Abstract
Genes can be transcribed from within chromosome territories; however, the major histocompatibilty complex locus has been reported extending away from chromosome territories, and the incidence of this correlates with transcription from the region. A similar result has been seen for the epidermal differentiation complex region of chromosome 1. These data suggested that chromatin decondensation away from the surface of chromosome territories may result from, and/or may facilitate, transcription of densely packed genes subject to coordinate regulation.
To investigate whether localization outside of the visible confines of chromosome territories can also occur for regions that are not coordinately regulated, we have examined the spatial organization of human 11p15.5 and the syntenic region on mouse chromosome 7. This region is gene rich but its genes are not coordinately expressed, rather overall high levels of transcription occur in several cell types. We found that chromatin from 11p15.5 frequently extends away from the chromosome 11 territory. Localization outside of territories was also detected for other regions of high gene density and high levels of transcription. This is shown to be partly dependent on ongoing transcription. We suggest that local gene density and transcription, rather than the activity of individual genes, influences the organization of chromosomes in the nucleus.
Keywords: gene density; chromatin; chromosome territories; nuclear organization; transcription
Introduction
Human nuclei have a radial organization. Chromosomes with the highest gene density are preferentially disposed toward the nuclear interior, and gene-poor chromosomes locate towards the nuclear periphery (Croft et al., 1999; Boyle et al., 2001; Cremer et al., 2001). This organization is conserved in other vertebrates (Habermann et al., 2001; Tanabe et al., 2002), suggesting that the nuclear interior may facilitate, or create a permissive environment for, transcription. However, many human chromosomes are a patchwork of domains with varying gene density and so some very gene-rich regions of the human genome are contained on chromosomes located close to the nuclear periphery.
We have previously shown that individual human genes can be transcribed from within the interior of chromosome territories that are not located in the nuclear center (Mahy et al., 2002). This showed that genes do not need to be either at the visible surface of interphase chromosome territories, or at the centre of the nucleus, in order to be transcribed. These genes were located in regions of moderate gene-density (the R-band 11p13). In contrast, the gene-dense major histocompatibilty complex (MHC)* locus is frequently observed on loops of chromatin that extend away from the human chromosome 6 territory that is detected by FISH with a chromosome paint, particularly when transcription of genes from this region is induced (Volpi et al., 2000). Similarly, the epidermal differentiation complex (EDC) at 1q21 is frequently located outside of the chromosome 1 territory in keratinocytes, cells in which the genes of the EDC are highly expressed (Williams et al., 2002). It was not clear whether localization outside of chromosome territories was a particular feature of regions of the genome that contain genes with related functions, and that are coordinately expressed, or whether it might represent a more general facet of genome organization wherever genes are particularly clustered together, or where the overall levels of transcription from a large number of genes across a region is high.
To address this, we have used FISH to examine territory organization of regions of the human genome with high gene densities and generally high levels of transcription. The T-band 11p15.5 contains at least 47 known genes within the most distal 4.5 megabase (Mb) of DNA. We found that many megabases of this chromatin is frequently found outside of the visible confines of the 11p territory. By extending this observation to other gene-dense parts of the human genome including; 11q13 and 16p13.3 and gene-dense regions of chromosomes 21 and 22, we suggest that there is a correlation between domains of high gene density and localization outside of chromosome territories. We show that the frequency of extraterritory localization decreases, but is not eliminated, when transcription is inhibited. This level of higher-order genome organization is conserved in the mouse, indicating that it likely has functional significance. We suggest that the propagation of a decondensed chromatin fibre outside of the confines of chromosome territories creates an environment that is permissive to transcription increasing the overall transcriptional potential of the domain (Tumbar et al., 1999), and that the structure of chromosome territories is, in part, driven by transcription.
Results
Distal 11p15.5 can locate outside of the chromosome 11 territory
It has been suggested that genes are positioned at the surface of chromosome territories (Kurz et al., 1996; for review see Cremer and Cremer, 2001). However, we recently reported that actively transcribing genes from the moderately gene-rich band Homo sapiens chromosome 11 p arm (HSA11p)13, or the region of conserved synteny on mouse chromosome (MMU)2 are located within the interior of the chromosome territory (Mahy et al., 2002). Conversely, the very gene-dense MHC locus is frequently observed on chromatin loops that extend away from the HSA6 chromosome territory (Volpi et al., 2000). These data suggest that the human genome is subject to different constraints of spatial organization, and that gene density or density of transcribed genes, rather than the activity of individual genes may influence chromosome organization.
To address this, we have analyzed the organization of distal 11p15, including the very gene-rich, subtelomeric T-band 11p15.5. The most distal 4.5 Mb of HSA11p is well characterized due to its association with Beckwith-Wiedemann Syndrome, and because of the cluster of imprinted genes located there. The region contains at least 47 known genes (Redeker et al., 1994; Alders et al., 1997; Hu et al., 1997; Reid et al., 1997; Lee et al., 1999; Engemann et al., 2000; Onyango et al., 2000; Paulsen et al., 2000; Fig. 1). In addition, it has a density of CpG islands that is much higher than that of 11p13 (Craig and Bickmore, 1994).
Figure 1.

Map of HSA11p15 and the region of conserved synteny from MMU7. (Left) Map of the distal 6 Mb of human HSA11p15 from the 11p telomere (tel) encompassing 11p15.5 and 11p15.4 and extending down to 11p15.3, showing the relative position of genes and loci used in this study. Genes are indicated in italics. Asterisks indicate the positions of loci used in this study. (Right) Map of the region of MMU7 in conserved synteny to the BWS-associated region of human 11p15. Gene locations were taken from NCBI online sequencing data (http://www.ncbi.nlm.nih.gov/cgi-bin/Entrez/maps.cgi?org=hum&chr=11), and from published maps (Alders et al., 1997; Hu et al., 1997; Reid et al., 1997; Buettner et al., 1998; Bepler et al., 1999; Kato et al., 1999; Lee et al., 1999; Engemann et al., 2000; Onyango et al., 2000; Paulsen et al., 2000).
