Abstract
Diverse stimuli initiate the activation of apoptotic signaling pathways that often causes nuclear DNA fragmentation. Here, we report a new antiapoptotic protein, a caspase-activated DNase (CAD) inhibitor that interacts with ASK1 (CIIA). CIIA, by binding to apoptosis signal-regulating kinase 1 (ASK1), inhibits oligomerization-induced ASK1 activation. CIIA also associates with CAD and inhibits the nuclease activity of CAD without affecting caspase-3–mediated ICAD cleavage. Overexpressed CIIA reduces H2O2- and tumor necrosis factor-α–induced apoptosis. CIIA antisense oligonucleotides, which abolish expression of endogenous CIIA in murine L929 cells, block the inhibitory effect of CIIA on ASK1 activation, deoxyribonucleic acid fragmentation, and apoptosis. These findings suggest that CIIA is an endogenous antagonist of both ASK1- and CAD-mediated signaling.
Keywords: apoptosis; apoptosis signal-regulating kinase 1; caspase-activated DNase; stress-activated protein kinase; c-Jun NH2-terminal kinase
Introduction
Apoptosis, or programmed cell death, is a fundamental cellular process for the self-elimination of unwanted or damaged cells. Apoptosis occurs during a variety of biological processes in multicellular organisms including development and pathogenesis of many diseases. Apoptotic cells undergo morphological changes such as chromosomal condensation, loss of mitochondrial membrane potential, and formation of apoptotic bodies. These morphological events follow biochemical events that include activation of caspases, changes in plasma membrane composition, and DNA fragmentation (Nagata, 1997; Green and Reed, 1998).
Caspases-3 and -7 have been shown to stimulate the fragmentation of chromosomal DNA through the activation of caspase-activated DNase (CAD), also known as caspase-activated nuclease and DNA fragmentation factor (DFF) 40 (Liu et al., 1997, 1998; Enari et al., 1998; Sakahira et al., 1998). In healthy cells, CAD–caspase-activated nucleus–DFF40 exists as a complex with its inhibitor, ICAD–DFF45. When the apoptotic pathway is activated, caspases cleave ICAD–DFF45, which results in the release of CAD–DFF40 from the complex and stimulation of its nuclease activity (Liu et al., 1997; Sakahira et al., 1998).
The MAPK signaling pathways mediate a variety of cellular events, including cell proliferation, differentiation, and death (Minden and Karin, 1997; Ip and Davis, 1998; Schaeffer and Weber, 1999). The mammalian MAPK subfamilies include extracellular signal-regulated kinase, c-Jun NH2-terminal kinase/stress-activated protein kinase (JNK/SAPK), and p38 (Minden and Karin, 1997; Ip and Davis, 1998; Schaeffer and Weber, 1999). SAPK/JNK and p38 are stimulated in response to a variety of cellular stresses, including UV light, osmotic and heat shock, DNA damaging agents, and proinflammatory cytokines (Derijard et al., 1994; Galcheva-Gargova et al., 1994; Han et al., 1994; Kyriakis et al., 1994). The JNK–SAPK and p38 pathways have been shown to be associated with mechanisms of apoptotic cell death under certain conditions (Xia et al., 1995; Verheij et al., 1996; Minden and Karin, 1997; Ip and Davis, 1998).
Apoptosis signal-regulating kinase 1 (ASK1) is a MAPK kinase kinase that activates the JNK/SAPK and the p38 signaling cascades (Ichijo et al., 1997). ASK1 is shown to be involved in apoptosis, induced by tumor necrosis factor (TNF)-α, Fas, and cellular stresses (Ichijo et al., 1997). Here, we report the identification of an antiapoptotic protein that physically interacts with both ASK1 and CAD. This protein is named a CAD inhibitor that interacts with ASK1 (CIIA). CIIA inhibits stress- or TNF-α–induced ASK1 activation. Furthermore, CIIA inhibits CAD-mediated DNA fragmentation. Thus, our data suggest that CIIA functions as an endogenous antagonist of ASK1 and CAD activities.
Results
Isolation of a cDNA clone of CIIA
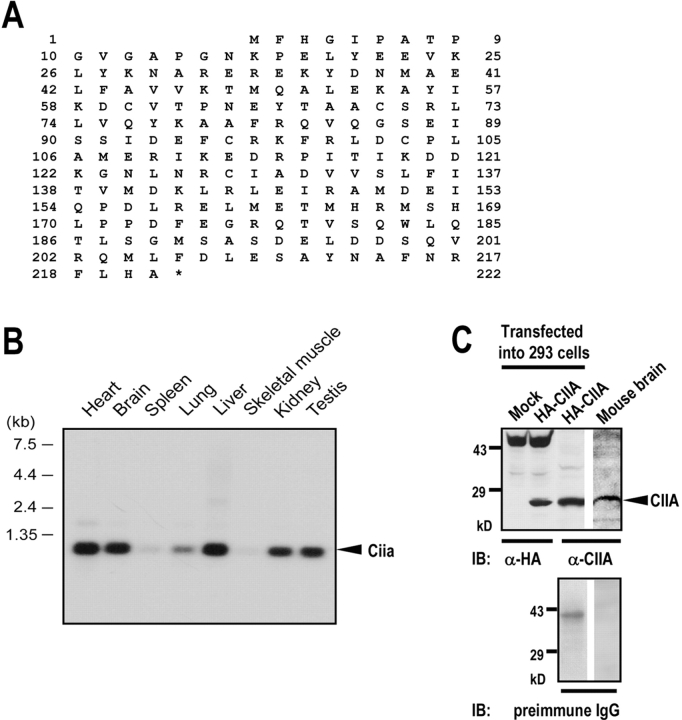
To better understand the mechanism underlying the regulation of ASK1 function, we searched initially for ASK1-binding proteins using the yeast two-hybrid screening method with a mouse adult brain cDNA library. From the two-hybrid study, we identified a new gene that encoded a 221-aa protein (Fig. 1 A). Surprisingly, the same gene was also identified in an independent yeast two-hybrid screening experiment using CAD as the bait. On the basis of the two-hybrid results, we named the protein encoded by this gene CIIA. We also found a human EST clone (GenBank/EMBL/DDBJ accession no. AA432040) by BLAST searches that has a high level of homology with the mouse CIIA gene. CIIA has no known conserved protein motif. A mouse multiple-tissue Northern blot analysis using a mouse CIIA cDNA probe revealed that a transcript of 1 kb was highly expressed in many adult tissues including heart, brain, liver, and kidney (Fig. 1 B). Immunoblot analysis using rabbit polyclonal anti-CIIA antibody identified a 26-kD protein in mouse brain tissue (Fig. 1 C) and in various mammalian cell lines, including murine fibrosarcoma L929, human neuroglioma H4, and human cervical cancer HeLa cells (not depicted). Immunostaining using anti-CIIA antibody showed that CIIA protein was present in both the nucleus and cytoplasm in cultured cells (unpublished data).
Figure 1.
Primary structure of CIIA. (A) Predicted amino acid sequences of the mouse CIIA. The cDNA sequence data of the mouse CIIA are available from GenBank/EMBL/DDBJ under accession no. AF373710. (B) Distribution of the CIIA transcript in mouse tissues. A multiple-tissue Northern blot membrane was probed with 32P-labeled BamHI–AccI fragment (714 bp) of mouse CIIA cDNA. (C) Immunodetection of CIIA protein in adult mouse brain tissue. 50 μg cell lysates were analyzed by immunoblot (IB) using a rabbit preimmune IgG or anti-CIIA antibody. The anti-CIIA antibody was produced from rabbits immunized with hexahistidine (His)-tagged CIIA and affinity-purified. Immunoblots with anti-HA antibody of the lysates from 293T cells transfected with pcDNA3-HA empty vector or pcDNA3-HA-CIIA were also shown.
