Abstract
CVaspase activation is a key event in apoptosis execution. In stress-induced apoptosis, the mitochondrial pathway of caspase activation is believed to be of central importance. In this pathway, cytochrome c released from mitochondria facilitates the formation of an Apaf-1 apoptosome that recruits and activates caspase-9. Recent data indicate that in some cells caspase-9 may not be the initiator caspase in stress-mediated apoptosis because caspase-2 is required upstream of mitochondria for the release of cytochrome c and other apoptogenic factors. To determine how caspase-2 is activated, we have studied the formation of a complex that mediates caspase-2 activation. Using gel filtration analysis of cell lysates, we show that caspase-2 is spontaneously recruited to a large protein complex independent of cytochrome c and Apaf-1 and that recruitment of caspase-2 to this complex is sufficient to mediate its activation. Using substrate-binding assays, we also provide the first evidence that caspase-2 activation may occur without processing of the precursor molecule. Our data are consistent with a model where caspase-2 activation occurs by oligomerization, independent of the Apaf-1 apoptosome.
Keywords: apoptosis; caspase activation; apoptosome; caspase-9; initiator caspase
Introduction
The execution of apoptosis is performed by a family of cysteine proteases known as caspases (for review see Nicholson, 1999). Caspases are present as zymogens in most animal cells and are activated in response to apoptotic signals. The apoptotic signals first lead to the activation of so called “initiator caspases,” such as caspase-8 and caspase-9. These caspases contain specific protein–protein interaction domains in their NH2 termini that allow them to be recruited to death-signaling complexes promoting their activation by an autocatalytic mechanism (for review see Kumar, 1999). Once the initiator caspases are activated, they process and activate downstream effector caspases, such as caspase-3, -6 and -7, which are responsible for the cleavage of several proteins in dying cells (Nicholson, 1999).
In mammals, there are two main caspase activation pathways: the death receptor pathway and the mitochondrial pathway (Kumar, 1999; Nicholson, 1999). In the first pathway, TNF receptor family members recruit Fas-associated protein with death domain (FADD),* an adaptor molecule that in turn recruits caspase-8 to the death-inducing signaling complex (DISC). Recruitment of caspase-8 to DISC results in its activation. The mitochondrial pathway of caspase activation is regulated by the Bcl-2 family of proteins and involves cytochrome c and Apaf-1–mediated activation of caspase-9. During stress signaling, cytochrome c released from mitochondria binds Apaf-1, resulting in a conformational change that facilitates oligomerization of Apaf-1 into an apoptosome and subsequent caspase recruitment domain (CARD)–mediated recruitment and activation of caspase-9 (Acehan et al., 2002). Interestingly, initiator caspase DRONC activation in Drosophila cells does not seem to require release of cytochrome c from mitochondria (Dorstyn et al., 2002).
Although caspase-9 is generally believed to be the initiator caspase in stress-induced apoptosis, recent studies suggest that caspase-2 can act upstream of mitochondria (Lassus et al., 2002; Robertson et al., 2002). The new findings indicate that caspase-2 is the initiator caspase in stress-induced apoptosis, whereas apoptosome mediated caspase-9, and subsequently caspase-3, activation may mainly serve as an amplification loop (Kumar and Vaux, 2002). Caspase-2, the first discovered apoptotic mammalian caspase, has been implicated in a variety of cell death pathways, and consistent with an initiator role it is activated rapidly in response to diverse death stimuli (Kumar et al., 1994, 1997; Wang et al., 1994; Kumar, 1995; Harvey et al., 1997). Since caspase-2−/− mice have only a subtle phenotype, it is likely that in the absence of this caspase other similar caspases functionally replace caspase-2 (Bergeron et al., 1998).
Although the process of caspase-9 activation via Apaf-1 apoptosome has been well studied, the mechanism of caspase-2 activation is not known. Caspase-2 can homodimerize and autoprocess in yeast and mammalian cells, but an Apaf-1-like adaptor has not been found for caspase-2 and it is not known whether an apoptosome-like complex is required for its activation (Butt et al., 1998). Because caspase-2 can be processed into subunits by caspase-3 and this processing is blocked in caspase-3–depleted extracts and cells derived from Apaf-1 and caspase-9–deficient mice, caspase-2 has often been suggested to be a downstream caspase (Harvey et al., 1996; Slee et al., 1999; O'Reilly et al., 2002). However, since caspase-9 activation can occur without processing (Stennicke et al., 1999) it is possible that initial caspase-2 activation may also occur without its cleavage and that caspase-3–mediated caspase-2 processing is simply a downstream amplification mechanism. In this paper, we report that caspase-2 is recruited to a large protein complex independent of cytochrome c and Apaf-1 and that this recruitment is sufficient for caspase-2 activation. We also provide data to suggest that initial caspase-2 activation is likely to occur without any processing of the precursor.