Cosmids encompassing the most distal part of 11p (11p15.3–p15.5), and a PAC to the 11p telomere itself, were first cohybridized with an 11p paint to methanol:acetic acid (MAA; 3:1 vol/vol)-fixed nuclei from lymphoblasts (Fig. 2 a) and primary fibroblasts (Fig. 2 b). For each probe, the mean distance (μm) between the probe signal and the nearest chromosome territory edge was calculated as described previously (Mahy et al., 2002). In contrast to the expressed RCN gene in 11p13, for which 76% of signals were well within (≥0.2 μm from the territory edge) the 11p territory (Mahy et al., 2002), all 11p15 loci were closer to the territory surface (Fig. 3 a). Only 48% of signals from probe cq26 in 11p15.3 were well within the 11p territory, decreasing to 32% of D11S12 (11p15.4) signals, 25% of signals from the IGF2 gene in 11p15.5, and only 11% of signals from the 11p telomere. Moreover, for the more distal loci, an increasing proportion of signals from these probes was found outside of the limits of the territory detectable by FISH with a chromosome paint. 53% of IGF2 signals were located >0.2 μm beyond the 11p territory in this experiment rising to 80% of signals from 11pter (Fig. 3 a). The null hypothesis that the location of a distal 11p15.5 probe (e.g., cI-11p15-25) was the same as cq26 in 11p15.3, was rejected using a 2 sample t-test, P < 0.000, but cI-11p15-25 has the same location as an adjacent probe cI-11p15-46, P = 0.43. Volpi et al. (2000) similarly observed a high proportion of signals (up to 36%) from the MHC locus located outside of the painted human chromosome 6 territory in MAA-fixed lymphoblastoid nuclei, though the distance from the territory edge was not established in that case. The mean distance of 11p15 probes from the territory edge confirmed this trend (Fig. 3 b). All 11p15.4 probes analyzed lie within the territory but close to the edge, whereas the mean positions of 11p15.5 probes are outside of the chromosome territory. The mean location of 11p15.5 distal markers is >1 μm beyond the chromosome territory (Fig. 3 b).
Figure 2.

Visualizing the spatial organization of 11p15 relative to the 11p chromosome territory. FISH of selected probes from 11p15.3, 11p15.4, or 11p15.5 (red) together with an HSA11p paint (green) to MAA-fixed lymphoblast (a) or primary fibroblast (b) nuclei. Known genes are italicized. (c) Maximal intensity projection of image stacks after three-dimensional FISH with probes from 11p15.5 (red) and a paint for 11p (green) on pFa-fixed fibroblast nuclei. Cells were counterstained with DAPI (blue). (d and e) Views of MAPaint reconstructions of pFa fixed fibroblast nuclei after three-dimensional FISH with probes (red) for IGF2 (d) and 80N22 (e) together with paints (green) for 11p (d) and 11q (e). Bars, 5 μm.
Figure 3.
Quantifying the spatial organization of 11p15. (a) Histogram showing the distribution of signals from 11p15 probes relative to the 11p territory edge (μm) after FISH to MAA fixed lymphoblast nuclei. An 11p13 probe is shown for comparison. Negative distances indicate probe signals located beyond the visible limits of the detectable chromosome territory. The localization of cI-11p15–25 is significantly different from that of cq26 (P < 0.000). (b) Mean distance (±95% confidence interval/CI) of 11p15 probes from the edge of the 11p territory of MAA-fixed lymphoblast (•) and fibroblast (▴) nuclei. The number of territories analyzed (n) =100. The mean position of a probe for the PAX6 gene in 11p13 (Mahy et al., 2002) and the chromosome 11 centromere (11pcen) are shown for comparison. cI-11p15.25 and cI-11p15–46 have the same location outside of the 11p territory (P = 0.43); however, cI-11p15–25 is significantly more distant from the territory than cq26 (P < 0.000). (c) Mean distance (±95% CI) in μm of probes from the edge of the 11p territory of pFa-fixed fibroblast nuclei (n ≥ 35).
MHC class II genes are more frequently observed outside of chromosome 6 territories in expressing cells (lymphoblastoid) than in cells that do not express class II genes (e.g., keratinocytes). Conversely, the localization of the EDC complex outside of chromosome 1 territories is more frequent in keratinocytes than lymphoblasts (Table I) (Williams et al., 2002). This suggested that localization outside of chromosome territories may relate to the levels of transcription emanating from these regions. Unlike the MHC or EDC, the genes in distal 11p15.5 are not functionally related nor is there any evidence that they are coordinately regulated. Analysis of the organization of this region in MAA-fixed primary fibroblasts gave a similar result to that seen in lymphoblast nuclei (Fig. 3 b). Hence, localization of distal 11p15.5 outside of the chromosome territory is not specific to any one cell type with a specific pattern of gene expression.
