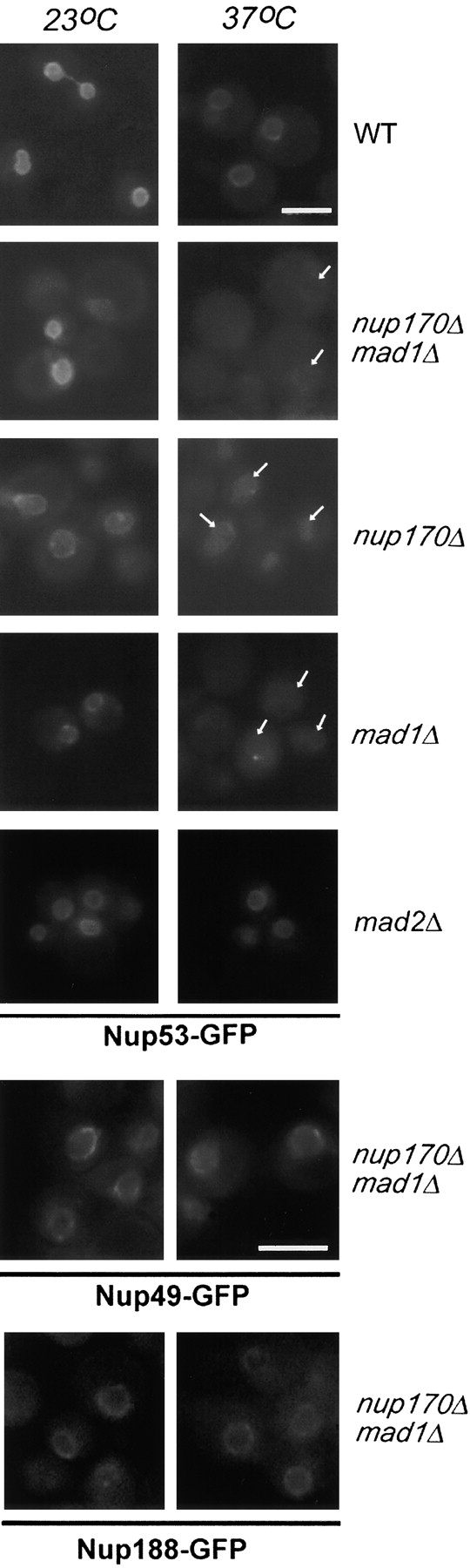
Mad1p is required for the stable association of Nup53p with the NE. WT, nup170Δmad1Δ, nup170Δ, mad1Δ, and mad2Δ strains expressing a plasmid borne copy of NUP53-GFP (pNP53) were grown to logarithmic phase and either maintained at 23°C or shifted to 37°C for 3 h. The localization of Nup53p-GFP was examined by fluorescence microscopy. The results of similar experiments examining the localization of two other nucleoporins, Nup49-GFP and Nup188-GFP, in the nup170Δ mad1Δ strain are shown in the bottom two rows. Note, arrows point to the NE. Bars, 5 μm.
Discussion
We have identified a functional link between components of the S. cerevisiae NPC and the mitotic checkpoint machinery. By a variety of criteria, we have shown that the checkpoint proteins Mad1p and Mad2p reside largely at the NPC throughout the cell cycle. Their association with the NPC occurs through a previously identified nup subcomplex containing Nup53p, Nup59p, and Nup170p (Marelli et al., 1998). The interactions between these nups and mads were detected by reciprocal affinity purification of the nup complex, Mad1p (Fig. 3), or Mad2p (unpublished data). Our data are consistent with a model in which the association of Mad2p with these nups is dependent on Mad1p (Fig. 10). In the absence of Mad1p, Mad2p fails to associate with NPCs and is dispersed throughout the cell. Nup53p plays an important role in either directly or indirectly tethering the Mad1p–Mad2p complex to the NPC. Removal of Nup53p causes a reduction in levels of Mad1p and Mad2p associated with NPCs (Fig. 4), but no significant changes in the cellular levels of either protein (unpublished data). It remains to be determined what nups, in the absence of Nup53p, would support the reduced levels of Mad1p and Mad2p at the NPC. One explanation is that their association is partially maintained by lower affinity interactions with other members of the Nup53p-containing complex. However, this remains an open question because null mutations of NUP59, NUP157, and NUP170 (Fig. 4) have no readily visible effects on the NPC association of Mad1p. Another candidate is the karyopherin Kap121p, which is also associated with Nup53p-containing complex. However, we fail to detect any effect on the localization of Mad1p and Mad2p in strains containing temperature-sensitive alleles of KAP121 (Anderson, A., and R. Wozniak, personal communication).