CIIA physically interacts with ASK1 and CAD
In our initial studies, NIH 3T3 cells stably expressing HA-tagged CIIA (HA-CIIA) were subjected to glycerol gradient fractionation. After cell lysates were subjected to centrifugation at 1,000 g, the resulting soluble fraction was subjected to glycerol gradient centrifugation. Immunoblot analysis of the glycerol gradient fractions revealed that the majority of ASK1 and CIIA were present in the same fractions (Fig. 2, fractions 10–16). CIIA and CAD also resided in the same fractions (Fig. 2, fractions 10–17). These fractionation results suggest that CIIA is in close proximity to ASK1 and to CAD. Next, we transiently transfected NIH 3T3 cells with CIIA plus CAD or CIIA plus ASK1 constructs and examined subcellular distribution of the proteins by immunofluorescence microscopy. Double-labeled immunostaining data revealed that ectopic CIIA was located in both the nucleus and the cytoplasm and that its distribution was overlapped with that of CAD or ASK1 (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200303003/DC1).
Figure 2.
Glycerol gradient analysis of CIIA, ASK1, and CAD proteins in NIH 3T3 cells. Lysates from NIH 3T3 cells stably expressing HA-CIIA were subjected to centrifugation at 1,000 g for 10 min. The resulting soluble fraction was subjected to 15–35% (wt/wt) linear glycerol gradient centrifugation. Equal volumes of the 22 collected fractions were analyzed by SDS-PAGE and immunoblotting with anti-HA, anti-ASK1, anti-CAD, and anti-ICAD antibodies.
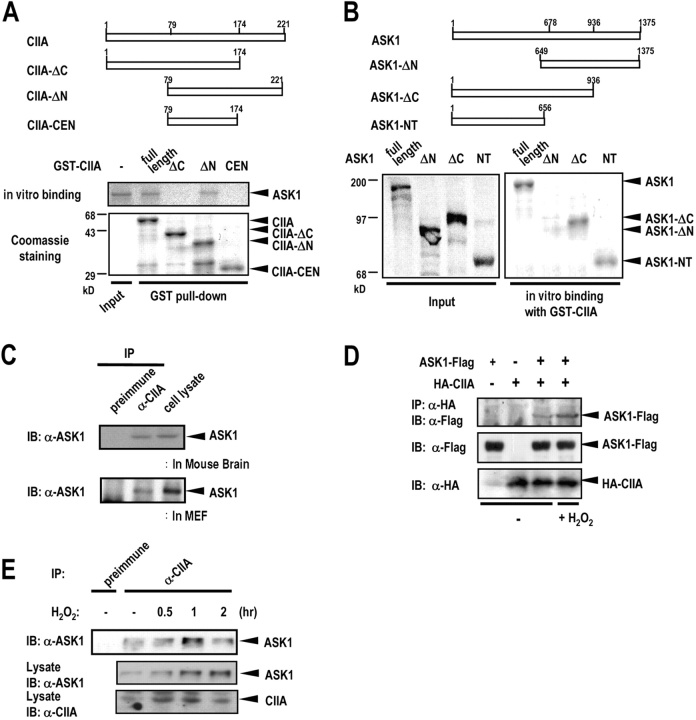
To test whether CIIA interacted directly with ASK1, we performed an in vitro binding study using recombinant GST-fused CIIA variants and in vitro–translated 35S-labeled ASK1 (Fig. 3 A). Both full-length CIIA and CIIA-ΔN associated with ASK1, whereas CIIA-ΔC and CIIA-CEN failed to bind to ASK1 (Fig. 3 A). In separate in vitro binding experiments, GST-CIIA interacted with in vitro–translated 35S-labeled full-length ASK1, ASK1-ΔC, and ASK1-NT, but not with ASK1-ΔN (Fig. 3 B). Thus, these data suggest that CIIA binds the NH2-terminal half region of ASK1. Other ASK1-interacting proteins such as TRAF2, GSTM1, and Daxx have been also shown to bind the NH2-terminal region of ASK1 (Chang et al., 1998; Liu et al., 2000; Cho et al., 2001). Therefore, we examined whether CIIA could affect the binding of ASK1 with TRAF2, GSTM1, or Daxx. A coimmunoprecipitation study revealed that CIIA inhibits the physical interaction of ASK1 with TRAF2, GSTM1, or Daxx (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200303003/DC1).
Figure 3.
CIIA physically interacts with ASK1. (A) 35S-Labeled ASK1 was produced by in vitro translation and incubated at 4°C for 3 h with GST-fused CIIA variants immobilized on glutathione-agarose beads. The bead-bound proteins were eluted and analyzed by SDS-PAGE and autoradiography. A lower part of the polyacrylamide gel was cut and stained with Coomassie brilliant blue to show the amount of GST-fused CIIA variants bound on the beads. (B) Binding of in vitro–translated 35S-labeled ASK1 variants to GST-CIIA was examined as in A. In A and B, the input 35S-labeled proteins (10%) were also shown. (C) The soluble fraction of mouse brain tissue or MEF homogenates was precleared with rabbit preimmune IgG and subjected to immunoprecipitation (IP) with rabbit anti-CIIA antibody, or rabbit preimmune IgG. The resulting precipitates were subjected to SDS-PAGE and analyzed by immunoblotting (IB) with anti-ASK1 antibody. Immunoblotting of cell lysates (5% of total) with anti-ASK1 antibody was also shown. (D) 293T cells were transfected with expression vectors encoding ASK1-Flag and HA-CIIA as indicated. After 48 h of transfection, the cells were untreated or treated with 500 μM H2O2 for 1 h. Cell lysates were subjected to immunoprecipitation with anti-HA antibody, and the resulting immunoprecipitates were subjected to immunoblot analysis with anti-Flag antibody. Cell lysates were also subjected to immunoblot analysis with the indicated antibodies. (E) L929 cells were untreated or treated with 500 μM H2O2 for indicated time periods. Cell lysates were subjected to immunoprecipitation and the resulting immunoprecipitates were analyzed by immunoblotting as in C. Cell lysates (5% of total) were also subjected to immunoblot analysis with anti-ASK1 or anti-CIIA antibody.
Next, we tested a physical interaction between two endogenous CIIA and ASK1 proteins in intact cells by coimmunoprecipitation. Lysates of mouse embryonic fibroblasts (MEFs) or mouse brain tissue were subjected to immunoprecipitation using anti-CIIA antibody, and the resulting immunoprecipitates were analyzed by immunoblotting with anti-ASK1 antibody. Immunoblot data revealed that CIIA physically associates with ASK1 in MEFs and cells from mouse brain (Fig. 3 C). Physical association between CIIA and ASK1 was also confirmed by coimmunoprecipitation in 293T cells transfected with plasmids encoding HA-tagged CIIA and Flag-tagged ASK1 (ASK1-Flag; Fig. 3 D). Interestingly, the interaction between ectopic CIIA and ASK1 was increased by H2O2 treatment. Subsequently, we examined a time course of the H2O2 action on the physical association of endogenous CIIA and ASK1 proteins in L929 cells (Fig. 3 E). Coimmunoprecipitation results indicated that H2O2-induced enhancement of the interaction between the two endogenous proteins was maximal at 1 h.