Table I. The proportion of signals external to chromosome territories.
| Gene/locus (cytogenetic location) | Percentage of signals outside of chromosome territory
|
|
|---|---|---|
| Two-dimensional (MAA-fixed) | Three-dimensional (pFa-fixed) | |
| WAGR (11p13) | 11 | 1 |
| Wt1 (MMU2) | 3 | ND |
| D11S12 (11p15.4) | 43 | 30 |
| D11S679 (11p15.5) | 68 | 31 |
| IGF2 (11p15.5) | 72 | 28 |
| D11S483 (11p15.5) | 88 | 43 |
| 11p tel (11pter) | 87 | 58 |
| 245N5 (MMU7) | 40 | ND |
| 80N2 (11q13) | 69 | 57 |
| MHC Class II (6p21.3) fibroblasts | 13 | 12 |
| MHC Class II (6p21.3) fibroblasts + IFN-γ | 26 | 19 |
| MHC Class II (6p21.3) keratinocytes | 19 | 12 |
| MHC Class II (6p21.3) lymphoblasts | 34 | 36 |
| EDC (1q21) keratinocytes | 25 | 22 |
| EDC (1q21) lymphoblasts | 8 | 6 |
The proportion (%) of signals scored as outside of chromosome territories in two-dimensional FISH (MAA-fixed) and three-dimensional FISH (fixed in pFa). All human chromosome 11 loci were analysed in fibroblasts. Mouse loci (MMU) were examined in embryonic stem cells. The analysis of the human MHC Class II (6p21.3) and EDC (1q21) of Volpi et al. (2000) and Williams et al. (2002) in fibroblasts, keratinocytes, and lymphoblasts is shown for comparison. For two-dimensional analysis, n = 100; for three-dimensional analysis, n = 35.
Our previous analyses of intraterritory organization of 11p13 gave similar results whether the analysis was carried out on flattened specimens fixed in MAA or on three-dimensional preserved nuclei fixed with para-Formaldehyde (pFa; Mahy et al., 2002). However, Volpi et al. (2000) noted that the proportion of MHC FISH signals scored as external to the chromosome territory was higher in cells fixed in MAA than in three-dimensional preserved pFa-fixed cells (Table I). This suggests that MAA fixation may preferentially loosen or decondense chromatin located at the surface of chromosome territories, or that pFA fixation condenses these chromatin regions. Three-dimensional FISH analysis of 11p15.5 on pFa fixed primary fibroblasts confirms this. Signals from distal 11p15 could still be seen outside of chromosome 11 territories (Figs. 2, c and d, and 3 c) in three-dimensional preserved nuclei; however, this was seen less frequently than with MAA-fixed samples (Table I).
It was possible that distal loci appeared outside of the chromosome territory because the HSA11p paint did not include sequences near the end of the chromosome. However, examination of metaphase chromosomes showed that chromosome paint FISH signal extended right to the end of the chromosome arm, ending at a point coincident with a probe to the 11p telomere (unpublished data). Furthermore, the most proximal sequence on HSA11p, the centromere, was positioned at the visible edge (but inside of) the HSA11p territory (Fig. 3 b).
Location of chromatin outside of chromosome territories is not common to telomeric or imprinted regions
11p15.5 may locate outside of the chromosome territory because of its distal position close to the 11p telomere. There is some evidence for associations between different telomeres and gene-rich subtelomeric regions in human nuclei (Stout et al., 1999; Nagele et al., 2001). However, other telomeric sequences that we examined (e.g., 18pter and 18qter) were located at the edge of, but not outside of, their respective chromosome territories (unpublished data).
11p15 contains clusters of imprinted genes. The mechanistic basis of imprinting is yet to be fully defined, but aspects of higher-order chromatin structure have been implicated, and homologous loci of human and mouse genes subject to imprinting have been reported to be transiently associated during late S-phase (LaSalle and Lalande, 1996). We considered it unlikely that the nuclear organization of 11p15.5 was linked to the imprinted state of genes in this region, as both alleles were found outside of chromosome territories in many nuclei. However, we wished to determine whether other imprinted regions of the human genome were also located outside of chromosome territories. There is a large cluster of imprinted genes, associated with Prader-Willi and Angelmann syndromes, located at 15q11–13. We found that loci from the imprinted region of 15q11–13 are positioned within the HSA15q territory (Fig. 4) indicating that localization outside of chromosome territories is not a common feature of imprinted regions.
Figure 4.
Correlation between gene density and localization relative to chromosome territories. Mean probe positions (normalized for territory radius) relative to the edge of chromosome territories measured in hybridizations to two-dimensional MAA-fixed lymphoblast nuclei, plotted against gene density (Genes/Mb) for all of the regions considered in this analysis. A value of 0 on the y-axis represents the edge of the chromosome territory and negative values indicate that the mean locus position is outside of the chromosome territory. The best-fit line was determined using Microsoft Excel. The equation for the line is y = −0.0242x + 0.315 and r2 = 67%.
Localization outside of a chromosome territory is common to gene-rich regions of the human genome
The relative intraterritory position of loci from 15q12 was intermediate to that of 11p13 (Mahy et al., 2002) and 11p15.5 loci (Fig. 4). Interestingly, the estimated gene densities of these regions follow the same trend. 14 genes have been mapped to a 2.5-Mb region within 15q11–13, equivalent to 5.6 genes/Mb (Gabriel et al., 1998), whereas there are only four genes per Mb in distal 11p13 (Mahy et al., 2002), and >10 genes per Mb in 11p15.5 (Fig. 1). These data suggest that gene density may be a determining factor in whether chromatin fibres extend outside of chromosome territories. In support of this, hybridization signals from BAC 80N22, located in the interstitial very gene-rich region 11q13 (http://www.chori.org/bacpac) were frequently seen outside of 11q chromosome territories in both two-dimensional and three-dimensional specimens (Figs. 2 e and 4; Table I). Localization outside of chromosome territories was also seen for a locus just proximal of the α-globin cluster at 16p13.3 (Flint et al., 1997; Fig. 4).
To assess the long-range effects of gene density on intraterritory position, we used the published sequences of HSA21 and HSA22 (The chromosome 21 mapping and sequencing consortium, 2000; Dunham et al., 1999; Saccone et al., 2001) to identify gene-rich (and GC-rich) and gene-poor 1 Mb regions across the long arms of both chromosomes. In two-dimensional FISH to lymphoblast nuclei, the localization of each probe relative to the edge of the chromosome 21 or 22 territories corresponded well to the local estimated gene-density (Fig. 4), except for the gene-rich probe 154H4, which is close to the centromere of chromosome 22. In three-dimensional analysis the borders of the chromosome 21 and 22 territories were too indistinct to be able to reliably measure locus position. Linear regression of the data in Fig. 4 confirms the correlation between gene density and localization relative to the edge of chromosome territories (r2 = 67%).