Our observation that S. cerevisiae Mad1p and Mad2p remain associated with NPCs throughout the cell cycle differs from the events described in vertebrate cells. Recent data have shown that vertebrate homologues of these proteins are also associated with NPCs during interphase (Campbell et al., 2001). However, in these cells, upon entering mitosis, the NE and NPCs are disassembled and Mad1p and Mad2p accumulate at unattached kinetochores during prometaphase. This situation does not occur in S. cerevisiae where the NE and NPCs remain intact during mitosis. Moreover, after replication of centromeric DNA, kinetochores are assembled and rapidly engaged by microtubules (for review see Winey and O'Toole, 2001). This may partially explain why Mad1p and Mad2p remain at the NPC during mitosis. Consistent with these predictions, only on activation of the spindle assembly checkpoint is Mad2p released from the NPCs (Fig. 2). In contrast, the bulk of Mad1p remains at the NPC and we fail to detect a discernible accumulation of Mad1p at the kinetochores. It is possible that a portion of Mad1p is also recruited to kinetochores but that it is either below the level of detection, obscured by the NPC signal, or its association with the kinetochores is transient, with Mad1p being quickly recycled back to the NPCs.
In addition to the release of Mad2p, checkpoint arrest induced by nocodazole also results in profound effects on the molecular interactions between members of the Nup53p-containing complex and Mad1p–Mad2p. In nocodazole-arrested cells, Nup53p, Mad1p, and Mad2p are no longer detected in association with Nup157p or Nup170p (Fig. 3). These changes are accompanied by the phosphorylation of Nup53p (Marelli et al., 1998; unpublished data) and Mad1p (Fig. 3; Hardwick and Murray, 1995). However, Nup53p and Mad1p remain associated, and this interaction is likely important for maintaining WT levels of Mad1p at the NPC after spindle checkpoint activation (Fig. 4). By analogy to higher eukaryotes where extensive changes in protein–protein interactions between nups occur during mitosis, these experiments suggest that distinct molecular rearrangements also occur within the yeast NPC. However, in yeast, these specific changes do not lead to NPC disassembly. Ourselves and others have not observed changes in the NPC localization of any nups during mitosis, including those that are part of the Nup53p-containing complex (Copeland and Snyder, 1993; Aitchison et al., 1995b; Marelli et al., 1998; Kerscher et al., 2001). We propose that in yeast, specific changes in protein–protein interactions occur during mitosis that alter the functional properties of the NPC, including its association with the checkpoint machinery.
Several observations suggest that the association of the Mad1p–Mad2p complex with the NPC plays an important role in the function of these checkpoint proteins and the NPC. We have shown that the checkpoint-induced hyperphosphorylation of Mad1p does not occur in the absence of Nup53p, suggesting that the physical association of Mad1p with Nup53p promotes the phosphorylation of Mad1p, an event that is believed to be mediated by the kinase Mps1p in response to spindle damage (Hardwick et al., 1996). This observation reiterates that the phosphorylation of Mad1p is not required for spindle checkpoint function (Farr and Hoyt, 1998), as the nup53Δ strain does not exhibit a readily detectable checkpoint defect, and raises the question of whether phosphorylation of Mad1p plays a role in NPC function.
The association of Mad2p with the NPC is dependent on Mad1p and consistent with this idea, both proteins show a decreased association with the NPC in asynchronous cultures of a nup53Δ strain (Fig. 4). We also detected a decrease in kinetochore associated Mad2p in the nup53Δ strain after checkpoint activation. Because the cellular levels of Mad1p are not altered in the nup53Δ strain, these results suggest that the association of the Mad1p–Mad2p complex with the NPC is an important prerequisite for the association of Mad2p with the kinetochores. Moreover, our results would suggest the checkpoint functions of Mad2p may not be tightly linked to the amount of Mad2p bound to kinetochores, as the nup53Δ strain exhibits no checkpoint defects.
Because Nup53p and Mad1p do not appear to leave the NPC or accumulate at the kinetochores during checkpoint activation, it seems unlikely that they play a direct role in the association of Mad2p to the kinetochores. A role of Mad1p and Nup53p in this process could be explained if we consider a model in which the NPC acts as a platform for regulating the assembly of checkpoint complexes that then associate with kinetochores. Our data, showing that Mad2p (but not Mad1p) is present in a cytoplasmic fraction, are consistent with the idea that a free pool of Mad2p exists in the cytoplasm and that formation of the Mad1p–Mad2p complex may occur at the NPC. Recent reports also place Schizosaccharomyces pombe Mad2p at the nuclear periphery (Ikui et al., 2002). It would also seem possible that the formation of other complexes that have been shown to be dependent on Mad1p, including, for example, the Mad2p–Mad3p–Bub3p–Cdc20p complex (Hardwick et al., 2000, Fraschini et al., 2001), could occur at the NPC. Such complex formation involving Mad2p could contribute to its function at kinetochores. Similar events may instead occur at kinetochores in vertebrate cells. For example, Chung and Chen (2002) have recently shown that a Mad1p-free pool of Mad2p is required for checkpoint function, and that this pool may cycle through a kinetochore-bound Mad1p intermediate. Conceptually, one could envision that these latter events occur at the NPC in yeast. It is possible that in vertebrates, kinetochore-bound Mad1p may also be associated with NPC components that are recruited there after NE disassembly, perhaps as a defined subcomplex similar to that observed in yeast. This idea is supported, in theory, by recent data showing that several NPC proteins are detectable at kinetochores after disassembly of the NPCs during mitosis (Belgareh et al., 2001).
