Abstract
The subnucleolar structure that is involved in rDNA transcription has been controversial. A report by Koberna et al. (2002)(this issue, page 743) adds significant weight toward the idea that dense fibrillar components (DFCs)* and fibrillar center (FC)/DFC borders are the sites of pre-rRNA synthesis.
Keywords: nucleolus; sites of DNA transcription; Christmas tree; Br-U labeling; dense fibrillar components
The nucleolus of an average mammalian cell can produce up to 10,000 ribosomes per min. Each cell contains hundreds of copies of rDNA in the form of tandem repeats, and ∼400 rDNA repeats are spread out on five pairs of chromosomes in human cells. At any given time, only a subset of rDNA repeats is actively being transcribed. Miller spreads (electron microscopic visualization of RNA being synthesized from DNA templates) of transcriptionally active rDNA repeats demonstrate Christmas tree–like transcription units. Transcription of rDNA generates 45S pre-rRNAs that are subsequently cleaved and processed into 28S, 18S, and 5.8S rRNA concomitant with their assembly into large and small subunits. The transcription, processing, and assembly of preribosomal particles take place in the nucleolus.
As one of the most prominent cellular organelles, the nucleolus provides an opportunity to investigate how different steps in this complex RNA metabolic process are structurally arranged to form a highly efficient macromolecular machine. High-resolution electron microscopic studies demonstrated that the nucleolus contains three distinct and highly conserved substructural features, the FCs, DFCs, and the granular components (GCs) (for reviews see Busch and Smetana, 1970; Hadjiolov, 1985). FCs are pale fibrillar regions that are enriched with RNA polymerase I (Pol I)–specific transcription factors including Pol I, DNA topoisomerase I, and UBF. DFCs are highly electron dense fibrillar regions that partially or completely surround FCs. DFCs are heavily labeled with antibodies specifically recognizing pre-rRNA processing factors including fibrillarin, a factor of the U3 complex primarily involved in early steps of preribosomal RNA processing. In addition, transcription factors are also localized to DFCs. GCs, the granular regions outside of FCs and DFCs, constitute the rest of the nucleolus. GCs are enriched with assembly factors and ribosomal proteins. Each nucleolus can contain multiple FCs surrounded by DFCs. Studies using pulse–chase labeling, in situ hybridization to pre-rRNAs, and immunolabeling of trans-acting factors involved in ribosome biogenesis suggest a vectorial process by which pre-rRNAs migrate from fibrillar regions to granular regions, and subsequently into the nucleoplasm while maturing into preribosomal particles (Granboulan and Granboulan, 1965, for review see Shaw and Jordan, 1995; Scheer and Hock, 1999). However, the precise structure and functional relationship between the steps of preribosome synthesis and specific nucleolar substructures remains unclear. In particular, the site of pre-rRNA transcription has been controversial.
In an attempt to clarify the nucleolar substructure that is involved in pre-rRNA transcription, a report by Koberna et al. (2002) elegantly depicts the sites of labeled nucleotide incorporation in the nucleolus using electron microscopy. HeLa cells underwent either hypotonic shift or detergent permeabilization to allow the uptake of Br-U for 5 min and were then fixed and embedded in lowicryl. The cellular sites of Br-U labeling represent RNA newly synthesized within the labeling period. Immunogold detection of Br-U epitopes on 50–70-nm sections reveals clusters of intense signals in DFCs as observed by transmission electron microscopy. Some signals are observed on FC/DFC borders and very little is seen in FCs. In addition, Koberna et al. (2002) detected concomitantly the Br-U incorporation signals in the same nucleolus by both light and electronic microscopy, demonstrating that in these mammalian cells the signals seen as intense dots by light microscopy, apparently covering both the FC and the DFC, can be resolved to DFC-specific localization with electron microscopy. With the short duration of labeling and slower transcriptional efficiency inherent to the protocol, most of the sites of Br-U incorporation are considered to be at or near the sites of transcription. The high concentration of Br-U labeling observed in DFCs, therefore, suggests that DFCs are the primary sites of active pre-rRNA synthesis. Three-dimensional reconstructions of serial sections further support this suggestion by demonstrating multiple clusters of Br-U signals in DFCs, implicating each cluster as one (or several spatially associated) units of actively transcripting rDNA (Christmas trees).