Transcription has a role in localizing chromatin outside of chromosome territories
Localization of chromatin outside of territories could reflect an “open” chromatin structure across large regions poised for transcription, and/or could be due to the process of transcription itself. Not only are 11p15.5, 11q13, and 16p13 regions of high gene- and CpG island density but they are also domains where there is a high density of transcribed genes and where the levels of expression from many genes is high in many cell types (Caron et al., 2001).
To investigate this we analysed the nuclear organization of 11p15 and 11q13 loci after transcription had been inhibited with Actinomycin D (ActD) or 5,6-dichloro-b-ribofuranosylbenzimidazole (DRB; Chodosh et al., 1989; Croft et al., 1999). The proportion of signals from probe cI-11p15-46 (11p15.5) located >0.2 μm beyond the territory edge decreased from 42% in control cells to 35 and 28% in ActD or DRB-treated cells, respectively (Fig. 5 a). The 72% of signals from 80N22 in 11q13 observed outside of the 11q territory dropped to 60% in ActD treated cells (Fig. 5 b). Even the modest 20% of signals from the β-globin locus (11p15.4) that could bee seen outside of the 11p territory in control cells was decreased to 15% of treated cells (Fig. 5 c). Hence ongoing transcription likely contributes to localization outside of chromosome territories, but gene dense domains can still locate outside of chromosome territories in the absence of transcription.
Figure 5.

Influence of transcription inhibitors on localization outside of chromosome territories. (a–c) Histograms showing the distribution of signals relative to the 11p (a and c), or 11q (b) territory edge (μm) after FISH with probes cI-11p15–46 in 11p15.5 (a), 80N22 (b), and β-globin (c), to MAA fixed lymphoblast nuclei from untreated cells (open bars), and from cells treated with DRB (filled bars) or ActD (hatched bars). n ≥ 75. (d) The proportion of territories in untreated cells (open bars), or ActD treated cells with both IGF2 and cI-11p15–25 loci contained within the 11p territory (in–in), both loci outside of the territory (out–out), or with one locus in and one locus out.
We did not find any locus in our studies that always located outside of its chromosome territory. In each case there were two peaks of localization, one within the territory, and the other some distance (>1 μm) beyond the detectable territory edge. Hence, the histograms in Figs. 3 and 5 are bimodal. This suggests a dynamic organization, with stochastic looping of very large domains of chromatin outside of the chromosome territory, rather than the presence of many smaller loops of chromatin emanating independently from each region. To investigate this further, we asked whether the extraterritory localization of two loci from the same chromosomal region was concerted. We compared the localization of the 11p15.5 IGF2 gene with cI-11p15–25 located 1 Mb more distally (Fig. 1). There was a high concordance in the behavior of the two loci. In 60% of chromosomes examined (n = 100), both loci were observed outside of the 11p territory, and in 23% of cases both of the loci were contained within the territory. In only 17% of cases could one probe be found within the territory with the other probed located outside of the territory (Fig. 5 d). It was this category of organization that proportionately was most affected by ActD treatment, their incidence being halved in treated cells.
Conservation of chromatin organization in the mouse
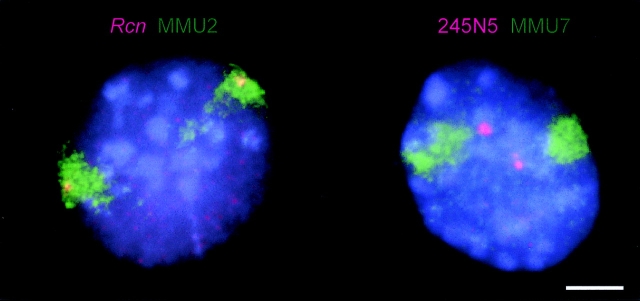
We have previously shown that the relative position of loci within chromosome territories is conserved between mouse and human (Mahy et al., 2002). To determine whether conservation of spatial organization extends to include loci that locate outside of chromosome territories, we examined the organization of loci from the region of the mouse chromosome 7 (MMU7) that is in conserved synteny with the BWS-associated region on HSA11p15.5 (Fig. 1). BACs 300P2 (Paulsen et al., 2000) and 245N5 (Engemann et al., 2000) were used in combination with an MMU7 chromosome paint (Jentsch et al., 2001) in two-dimensional FISH to MAA-fixed ES cell nuclei. 40% of 245N5 signals and 29% of 300P2 signals were outside of the MMU7 territory (Table I; Fig. 6). This contrasts with only 3% of signals from the Wt1 gene on MMU2, the region of conserved synteny to human 11p13 (Mahy et al., 2002). We conclude that the localization of gene-rich chromatin external to chromosome territories is conserved between human and mouse cells and that this indicates its functional importance.
Figure 6.
Conservation of intranuclear organization in mouse cells. FISH of BACs (red) and MMU2 and 7chromosome paints (green) hybridized to MAA-fixed ES cell nuclei and counterstained with DAPI. Murine Rcn is in conserved synteny with human 11p13. (Mahy et al., 2002). BAC 245N5 contains the genes from Obph1 to Tssc5 in conserved synteny with human 11p15.5 (Fig. 1). Bar, 5 μm.