Results and discussion
Recruitment of caspase-2 to a large protein complex
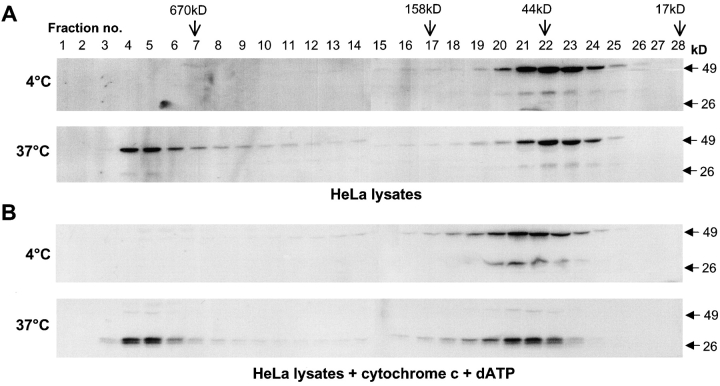
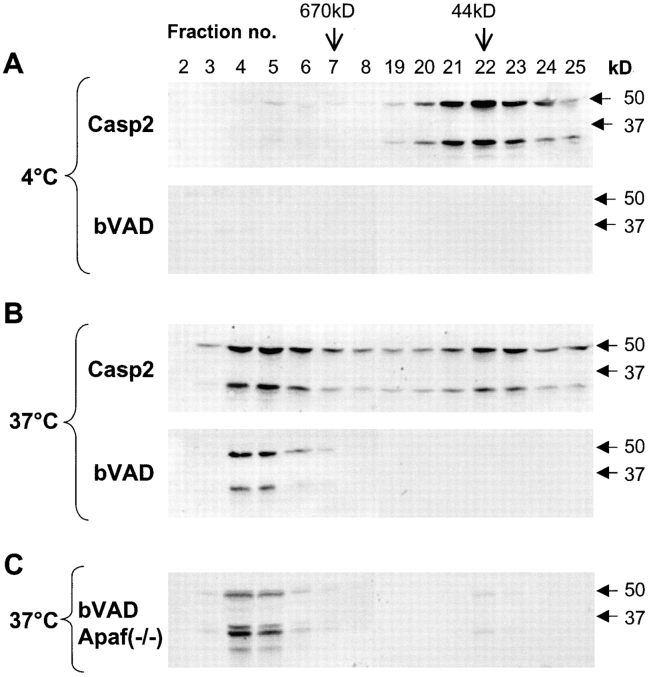
We have shown previously that caspase-2 overexpression in mammalian cell lines results in the formation of elaborate filamentous structures that are dependent on the prodomain of caspase-2 (Colussi et al., 1998a,b). To further dissect the ability of caspase-2 to form large complexes, we subjected whole cell lysates from HeLa cells to size exclusion chromatography. Size exclusion chromatography has been used successfully by several groups to show the formation of the Apaf-1 apoptosome (Cain et al., 1999, 2000; Zou et al., 1999). Upon the addition of cytochrome c and dATP to cell lysates containing monomeric Apaf-1 and caspase-9, these proteins were found to interact via their CARDs to form a complex >700 kD in size (Cain et al., 1999, 2000; Zou et al., 1999). Our results showed that in untreated HeLa lysates, caspase-2 eluted mainly as monomeric zymogen (Fig. 1 A). Interestingly, incubation of the cell lysates at 37°C for 1 h without the addition of cytochrome c and dATP resulted in a dramatic change in the elution behavior of caspase-2 (Fig. 1 A). Approximately 50% of the caspase-2 was now associated with fractions corresponding to a molecular mass of >670 kD (Fig. 1).
Figure 1.
Caspase-2 is recruited to a large protein complex. HeLa cell lysates were subjected to size exclusion chromatography (A and B). Aliquots from the fractions were analyzed by immunoblotting using the monoclonal antibody 11B4 that recognizes the p19 subunit and full-length caspase-2. Cells were lysed in buffer A and incubated at 4 or 37°C for 60 min before loading onto the column. In B, lysates were incubated at 4 or 37°C for 2 h with 2 μg cytochrome c and 2 mM dATP. The elution positions of the markers on the Superdex 200 column are indicated. The positions of the SDS-PAGE prestained M r standards are indicated on the right of the blots. The smaller caspase-2 immunoreactive band is prodomain+p19 processing intermediate that is often detected in cell lysates, probably due to some processing of procaspase-2 during cell harvesting and lysis.