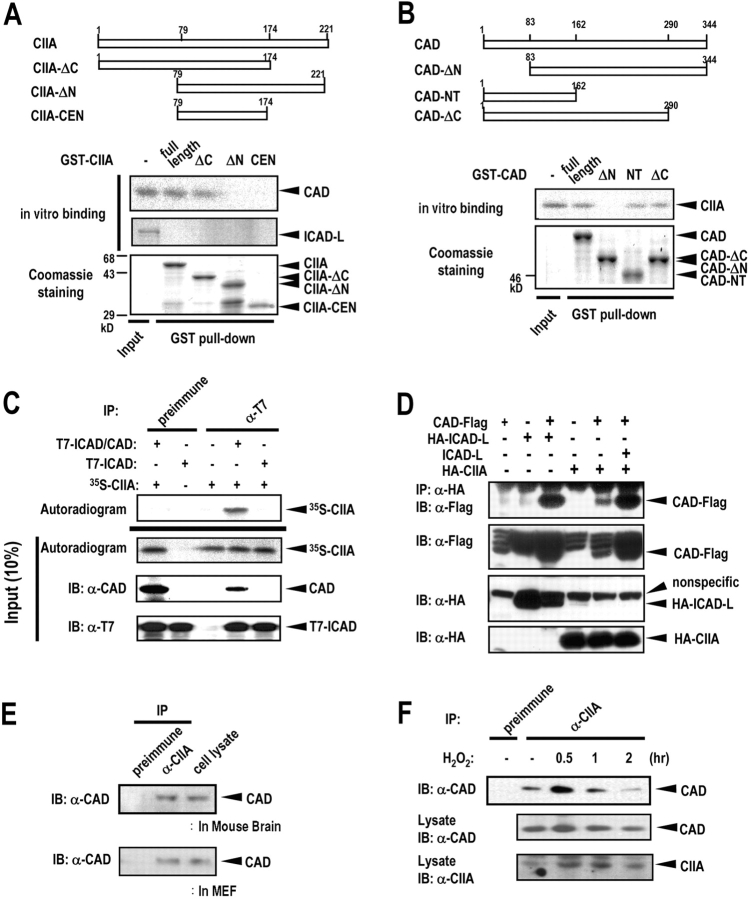
Next, we examined in vitro binding between CIIA and CAD using recombinant GST-CIIA variants and in vitro–translated 35S-Labeled CAD. 35S-labeled CAD bound to CIIA and CIIA-ΔC, but not to CIIA-ΔN or CIIA-CEN (Fig. 4 A). In comparison, 35S-labeled ICAD-L did not interact with CIIA in vitro. We also conducted in vitro binding studies using 35S-labeled CIIA and GST-fused CAD variants. 35S-Labeled CIIA bound to CAD, CAD-NT, and CAD-ΔC, but not to CAD-ΔN (Fig. 4 B). Interestingly, CAD-ΔN lacks amino acid residues 1–83, which is homologous to a NH2-terminal domain of CIDE proteins (the CIDE-N domain; Inohara et al., 1998). It was reported previously that the NH2-terminal domain of CAD is involved in binding to ICAD (Inohara et al., 1999). Therefore, we examined whether in vitro–translated 35S-labeled CIIA can form a tertiary complex with the CAD–ICAD complex. 35S-Labeled CIIA bound to the recombinant CAD–ICAD complex, but not to ICAD (Fig. 4 C).
Figure 4.
CIIA physically interacts with CAD. (A) 35S-Labeled CAD was produced by in vitro translation and applied to GST-fused CIIA variants immobilized on glutathione-agarose beads. The bead-bound proteins were eluted, separated by SDS-PAGE, and autoradiography. The polyacrylamide gel was also stained with Coomassie brilliant blue. (B) 35S-labeled CIIA was produced by in vitro translation and examined for in vitro binding to GST-fused CAD variants as in A. In A and B, the input 35S-labeled proteins (10%) were also shown. (C) Recombinant T7-ICAD–CAD complex or T7-ICAD alone was incubated at 4°C for 3 h with in vitro–translated 35S-labeled CIIA, and immunoprecipitated with mouse preimmune IgG or mouse monoclonal anti-T7 antibody. The resulting precipitates were analyzed by SDS-PAGE and autoradiography. The input (10%) of 35S-labeled CIIA, CAD, and T7-ICAD was also shown. (D) 293T cells were transfected for 48 h with indicated combinations of plasmids encoding CAD-Flag, HA-ICAD-L, ICAD-L, and HA-CIIA. Cell lysates were subjected to immunoprecipitation with anti-HA antibody, and the resulting HA immunoprecipitates were subjected to immunoblot analysis with anti-Flag antibody. Cell lysates (10% of total) were also subjected to immunoblot analysis with indicated antibodies. (E) Physical interaction between endogenous CIIA and CAD. The lysates of mouse brain tissue and MEFs were subjected to immunoprecipitation with rabbit preimmune IgG or anti-CIIA antibody, and the resulting precipitates were subjected to immunoblot analysis with rabbit anti-CAD antibody. Immunoblotting of the cell lysates (5% of total) was also shown. (F) L929 cells were untreated or treated with 500 μM H2O2 for indicated time periods. Cell lysates were subjected to immunoprecipitation, and the immunoprecipitates were subjected to immunoblot analysis as in E. Cell lysates (5% of total) were also subjected to immunoblot analysis with anti-CAD or anti-CIIA antibody.
Next, we examined the physical interaction between CIIA and CAD in 293T cells expressing HA-CIIA and CAD-Flag (Fig. 4 D). Immunoblot analysis of the HA immunoprecipitates revealed a physical association of CIIA with CAD in the cotransfected cells. We also examined whether ICAD could inhibit the interaction between CAD and CIIA. Coexpression of ICAD-L did not interfere with the interaction between HA-CIIA and CAD-Flag (Fig. 4 D). In fact, coexpression of ICAD-L resulted in an increase in the amount of CAD-Flag coimmunoprecipitated with HA-CIIA, presumably because of an increase in the expression of functional CAD-Flag. ICAD-L has been shown not only to inhibit CAD activity but also to enhance the expression of functional CAD (Enari et al., 1998; Sakahira et al., 1999). Physical association of two endogenous CIIA and CAD proteins was also confirmed in MEFs and cells from mouse brain by coimmunoprecipitation (Fig. 4 E). Next, we examined a possible effect of H2O2 on the physical interaction between endogenous CIIA and CAD in L929 cells. L929 cells were unexposed or exposed to H2O2 for various times, and cell lysates were analyzed by coimmunoprecipitation. Physical interaction between CIIA and CAD was maximal after 30 min of an exposure of L929 cells to H2O2 (Fig. 4 F).
CIIA inhibits the kinase activity of ASK1
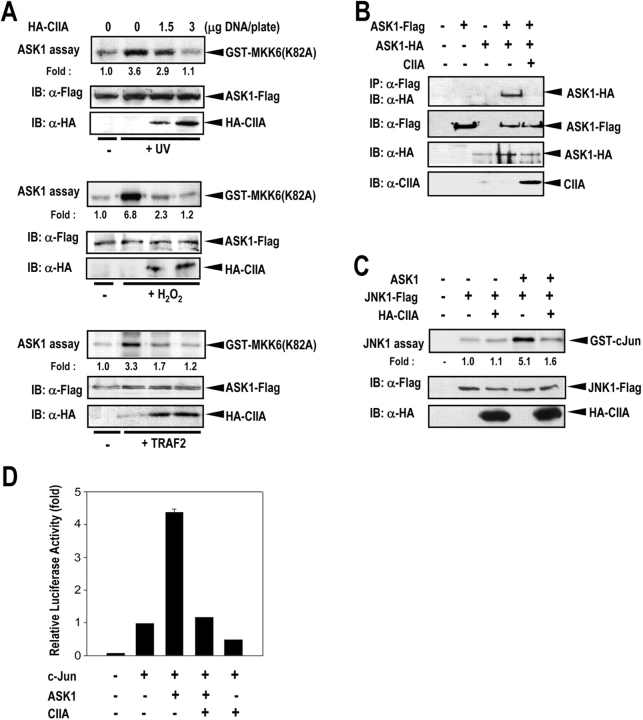
Next, we investigated whether CIIA could affect the kinase activity of ASK1. Ectopic ASK1 was stimulated by an exposure of the transfected cells to UV light or H2O2 or by coexpression of TRAF2 (Fig. 5 A). Expression of CIIA suppressed UV-, H2O2-, and TRAF2-stimulated ASK1 activities in the cells. In comparison, CIIA neither bound to nor inhibited MAPK/extracellular signal–regulated kinase kinase kinase 1 (MEKK1), another MAPK kinase kinase that stimulates the JNK–SAPK pathway (Figs. S3 and S4, available at http://www.jcb.org/cgi/content/full/jcb.200303003/DC1).
Figure 5.