Discussion
Gene density correlates with localization outside of chromosome territories
It has been suggested that genes are positioned at the surface of chromosome territories, so as to be accessible to transcription factors and the transcription machinery residing within an interchromosome compartment (Zirbel et al., 1993; Kurz et al., 1996; for review see Cremer and Cremer, 2001). However, poly(A) RNA, nascent RNA, and transcription factors are not excluded from chromatin territories (Abranches et al., 1998; Verschure et al., 1999), and we have recently shown that both housekeeping and tissue-specific genes can be transcribed from within the territory interior (Mahy et al., 2002). In contrast, the MHC and EDC loci have been observed on chromatin loops extending away from the surface of their chromosome territories (Volpi et al., 2000; Williams et al., 2002). Because the MHC and EDC are regions where genes with related function and with coordinated patterns of expression are clustered, localization outside of territories might reflect their specialized regulation. Alternatively, there may be many other regions of the human genome that also locate outside of chromosome territories.
To establish if localization outside of territories is not uncommon, we have analysed the organization of several gene-dense regions of the human genome. Here we show that one of the sites of highest gene density on chromosome 11p (11p15.5), but where the genes are functionally unrelated and have different patterns of gene expression, can also be found outside of the HSA11p chromosome territory (Table I; Figs. 2 and 3). We saw similar “extra-territory” localizations for two other gene-rich T-bands of the human genome (11q13 and 16p13; Figs. 2 e and 4). A strong correlation (r2 = 67%) between gene-density and chromosome territory organization was confirmed by a systematic analysis along the long arms of chromosomes 21 and 22 (Fig. 4). It is interesting to note that even though the cell types that we examined do not express globin genes, the localization of the β-globin locus close to the surface of, but within, the chromosome 11 territory (Fig. 2 a; Kurz et al., 1996) contrasts with the localization of the α-globin region outside of the chromosome 16 territory (Fig. 4). This adds to the growing list of differences in chromatin structure and nuclear organization that have been described for α-and β globin genes (Brown et al., 2001).
Transcriptional activity can influence localization outside of territories
Localization of chromatin outside of the confines of chromosome territories could result from the process of transcription itself, or could reflect the structure of the chromatin fibre (e.g., histone modifications) in domains poised for transcription. Extrusion of the MHC and EDC loci from their chromosome territories is clearly related to the levels of transcription from these complexes (Volpi et al., 2000; Williams et al., 2002). Even though the loci that we have identified here as being frequently located outside of territories do not contain genes whose expression is coordinately switched on in the cell types that we studied, there are high levels of gene expression emanating from these regions. Genome-wide expression profiling using human ESTs highlighted the distal part of 11p, and regions likely corresponding to 11q13 and 16p13.3, as large regions of increased gene expression in many different cell and tissue types, including primary fibroblasts used in our study (Caron et al., 2001). Many of the genes in 16p13 are also known to be widely expressed (Daniels et al., 2001). Together with the results of Volpi et al. (2000) and Williams et al. (2002) this suggests that it may indeed be high levels of gene expression over a large genomic region, rather than just the density of genes per se, that determines whether chromatin domains will locate outside of chromosome territories.
To investigate this we examined the localization of loci outside of territories after treatment of cells with agents (ActD and DRB) that inhibit transcription. We did observe a decrease in the number of signals seen outside of chromosome territories in treated cells compared to controls and a concomitant increase in the signals located within the bulk chromosome territory (Fig. 5). Retraction of gene-dense domains into the confines of condensed chromosome territories in the absence of transcription is consistent with the compaction of the territory of (gene-dense) human chromosome 19 after treatment with ActD or DRB (Croft et al., 1999), and with the failure of an mouse mammary tumor virus promoter array to decondense upon steroid hormone addition in DRB-treated cells (Müller et al., 2001).
However, even in DRB or ActD-treated cells most signals from 11p15.5 and 11q13 loci are still outside of territories. ActD had most effect on territories in which the loops of chromatin extending beyond the territory edge contain only some of the loci from the region (Fig. 5 d) i.e., they may not be fully extended. The role of transcription may be during formation of the loops, whereas other factors, e.g., chromatin structure, may maintain them. The visible decondensation of a 90-Mb lacO array can be induced by a transcriptional activator, even in the absence of transcription itself, and is accompanied by increased levels of histone acetylation (Tumbar et al., 1999). 11p15.5, 11q13, and 16p13 are all regions of the human genome with hyperacetylated histones (Jeppesen, 1997), and are identified here as regions that frequently locate outside of chromosome territories.
Reconsidering the concept of chromosome territories
Volpi et al. (2000) suggested that FISH signals located outside of chromosome territories are the visual manifestation of chromatin decondensation over large regions. Here we have shown that this phenomenon is quite widespread, and not limited to clusters of coordinately regulated genes. Previous studies of long-range chromatin decondensation as the result of transcriptional activator binding (Tumbar et al., 1999) or steroid hormone recruitment and transcription (Müller et al., 2001) on artificial reporter arrays have tried to quantify the level of chromatin compaction. In our study of endogenous loci in human cells we observe a maximal distance between an extended 11p15.5 locus and the 11p territory of 2 μm in pFa-fixed cells, the mean distances being ∼1 μm (Fig. 3 c). Based on the human genome sequence, the genomic distance between 11ptel and IGF2 is ∼1.5 Mb (Fig. 1). This represents a fourfold higher level of compaction than that seen in the presence of transcription from a reporter array (2 Mb extending over an average of 6 μm; Müller et al., 2001). However, it is a similar level of decondensation to that reported by Tumbar et al. (1999)(90 Mb extended across a 30 μm fiber). Hence, there is still a large degree of higher-order structure, beyond a 30-nm fiber, in regions that extend out from chromosome territories
The data presented here, and previously published (Volpi et al., 2000; Williams et al., 2002) lead us to suggest that the organization of chromosomes within the nucleus is probably somewhere in between the complete decondensation of chromatin fibres like spaghetti on a plate suggested >30 y ago and the model of a discrete territorial organization favored recently (for review see Cremer and Cremer, 2001). Although the chromosome territory is a useful term to describe the appearance of hybridization signals from complex chromosome paints at the light microscope level, does it have any biological significance if there are many genomic regions contained outside of these domains and “invisible” by chromosome painting?