The lack of requirement for cytochrome c and dATP addition for the formation of this complex suggests that either there is sufficient cytochrome c and dATP in the cell extracts or that these components are not required for the generation of the caspase-2 complex. In all experiments described in this paper, we found that it was unnecessary to add cytochrome c and dATP for the formation of caspase-2 or Apaf-1 complex. Similar results were obtained with extracts prepared from the murine neuroblastoma cell line N18 (unpublished data) and human embryonic kidney 293T cells (see below), suggesting that the recruitment of caspase-2 to a putative high molecular weight complex is not cell specific. In experiments where the lysates supplemented with cytochrome c and dATP were incubated at 37°C for >2 h, processing of procaspase-2, likely to be mediated by active caspase-3, was clearly evident in both low and high M r fractions (Fig. 1 B).
Caspase-2 complex is sensitive to ionic concentration and requires the prodomain
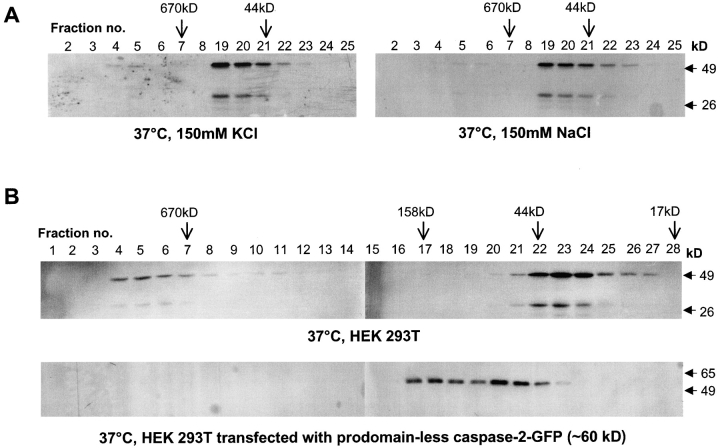
A time course demonstrated that the formation of caspase-2 complex is rapid and complete. Within 30 min of incubation at 37°C, >30% of the monomeric caspase-2 was recruited to the large complex, whereas a 2-h incubation was sufficient for >75% of caspase-2 to shift to the high molecular mass complex in fractions 4–6 (unpublished data). Previous studies have shown that physiological levels of KCl or NaCl prevent the formation of the apoptosome in vitro, suggesting that this may be a mechanism to ensure that inadvertent caspase activation does not occur in healthy cells (Cain et al., 2001). In similar experiments, we prepared HeLa cell lysates in salt-free buffer and supplemented them with either 150 mM KCl or 150 mM NaCl. The addition of the 150 mM salt to HeLa lysates strongly inhibited the caspase-2 complex formation (Fig. 2 A). Thus, as is the case for the apoptosome, inadvertent caspase-2 complex formation may be prevented by normal cytoplasmic salt concentrations.
Figure 2.
Salt and prodomain dependence of caspase-2 complex. (A) HeLa cell lysates in 20 mM Hepes, pH 7.5, were supplemented with either 150 mM KCl or NaCl as indicated before incubation at 37°C for 60 min. Only selected fractions 2–8 and 19–25 were analyzed by immunoblotting. (B) Lysates from control HEK 293T cells or those transfected with a GFP-tagged prodomain-less caspase-2 expression construct were incubated at 37°C for 1 h and subjected to size exclusion chromatography. Fractions were immunoblotted with the caspase-2 antibody (top) or a GFP antibody (bottom). Note that prodomain-less caspase-2–GFP fusion protein (∼60 kD) appears as monomer and dimer but was not detected in the high M r fractions. Full-length caspase-2–GFP (Cys mutant) protein expressed in HEK 293T cells was largely insoluble and therefore could not be used in fractionation experiments.
To test if caspase-2 CARD was necessary for its recruitment to the large complex, we fractionated extracts prepared from cells transfected with a caspase-2 construct lacking the prodomain and found that upon incubation at 37°C this form of caspase-2 was not recruited to the large complex (Fig. 2 B). This result suggests that CARD is necessary for procaspase-2 recruitment into the complex.