CIIA inhibits ASK1 activation. (A) CIIA inhibits ASK1 activation in transfected cells. 293T cells were transfected for 48 h with the combinations of ASK1-Flag, HA-CIIA, and TRAF2 constructs as indicated. Where noted, cells were treated with 60 J/m2 UV light or 500 μM H2O2 and incubated further for 20 min. Cell lysates were subjected to immunoprecipitation with anti-Flag antibody, and the resulting precipitates were examined for ASK1 activity by the immunocomplex kinase assay with GST-MKK6(K82A) as the substrate. Cell lysates were also examined by immunoblot analysis with the indicated antibodies. (B) CIIA inhibits ASK1 homo-oligomerization. 293T cells were transfected for 48 h with expression vectors encoding ASK1-Flag, HA-ASK1, and CIIA, as indicated. Cell lysates were subjected to immunoprecipitation with anti-Flag antibody, and the resulting Flag immunoprecipitates were subjected to immunoblot analysis with anti-HA antibody. Cell lysates were also immunoblotted with the indicated antibodies. (C) CIIA inhibits ASK1-dependent JNK activation. 293T cells were transfected for 48 h with expression vectors for JNK1-Flag, ASK1, or HA-CIIA as indicated. Cell lysates were subjected to immunoprecipitation with anti-Flag antibody, and the resulting Flag immunoprecipitates were examined for JNK1 activity by the immunocomplex kinase assay. (D) CIIA inhibits the ASK1-induced stimulation of the transcriptional activity of c-Jun. 293T cells were transfected with pFR-Luc and pcDNA3-β-gal and the indicated combinations of plasmids for c-Jun, ASK1, and CIIA. After 48 h of transfection, the cells were lysed and the soluble fraction of cell lysates was assayed for both luciferase and β-galactosidase activities. Luciferase activity was normalized to β-galactosidase activity. Data are the mean of triplicate determinations ± SEM.
Homo-oligomerization of ASK1 is one mechanism for ASK1 activation (Gotoh and Cooper, 1998; Liu et al., 2000). Therefore, we examined whether CIIA interferes with ASK1 oligomerization (Fig. 5 B). 293T cells were transfected with ASK1-Flag and ASK1-HA constructs in the absence or presence of the CIIA construct. Coimmunoprecipitation analysis indicated that ASK1-HA was found to be associated with ASK1-Flag in the transfected cells. The ASK1 homo-oligomerization was inhibited by coexpression of CIIA. These results suggest that CIIA may inhibit ASK1 activation, at least in part, by the suppression of ASK1 homo-oligomerization.
Next, we examined the effect of CIIA on the signaling events downstream of ASK1. ASK1 activation can induce stimulation of JNK, which in turn enhances the transcription-stimulating activity of c-Jun (Ichijo et al., 1997; Ip and Davis, 1998). Overexpressed ASK1-induced JNK1 activation and this ASK1-dependent JNK1 activation was blocked by CIIA expression in transfected 293T cells (Fig. 5 C). CIIA also inhibited the ASK1-induced stimulation of the transcription-stimulating activity of c-Jun (Fig. 5 D). CIIA did not interact physically with JNK1 or c-Jun (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200303003/DC1).
CIIA inhibits the DNase activity of CAD
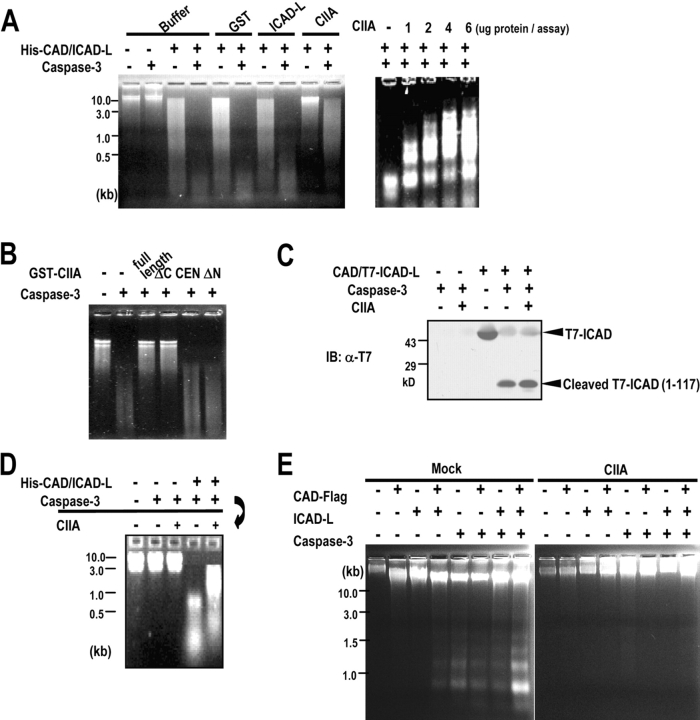
Next, we investigated whether CIIA could modulate the DNase activity of CAD. An in vitro CAD assay showed that treatment of a recombinant CAD–ICAD-L complex with caspase-3 resulted in the stimulation of the nuclease activity of CAD (Fig. 6 A). CIIA, in the form of a GST fusion protein, inhibited the caspase-3–induced stimulation of CAD activity, whereas GST alone or GST-ICAD-L did not. Caspase-3–dependent CAD activation was also inhibited by CIIA-ΔC, but not by CIIA-ΔN or CIIA-CEN (Fig. 6 B). To further understand the mechanism underlying the inhibitory action of CIIA on CAD activation, we examined whether CIIA could block a caspase-3–catalyzed cleavage of ICAD. In vitro cleavage results indicated that CIIA did not inhibit a cleavage of ICAD by caspase-3 (Fig. 6 C). Next, recombinant CAD–ICAD complex was pretreated with caspase-3 and examined for the nuclease activity in the absence or presence of CIIA. CIIA inhibited DNA fragmentation mediated by caspase-3–pretreated CAD–ICAD complex (Fig. 6 D). Collectively, these results suggest that CIIA blocks the CAD-dependent DNA fragmentation through an inhibition of the DNase activity of CAD.
Figure 6.
CIIA inhibits the nuclease activity of CAD. (A) In vitro nuclease activity of CAD was examined by incubating 1 μg His-CAD–ICAD-L complex and 5 μg Jurkat chromosomal DNA with 2 μg GST, 2 μg GST-ICAD-L, or 2 μg in left panel or the indicated amounts in right panel of GST-CIIA in the absence or presence of caspase-3. (B) In vitro nuclease activity of CAD was examined in the presence of 2 μg GST-fused CIIA variants as in A. (C) 1 μg CAD–T7-ICAD complex was treated with 200 ng recombinant caspase-3 in the absence or presence of 4 μg GST-CIIA for 2 h at 37°C. The reaction mixture was analyzed by SDS-PAGE and immunoblotting with mouse anti-T7 mAb. (D) 1 μg His-CAD–ICAD-L complex was pretreated with 200 ng caspase-3 for 2 h at 37°C, where indicated, and examined for the nuclease activity of CAD in the absence or presence of 2 μg GST-CIIA. (E) Overexpressed CIIA inhibits the nuclease activity of CAD in 293T cells. 293T cells were transfected for 48 h with indicated combinations of expression vectors encoding CAD-Flag, ICAD-L, constitutively active caspase-3, and HA-CIIA. Chromosomal DNA from the transfected cells was examined for DNA fragmentation (Liu et al., 1998).
Next, the effect of CIIA on CAD-mediated DNA fragmentation was examined in 293T cells. The cells were transfected with various combinations of plasmids encoding CAD-Flag, ICAD-L, a constitutively active caspase-3, and HA-CIIA (Fig. 6 E). Expression of CAD and ICAD-L with the caspase-3 enhanced DNA fragmentation in the transfected cells. Coexpression of CIIA inhibited the caspase-3/CAD-dependent DNA fragmentation. Together, our data suggest that CIIA suppresses the DNase activity of CAD.