To answer this question it will be important to determine whether extended chromatin fibers are located in a “space” between chromosome territories, or whether they are embedded with the territories of other chromosomes. A light microscopy study of in vivo labelled chromatin has demonstrated that, although in general the borders between chromosome territories are well defined, fiber-like structures can be observed embedded in other chromosome territories (Visser and Aten, 1999).
Materials and methods
Cell culture and FISH
All cells were grown and prepared for FISH as previously described (Mahy et al., 2002). Treatment of cells with ActD or DRB to inhibit transcription was as previously described (Croft et al., 1999). Paints for human chromosomes 11p (HSA11p), HSA11q, HSA15q, HSA16p, gifts of Michael Bittner (National Institutes of Health, Bethesda, MD), were labelled with biotin-16-dUTP by PCR amplification (Guan et al., 1996; Croft et al., 1999). Paints for HSA21q and HSA22q and MMU7 (Jentsch et al., 2001) were amplified by PCR, and then biotin labelled by nick translation. Human cosmids from 11p15 (Alders et al., 1997), PAC 44h16 (Genome Systems) mapping to the 11p telomere, BACs from 11q13 (80N22), and 15q (80H14 and 88P10; BACPAC Resources, http://www/chori.org/bacpac/), cos 12 from 16p13 (Flint et al., 1997), BACs from HSA21 (The chromosome 21 mapping and sequencing consortium, 2000), and HSA22 (Sanger Centre, http://www.sanger.ac.uk/HGP/Cytogenetics/Bacset.shtml) were labelled with digoxigenin by nick translation. Mouse BACs 245N5 (Engemann et al., 2000) and 300P2 (Paulsen et al., 2000) were also digoxigenin labelled. This labelling scheme was adopted because chromosome territories detectable with biotin-labelled paint were brighter and more reproducible than those seen using digoxigenin-labeled paints (Mahy et al., 2002). ∼200 ng of paint and 50 ng cosmid or 100 ng BAC/PAC were used per slide, together with 6 μg human CotI, or 14 μg mouse CotI DNA (GIBCO BRL) as competitor.
FISH performed on MAA-fixed cells (two-dimensional analysis) or on three-dimensional preserved cells fixed with 4% pFa buffered in PBS, probe detection, examination of slides, and image capture, were as previously described (Mahy et al., 2002).
Image analysis
Analysis of probes located within chromosome territories in two-dimensional samples was as previously described (Mahy et al., 2002). Where probe signals appeared outside of the chromosome territory, the following script was used. Nuclear area was calculated from the segmented DAPI image. Locus-specific hybridization signals were segmented and a region of interest was manually defined around them. Hybridization signal from the chromosome territory was then segmented by thresholding, without knowledge of the locus signal, and a region of interest manually defined around the detectable territory. The area of the territory was calculated. A segmentation disc was dilated out from the locus signal centroid, and then eroded until a pixel containing territory signal was found. This was taken to be the nearest edge of the territory to the locus, and the radius of the disc was calculated, representing the distance (μm) from the centre of the locus to the nearest edge of the territory. A similar procedure was used to determine the distance between the territory centroid and territory edge. To verify the reproducibility of this analysis, the localization of 11p15 probes cI-11p15-46 and β-globin LCR, as well as an 11q13 probe were assessed in lymphoblastoid cells in separate experiments by independent investigators. For the 11p15.5 probe cI-11p15-46, both investigators scored the mean position of the locus as outside of the chromosome 11p territory and >0.7 μm away from the territory edge (−1.0 ± 0.34; −0.70 ± 0.2). β-Globin was measured within the 11p territory and close to the chromosome territory edge (0.29 ± 0.1; 0.1 ± 0.2).
Because actual territory size varied between chromosomes, between cell types, and between species, the locus to territory edge distance was normalized by dividing it by the radius of a circle of equal area to that of the territory. Thus, a value of 0 denotes a locus at the edge of a territory and negative values describe loci that locate outside of the detectable limits of the chromosome territory. A value of 1.0 denotes a locus at the theoretical territory center, but in practice values of 1.0 are not seen because territories are not circular. On this scale, the actual mean territory centroids were at 0.64±0.02 and 0.63±0.03 (HSA11p and q, respectively), 0.59±0.03 (HSA16p), and 0.66±0.03 (MMU7).
Three-dimensional images were analyzed as previously described (Mahy et al., 2002), using the program MAPaint (Mouse Atlas Project, http://genex.hgu.mrc.ac.uk/).
Statistical analyses of data by linear regression, and by Students t test, were carried out using Microsoft Excel.
Acknowledgments
We thank Irina Solovei (University of Munich, Munich, Germany) and Nigel Carter (Sanger Centre, Cambridge, UK) for MMU2 and 7 paints and for chromosome 22 BACs. Various probes used in this study were gifts of Doug Higgs (MRC Molecular Haematology Unit, Oxford, UK), Peter Fraser (Babraham Institute, Cambridge, UK), Hans Lehrach and Sabine Engemann (Max-Planck-Institute fur Molekulare Genetik, Berlin, Germany), and Mariette Alders (Academic Medical Centre, University of Amsterdam, The Netherlands).
N. Mahy was supported by a studentship from the Medical Research Council, and W.A. Bickmore is a Centennial Fellow of the James S. McDonnell Foundation.
Footnotes
Abbreviations used in this paper: ActD, Actinomycin D; DRB, 5,6-dichloro-β-ribofuranosylbenzimidazole; EDC, epidermal differentiation complex; HSA11p, Homo sapiens chromosome 11 p arm; MAA, methanol:acetic acid; Mb, megabase; MHC, major histocompatibilty complex; MMU, Mus musculus chromosome; pFa, para-Formaldehyde.