The elution profile of caspase-2 complex is similar to that of the Apaf-1 apoptosome
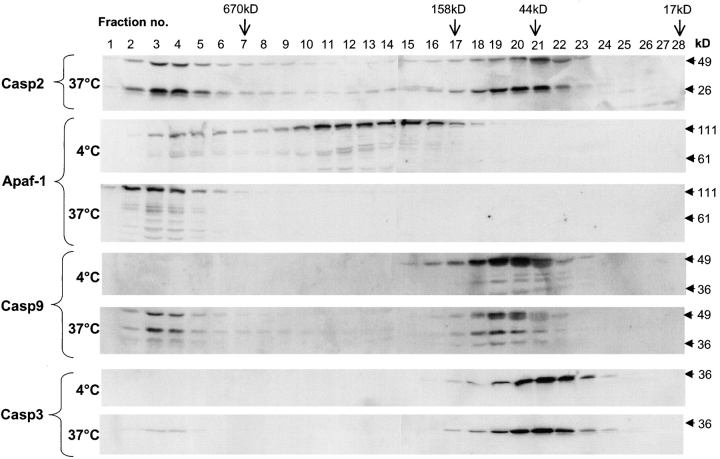
The size of the caspase-2 complex corresponds to that of the Apaf-1 apoptosome. To test if the apoptosome and the caspase-2 complex coelute in the same fractions, we probed the fractions obtained from the size exclusion column with Apaf-1, caspase-9, and caspase-3 antibodies. As shown in Fig. 3, in untreated extracts all three proteins were present in the fractions corresponding to their relative monomeric molecular mass as has been shown previously (Cain et al., 1999, 2000; Zou et al., 1999). However, after incubation of extracts at 37°C for 1 h all of the Apaf-1 and ∼50% of the endogenous caspase-9 was recruited to a high M r complex in fractions that also contained caspase-2. A small fraction of procaspase-3 was also present in the same high molecular mass fractions. These data suggest that caspase-2 is either a part of the Apaf-1 and caspase-9 apoptosome or a complex of similar size to the apoptosome.
Figure 3.
The caspase-2 complex is similar to the apoptosome. Treated HeLa lysates were subjected to size exclusion chromatography. Aliquots of the fractions were immunoblotted using antibodies to caspase-2 (Casp2), Apaf-1, caspase-9 (Casp9), and caspase-3 (Casp3).
Caspase-2 complex formation occurs independent of cytochrome c and Apaf-1
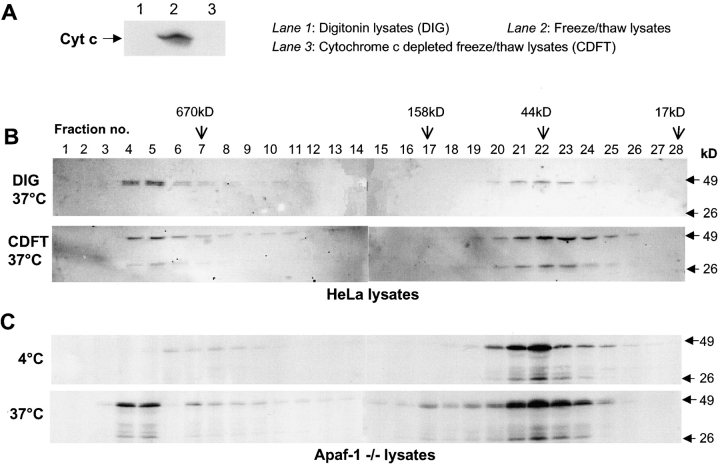
Although caspase-2 was detected in the same fractions as Apaf-1/cytochrome c apoptosome, we were unable to detect caspase-2 in Apaf-1 immunoprecipitations from fractions 2–6, which suggested that caspase-2 is unlikely to directly interact with Apaf-1 (unpublished data). To test if caspase-2 complex formation requires cytochrome c, we used HeLa lysates depleted of cytochrome c or prepared cell extracts by digitonin treatment to prevent lysis of mitochondria. As shown in Fig. 4 B, the lack of cytochrome c had no apparent effect on caspase-2 complex formation. To analyze if Apaf-1 was necessary for caspase-2 complex, we used a myeloid cell line derived from Apaf-1 knock-out mice. Extracts prepared from Apaf-1–deficient cells were analyzed by gel filtration, and as shown in Fig. 4 B caspase-2 recruitment to the high molecular weight fractions was unaffected in these cells. These results indicate that caspase-2 is not recruited to the high molecular weight complex via an interaction with Apaf-1 and that caspase-2 complex forms independently of the Apaf-1/cytochrome c apoptosome. One possibility is that caspase-2 is recruited to this high molecular weight complex by interacting with an adaptor molecule functionally similar to Apaf-1. One candidate adaptor molecule for caspase-2 is RAIDD, which is known to interact with caspase-2 via a CARD (Duan and Dixit, 1997; Shearwin-Whyatt et al., 2000). However, we were unable to detect any RAIDD in the high molecular weight fractions (unpublished data), ruling out the involvement of RAIDD in caspase-2 complex formation.