CIIA reduces TNF-α– and H2O2-induced apoptosis
To further assess the function of CIIA, HA-CIIA construct was stably transfected into 293T cells, in which a level of endogenous CIIA was quite low (unpublished data). Expression of HA-CIIA did not affect endogenous levels of ASK1, CAD, or ICAD in 293 cells (unpublished data). TNF-α treatment of 293-neo control cells resulted in ASK1 activation, however, TNF-α–induced ASK1 activation was impaired in the cells expressing HA-CIIA (293-CIIA cells; Fig. 7 A). CIIA overexpression also inhibited TNF-α–induced activation of JNK1, a downstream kinase of ASK1. Furthermore, TNF-α–induced DNA fragmentation was lowered in 293-CIIA cells, compared with that of 293-neo cells (Fig. 7 B). ASK1 plays a crucial role in the mechanisms of TNF-α– and stress-induced apoptosis through the activation of stress-activated MAPKs, including JNK/SAPK (Ichijo et al., 1997), and the mitochondria-dependent activation of caspases (Hatai et al., 2000). Therefore, we examined the effect of CIIA on apoptotic cell death induced by TNF-α or H2O2 (Fig. 7 C). 293-CIIA cells were more resistant to both TNF-α– and H2O2-induced apoptosis, compared with 293-neo cells. CIIA also protected 293-CIIA cells against apoptosis induced by Daxx, an activator of ASK1 (Chang et al., 1998; unpublished data).
Figure 7.
CIIA inhibits apoptosis. (A) CIIA inhibits TNF-α–stimulated ASK1 activity in 293-CIIA cells. 293T cells were stably transfected with pcDNA3 empty vector or pcDNA3-HA-CIIA, yielding 293-neo control or 293-CIIA cells, respectively. Heterogeneous populations of the stably transfected cells were used to avoid clonal variations. The cells were untreated or treated with 20 ng/ml TNF-α for 20 min. Cell lysates were subjected to immunoprecipitation with anti-ASK1 or anti-JNK1 antibodies, and the resulting immunoprecipitates were assayed for the kinase activities of endogenous ASK1 or JNK1 by immunocomplex kinase assay. Cell lysates were also examined by immunoblot analysis with anti-ASK1, anti-JNK1, or anti-HA antibody. (B) CIIA decreases TNF-α–induced DNA fragmentation in 293-CIIA cells. 293-neo and 293-CIIA cells were treated with 20 ng/ml TNF-α plus 1 μg/ml actinomycin D for the indicated time periods. Chromosomal DNA was obtained from the cells and examined for DNA fragmentation. (C) CIIA suppresses TNF-α– and H2O2-induced apoptosis. 293-neo and 293-CIIA cells were exposed to 20 ng/ml TNF-α plus 1 μg/ml actinomycin D (TNF-α/ActD) or 500 μM H2O2, incubated overnight, and assayed for apoptotic cell death by flow cytometry with annexin V staining. Data are mean ± SEM from three experiments. (D and E) 293T cells were transfected with plasmids encoding (D) HA-ASK1 or (E) HA-JNK1 alone or together with full-length CIIA, CIIA-ΔN, or CIIA-ΔC. After 48 h of transfection, the cells were untreated or treated with 500 μM H2O2 or 20 ng/ml TNF-α for 20 min. Cell lysates were assayed for (D) ASK1 or (E) JNK1 activity by immunocomplex kinase assay with the use of anti-HA antibody. Cell lysates were also examined by immunoblot analysis with anti-HA and anti-Flag antibodies. (F) 293T cells were transiently transfected for 50 h with plasmids encoding full-length CIIA, CIIA-ΔN, or CIIA-ΔC. The cells were untreated or treated with 20 ng/ml TNF-α plus 1 μg/ml actinomycin D (TNF-α/ActD) or 500 μM H2O2, further incubated overnight, and assayed for apoptosis by DAPI staining. The data are presented as the mean ± SEM. The statistical significance is designated by an asterisk. P < 0.01 versus control (mock transfection).
To further investigate the inhibitory effect of CIIA on apoptosis, we transiently transfected 293T cells with expression vectors for full-length CIIA, CIIA-ΔN, and CIIA-ΔC, and measured TNF-α– and H2O2-induced apoptosis as well as ASK1 and JNK1 activation in the transfected cells. CIIA-ΔN is a CIIA mutant that interacted with ASK1 (Fig. 3 A), but not with CAD (Fig. 4 A). CIIA-ΔC is a CIIA mutant that bound and inhibited CAD (Figs. 4 A and 6 B), but it did not interact with ASK1 (Fig. 3 A). CIIA and CIIA-ΔN inhibited the TNF-α– and H2O2-stimulated activities of ASK1 (Fig. 7 D) and JNK1 (Fig. 7 E), whereas CIIA-ΔC had little effect, if any, on the TNF-α– and H2O2-stimulated activities of ASK1 (Fig. 7 D) and JNK1 (Fig. 7 E). CIIA and CIIA-ΔN inhibited TNF-α– and H2O2-induced apoptosis in the transfected cells (Fig. 7 F). In contrast, the effect of CIIA-ΔC was marginal. Together, these results suggest that the inhibition of ASK1 activity by CIIA is crucial for the antiapoptotic function of CIIA.
CIIA antisense oligonucleotides prevent CIIA from inhibiting ASK1 activation and DNA fragmentation
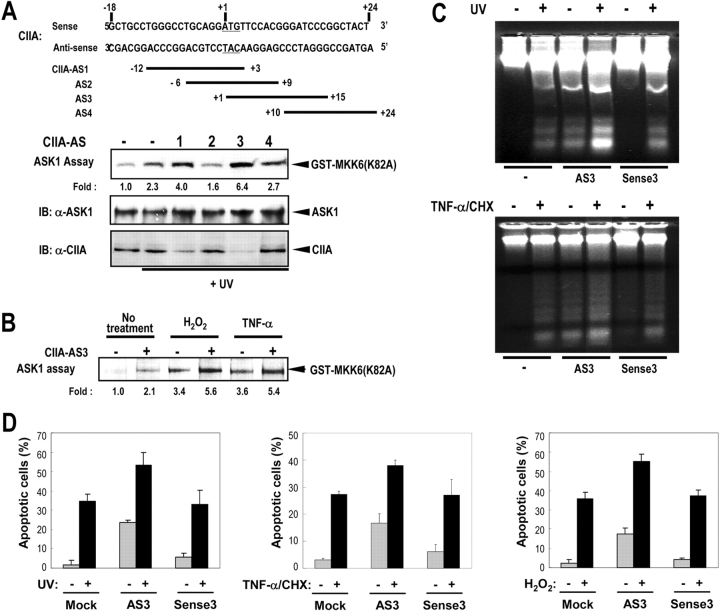
To test the role of endogenous CIIA protein in stress-induced ASK1 activation and DNA fragmentation, CIIA antisense oligonucleotides were transfected into L929 cells (Fig. 8 A). Among the antisense oligonucleotides used, CIIA-AS3 most effectively blocked the expression of endogenous CIIA in L929 cells. Inhibition of CIIA expression by CIIA-AS3 resulted in an increase in the UV-stimulated activity of endogenous ASK1 in L929 cells. Transfection of L929 cells with antisense oligonucleotides CIIA-AS3 also resulted in an increase in the H2O2- and TNF-α–stimulated activity of endogenous ASK1 (Fig. 8 B). Moreover, treatment of cells with CIIA-AS3 markedly enhanced DNA fragmentation and apoptosis even in the absence of any apoptotic stimuli (Fig. 8, C and D). Either UV- or TNF-α–induced DNA fragmentation also increased in L929 cells transfected with CIIA-AS3, compared with the mock-transfected cells or cells transfected with the corresponding sense oligonucleotides (Fig. 8 C). Furthermore, transfection of the cells with CIIA-AS3 enhanced UV-, TNF-α–, and H2O2-induced apoptosis, compared with the mock-transfected cells or cells transfected with the sense oligonucleotides (Fig. 8 D). Collectively, these data suggest that CIIA functions as a natural antagonist against ASK1-mediated signaling and DNA fragmentation.
Figure 8.