It still remains to be determined whether the interactions between the Nup53p-containing complex and Mad1p and Mad2p are critical for their checkpoint functions or whether this association plays another role, such as regulating the level of the checkpoint response. The functional redundancy between members of the Nup53p-containing complex makes this difficult to test directly. However, a potential functional link is the benomyl-resistant phenotype of the nup null mutants, which is dependent on a functional spindle checkpoint, as the deletion of MAD1 or MAD2 suppressed this phenotype (Fig. 6). The mechanistic basis for this phenotype is not clear, but one possibility is that deletion of any one of its members alters the ability of the Nup53p-containing complex to regulate the level of the checkpoint response. The benomyl resistant phenotype of the nup mutants could reflect an up-regulation of the checkpoint response that prolongs mitotic arrest and increases cell survival in the presence of elevated levels of benomyl. Alternatively, the benomyl resistance of these mutants may reflect a separate, as yet undefined, role for these nups in regulating spindle dynamics.
Our data also support the idea that Mad1p plays an active role in NPC function. Import assays using a reporter protein imported by Kap121p show that both the mad1Δ and mad1Δ nup170Δ strains exhibit import defects (Fig. 8). The functional basis for this defect remains to be determined, as does the extent to which other karyopherin-mediated pathways are affected. One possibility is that the function of Mad1p is linked to Nup170p and its role in establishing the size limits of the diffusion channel through the NPC (Shulga et al., 2000). Changes in the diffusion channel may explain the defects in reporter accumulation we observe in the nup170Δ stain. Alternatively, Mad1p may play a role in active transport. In addition to this function, we show that in the absence of Mad1p, the association of Nup53-GFP with the NPC becomes thermolabile, with levels of the protein at the NE being greatly reduced at 37°C (Fig. 9). A similar phenomenon was also observed in strains lacking Nup170p. These data lead us to conclude that Mad1p, like Nup170p, contributes to the stable association of Nup53p with the NPC, either by stabilizing its association with other nups or influencing its assembly into the NPC. The latter scenario could be linked to Mad1p's effects on transport, as we have recently shown that Kap121p plays a role in assembly of Nup53p into the NPC (Lusk et al., 2002).
In conclusion, we have shown that in yeast specific nups and mads physically and functionally interact. This functional relationship between checkpoint protein complexes and the NPC sets the stage for our further understanding of the molecular role of these evolutionarily conserved proteins in genome stability in model organisms and humans.
Materials and methods
Yeast media, strains, and plasmids
Media for yeast growth and sporulation were as described previously (Adams et al., 1997) except where indicated. Benomyl-containing plates were made as described previously (Hyland et al., 1999). Procedures for yeast manipulation were performed as described previously (Sherman et al., 1986). All yeast strains are listed in Table I.
Table I.
Strains used in this study
| Strain | Genotype | Source, derivation, or reference |
|---|---|---|
| W303 | Mat a/α ade2-1/ade2-1 ura3-1/ura3-1 his3-11,15/his3-11,15 trp1-1/trp1-1 leu2-3, 112 can 1-100/112 can1-100 | |
| DF5a | Mat a ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 | |
| DF5α | Matα ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 | |
| DF5 | Mat a/α ura3-52/ura3-52 lys2-801/52/lys2-801 his3-200/his3-200 leu2-3,112/leu2-3,112 trp1-1/trp1-1 nup157Δ::URA3 | |
| NP157-2.1 | Mat a ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 | Aitchison et al.,1995a |
| NP120-25-3 | Matα ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 nup120Δ::URA3 | Aitchison et al.,1995a |
| NP120-25-4 | Mat a ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 nup120Δ::URA3 | Aitchison et al.,1995a |
| NP53/NP59 | Matα ade2 ura3 his3 trp1 leu2 nup53Δ::HIS3 nup59Δ::HIS3 | Marelli et al., 1998 |
| NP53-B1 | Mat a ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 nup53Δ::HIS3 | Marelli et al., 1998 |
| NP59-23 | Mat a ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 nup59Δ::HIS3 | Marelli et al., 1998 |
| NP170pA | Mata ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 Nup170-protA (URA3-HIS3)/+ | Rout et al., 2000 |