Figure 4.
Formation of caspase-2 complex is independent of cytochrome c and Apaf-1. In A and B, HeLa cell lysates were prepared by digitonin lysis or freeze/thawing. Immunoblotting in A, lane 1, demonstrated that digitonin lysates do not have any detectable cytochrome c (Cyt c). Cytochrome c from the freeze/thaw lysates was removed by immunodepletion. As shown in A, lane 3, immunodepleted cell extracts do not have any detectable cytochrome c. In B, digitonin lysates (DIG) and cytochrome c–depleted lysates (CDFT) were incubated at 37°C for 60 min, subjected to size exclusion chromatography, and analyzed by immunoblotting using a caspase-2 antibody. (C) Lysates from Apaf-1−/− cells were incubated at 4 or 37°C for 60 min and subjected to size exclusion chromatography. Fractions were precipitated with 10% TCA/0.07% β-mercaptoethanol/acetone at −20°C. Protein pellets were washed with cold 0.07% β-mercaptoethanol/acetone and resuspended in 1× SDS protein loading buffer. The entire sample was resolved by SDS-PAGE and analyzed by immunoblotting with the caspase-2 antibody.
Recruitment to the large complex results in activation of caspase-2
Although caspase-2 is structurally similar to the initiator caspases, previous studies have shown that procaspase-2 is a substrate for caspase-3–mediated activation and depletion of caspase-3, or its upstream regulators caspase-9 and Apaf-1, blocks processing of procaspase-2 (Harvey et al., 1996; Slee et al., 1999; O'Reilly et al., 2002). How then can we reconcile the role of caspase-2 upstream of mitochondria? One possibility is that the initial activation of caspase-2 occurs without any significant processing of procaspase-2, and caspase-2 zymogen cleavage by caspase-3 is a downstream event that acts as an amplification mechanism to fully and irreversibly activate all cellular caspase-2. Some support for this hypothesis comes from the observation that most of the caspase-2 in the large complex is in its zymogen form; however, longer incubation (>2 h) of cell extracts at 37°C with cytochrome c and dATP resulted in mostly processed caspase-2 (Fig. 1 B), suggesting that this required activation of downstream caspases, such as caspase-3, which was fully activated by 2 h (unpublished data).
Since there are no specific substrates for caspase-2, and the most widely used caspase-2 substrate VDVAD is also readily cleaved by several other caspases including caspase-3 and caspase-7 (unpublished data), we tested if we can distinguish active caspase-2 from inactive form by its ability to directly bind a substrate/inhibitor. As suggested for caspase-9 zymogen activation (Stennicke et al., 1999; Renatus et al., 2001), if caspase-2 is initially activated by a conformational change, the active form might be detected by its ability to bind biotin–VAD. To this end, fractions from HeLa lysates separated by size exclusion chromatography were incubated with biotin–VAD-fmk, precipitated with streptavidin-coupled Sepharose, and the precipitate was subjected to immunoblotting using anti–caspase-2 antibody. As shown in Fig. 5 A, fractions collected from control lysates did not show any biotin–VAD-fmk binding. However, fractions 4–7 from HeLa lysate incubated at 37°C contained caspase-2 with the ability to bind biotin–VAD-fmk (Fig. 5 B). This result suggests the recruitment of caspase-2 to the high molecular weight complex enables the activation of caspase-2 as measured by substrate binding. Interestingly, the fractions containing monomeric caspase-2 did not show any biotin–VAD-fmk binding. We repeated this experiment with column fractions collected from Apaf-1−/− lysates. Again, the high molecular weight fractions contained caspase-2 protein capable of binding biotin–VAD-fmk (Fig. 5 C). These results show that although the monomeric procaspase-2 is largely inactive, caspase-2 in the high molecular weight complex is catalytically active. Furthermore, whereas the caspase-2 complex resembles the Apaf-1/caspase-9 apoptosome, our data suggest that the complex forms independently of Apaf-1 and that Apaf-1 is not required for caspase-2 activation.
Figure 5.