These findings build on previous studies from the same group (Dundr and Raska, 1993; Stanek et al., 2001) and are in agreement with reports by Hozak et al. (1994) and Cmarko et al. (2000), in which short pulse labeling of Br-U resulted in the detection of Br-U in DFCs and at DFC/FC borders, and prolonged labeling generated signals in the surrounding GCs. The overwhelming intensity of the Br-U labeling signals in DFCs in the current report makes it much more believable that pre-rRNAs are transcribed mainly in DFCs and sometimes at DFC/FC borders (Koberna et al., 2002). Furthermore, this idea is also consistent with the findings of transcriptionally active rDNA and Br-U labeled “Christmas trees” in plant cell DFCs (Melcak et al., 1996; Gonzalez-Melendi et al., 2001). However, these observations are in contrast to findings by at least two groups where the Br-U pulse labeling resulted in signals in FCs viewed by light and electron microscopy. Mais and Scheer (2001) analyzed the localization of Pol I transcription factors, fibrillarin, and Br-U pulse labeling in amplified Xenopus oocytes nucleoli where FCs and DFCs can be distinguished at the light microscopic level. They found that the labelings of Br-U, UBF, and Pol I are surrounded by the labeling of fibrillarin (indicative of DFCs), suggesting that transcription takes place in FCs. Furthermore, Thiry et al. (2000) demonstrated a time-dependent migration of Br-U labeling from FCs to DFCs, to GCs, and finally to the cytoplasm. The signals are observed exclusively in DFCs after a 5-min labeling and a 10-min chase. These observations imply that transcription of rRNA primarily takes place in FCs. It is curious that the use of Br-U pulse labeling technique has resulted in different observations from different groups. The key difference is the positive versus the negative labeling of FCs with Br-U after a short pulse labeling. Could it be that some of the nascent pre-rRNAs may be synthesized in FCs and are transported or extended so rapidly into DFCs that a slight delay in fixation could result in loss of Br-U labeling in FCs? Or might the density of the DFC reduce its labeling relative to the FC in some preparations? It may be constructive for groups studying this subject to discuss technical details that may contribute to the discrepancies in the detection of Br-U labeling in the nucleolus.
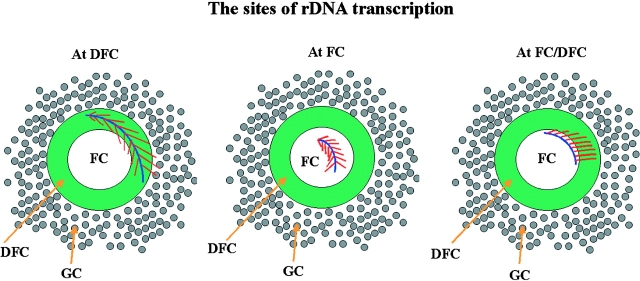
Most recently, using the Miller chromatin spread method, Osheim and Beyer (personal communication) demonstrated that U3 complex forms terminal nodules on the 5′-end of transcribing pre-rRNA, suggesting that transcription and pre-rRNA processing proceed simultaneously. This finding supports that transcription occurs in DFCs since fibrillarin, a component of the U3 complex, is observed mostly in DFCs and is not present in FCs. On the other hand, it is also possible that some of the active rDNA templates are in FCs, and the elongating RNAs extend into DFCs. The studies by Koberna et al. (2002) provide compelling evidence that the DFC, with the DFC/FC boundary, is a strong contender for the primary site of rRNA transcription in the robust nucleolar factory of mammalian cells. However, the subject is likely to remain controversial. Therefore, as of now, there remain several possibilities concerning the sites of rRNA transcription, including the DFCs, FCs, FC/DFC borders, or some combination of these (Fig. 1). An unambiguous resolution may have to await convincing labeling of active rDNA clusters in well-preserved nucleoli using electron microscopy.