| NP157pA | Mat a ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 Nup157-protA (URA3-HIS3)/+ | Rout et al., 2000 |
| NP60pA | Mat a ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 Nup60-protA (HIS3)/+ | Rout et al., 2000 |
| M1GFP | Matα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 MAD1-GFP/HIS5 | This work |
| M2GFP | Matα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 MAD2-GFP/HIS5 | This work |
| YMB1296 | Matα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 MAD2-GFP/HIS5 CFIII (CEN3L.YPH278)URA3 SUP11 | This work |
| YMB1299 | Matα ura3-52 lys2-801 ade2-101 his3Δ200 leu2Δ1 MAD1-GFP/HIS5 CFIII (CEN3L.YPH278)URA3 SUP11 | This work |
| YMB1451 | Mat a ura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ1 nup170Δ::KAN | This work |
| YMB1482 | Matα ura3-52 lys2-801 his3Δ-200 leu2-3,112 trp1-1 nup170Δ::KAN | This work |
| YMB1900 | Mat a ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 mad2Δ::HIS3 [pMAD2/URA3] | This work |
| YMB1906 | Matα ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 mad2Δ::HIS3 | This work |
| YMB1908 | Matα ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 mad1Δ::KAN | This work |
| YMB1911 | Mat a ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 mad1Δ::KAN [pMAD1/URA3] | This work |
| YMB1979 | Matα ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 nup170Δ::HIS3 mad1Δ::KAN | Segregant of sporulated YMB1482/1911 |
| YMB2008 | Mat a ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 nup170Δ::HIS3 mad2Δ::HIS3 | Segregant of sporulated YMB1482/1900 |
| YMB2018 | Matα ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 MAD1-GFP/HIS5 | This work |
| YMB2020 | Mat a ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 MAD1-GFP/HIS5 | This work |
| YMB2022 | Mat a ura3 his3 nup53Δ::HIS3 mad2Δ::HIS | Segregant of sporulated NP53/NP15782/YMB1906 |
| YMB2029 | Matα ura3 his3 nup157Δ::URA3 mad2Δ::HIS3 | Segregant of sporulated NP53/NP1578-2/YMB1906 |
| YMB2032 | Matα ura3 his3 nup157Δ::URA3 mad1Δ::KAN | Segregant of sporulated NP53/NP1578-3/YMB1908 |
| YMB2035 | Mat a ura3 his3 nup53Δ::HIS3 mad1Δ::KAN | Segregant of sporulated NP53/NP1578-3/YMB1908 |
| YMB2064 | Matα ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 nup170Δ::HIS3 MAD1-GFP/HIS5 | Segregant of sporulated YMB1482/YMB2020 |
| YMB2067 | Matα ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 nup53Δ::HIS3 MAD1-GFP/HIS5 | Segregant of sporulated NP53-B1/YMB2018 |
| YMB2081 | Matα ura3-52 lys2-801 his3-200 leu2-3,112 trp1-1 nup157Δ::URA3 MAD1-GFP/HIS5 | Segregant of sporulated NP157-2.1/YMB2018 |
| YMB2086 | Matα ura3-52 lys2-801 ade2-101 his3-200 leu2Δ1 nup170Δ::KAN MAD2-GFP/HIS5 CFIII (CEN3L.YPH278)URA3 SUP11 | This work |
| YMB2087 | Matα ura3-52 lys2-801 ade2-101 his3-200 leu2Δ1 nup53Δ::KAN MAD2-GFP/HIS5 CFIII (CEN3L.YPH278)URA3 SUP11 | This work |
| YMB2088 | Matα ura3-52 lys2-801 ade2-101 his3-200 leu2Δ1 mad1Δ::KAN MAD2-GFP/HIS5 CFIII (CEN3L.YPH278)URA3 SUP11 | This work |
| YMB3048 | Mat a ura his3 trp1-1 leu2-3,112 nup59Δ::HIS3 mad2Δ::HIS3 | Segregant of sporulated NP59-23/YMB1906 |
| YMB3051 | Mat a ura his3 trp1-1 leu2-3,112 nup59Δ::HIS3 mad2Δ::KAN | Segregant of sporulated NP59-23/YMB1908 |
| NP120M1G | Matα ura3 his3 trp1 leu2 nup120Δ::URA3 MAD1-GFP/HIS5 | Segregant of sporulated NP120-25-3/YMB2020 |
| NP120M2G | Matα ura3 his3 trp1 leu2 nup120Δ::URA3 MAD1-GFP/HIS5 | Segregant of sporulated NP120-25-4/YMB1296 |
| YPH278 | Matα ura3-52 lys2-801 ade2-101 his3-200 leu2Δ1 CFIII (CEN3L.YPH278)URA3 SUP11 | Spencer et al., 1990 |
Production of NUP and MAD deletions
Unless otherwise noted, complete chromosomal deletions were constructed in WT strains DF5a, DF5α, or DF5. Deletions were confirmed by PCR and marker segregation in subsequent tetrad analysis.
Deletions of nup and checkpoint genes were produced by two similar PCR-mediated gene deletion techniques (Baudin et al., 1993). In the first technique, we used a PCR product derived from 40 bp of sequences immediately upstream of the start and downstream of the stop codon of the gene to be deleted and 20 bp of sequence homologous to pRS303 (HIS3) or pRS400 (KAN; Sikorski and Hieter, 1989; Brachmann et al., 1998) immediately adjacent to the vector selectable marker. The second technique utilizes genomic DNA of an existing deletion strain to PCR amplify a deletion cassette module containing ∼200 bp upstream of the start and downstream of the stop codon of the deleted gene of interest plus the deletion marker.