Recruitment of caspase-2 to the large complex results in its activation. Lysates from HeLa (A and B) or Apaf-1−/− (C) cells were subjected to size exclusion chromatography. Aliquots from fractions were analyzed by immunoblotting using caspase-2 antibody (Casp2) or 200 μl of the HeLa fractions, or all of the Apaf-1−/− fractions were incubated with biotin–VAD-fmk (bVAD) for 60 min at RT followed by overnight incubation with streptavidin-Sepharose. Sepharose pellets were washed, and caspase-2 was detected by immunoblotting. HeLa lysates incubated at 4°C for 60 min (A); HeLa lysates incubated at 37°C for 60 min (B); Apaf-1−/− lysates incubated at 37°C for 60 min (C). Only caspase-2–containing fractions (2–8 and 19–25) are shown.
Although it is possible to demonstrate the formation of the Apaf-1/caspase-9 apoptosome in cell lysates, it has been difficult to isolate this complex from intact cells in response to a cell death stimulus. In some studies, Apaf-1 was shown to migrate to fractions of >700 kD in size in extracts prepared from cells treated with cytotoxic drugs (Zou et al., 1999; Almond et al., 2001). However, to our knowledge no one has demonstrated the recruitment of caspases to the putative Apaf-1 complex in dying cells. This may indicate the transient nature of the apoptosome in vivo and the fact that only a small fraction of cellular caspase-9 is recruited to the apoptosome at a given time, making it technically difficult to experimentally observe it. Similarly, we were unable to see any caspase-2 in high M r fractions of cell extracts prepared from etoposide-treated HeLa cells, although these cells did show recruitment of a part of the monomeric Apaf-1 pool to the large complex (unpublished data). Since the putative adaptor required for caspase-2 activation is currently unknown, it was not possible to see whether an Apaf-1–like complex specific for caspase-2 activation was being formed in vivo. We are currently in the process of identifying the components of the caspase-2 complex using proteomics-based approaches. This will help us delineate fully the molecular mechanism of caspase-2 activation.
Materials and methods
Cell culture and preparation of lysates
HeLa cells were maintained in DME supplemented with 10% FBS and 1 mM glutamine. Apaf-1 wild-type and Apaf-1−/− cells (retrovirally immortalized murine fetal liver cells) were maintained in DME with 10% FBS, 1 mM glutamine and 1/4,000 recombinant murine IL-3 (a gift from A.F. Lopez, Hanson Institute). To prepare lysates, cells were washed twice with PBS and resuspended in buffer A (20 mM Hepes-KOH, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, pH 7.5) supplemented with protease inhibitors (Complete™; Roche). Resuspended cells were subjected to three rounds of freeze thawing using liquid nitrogen and ice water. Cellular debris was removed by centrifugation at 9,000 g, 20 min at 4°C, followed by 100,000 g, 30 min at 4°C. Cleared lysates were stored at −20°C until required. Where indicated, depletion of cytochrome c from cell extracts was performed as described (Dorstyn et al., 2002). To maintain mitochondria integrity in some experiments, cells were lysed by homogenization in a buffer containing 0.1 mg/ml digitonin (Bourgeron et al., 1992). HEK 293T cells were grown in RPMI1640 with 10% FBS. Cell were transfected with a catalytically inactive caspase-2–GFP expression construct lacking the prodomain as described (Colussi et al., 1998a). Cell lysates from untransfected and transfected cells were prepared by freeze thawing as above.
Chromatographic methods
Treated lysates were fractionated using FPLC protein purification system on a Superdex 200 column (Amersham Biosciences) at 4°C. The column was equilibrated with buffer A, and lysates (5 mg) were applied and eluted from the column with buffer A at a flow rate of 0.4 ml/min and 400-μl fractions were collected. The column was calibrated with Bio-Rad Laboratories gel filtration standards containing bovine thyroglobulin (670 kD), horse gamma globulin (158 kD), chicken ovalbumin (44 kD), horse myoglobin (17 kD), and vitamin B-12 (1.35 kD).
Immunoblotting
25 μl of each fraction from gel filtration column were resolved on 8–15% SDS–polyacrylamide gels and transferred to polyvinylidine difluoride membrane (Hybond-P; Amersham Biosciences). Membranes were blocked in 5% blocking solution (Amersham Biosciences) before probing with antibodies to caspase-2 (11B4) (O'Reilly et al., 2002), Apaf-1 (12F11) (O'Reilly et al., 2002), caspase-9 (B40) (BD PharMingen), cytochrome c (BD PharMingen), GFP (Roche), and caspase-3 (BD PharMingen). Antibody binding was detected using goat anti–rat IgG (Pierce Chemical Co.), sheep anti–mouse IgG (Silenus), or sheep anti–rabbit IgG (Silenus), all conjugated to alkaline phosphatase, and ECF (Amersham Biosciences) using the Typhoon 9410 and ImageQuant software (Amersham Biosciences).