CIIA antisense oligonucleotides block the inhibitory effect of CIIA on ASK1 activation and DNA fragmentation. (A) CIIA antisense oligonucleotides block the expression of endogenous CIIA and enhance UV-induced ASK1 activation in L929 cells. L929 cells were transfected for 48 h with CIIA antisense oligonucleotides (CIIA-AS1 to 4). The cells were treated with 80 J/m2 UV light and incubated further for 20 min. The cell lysates were immunoblotted to monitor the intracellular levels of CIIA and ASK1, and also examined for ASK1 activity by immunocomplex kinase assay using anti-ASK1 antibody. (B) CIIA-AS3 enhances ASK1 activity stimulated by H2O2 and TNF-α in L929 cells. L929 cells were transfected with CIIA-AS3 (+) or its complementary sense oligonucleotides (−). After 48 h of transfection, the cells were untreated or treated with 500 μM H2O2 or 20 ng/ml TNF-α for 20 min. The cell lysates were assayed for ASK1 activity as in A. (C and D) CIIA antisense oligonucleotides enhance DNA fragmentation and apoptosis. L929 cells were mock-transfected or transfected with CIIA-AS3 (AS3) or its complementary sense oligonucleotides (Sense3). After 48 h of transfection, the cells were untreated (−) or treated (+) with 80 J/m2 UV light, 20 ng/ml TNF-α plus 10 μg/ml cycloheximide (TNF/CHX), or 500 μM H2O2 and incubated overnight. The cells were then examined for (C) DNA fragmentation or for (D) apoptosis by DAPI staining. Data are the mean of triplicate determinations ± SEM. The data represent results from three independent experiments.
Discussion
Here, we identified a new protein, CIIA, which functions as an antagonist of both ASK1 and CAD activities. ASK1 has been shown to function in apoptosis triggered by a variety of cellular stresses and proinflammatory cytokines (Ichijo et al., 1997; Chang et al., 1998; Chen et al., 1999). Oligomerization of ASK1 is shown to be associated with the mechanism for ASK1 activation (Gotoh and Cooper, 1998; Nishitoh et al., 1998; Hoeflich et al., 1999; Liu et al., 2000). Our results suggest that CIIA, by binding ASK1, interferes with ASK1 oligomerization and blocks ASK1 activation. Interestingly, other ASK1 inhibitors, including thioredoxin and GSTμ, also block ASK1 oligomerization (Saitoh et al., 1998; Cho et al., 2001), suggesting that the mitigation of ASK1 oligomerization may be a general mechanism underlying the negative regulation of ASK1. It is also noteworthy that ASK1 does not phosphorylate CIIA (unpublished data).
The NH2-terminal region of CAD contains a CIDE-N domain, which is conserved in ICAD/DFF45 and CIDE-B (Inohara et al., 1998; Sakahira et al., 1998). This domain of CAD is essential for interaction with ICAD-L, and thus, for regulation of the nuclease activity of CAD (McCarty et al., 1999a,b; Sakahira et al., 1999). Our data show that CIIA associates with the CIDE-N domain of CAD. Interestingly, CIIA does not associate with ICAD-L even though ICAD-L also contains a CIDE-N domain. Furthermore, CIIA can associate with CAD regardless of the presence of ICAD-L. The physical association of CIIA with CAD leads to the inhibition of the nuclease activity of CAD.
Upon exposure of cells to a variety of apoptotic stimuli, CIIA appears to suppress ASK1 activation and CAD-mediated DNA fragmentation. In this regard, it was reported recently that ASK1 activation enhances cytochrome c release from the mitochondria into the cytoplasm, as well as the subsequent activation of caspase-9 and downstream caspases (Hatai et al., 2000). The activated caspase cascade may stimulate downstream apoptotic pathways including CAD-mediated DNA fragmentation. Thus, CIIA may antagonize the ASK1-mediated apoptotic pathway with high efficiency by inhibiting ASK1 activation at an early stage of apoptosis and induction of ASK1-induced DNA fragmentation at a later stage.
On the basis of our findings, we propose that CIIA functions as a natural inhibitor of both ASK1 and CAD. The dual function of CIIA may constitute an integral part of the mechanism by which the apoptotic pathways in the cytoplasm and in the nucleus are controlled. In addition, while the present paper was in preparation, a human counterpart of mouse CIIA was reported. This human protein, named hVPS28, interacts with the Tsg101 protein and appears to be involved in endosomal sorting (Bishop and Woodman, 2001). Tsg101, originally discovered as a tumor susceptibility gene, has been implicated in transcriptional regulation and neoplastic transformation (Li and Cohen, 1996). Therefore, it is tempting to propose that CIIA may be a multifaceted regulator that associates with intracellular signaling networks for apoptosis, cellular stress, and tumorigenesis.
Materials and methods
Antibodies
Rabbit anti-ASK1 and anti-hexahistidine (His) pAbs were purchased from Santa Cruz Biotechnology. Rabbit anti-CAD and anti-ICAD pAbs were purchased from Oncogene Research Products. Affinity-purified rabbit anti-CIIA pAb was produced from rabbits immunized with His-CIIA protein. Mouse anti-HA, anti-Flag, anti-Myc, and anti-H7 mAbs were purchased from Roche Molecular Biochemicals, Stratagene, Cell Signaling Biotechnology, and Novagen, respectively.
DNA constructs
ASK1 deletion mutants, ASK-ΔN, ASK1-ΔC, and ASK1-NT, were generated by PCR, and subcloned into pcDNA3 vector (Invitrogen; Cho et al., 2001). Daxx(498–740) was a gift from S.H. Kim (Sungkyunkwan University, Suwon, Korea). JNK1, TRAF2, and caspase-3 cDNA clones were from R.J. Davis (University of Massachusetts, Worchester, MA), D.V. Goeddel Tularik Inc., South San Francisco, CA), and Dr. Y.K. Chung (Kwangju Institute of Science and Technology, Kwangju, Korea), respectively. CAD and ICAD-L cDNAs were obtained by RT-PCR (Park et al., 2000). The cDNA of the mouse CIIA gene was obtained from the screening of a mouse adult brain cDNA library (CLONTECH Laboratories, Inc.) and 5′-RACE. Human CIIA cDNA was obtained from human fetal brain total RNA (CLONTECH Laboratories, Inc.) by RT-PCR using primers CIIA-H1 (5′-CCCAGAGCCTAGAGGATGTTTCATGG-3′) and CIIA-H2 (5′-CCGGGCTCAGGCATGCAGGAAGCG-3′), whose nucleotide sequences were determined from the human cDNA EST clones (zw 80e08.s1; zw 80e08.r1).
Yeast two-hybrid screening
Yeast two-hybrid screening was performed according to the manufacturer's protocol (CLONTECH Laboratories, Inc.). In brief, a full-length cDNA of either ASK1 or CAD was inserted adjacent to the LexA DNA-binding domain in the pLexA bait vector. About 2 × 106 clones of a mouse adult brain cDNA library (CLONTECH Laboratories, Inc.) were screened using Saccharomyces cerevisiae EGY48[p8op-lacZ]. Positive clones were rescued from yeast cotransformants using Escherichia coli KC8 cells, and the cDNA inserts in the rescued plasmids were sequenced.
Isolation of the mouse CIIA cDNA
A 714-bp fragment of CIIA cDNA that had been isolated from the yeast two-hybrid screening using ASK1 as the bait was used as a probe to isolate cDNA clones from a mouse brain Lambda ZAPII cDNA library (Stratagene). Plaque hybridization was performed at 42°C for 12 h in 5× SSPE, 0.1% SDS, 5× Denhardt's solution, 50% formamide, and 100 μg/ml denatured salmon sperm DNA. Positive cDNA inserts were in vivo excised, recovered in a pBluescript SK(−) plasmid, and sequenced. To obtain the 5′ region of mouse CIIA, 5′-RACE was performed using mouse brain total RNA (CLONTECH Laboratories, Inc.) and a 5′-RACE kit (Roche Molecular Biochemicals). The gene-specific antisense primer sequences used for 5′-RACE were 5′-GGGAGCGGGAGAAGTATGACAACATGG-3′ and 5′-GGATGTTCCACGGGATCCCGGCTAC-3′.
