Abstract
The integrin α3β1 mediates cellular adhesion to the matrix ligand laminin-5. A second integrin ligand, the urokinase receptor (uPAR), associates with α3β1 via a surface loop within the α3 β-propeller (residues 242–246) but outside the laminin binding region, suggesting that uPAR–integrin interactions could signal differently from matrix engagement. To explore this, α3−/− epithelial cells were reconstituted with wild-type (wt) α3 or α3 with Ala mutations within the uPAR-interacting loop (H245A or R244A). Wt or mutant-bearing cells showed comparable expression and adhesion to laminin-5. Cells expressing wt α3 and uPAR dissociated in culture, with increased Src activity, up-regulation of SLUG, and down-regulation of E-cadherin and γ-catenin. Src kinase inhibition or expression of Src 1–251 restored the epithelial phenotype. The H245A and R244A mutants were unaffected by coexpression of uPAR. We conclude that α3β1 regulates both cell–cell contact and matrix adhesion, but through distinct protein interaction sites within its β-propeller. These studies reveal an integrin- and Src-dependent pathway for SLUG expression and mesenchymal transition.
Keywords: integrin α3β1; urokinase receptor; Src; SLUG; epithelial and mesenchymal transition
Introduction
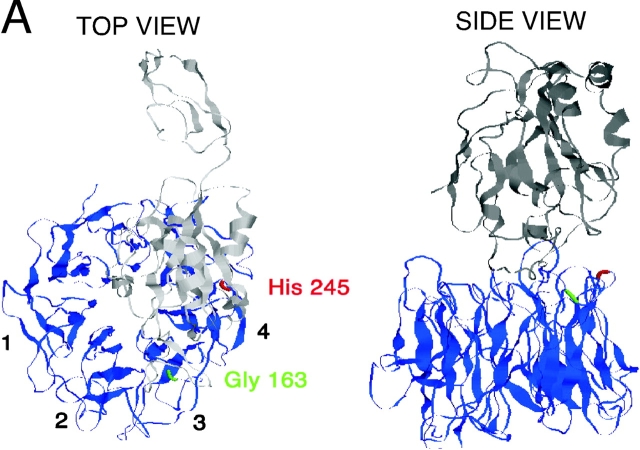
The integrin α3β1 is highly expressed in epithelial cells and associated with invasiveness of malignant carcinomas (Morini et al., 2000). The major endogenous ligand for α3β1 has proven to be laminin-5 (Eble et al., 1998). The binding of laminin-5 to α3β1 is non-RGD dependent but, like RGD ligands, involves the upper surface of the integrin α chain β-propeller. Mutation of glycine 163, located within an upper surface loop between blades 2 and 3 of the propeller (highlighted as green in Fig. 1), completely abrogates laminin-5 interaction with α3β1 (Zhang et al., 1999).
Figure 1.
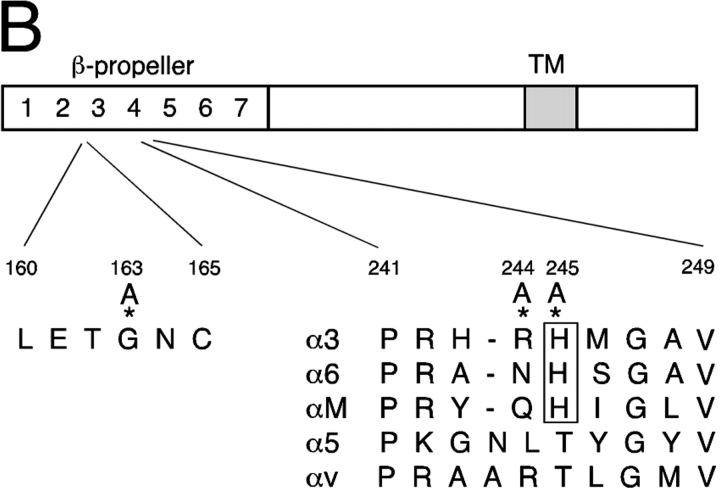
Model of α3 β-propeller structure. (A) Top and side views of the predicted β-propeller structure of the α3 chain (blue) and the NH2-terminal part of its associated β1 chain (gray) modeled on the crystal structure of integrin αvβ3 (Xiong et al., 2001). The positioning of Gly163 and His245 in the β-propeller are indicated in green and red, respectively. (B) A schematic of the integrin α3 chain showing sequence homologies between the reported α3 interaction site with uPAR (repeat 4 BC loop of the β-propeller structure) and αM as well as other β1-integrin–coupled α chains. Three mutations (*) were made in repeat 2 or repeat 4 of the α3 β-propeller region, as indicated in the figure.