Detection of active caspase-2
Whole cell lysates (50 μg) or 100–200 μl of Superdex 200 column fractions were incubated with 2 μM biotin–VAD-fmk (Enzyme Systems) for 60 min at RT. Reaction volumes were made up to 1 ml in buffer A and the biotin–VAD-fmk–reacting proteins were precipitated by incubation with streptavidin-coupled Sepharose overnight at 4°C on a rotator. After washing with buffer A plus 0.1% Tween 20, Sepharose bound proteins were resolved by SDS-PAGE and immunoblotting with the caspase-2 antibody.
Acknowledgments
We thank Paul Colussi and Guy Salvesen for helpful suggestions.
This work was supported by the National Health and Medical Research Council and the Cancer Council.
Footnotes
Abbreviations used in this paper: CARD, caspase recruitment domain; DISC, death-inducing signaling complex; FADD, Fas-associated protein with death domain.
References
- Acehan, D., X. Jiang, D.G. Morgan, J.E. Heuser, X. Wang, and C.W. Akey. 2002. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol. Cell. 9:423–432. [DOI] [PubMed] [Google Scholar]
- Almond, J.B., R.T. Snowden, A. Hunter, D. Dinsdale, K. Cain, and G.M. Cohen. 2001. Proteasome inhibitor-induced apoptosis of B-chronic lymphocytic leukaemia cells involves cytochrome c release and caspase activation, accompanied by formation of an approximately 700 kDa Apaf-1 containing apoptosome complex. Leukemia. 15:1388–1397. [DOI] [PubMed] [Google Scholar]
- Bergeron, L., G.I. Perez, G. Macdonald, L. Shi, Y. Sun, A. Jurisicova, S. Varmuza, K.E. Latham, J.A. Flaws, J.C. Salter, et al. 1998. Defects in regulation of apoptosis in caspase-2-deficient mice. Genes Dev. 12:1304–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeron, T., D. Chretien, A. Rotig, A. Munnich, and P. Rustin. 1992. Isolation and characterization of mitochondria from human B lymphoblastoid cell lines. Biochem. Biophys. Res. Commun. 186:16–23. [DOI] [PubMed] [Google Scholar]
- Butt, A.J., N.L. Harvey, G. Parasivam, and S. Kumar. 1998. Dimerization and autoprocessing of the Nedd2 (caspase-2) precursor requires both the prodomain and the carboxyl-terminal regions. J. Biol. Chem. 273:6763–6768. [DOI] [PubMed] [Google Scholar]
- Cain, K., D.G. Brown, C. Langlais, and G.M. Cohen. 1999. Caspase activation involves the formation of the aposome, a large (approximately 700 kDa) caspase-activating complex. J. Biol. Chem. 274:22686–22692. [DOI] [PubMed] [Google Scholar]
- Cain, K., S.B. Bratton, C. Langlais, G. Walker, D.G. Brown, X.M. Sun, and G.M. Cohen. 2000. Apaf-1 oligomerizes into biologically active approximately 700-kDa and inactive approximately 1.4-MDa apoptosome complexes. J. Biol. Chem. 275:6067–6070. [DOI] [PubMed] [Google Scholar]
- Cain, K., C. Langlais, X.M. Sun, D.G. Brown, and G.M. Cohen. 2001. Physiological concentrations of K+ inhibit cytochrome c-dependent formation of the apoptosome. J. Biol. Chem. 276:41985–41990. [DOI] [PubMed] [Google Scholar]
- Colussi, P.A., N.L. Harvey, and S. Kumar. 1998. a. Prodomain-dependent nuclear localization of the caspase-2 (Nedd2) precursor. A novel function for a caspase prodomain. J. Biol. Chem. 273:24535–24542. [DOI] [PubMed] [Google Scholar]
- Colussi, P.A., N.L. Harvey, L.M. Shearwin-Whyatt, and S. Kumar. 1998. b. Conversion of procaspase-3 to an autoactivating caspase by fusion to the caspase-2 prodomain. J. Biol. Chem. 273:26566–26570. [DOI] [PubMed] [Google Scholar]