References
- Abranches, R., A.F. Beven, L. Aragon-Alcaide, and P.J. Shaw. 1998. Transcription sites are not correlated with chromosome territories in wheat nuclei. J. Cell Biol. 143:5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alders, M., M. Hodges, A.K. Hadjantonakis, J. Postmus, I. van Wijk, J. Bliek, M. de Meulemeester, A. Westerveld, F. Guillemot, C. Oudejans, P. Little, M. Mannens. 1997. The human Achaete-Scute homologue 2 (ASCL2,HASH2) maps to chromosome 11p15.5, close to IGF2 and is expressed in extravillus trophoblasts. Hum. Mol. Genet. 6:859–867. [DOI] [PubMed] [Google Scholar]
- Bepler, G., K.C. O'Briant, Y.C. Kim, G. Schreiber, and D.M. Pitterle. 1999. A 1.4-Mb high-resolution physical map and contig of chromosome segment 11p15.5 and genes in the LOH11A metastasis suppressor region. Genomics. 55:164–175. [DOI] [PubMed] [Google Scholar]
- Boyle, S., S. Gilchrist, J.M. Bridger, N.L. Mahy, J.A. Ellis, and W.A. Bickmore. 2001. The spatial organization of human chromosomes in the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211–219. [DOI] [PubMed] [Google Scholar]
- Brown, K.E., S. Amoils, J.M. Horn, V.J. Buckle, D.R. Higgs, M. Merkenschlager, and A.G. Fisher. 2001. Expression of alpha- and beta-globin genes occurs within different nuclear domains in haemopoietic cells. Nat. Cell Biol. 3:602–606. [DOI] [PubMed] [Google Scholar]
- Buettner, J.A., G. Glusman, N. Ben-Arie, P. Ramos, D. Lancet, and G.A. Evans. 1998. Organization and evolution of olfactory receptor genes on human chromosome 11. Genomics. 53:56–68. [DOI] [PubMed] [Google Scholar]
- Caron, H., B. van Schaik, M. van der Mee, F. Baas, G. Riggins, P. van Sluis, M-C. Hermus, R. van Asperen, K. Boon, P.A. Voute, S. Heisterkamp, A. van Kampen, and R. Versteeg. 2001. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 291:1289–1292. [DOI] [PubMed] [Google Scholar]
- Chodosh, L.A., A. Fire, M. Samuels, and P.A. Sharp. 1989. 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J. Biol. Chem. 264:2250–2257. [PubMed] [Google Scholar]
- Craig, J.M., and W.A. Bickmore. 1994. The distribution of CpG islands in mammalian chromosomes. Nat. Genet. 7:376–382. [DOI] [PubMed] [Google Scholar]
- Cremer, T., and C. Cremer. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292–301. [DOI] [PubMed] [Google Scholar]
- Cremer, M., J. von Hase, T. Volm, A. Brero, G. Kreth, J. Walter, C. Fischer, I. Solovei, C. Cremer, and T. Cremer. 2001. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 9:541–567. [DOI] [PubMed] [Google Scholar]
- Croft, J.A., J.M. Bridger, S. Boyle, P. Perry, P. Teague, and W.A. Bickmore. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145:1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, R.J., J.F. Peden, C. Lloyd, S.W. Horsley, K. Clark, C. Tufarelli, L. Kearney, V.J. Buckle, N.A. Doggett, J. Flint, and D.R. Higgs. 2001. Sequence, structure and pathology of the fully annotated terminal 2 Mb of the short arm of human chromosome 16. Hum. Mol. Genet. 10:339–352. [DOI] [PubMed] [Google Scholar]
- Dunham, I., N. Shimizu, B.A. Roe, S. Chissoe, A.R. Hunt, J.E. Collins, R. Bruskiewich, D.M. Beare, M. Clamp, L.J. Smink, et al. 1999. The DNA sequence of human chromosome 22. Nature. 402:489–495. [DOI] [PubMed] [Google Scholar]
- Engemann, S., M. Strodicke, M. Paulsen, O. Franck, R. Reinhardt, N. Lane, W. Reik, and J. Walter. 2000. Sequence and functional comparison in the Beckwith-Wiedemann region: implications for a novel imprinting centre and extended imprinting. Hum. Mol. Genet. 9:2691–2706. [DOI] [PubMed] [Google Scholar]
- Flint, J., K. Thomas, G. Micklem, H. Raynham, K. Clark, N.A. Doggett, A. King, and D.R. Higgs. 1997. The relationship between chromosome structure and function at a human telomeric region. Nat. Genet. 15:252–257. [DOI] [PubMed] [Google Scholar]
- Gabriel, J.M., M.J. Higgins, T.C. Gebuhr, T.B. Shows, S. Saitoh, and R.D. Nicholls. 1998. A model system to study genomic imprinting of human genes. Proc. Natl. Acad. Sci. USA. 95:14857–14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, X.Y., H. Zhang, M. Bittner, Y. Jiang, P. Meltzer, and J. Trent. 1996. Chromosome arm painting probes. Nat. Genet. 12:10–11. [DOI] [PubMed] [Google Scholar]
- Habermann, F.A., M. Cremer, J. Walter, G. Kreth, J. von Hase, K. Bauer, J. Wienberg, C. Cremer, T. Cremer, and I. Solovei. 2001. Arrangements of macro- and microchromosomes in chicken cells. Chromosome Res. 9:569–584. [DOI] [PubMed] [Google Scholar]
- Hu, R.J., M.P. Lee, T.D. Connors, L.A. Johnson, T.C. Burn, K. Su, G.M. Landes, and A.P. Feinberg. 1997. A 2.5-Mb transcript map of a tumor-suppressing subchromosomal transferable fragment from 11p15.5, and isolation and sequence analysis of three novel genes. Genomics. 46:9–17. [DOI] [PubMed] [Google Scholar]