Northern blot analysis
A 714-bp fragment of mouse CIIA cDNA excised by BamHI and AccI was labeled with α-[32P]ATP by a random priming method and hybridized with a mouse multiple-tissue mRNA blot (CLONTECH Laboratories, Inc.).
Cell culture, transfection, and apoptotic cell death
293T, L929, and HeLa cells were cultured in DME supplemented with 10% FBS. DNA transfections were performed with the LipofecAMINETM (GIBCO BRL), GenePorter 2 (Gene Therapy Systems, Inc.), calcium phosphate, or electroporation method. Apoptotic cell death was measured by flow cytometry (Facs®Calibur; Becton Dickinson) with annexin V staining or by DAPI staining. For annexin V staining, cultured cells were resuspended in binding buffer (10 mM Hepes/NaOH, pH 7.4, 140 mM NaCl, and 2.5 mM CaCl2) and stained with FITC–annexin V and propidium iodide. Apoptotic cells (Annexin V–FITC positive, propidium iodide negative) were analyzed by flow cytometry (Facs®Calibur; Becton Dickinson).
For DAPI staining, cultured cells were transfected with pEGFP (CLONTECH Laboratories, Inc.) and expression vectors for the indicated proteins. After transfection, the cells were washed twice with PBS solution. Next, the cells were fixed with 0.25% glutaraldehyde, permeabilized with 0.1% Triton X-100, and stained with DAPI. The DAPI-stained nuclei in GFP-positive cells were examined for apoptotic morphology by fluorescence microscopy. The percentage of GFP-expressing cells that were apoptotic was determined from three independent dishes.
Coimmunoprecipitation analysis
Cells were lysed in buffer A that contained 20 mM Tris-HCl, pH 7.4, 150 mM sodium chloride, 1% Triton X-100, 1% deoxycholate, 12 mM β-glycerphosphate, 10 mM sodium fluoride, 5 mM EGTA, and 1 mM PMSF. The cell lysates were subjected to immunoprecipitation using the appropriate antibodies. The resulting immunoprecipitates were analyzed by SDS-PAGE and immunoblotting with the use of an ECL detection method (Amersham Biosciences).
Immunocomplex kinase assays
Cell lysates were subjected to immunoprecipitation using the proper antibody, and the resulting immunopellets were assayed for the indicated protein kinases as described previously (Park et al., 2001). Phosphorylated substrates were resolved by SDS-PAGE, and phosphorylation was quantified using a phosphoimager (model BAS2500; Fuji). GST-MKK6(K82A) and GST-c-Jun(1–79) were used as substrates for ASK1 and JNK/SAPK.
In vitro binding assay
CIIA, ASK1, CAD, or their variants were in vitro translated in the presence of [35S]methionine using the TNT reticulocyte lysate system (Promega). The 35S-labeled proteins were incubated at 4°C for 3 h with GST-fused proteins immobilized on glutathione-Sepharose beads or with T7-tagged proteins immobilized on anti-T7 antibody/protein G–Sepharose beads in a buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1 mM DTT, 0.1% NP-40, and 5 mg/ml BSA. The bound 35S-labeled proteins were eluted from the beads and analyzed by SDS-PAGE and autoradiography.
Luciferase reporter assay of c-Jun–dependent transcription
The transcription-stimulating activity of c-Jun was measured with the PathDetect luciferase reporter kit (Stratagene). 293T cells were transfected for 48 h with luciferase reporter plasmid pFR-Luc, pFA2-c-Jun, and pcDNA3-β-gal and the indicated combinations of plasmids for ASK1 and CIIA. The soluble fraction of the cell lysates was assayed for luciferase activity using a luciferase assay kit (Promega) and for β-galactosidase activity. The luciferase activities in the transfected cells were normalized with reference to the β-galactosidase activities in the same cells.
CAD and DNA fragmentation assays
A His-tagged CAD–ICAD-L complex was bacterially expressed using pET23b (Novagen) and purified with Ni2+-NTA–agarose (QIAGEN). 1 μg of the His-CAD–ICAD-L complex protein was incubated for 2 h at 37°C with 2 μg GST-CIIA or its various deletion mutants in the absence or presence of 200 ng of recombinant caspase-3 in 50 μl of a nuclease reaction buffer containing 10 mM Hepes, pH 7.5, 1 mM EGTA, 5 mM MgCl2, 50 mM NaCl, 1 mg/ml BSA, and 0.1 mg/ml chromosomal DNA extracted from Jurkat cells (Halenbeck et al., 1998). CAD-mediated DNA fragmentation was analyzed by electrophoresis on a 2% agarose gel and staining with ethidium bromide. DNA fragmentation in 293T cells was measured as described previously (Liu et al., 1998) after 48 h of transfection with expression vectors encoding CIIA, CAD, ICAD-L, and prodomain-deleted active caspase-3 (Srinivasula et al., 1998).
Glycerol gradient centrifugation
NIH 3T3 cells stably expressing HA-CIIA were homogenized using a Dounce homogenizer in PBS solution containing 1 mM PMSF, 2 μg/ml leupeptin, and 2 μg/ml aprotinin. Cell extracts were subjected to centrifugation at 1,000 g for 10 min, and the resulting soluble fraction was layered on the top of linear 15–35% (wt/wt) glycerol gradient adjusted to 20 mM Tris-HCl, pH 6.7, 150 mM MgCl2, and 10 mM KCl. Centrifugation was performed at 39,000 rpm for 18 h at 4°C using a rotor (model SW40Ti; Beckman Coulter). 22 fractions of the soluble fraction were collected sequentially from the bottom and equal volumes were analyzed by SDS-PAGE and immunoblotting with the use of anti-HA, anti-ASK1, anti-CAD, and anti-ICAD antibodies.
Online supplemental material
Fig. S1 shows subcellular distribution of ectopic CIIA, ASK1, and CAD in NIH 3T3 cells. Fig. S2 shows the effect of CIIA on the binding of ASK1 with TRAF2, GSTM1, or Daxx. Fig. S3 shows coimmunoprecipitation data indicating that CIIA does not bind JNK1, MEKK1, or c-Jun. Fig. S4 shows an effect of CIIA on MEKK1 activity. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200303003/DC1.
Acknowledgments
We thank Dr. G. Hoschek for critical reading of the manuscript.
This work was supported by the Creative Research Initiatives Program of the Korean Ministry of Science and Technology (to E.-J. Choi).
S.-G. Cho, J.W. Kim, and Y.H. Lee contributed equally to this work.
The online version of this paper contains supplemental material.
Abbreviations used in this paper: ASK1, apoptosis signal-regulating kinase 1; CAD, caspase-activated DNase; CIIA, CAD inhibitor that interacts with ASK1; DFF, DNA fragmentation factor; His, hexahistidine; JNK, c-Jun NH2-terminal kinase; MEF, mouse embryonic fibroblast; MEKK1, MAPK/extracellular signal–regulated kinase kinase kinase 1; SAPK, stress-activated protein kinase.