- Dorstyn, L., S. Read, D. Cakouros, J.R. Huh, B.A. Hay, and S. Kumar. 2002. The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J. Cell Biol. 156:1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, H., and V.M. Dixit. 1997. RAIDD is a new ‘death’ adaptor molecule. Nature. 385:86–89. [DOI] [PubMed] [Google Scholar]
- Harvey, N.L., J.A. Trapani, T. Fernandes-Alnemri, G. Litwack, E.S. Alnemri, and S. Kumar. 1996. Processing of the Nedd2 precursor by ICE-like proteases and granzyme B. Genes Cells. 1:673–685. [DOI] [PubMed] [Google Scholar]
- Harvey, N.L., A.J. Butt, and S. Kumar. 1997. Functional activation of Nedd2/ICH-1 (caspase-2) is an early process in apoptosis. J. Biol. Chem. 272:13134–13139. [DOI] [PubMed] [Google Scholar]
- Kumar, S. 1995. Inhibition of apoptosis by the expression of antisense Nedd2. FEBS Lett. 368:69–72. [DOI] [PubMed] [Google Scholar]
- Kumar, S. 1999. Mechanisms mediating caspase activation in cell death. Cell Death Differ. 6:1060–1066. [DOI] [PubMed] [Google Scholar]
- Kumar, S., and D.L. Vaux. 2002. A cinderella caspase takes center stage. Science. 297:1290–1291. [DOI] [PubMed] [Google Scholar]
- Kumar, S., M. Kinoshita, M. Noda, N.G. Copeland, and N.A. Jenkins. 1994. Induction of apoptosis by the mouse Nedd2 gene, which encodes a protein similar to the product of the Caenorhabditis elegans cell death gene ced-3 and the mammalian IL-1 beta-converting enzyme. Genes Dev. 8:1613–1626. [DOI] [PubMed] [Google Scholar]
- Kumar, S., M. Kinoshita, L. Dorstyn, and M. Noda. 1997. Origin, expression and possible functions of the two alternatively spliced forms of the mouse Nedd2 mRNA. Cell Death Differ. 4:378–387. [Google Scholar]
- Lassus, P., X. Opitz-Araya, and Y. Lazebnik. 2002. Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science. 297:1352–1354. [DOI] [PubMed] [Google Scholar]
- Nicholson, D.W. 1999. Caspase structure, proteolytic substrates, and function during apoptotic cell death. Cell Death Differ. 6:1028–1042. [DOI] [PubMed] [Google Scholar]
- O'Reilly, L.A., P. Ekert, N. Harvey, V. Marsden, L. Cullen, D.L. Vaux, G. Hacker, C. Magnusson, M. Pakusch, F. Cecconi, et al. 2002. Caspase-2 is not required for thymocyte or neuronal apoptosis even though cleavage of caspase-2 is dependent on both Apaf-1 and caspase-9. Cell Death Differ. 9:832–841. [DOI] [PubMed] [Google Scholar]
- Renatus, M., H.R. Stennicke, F.L. Scott, R.C. Liddington, and G.S. Salvesen. 2001. Dimer formation drives the activation of the cell death protease caspase 9. Proc. Natl. Acad. Sci. USA. 98:14250–14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, J.D., M. Enoksson, M. Suomela, B. Zhivotovsky, and S. Orrenius. 2002. Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J. Biol. Chem. 277:29803–29809. [DOI] [PubMed] [Google Scholar]
- Shearwin-Whyatt, L.M., N.L. Harvey, and S. Kumar. 2000. Subcellular localization and CARD-dependent oligomerization of the death adaptor RAIDD. Cell Death Differ. 7:155–165. [DOI] [PubMed] [Google Scholar]
- Slee. E.A., C. Adrain, and S.J. Martin. 1999. Serial killers: ordering caspase activation events in apoptosis. Cell Death Differ. 16:1067–1074. [DOI] [PubMed] [Google Scholar]
- Stennicke, H.R., Q.L. Deveraux, E.W. Humke, J.C. Reed, V.M. Dixit, and G.S. Salvesen. 1999. Caspase-9 can be activated without proteolytic processing. J. Biol. Chem. 274:8359–8362. [DOI] [PubMed] [Google Scholar]
- Wang, L., M. Miura, L. Bergeron, H. Zhu, and J. Yuan. 1994. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 78:739–750. [DOI] [PubMed] [Google Scholar]
- Zou, H., Y. Li, X. Liu, and X. Wang. 1999. An APAF-1-cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 274:11549–11556. [DOI] [PubMed] [Google Scholar]