- Jentsch, I., I.D. Adler, N.P. Carter, and M.R. Speicher. 2001. Karyotyping mouse chromosomes by multiplex-FISH (M-FISH). Chromosome Res. 9:211–214. [DOI] [PubMed] [Google Scholar]
- Jeppesen, P. 1997. Histone acetylation: a possible mechanism for the inheritance of cell memory at mitosis. Bioessays. 19:67–74. [DOI] [PubMed] [Google Scholar]
- Kato, R., H. Shirohzu, T. Yokomine, S. Mizuno, T. Mukai, and H. Sasaki. 1999. Sequence-ready 1-Mb YAC, BAC and cosmid contigs covering the distal imprinted region of mouse chromosome 7. DNA Res. 6:401–405. [DOI] [PubMed] [Google Scholar]
- Kurz, A., S. Lampel, J.E. Nickolenko, J. Bradl, A. Benner, R.M. Zirbel, T. Cremer, and P. Lichter. 1996. Active and inactive genes localize preferentially in the periphery of chromosome territories. J. Cell Biol. 135:1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle, J.M., and M. Lalande. 1996. Homologous association of oppositely imprinted chromosomal domains. Science. 272:725–728. [DOI] [PubMed] [Google Scholar]
- Lee, M.P., S. Brandenburg, G.M. Landes, M. Adams, G. Miller, and A.P. Feinberg. 1999. Two novel genes in the center of the 11p15 imprinted domain escape genomic imprinting. Hum. Mol. Genet. 8:683–690. [DOI] [PubMed] [Google Scholar]
- Mahy, N.L., P. Perry, S. Gilchrist, R.A. Baldock, and W.A. Bickmore. 2002. Spatial organization of active and inactive genes, and noncoding DNA, within chromosome territories. J. Cell Biol. 157:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, W.G., D. Walker, G.L. Hager, and J.G. McNally. 2001. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J. Cell Biol. 154:33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagele, R.G., A.Q. Velasco, W.J. Anderson, D.J. McMahon, Z. Thomson, J. Fazekas, K. Wind, and H. Lee. 2001. Telomere associations in interphase nuclei: possible role in maintenance of interphase chromosome topology. J. Cell Sci. 114:377–388. [DOI] [PubMed] [Google Scholar]
- Onyango, P., W. Miller, J. Lehoczky, C.T. Leung, B. Birren, S. Wheelan, K. Dewar, and A.P. Feinberg. 2000. Sequence and comparative analysis of the mouse 1-megabase region orthologous to the human 11p15 imprinted domain. Genome Res. 10:1697–1710. [DOI] [PubMed] [Google Scholar]
- Paulsen, M., O. El-Maarri, S. Engemann, M. Strodicke, O. Franck, K. Davies, R. Reinhardt, W. Reik and J. Walter. 2000. Sequence conservation and variability of imprinting in the Beckwith-Wiedemann syndrome gene cluster in human and mouse. Hum Mol Genet. 9:1829–1841. [DOI] [PubMed] [Google Scholar]
- Redeker, E., J.M. Hoovers, M. Alders, C.J. van Moorsel, A.C. Ivens, S. Gregory, L. Kalikin, J. Bliek, L. de Galan, R. van den Bogaard, J. Visser, R. van der Voort, et al. 1994. An integrated physical map of 210 markers assigned to the short arm of human chromosome 11. Genomics. 21:538–550. [DOI] [PubMed] [Google Scholar]
- Reid, L.H, C. Davies, P.R. Cooper, S.J. Crider-Miller, S.N. Sait, N.J. Nowak, G. Evans, E.J. Stanbridge, P. deJong, T.B. Shows, B.E. Weissman, and M.J. Higgins. 1997. A 1-Mb physical map and PAC contig of the imprinted domain in 11p15.5 that contains TAPA1 and the BWSCR1/WT2 region. Genomics. 43:366–375. [DOI] [PubMed] [Google Scholar]
- Saccone, S., A. Pavlicek, C. Federico, J. Paces, and G. Bernard. 2001. Genes, isochores and bands in human chromosomes 21 and 22. Chromosome Res. 9:533–539. [DOI] [PubMed] [Google Scholar]
- Stout, K., S. van der Maarel, R.R. Frants, G.W. Padberg, H.H. Ropers, and T. Haaf. 1999. Somatic pairing between subtelomeric chromosome regions: implications for human genetic disease? Chromosome Res. 7:323–329. [DOI] [PubMed] [Google Scholar]
- Tanabe, H., S. Muller, M. Neusser, J. von Hase, E. Calcagno, M. Cremer, I. Solovei, C. Cremer, and T. Cremer. 2002. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl. Acad. Sci. USA. 99:4424–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar, T., G. Sudlow, and A.S. Belmont. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschure, P.J., E.M. Manders, and R. van Driel. 1999. Spatial relationship between transcription sites and chromosome territories. J. Cell Biol. 147:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser, A.E., and J.A. Aten. 1999. Chromosomes as well as chromosomal subdomains constitute distinct units in interphase nuclei. J. Cell Sci. 112:3353–3360. [DOI] [PubMed] [Google Scholar]
- Volpi, E.V., E. Chevret, T. Jones, R. Vatcheva, J. Williamson, S. Beck, R.D. Campbell, M. Goldsworthy, S.H. Powis, J. Ragoussis, et al. 2000. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113:1565–1576. [DOI] [PubMed] [Google Scholar]
- Williams, R.R.E., S. Broad, D. Sheer, and J. Ragoussis. 2002. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 272:163–175. [DOI] [PubMed] [Google Scholar]
- Zirbel, R.M., U.R. Mathieu, A. Kurz, T. Cremer, and P. Lichter. 1993. Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chromosome Res. 1:93–106. [DOI] [PubMed] [Google Scholar]