The NUP170 ORF was replaced with the HIS3 marker in the DF5α strain (YMB1482) and with the KAN marker in the strain YMB1451. Heterozygous mad1Δ::KAN/MAD1 (YMB1488) and mad2Δ::HIS3/MAD2 (YMB1496) deletions were made in diploid WT strain DF5 using a mad1Δ::KAN strain (YFS1120; provided by F. Spencer, The Johns Hopkins School of Medicine, Baltimore, MD) or a mad2Δ::HIS3 strain (YKH447; provided by K. Hyland and P. Hieter, University of British Columbia, British Columbia, Canada) to PCR amplify the respective deletion module. The resulting strain mad1Δ::KAN/MAD1 (YMB1488) was transformed with a MAD1/URA3 plasmid (pKH130; Hardwick and Murray, 1995), and mad2Δ::HIS3/MAD2 (YMB1496) was transformed with a MAD2/URA3 containing plasmid (pCD2; Warren et al., 2002). pKH130 and pCD2 are gifts from F. Spencer. The diploid strains were sporulated and haploid mad1Δ::KAN (YMB1908 and YMB1911) and mad2Δ::HIS3 strains (YMB1900 and YMB1906) were used for subsequent crosses.
For double deletion mutants, mad1Δ::KAN (YMB1908 and YMB1911) or mad2Δ::HIS3 (YMB1900 and YMB1906) was mated to nup170Δ::HIS3 (YMB1482), nup157Δ::URA3 (NP157–2.1), nup53Δ::HIS3 (from NP53/NP157[8–2]), and nup59Δ::HIS3 (NP59–23). Haploid meiotic progeny of the diploids was analyzed. Similarly, nup120Δ::URA3 MAD1-GFP (NP120M1G) and nup120Δ::URA MAD2-GFP (NP120M2G) haploid strains were derived from diploids made by crossing NP120–25–4 with M1GFP or M2GFP, respectively. Strains with nup gene deletions containing MAD1-GFP or MAD2-GFP are described below.
Construction of tagged MAD1, MAD2, and MTW1 genes
MAD1 and MAD2 were tagged with GFP after their last amino acid codon using an integrative PCR-based transformation procedure. Primers and the GFP/HIS5 template plasmid pGFP (Wigge et al., 1998) were supplied by Dan Burke (University of Virginia, Charlottesville, VA). In brief, GFP and the HIS5 marker were PCR amplified from plasmid pGFP using a sense primer containing a region of MAD1 or MAD2 just before the termination codon, and an antisense primer containing a region of MAD1or MAD2 just past the termination codon. PCR products were transformed into a WT strain (YPH278) and His+ transformants were screened for in-frame fusions of MAD1-GFP and MAD2-GFP by PCR and Western blot analysis. Strains MAD1-GFP/HIS5 (YMB1299) and MAD2-GFP/HIS5 (YMB1296) were used for subsequent studies.
For the examination of Mad1-GFP protein in nup deletion mutants, a MAD1-GFP/HIS5 module (from strain YMB1299) was PCR-amplified and transformed into the WT strain DF5 to generate MAD1-GFP/HIS5 strains (YMB2018 or YMB2020). Subsequently, these MAD1-GFP/HIS5 strains were crossed with NP53-B1 (nup53Δ::HIS3), YMB1482 (nup170Δ::HIS3), NP157–2.1 (nup157Δ::URA3), and NP59–23 (nup59Δ::HIS3). Meiotic progeny was analyzed and appropriate haploid strains were chosen for further analysis (Table I).
For the examination of Mad2-GFP in nup deletion mutants, nup deletions were made in the MAD2-GFP strain (YMB1296) using a deletion technique described earlier. The nup170Δ::KAN deletion module was derived from YMB1451; the nup53Δ::KAN deletion module was derived from strain no. 10734 (Research Genetics); the nup59Δ::KAN deletion module was derived from strain no. 3785 (Research Genetics); and the mad1Δ::KAN deletion module was derived from YMB1979.
The gene encoding the centromere associated protein Mtw1p (Goshima and Yanagida, 2000) was tagged with the gene-encoding GFP or the CFP as follows: MTW1 and the adjoining promoter sequences were amplified from genomic DNA with the following primers: OMB338 (−600 to −581 of MTW1) and OMB339 (+846 to +867). The resulting PCR product was digested with XhoI and NotI and cloned into LEU2/CEN plasmid pAA3 (Sesaki and Jensen, 1999). In pAA3, MTW1 was placed in frame with GFP to form pOKMTW1-GFP. pOKMTW1-CFP was constructed by replacing a NotI-SacII GFP containing fragment of pOKMTW1-GFP with a corresponding segment from CFP derived from the plasmid pDH3 (pDH3 [pFA6a-CFP-KAN]) by PCR. pOKMTW1-GFP and pOKMTW1-CFP complement the lethal phenotype of an mtw1Δ::KAN deletion. Additionally, both Mtw1p-GFP and Mtw1p-CFP staining is lost in an ndc10–1 mutant 3 h after shift to 37°C.