In spite of its typical matrix binding capacity, among β1 integrins, α3β1 has unusual and distinctive features. This integrin does not localize to focal adhesion sites only (Dogic et al., 1998), but instead is frequently also found in or near epithelial adherens junctions (Nakamura et al., 1995). Newborn mice lacking α3β1 have disordered adherens junction formation in kidney epithelial cells (Wang et al., 1999). α3β1 has also been reported to associate tightly and specifically with the tetraspan protein CD151 (Berditchevski et al., 2001).
Although there is currently no mechanism linking α3β1 expression and E-cadherin function, there is evidence that integrins influence E-cadherin. In mammary epithelial cells, β1 integrin signaling promoted formation of adherens junctions with relocalization of E-cadherin to the lateral side of cells. Conversely, in colon carcinoma cells, overexpression of active Src leads to redistribution of E-cadherin away from adherens junctions. As judged by antibodies to either αv or β1, the Src responses required integrin signaling (Avizienyte et al., 2002). As mentioned, kidney epithelial cells deficient in α3β1 show loss of normal E-cadherin junctions (Wang et al., 1999). Together these data are consistent with the view that matrix–integrin associations regulate epithelial cell–cell interactions involving E-cadherin, and that in some way this cross-talk likely involves Src activation through integrins.
We recently reported that the urokinase receptor (uPAR), a glycosylphosphorylinositol (GPI)-linked surface protein, preferentially and physically associates with α3β1 (Wei et al., 2001). uPAR, by virtue of its ability to initiate a proteolytic cascade and its capacity to modify integrin function, is also implicated in cell migration (Blasi and Carmeliet, 2002). Numerous studies document the independent poor predictive value of uPAR expression on tumor metastasis (Yang et al., 2000). Several recent reports attest to the ability of uPAR to influence cell migration independently of urokinase protease activity (Ossowski and Aguirre-Ghiso, 2000). One such mechanism appears to be the ability of uPAR to influence integrin function, either by direct binding or through signaling. Based on sequence homology to a previously described uPAR-binding phage display peptide, we identified and provided evidence for an interaction site for uPAR within the repeat 4 BC loop of αM (CD11b), a β2-associating α chain (Simon et al., 2000). Among integrin α chains associating with β1 integrins, the homologous loop of α3 has the closest sequence homology to uPAR-binding site of αM (highlighted in Fig. 1 B). Unlike the association of CD151 with α3β1, the physical interaction of uPAR with this integrin is not constitutive. Binding of purified, soluble uPAR with purified soluble α3β1 is strongly promoted by concurrent binding of urokinase to uPAR and by agents that activate integrins (Wei et al., 2001). Similar patterns have been reported for uPAR–integrin interactions by several groups (Carriero et al., 1999; Tarui et al., 2001). Thus, the interaction of uPAR with α3β1 appears to be dynamic and influenced by multiple factors that affect the conformation of each interacting partner, typical for integrin ligands.
To probe further the interaction between uPAR and α3β1, we generated point mutants of the most conserved amino acids within the repeat 4 BC loop of the integrin α chains reported to interact with uPAR and expressed them in an α3-null background. Analysis of multiple clones of mutant and wild-type (wt) α3– and uPAR-expressing cells confirms the fourth repeat BC loop as an important interaction site for uPAR and reveals a mechanism linking α3β1 and E-cadherin.
Results
Selection and expression of integrin mutants
Recently reported studies of the crystal structure of αvβ3 confirm the earlier proposal that the NH2-terminal region (∼450 residues) of integrin α chains folds into a seven-bladed β-propeller (Springer, 1997; Xiong et al., 2001). Schematics of the NH2-terminal regions of α3β1, modeled on the atomic coordinates of αvβ3 (using Modeller v4.0), showing the numbered blades are illustrated in Fig. 1. Because His 245 is conserved among the three integrin α chains showing the most robust interaction with uPAR in coprecipitation studies (Fig. 1 B), and is contained in the original phage peptide used to block integrin–uPAR association, we first mutated this His to Ala. In subsequent experiments, we also mutated the adjacent Arg 244 to Ala. Similar point mutations of several additional mutations in this region, including Gly, Val, and Arg, were found to be either weakly expressed or not expressed at all. These mutants were not further pursued. In addition, we expressed a Gly 163 to Ala mutation in α3 (Fig. 1). This point mutant has previously been reported by Zhang et al. (1999) to be expressible and yet exhibit no binding to laminin-5 when expressed in K562 cells. Inspection of the top view of the modeled α3β1 indicates that G163 is accessible on the β-propeller upper surface, consistent with its role in laminin-5 binding, whereas H245 is not. Both G163 and H245 are on loops predicted to be accessible via the lateral surface.