Figure 1.
Possible models illustrate the nucleolus substructure that could be involved in pre-rRNA synthesis.
Acknowledgments
I would like to thank Drs. Ivan Raska, Ulrich Scheer, Ann Beyer, Steve Adam, Daniel Leary, and Jeanne Lawrence for their constructive comments.
Footnotes
Abbreviations used in this paper: DFC, dense fibrillar component; FC, fibrillar center; GC, granular component; Pol I, polymerase I.
References
- Busch, H., and K. Smetana. 1970. The Nucleolus. Academic Press, New York. 626 pp.
- Cmarko, D., P.J. Verschure, L.I. Rothblum, D. Hernandez-Verdun, F. Amalric, R. van Driel, and S. Fakan. 2000. Ultrastructural analysis of nucleolar transcription in cells microinjected with 5-bromo-UTP. Histochem. Cell Biol. 113:181–187. [DOI] [PubMed] [Google Scholar]
- Dundr, M., and I. Raska. 1993. Nonisotopic ultrastructural mapping of transcription sites within the nucleolus. Exp. Cell Res. 208:275–281. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Melendi, P., B. Wells, A.F. Beven, and P.J. Shaw. 2001. Single ribosomal transcription units are linear, compacted Christmas trees in plant nucleoli. Plant J. 27:223–233. [DOI] [PubMed] [Google Scholar]
- Granboulan, N., and P. Granboulan. 1965. Cytochmie Ultrasructurale du nucleole: II etude des sites de synthese du RNA dans le nucleole et le noyau. Exp. Cell Res. 38:605–619. [DOI] [PubMed] [Google Scholar]
- Hadjiolov, A.A. 1985. The nucleolus and ribosome biogenesis. Cell Biology Monographs. Springer-Verlag, New York. 1–263.
- Hozak, P., P.R. Cook, C. Schofer, W. Mosgoller, and F. Wachtler. 1994. Site of transcription of ribosomal RNA and intranucleolar structure in HeLa cells. J. Cell Sci. 107:639–648. [DOI] [PubMed] [Google Scholar]
- Koberna, K., J. Malínsky, A. Pliss, M. Masata, J. Vecerová, M. Fialová, J. Bednár, and I. Raska. 2002. Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees” in situ. J. Cell Biol. 157:743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mais, C., and U. Scheer. 2001. Molecular architecture of the amplified nucleoli of Xenopus oocytes. J. Cell Sci. 114:709–718. [DOI] [PubMed] [Google Scholar]
- Melcak, I., M.C. Risueno, and I. Raska. 1996. Ultrastructural nonisotopic mapping of nucleolar transcription sites in onion protoplasts. J. Struct. Biol. 116:253–263. [DOI] [PubMed] [Google Scholar]
- Scheer, U., and R. Hock. 1999. Structure and function of the nucleolus. Curr. Opin. Cell Biol. 11:385–390. [DOI] [PubMed] [Google Scholar]
- Shaw, P.J., and E.G. Jordan. 1995. The nucleolus. Annu. Rev. Cell Dev. Biol. 11:93–121. [DOI] [PubMed] [Google Scholar]
- Stanek, D., K. Koberna, A. Pliss, J. Malinsky, M. Masata, J. Vecerova, M.C. Risueno, and I. Raska. 2001. Non-isotopic mapping of ribosomal RNA synthesis and processing in the nucleolus. Chromosoma. 110:460–470. [DOI] [PubMed] [Google Scholar]
- Thiry, M., T. Cheutin, M.F. O'Donohue, H. Kaplan, and D. Ploton. 2000. Dynamics and three-dimensional localization of ribosomal RNA within the nucleolus. RNA. 6:1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]