Cell cycle arrest
For cell cycle arrests, we used early logarithmic phase cultures grown at 30°C. For G1 arrest, cells were incubated with 1 μg/ml α-factor (T-6901; Sigma-Aldrich) for 90 min. For G2/M arrest, cells were incubated with 15 μg/ml nocodazole (M-1404; Sigma-Aldrich) or 30 μg/ml benomyl (Sigma-Aldrich) for 90–120 min (Hardwick and Murray, 1995). Cell cycle arrest was monitored by microscopic examination of cells and FACS® analysis (Basrai et al., 1996).
Cell fractionation, affinity purification, and Western blotting procedures
Cell fractionation of the strain M1GFP was performed as described previously (Strambio-de-Castillia et al., 1995). The cytosol and crude nuclei fractions are the supernatant and pellet fractions, respectively, from a low speed centrifugation (10,000 g for 15 min) of lysed spheroplasts. Purified nuclei were used to produce a NE fraction. Nuclei were digested with 20 μg/ml DNase I-EP and then diluted with an equal volume of buffer to a final concentration of 200 mM NaCl, 10 mM Bis-Tris, pH 6.5, and 0.1 mM MgCl2 and incubated on ice for 15 min. Samples were then centrifuged at 200,000 g for 15 min at 4°C to produce a pellet fraction containing crude NEs and a supernatant fraction (containing histone proteins) designated as the nucleoplasmic fraction.
Immunoprecipitations and affinity purification of nup–pA fusion proteins were performed on nuclear extracts derived from crude nuclear fraction isolated from a 250-ml culture (Tcheperegine et al., 1999). The nuclear fraction was digested with 10μg/ml DNAase and then diluted with 1 vol of extraction buffer to a final concentration of 1% Triton X-100, 20% dimethyl sulfoxide, 150 mM NaCl, 0.1 mM MgCl2, 1 mM DTT, 50 mM Tris-HCl, pH 7.5, and containing a protease inhibitor cocktail (50 mM NaF, 0.2 mM PMSF, 2 μg/ml leupeptin, 2 μg/ml aprotinin, and 0.4 μg/ml pepstatin A). Samples were clarified by centrifugation at 200,000 g for 15 min at 4°C. The supernatant fraction containing the nuclear extract was incubated with IgG-Sepharose beads (Amersham Biosciences), using 10 μl of bead slurry per 1 ml of extract for 3 h at 4°C. Alternatively, extracts from M1GFP or M2GFP cells were incubated with rabbit polyclonal anti-GFP antibodies (provided by L. Berthiaume, University of Alberta, Edmonton, Alberta, Canada) or polyclonal anti-Nup53p antibodies (Marelli et al., 1998) followed by the addition of 20 μl protein G-Sepharose beads (Amersham Biosciences). Beads were washed extensively in wash buffer (0.1% Tween 20, 150 mM NaCl, 0.1 mM MgCl2, 1 mM DTT, 50 mM Tris-HCl, pH 7.5, and a protease inhibitor cocktail) and then with wash buffer containing 50 mM MgCl2. Bound proteins were eluted with a MgCl2 step gradient (0.1 M, 0.5 M, 1 M, and 2.0 M for protein A fusions and 0.1 M, 0.5 M, and 2 M for anti-Nup53p and anti-GFP) and a final wash with 0.5 M acetic acid, pH 3.4.
For the analysis of whole cell lysates, samples were prepared as described previously (Marelli et al., 2001) and proteins were separated by SDS-PAGE. For the examination of phosphorylated Mad1-GFP, proteins from whole cell lysates were separated by double-inverted gradient gel electrophoresis as described previously (Zardoya et al., 1994). Immunoblotting was performed as described previously (Marelli et al., 1998). Protein A fusions were detected with rabbit anti–mouse IgG (ICN Biomedicals). Polyclonal anti-GFP antibodies were a gift from Michael Rout (The Rockefeller University, New York, NY). Anti-Mad1 pAb was provided by Kevin Hardwick (University of Edinburgh, Edinburgh, UK). Anti-Mad2 polyclonal antibodies were a gift from Rei-Huei Chen (Cornell University, Ithaca, NY; Chen et al., 1999). Nup2p was detected using anti-Nup2p pAbs provided by John Aitchison (Institute for Systems Biology, Seattle, WA; Dilworth et al., 2001). Immune complexes were detected with HRP-conjugated secondary antibodies and the ECL system (Amersham Biosciences).