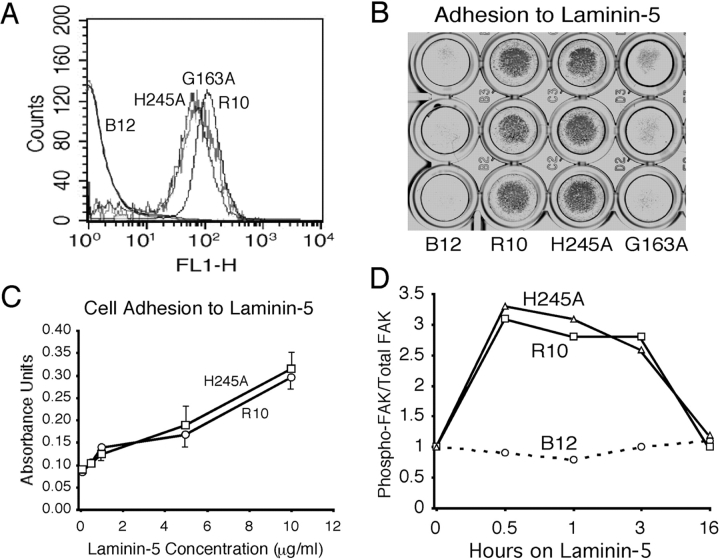
The wt, H245A, and G163A integrin α chains were stably expressed in a murine α3-null kidney epithelial cell line (Fig. 2 A). Multiple clones of each expression construct were isolated and frozen for subsequent experiments. To examine their matrix-binding function, the various integrin-expressing cells were tested for their capacity to adhere to purified laminin-5. The α3-null cells showed very weak attachment to laminin-5. The photograph in Fig. 2 B highlights robust adhesion of both wt and H245A integrins to 0.5 μg/ml of laminin-5. Both wt α3 and the H245A mutant adhered indistinguishably to all coating concentrations of laminin-5 tested (0.1–10 μg/ml) (Fig. 2 C). Strong adhesion of the H245A mutant was not unexpected, as the repeat 4 BC loop of integrins has not been implicated directly in matrix ligand engagement. Also as expected, cells bearing the G163A α3 mutant did not attach to laminin-5. Adhesion to laminin-5 by cells expressing either wt or the H245A mutant integrin was accompanied by transient FAK activation (Fig. 2 D). FAK activation initiated by either the wt or the H245A mutant integrin was identical, increasing within minutes of plating and returning to baseline by 16 h.
Figure 2.
Kidney epithelial cells expressing either wt α3 or the H245A mutant adhere to laminin-5. (A) FACS® analysis of wt (R10) and mutant α3 (H245A) expression on α3-null cells (B12). R10, H245A, or G163A α3 integrins were stably expressed in B12 cells and visualized by staining with P1B5. (B) Adhesion to laminin-5. Cells were seeded on laminin-5 (0.5 μg/ml) for 1 h at 37°C, washed, fixed, and stained with Giemsa. A representative of three independent experiments with triplicate wells is shown. (C) R10 and H245A cells adhere identically to laminin-5. A 96-well tissue culture plate was precoated with laminin-5 (0.1–10 μg/ml) and then blocked in 1% BSA. Adhesion was assayed as described in B. Data are expressed as mean ± SD of triplicate wells (n = 3). (D) FAK phosphorylation induced by laminin-5 engagement. α3-null (B12) or wt (R10) or H245A mutant α3–expressing cells were serum starved for 4 h and exposed to the immobilized laminin-5. Cells were lysed in RIPA buffer and immunoblotted for phospho-FAK and total FAK at various times as indicated. Data are expressed as ratio of phospho-FAK/total FAK. The ratio at time 0 for each cell line was made 1. This experiment was repeated three times with similar results.