References
- Bishop, N., and P. Woodman. 2001. Tsg101/mammalian vps23 and mammalian vps28 interact directly and are recruited to vps4-induced endosomes. J. Biol. Chem. 276:11735–11742. [DOI] [PubMed] [Google Scholar]
- Chang, H.Y., H. Nishitoh, X. Yang, H. Ichijo, and D. Baltimore. 1998. Activation of apoptosis signal-regulating kinase 1 (ASK1) by the adapter protein Daxx. Science. 281:1860–1863. [DOI] [PubMed] [Google Scholar]
- Chen, Z., H. Seimiya, M. Naito, T. Mashima, A. Kizaki, S. Dan, M. Imaizumi, H. Ichijo, K. Miyazono, and T. Tsuruo. 1999. ASK1 mediates apoptotic cell death induced by genotoxic stress. Oncogene. 18:173–180. [DOI] [PubMed] [Google Scholar]
- Cho, S.G., Y.H. Lee, H.S. Park, K. Ryoo, K.W. Kang, J. Park, S.J. Eom, M.J. Kim, T.S. Chang, S.Y. Choi, et al. 2001. Glutathione S-transferase mu modulates the stress-activated signals by suppressing apoptosis signal-regulating kinase 1. J. Biol. Chem. 276:12749–12755. [DOI] [PubMed] [Google Scholar]
- Derijard, B., M. Hibi, I.H. Wu, T. Barrett, B. Su, T. Deng, M. Karin, and R.J. Davis. 1994. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 76:1025–1037. [DOI] [PubMed] [Google Scholar]
- Enari, M., H. Sakahira, H. Yokoyama, K. Okawa, A. Iwamatsu, and S. Nagata. 1998. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 391:43–50. [DOI] [PubMed] [Google Scholar]
- Galcheva-Gargova, Z., B. Derijard, I.H. Wu, and R.J. Davis. 1994. An osmosensing signal transduction pathway in mammalian cells. Science. 265:806–808. [DOI] [PubMed] [Google Scholar]
- Gotoh, Y., and J.A. Cooper. 1998. Reactive oxygen species- and dimerization-induced activation of apoptosis signal-regulating kinase 1 in tumor necrosis factor-alpha signal transduction. J. Biol. Chem. 273:17477–17482. [DOI] [PubMed] [Google Scholar]
- Green, D.R., and J.C. Reed. 1998. Mitochondria and apoptosis. Science. 281:1309–1312. [DOI] [PubMed] [Google Scholar]
- Halenbeck, R., H. MacDonald, A. Roulston, T.T. Chen, L. Conroy, and L.T. Williams. 1998. CPAN, a human nuclease regulated by the caspase-sensitive inhibitor DFF45. Curr. Biol. 8:537–540. [DOI] [PubMed] [Google Scholar]
- Han, J., J.D. Lee, L. Bibbs, and R.J. Ulevitch. 1994. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 265:808–811. [DOI] [PubMed] [Google Scholar]
- Hatai, T., A. Matsuzawa, S. Inoshita, Y. Mochida, T. Kuroda, K. Sakamaki, K. Kuida, S. Yonehara, H. Ichijo, and K. Takeda. 2000. Execution of ASK1-induced apoptosis by the mitochondria-dependent caspase activation. J. Biol. Chem. 275:26576–26581. [DOI] [PubMed] [Google Scholar]
- Hoeflich, K.P., W.C. Yeh, Z. Yao, T.W. Mak, and J.R. Woodgett. 1999. Mediation of TNF receptor-associated factor effector functions by apoptosis signal-regulating kinase-1 (ASK1). Oncogene. 18:5814–5820. [DOI] [PubMed] [Google Scholar]
- Ichijo, H., E. Nishida, K. Irie, P. ten Dijke, M. Saitoh, T. Moriguchi, M. Takagi, K. Matsumoto, K. Miyazono, and Y. Gotoh. 1997. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 275:90–94. [DOI] [PubMed] [Google Scholar]
- Inohara, N., T. Koseki, S. Chen, X. Wu, and G. Nunez. 1998. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 17:2526–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara, N., T. Koseki, S. Chen, M.A. Benedict, and G. Nunez. 1999. Identification of regulatory and catalytic domains in the apoptosis nuclease DFF40/CAD. J. Biol. Chem. 274:270–274. [DOI] [PubMed] [Google Scholar]
- Ip, Y.T., and R.J. Davis. 1998. Signal transduction by the c-Jun N-terminal kinase (JNK)-from inflammation to development. Curr. Opin. Cell Biol. 10:205–219. [DOI] [PubMed] [Google Scholar]
- Kyriakis, J.M., P. Banerjee, E. Nikolakaki, T. Dai, E.A. Rubie, M.F. Ahmad, J. Avruch, and J.R. Woodgett. 1994. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 369:156–160. [DOI] [PubMed] [Google Scholar]
- Li, L., and S.N. Cohen. 1996. Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell. 85:319–329. [DOI] [PubMed] [Google Scholar]
- Liu, H., H. Nishitoh, H. Ichijo, and J.M. Kyriakis. 2000. Activation of apoptosis signal-regulating kinase 1 (ASK1) by tumor necrosis factor receptor-associated factor 2 requires prior dissociation of the ASK1 inhibitor thioredoxin. Mol. Cell. Biol. 20:2198–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., H. Zou, C. Slaughter, and X. Wang. 1997. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 89:175–184. [DOI] [PubMed] [Google Scholar]
- Liu, X., P. Li, P. Widlak, H. Zou, X. Luo, W.T. Garrard, and X. Wang. 1998. The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc. Natl. Acad. Sci. USA. 95:8461–8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty, J.S., S.Y. Toh, and P. Li. 1999. a. Study of DFF45 in its role of chaperone and inhibitor: two independent inhibitory domains of DFF40 nuclease activity. Biochem. Biophys. Res. Commun. 264:176–180. [DOI] [PubMed] [Google Scholar]
- McCarty, J.S., S.Y. Toh, and P. Li. 1999. b. Multiple domains of DFF45 bind synergistically to DFF40: roles of caspase cleavage and sequestration of activator domain of DFF40. Biochem. Biophys. Res. Commun. 264:181–185. [DOI] [PubMed] [Google Scholar]
- Minden, A., and M. Karin. 1997. Regulation and function of the JNK subgroup of MAP kinases. Biochim. Biophys. Acta. 1333:F85–F104. [DOI] [PubMed] [Google Scholar]
- Nagata, S. 1997. Apoptosis by death factor. Cell. 88:355–365. [DOI] [PubMed] [Google Scholar]
- Nishitoh, H., M. Saitoh, Y. Mochida, K. Takeda, H. Nakano, M. Rothe, K. Miyazono, and H. Ichijo. 1998. ASK1 is essential for JNK/SAPK activation by TRAF2. Mol. Cell. 2:389–395. [DOI] [PubMed] [Google Scholar]
- Park, H.S., S.H. Huh, Y. Kim, J. Shim, S.H. Lee, I.S. Park, Y.K. Jung, I.Y. Kim, and E.-J. Choi. 2000. Selenite negatively regulates caspase-3 through a redox mechanism. J. Biol. Chem. 275:8487–8491. [DOI] [PubMed] [Google Scholar]
- Park, H.S., J.S. Lee, S.H. Huh, J.S. Seo, and E.-J. Choi. 2001. Hsp72 functions as a natural inhibitory protein of c-Jun N-terminal kinase. EMBO J. 20:446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, M., H. Nishitoh, M. Fujii, K. Takeda, K. Tobiume, Y. Sawada, M. Kawabata, K. Miyazono, and H. Ichijo. 1998. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 17:2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakahira, H., M. Enari, and S. Nagata. 1998. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature. 391:96–99. [DOI] [PubMed] [Google Scholar]
- Sakahira, H., M. Enari, and S. Nagata. 1999. Functional differences of two forms of the inhibitor of caspase-activated DNase, ICAD-L, and ICAD-S. J. Biol. Chem. 274:15740–15744. [DOI] [PubMed] [Google Scholar]
- Schaeffer, H.J., and M.J. Weber. 1999. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol. Cell. Biol. 19:2435–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula, S.M., M. Ahmad, M. MacFarlane, Z. Luo, Z. Huang, T. Fernandes-Alnemri, and E.S. Alnemri. 1998. Generation of constitutively active recombinant caspases-3 and -6 by rearrangement of their subunits. J. Biol. Chem. 273:10107–10111. [DOI] [PubMed] [Google Scholar]
- Verheij, M., R. Bose, X.H. Lin, B. Yao, W.D. Jarvis, S. Grant, M.J. Birrer, E. Szabo, L.I. Zon, J.M. Kyriakis, et al. 1996. Requirement for ceramide-initiated SAPK/JNK signalling in stress-induced apoptosis. Nature. 380:75–79. [DOI] [PubMed] [Google Scholar]
- Xia, Z., M. Dickens, J. Raingeaud, R.J. Davis, and M.E. Greenberg. 1995. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 270:1326–1331. [DOI] [PubMed] [Google Scholar]