Nuclear transport assays
Nuclear import of a Pho4140–166-GFP3 reporter protein was assessed in the strains DF5, YMB1482, YMB1906, YMB1908, and YMB1979 transformed with the plasmid EB0836 (Kaffman et al., 1998; provided by E. O'Shea, University of California, San Francisco, San Francisco, CA) using a previously described assay (Shulga et al., 1996, 2000) with modifications. Cells were treated with a metabolic poison cocktail of 100 mM 2-deoxyglucose and 10 mM sodium azide for 45 min at 30°C. Cells were then washed and resuspended in CM-Ura media at RT. Recovery was allowed to proceed at RT on a slide and confocal images were captured at the indicated times and scored for nuclear accumulation of the reporter. For 37°C-treated cells, cultures were grown for 3 h at 37°C before metabolic poisoning.
Fluorescence microscopy
Strains containing both GFP- and CFP-tagged proteins were examined on a microscope (Axioscope 2; Carl Zeiss MicroImaging, Inc.) fitted with a Cooke Sensicam, a Chroma GFP filter set (model CZ909; Chroma Technology Corp.), an optical CFP filter set (model XF114–2; Omega Optical Inc.), and a Uniblitz Shutter assembly. For analysis of GFP-tagged Nup53p, Nup49p, and Nup188p in the indicated mutants, cultures were grown at 27°C to early logarithmic phase, split into two cultures, and then maintained at 23°C or shifted to 37°C for 3 h. Images were obtained using a microscope (model BX-50; Olympus) and a digital camera (Spot HRD060-NIC; Diagnostic Instruments). All confocal images discussed in the results were obtained using a confocal microscope (model LSM501; Carl Zeiss MicroImaging, Inc.).
Acknowledgments
We thank John Aitchison and Bob Skibbens for valuable criticisms of the manuscript. Numerous individuals have provided helpful discussions, including members of the Aitchison lab, D. Goldfard, R. Rachubinski, F. Spencer, C. Dunbar, M.S. Lee, C. Connelly and M. Nau, members of the Wozniak and Basrai laboratories. We are very grateful for all those cited in the text for reagents and Carole Carter for FACS® analysis.
Funding for this paper was provided by the Canadian Institutes for Health Research, the Alberta Heritage Foundation for Medical Research, and the National Institutes of Health. Salary support is provided to R.W. Wozniak from Canadian Institutes for Health Research and the Alberta Heritage Foundation for Medical Research.
T. Iouk and O. Kerscher contributed equally to this paper.
Footnotes
Abbreviations used in this paper: NE, nuclear envelope; NPC, nuclear pore complex; pA, protein A; WT, wild-type.
References
- Adams, A., D.E. Gottschling, C.A. Kaiser, and T. Stearns. 1997. Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 177 pp.
- Aitchison, J.D., G. Blobel, and M. Rout. 1995. a. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J. Cell Biol. 131:1659–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison, J.D., M.P. Rout, M.M. Marelli, G. Blobel, and R.W. Wozniak. 1995. b. Two novel related yeast nucleoporins Nup170p and Nup157p: Complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131:1133–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basrai, M.A., J. Kingsbury, D. Koshland, F. Spencer, and P. Hieter. 1996. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae Mol. Cell. Biol. 6:2838–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh, N., G. Rabut, S.W. Bai, M. van Overbeek, J. Beadouin, N. Daigle, O.V. Zatsepina, F. Pasteau, V. Labas, M. Fromont-Racine, et al. 2001. An evolutionary conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154:1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C.B., A. Davies, G.J. Cost, E. Caputo, J. Li, P. Hieter, and J.D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132. [DOI] [PubMed] [Google Scholar]
- Campbell, M.S., G.K. Chan, and T.J. Yen. 2001. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 114:953–963. [DOI] [PubMed] [Google Scholar]
- Chen, R.H., J.C. Waters, E.D. Salmon, and A.W. Murray. 1996. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 274:242–246. [DOI] [PubMed] [Google Scholar]
- Chen, R.H., D.M. Brady, D. Smith, A.W. Murray, and K.G. Hardwick. 1999. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell. 10:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, E., and R.-H. Chen. 2002. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol. Biol. Cell. 13:1501–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland, C.S., and M. Snyder. 1993. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. Yeast. 9:235–249. [DOI] [PubMed] [Google Scholar]
- Dilworth, D.J., A. Suprapto, J.C. Padovan, B.T. Chait, R.W. Wozniak, M.P. Rout, and J.D. Aitchison. 2001. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 153:1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, G. 2002. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell. 13:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr, K., and A. Hoyt. 1998. Bub1p kinase activates the Saccharomyces cerevisiae spindle checkpoint. Mol. Cell. Biol. 18:2738–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro, S.B., M.L. McCleland, P.T. Stukenberg, D.J. Burke, M.M. Ross, J. Shabanowitz, D.F. Hunt, and F.M. White. 2002. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20:301–305. [DOI] [PubMed] [Google Scholar]
- Fraschini, R., A. Beretta, L. Sironi, A. Musacchio, G. Lucchini, and S. Piatti. 2001. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 20:6648–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky, G.J., R.H. Chen, and A.W. Murray. 1998. Microinjection of antibody to Mad2 protein into mammalian cells in mitosis induces premature anaphase. J. Cell Biol. 142:1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and M. Yanagida. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle in budding yeast. Cell. 100:619–633. [DOI] [PubMed] [Google Scholar]
- Hardwick, K.G. 1998. The spindle checkpoint. Trends Genet. 1:1–4. Review. [DOI] [PubMed] [Google Scholar]
- Hardwick, K.G., and A.W. Murray. 1995. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 131:709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick, K.G., E. Weiss, F.C. Luca, M. Winey, and A.W. Murray. 1996. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 273:953–956. [DOI] [PubMed] [Google Scholar]
- Hardwick, K.G., R.C. Johnston, D.L. Smith, and A.W. Murray. 2000. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 148:871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, M.A. 2001. A new view of the spindle checkpoint. J. Cell Biol. 154:909–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland, K.M., J. Kingsbury, D. Koshland, and P. Hieter. 1999. Ctf19p: A novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 145:15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikui, A.E., K. Furuya, M. Yanagida, and T. Matsumoto. 2002. Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J. Cell Sci. 115:1603–1610. [DOI] [PubMed] [Google Scholar]
- Joseph, J., S.H. Tan, T.S. Karpova, J.C. McNally, and M. Dasso. 2002. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman, A., N.M. Rank, and E.K. O'Shea. 1998. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 12:2673–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher, O., P. Hieter, M. Winey, and M.A. Basrai. 2001. Novel role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation. Genetics. 157:1543–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., and R. Benezra. 1996. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 274:246–248. [DOI] [PubMed] [Google Scholar]
- Lusk, C.P., T. Makhnevych, M. Marelli, J.D. Aitchison, and R.W. Wozniak. 2002. Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes. J. Cell Biol. 159:267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara, I.G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli, M.M., J.D. Aitchison, and R.W. Wozniak. 1998. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143:1813–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli, M., C.P. Lusk, H. Chan, J.D. Aitchison, and R.W. Wozniak. 2001. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 153:709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millband, D.N., L. Campbell, and K.G. Hardwick. 2002. The awesome power of multiple model systems: interpreting the complex nature of spindle checkpoint signaling. Trends Cell Biol. 9:205–209. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., and J.D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593–16596. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., J.D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B.T. Chait. 2000. A comprehensive analysis of the yeast nuclear pore complex. J. Cell Biol. 148:635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki, H., and R.E. Jensen. 1999. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147:699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J.V., and D.W. Cleveland. 2000. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell. 103:997–1000. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1986. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 186 pp.
- Shulga, N., P. Roberts, Z. Gu, L. Spitz, M.M. Tabb, M. Nomura, and D.S. Goldfarb. 1996. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 135:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga, N., N. Mosammaparast, R. Wozniak, and D.S. Goldfarb. 2000. Yeast nucleoporins involved in passive nuclear envelope permeability. J. Cell Biol. 149:1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, F., S.L. Gerring, C. Connelly, and P. Hieter. 1990. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 124:237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia, C., G. Blobel, and M.P. Rout. 1995. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces. J. Cell Biol. 131:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin, V., G.K.T. Chan, and T.J. Yen. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, MAD2. J. Cell Biol. 154:925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z., R. Bharadwaj, B. Li, and H. Yu. 2001. Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 1:227–237. [DOI] [PubMed] [Google Scholar]
- Tcheperegine, S., M. Marelli, and R.W. Wozniak. 1999. Topology and functional domains of the yeast pore membrane protein Pom152p. J. Biol. Chem. 274:5252–5258. [DOI] [PubMed] [Google Scholar]
- Uetz, P., L. Giot, G. Cagney, T.A. Mansfield, R.S. Judson, J.R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 403:623–627. [DOI] [PubMed] [Google Scholar]
- Warren, C.D., D.M. Brady, R.C. Johnston, J.S. Hanna, K.G. Hardwick, and F.A. Spencer. 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell. 13:3029–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente, S.R. 2000. Gatekeepers of the nucleus. Science. 288:1374–1377. [DOI] [PubMed] [Google Scholar]
- Wigge, P.A., O.N. Jensen, S. Holmes, S. Soues, M. Mann, and J.V. Kilmartin. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141:967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., and E. O'Toole. 2001. The spindle cycle in budding yeast. Nat. Cell Biol. 3:E23–E27. [DOI] [PubMed] [Google Scholar]
- Wozniak, R.W., M.P. Rout, and J.D. Aitchison. 1998. Karyopherins and kissing cousins. Trends Cell Biol. 5:184–188. [DOI] [PubMed] [Google Scholar]
- Zardoya, R., A. Diez, P.J. Mason, L. Luzzatto, A. Garrido-Pertierra, and J.M. Bautista. 1994. High resolution of proteins by double-inverted gradient polyacrylamide gel electrophoresis (DG-PAGE). Biotechniques. 16:270–272. [PubMed] [Google Scholar]