Integrins affect cell–cell contact: influence of uPAR
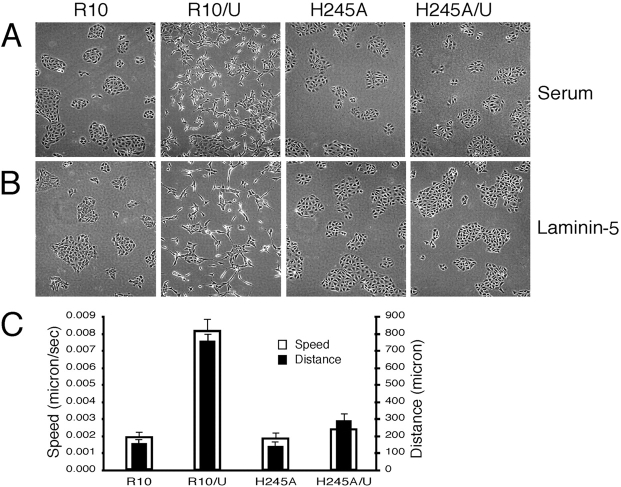
We next compared the morphology and cytoskeletal organization of cells expressing either wt or repeat 3 (G163A) or repeat 4 (H245A) mutants. Cells expressing wt α3 (R10 cells) illustrated a classical epithelial cell morphology in two-dimensional culture with clustering and formation of extensive cell–cell borders. This pattern was seen when cells were plated onto either serum- or laminin-5–coated surfaces (Fig. 3, A and B). The G163A mutant formed even more compact cell clusters, showing little tendency to spread either on vitronectin, fibronectin, or laminin-5 (not depicted). Although the H245A mutant formed clear cell–cell borders and clusters of epithelial cells, these clusters appeared somewhat less compact than those of R10 or G163A cells (Fig. 3, A and B).
Figure 3.
Expression of uPAR alters cell–cell contact and cytoskeleton organization. (A and B) Cells expressing wt or H245A α3 form clusters with extensive cell–cell contact when cultured either in 10% serum (A) or serum-free on purified laminin-5 (B). After uPAR transfection, wt α3–bearing cells scatter (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200304065/DC1), whereas cells expressing the H245A are unaffected (Video 2). Nearly identical changes in cellular morphology after uPAR transfection were seen with serum- or laminin-5–coated surfaces. (C) Cells expressing both uPAR and wt α3 are motile. R10, H245A, R10/U, or H245/U cells were maintained in a heated chamber, and images were collected every 10 min using a time-lapse imaging system (Spot Camera). Tracking of individual cells was done using SimplePCI software. Data (mean ± SD) of cell distance (μm) moved and speed are based on 18 cells in each movie tracked.
Morphological differences among the cell lines became more apparent upon transfection with uPAR. Epithelial cells coexpressing uPAR and wt α3 (R10/U) dissociate in culture and fail to form extensive cell–cell borders or clusters (Fig. 3 A). These findings were observed in at least five distinct clones of uPAR/wt α3–coexpressing cells and were critically dependent upon expression of both proteins. Periodic loss of expression of either α3 or uPAR upon passaging for months led to a reversion to the phenotype of α3-null or uPAR minus α3–bearing cells, respectively. Plating of cells on laminin-5 to ensure engagement of surface α3β1 also led to stable clusters and did not block the dissociative effect of concurrent uPAR expression (Fig. 3 B). In contrast to the striking phenotypic effect of uPAR overexpression on wt α3 cells, expression of uPAR had no discernible effect on cells expressing the H245A mutant. Again, multiple clones were examined, and no H245A α3 clone showed a morphological response to uPAR expression.
These morphological differences were reflected in altered motility as judged by 18-h time-lapse microscopy. Wt α3 cells coexpressing uPAR showed marked enhancement of random motility over that of cells coexpressing H245A α3 and uPAR (Fig. 3 C), with little tendency after cell division or contact to form stable cell–cell clusters. The H245A α3 cells coexpressing uPAR formed the clusters seen in Fig. 3 A largely by replication of cells within smaller two- to four-cell clusters, consistent with their largely stationary state during the observation period (Fig. 3 C; Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200304065/DC1).
To test whether these observations were unique to the H245A mutant, the adjacent Arg 244 was also point mutated to Ala (Fig. 1 B). This mutant, like the H245A mutant, was expressible and showed normal adhesion to laminin-5 (unpublished data). Coexpression of uPAR in these cells also failed to influence cell–cell border formation or organization in culture, consistent with the response of the H245A mutant to uPAR expression. Expression of uPAR in cells bearing the G163A integrin mutant also failed to show morphological changes; the cells maintained a tightly clustered phenotype (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200304065/DC1). Because of the striking differences between the phenotypes of wt and mutant α3 responses to coexpression of uPAR, we focused on the physical and molecular connections between α3β1 and uPAR that could explain these responses.
The H245A mutation impairs uPAR–integrin complex formation
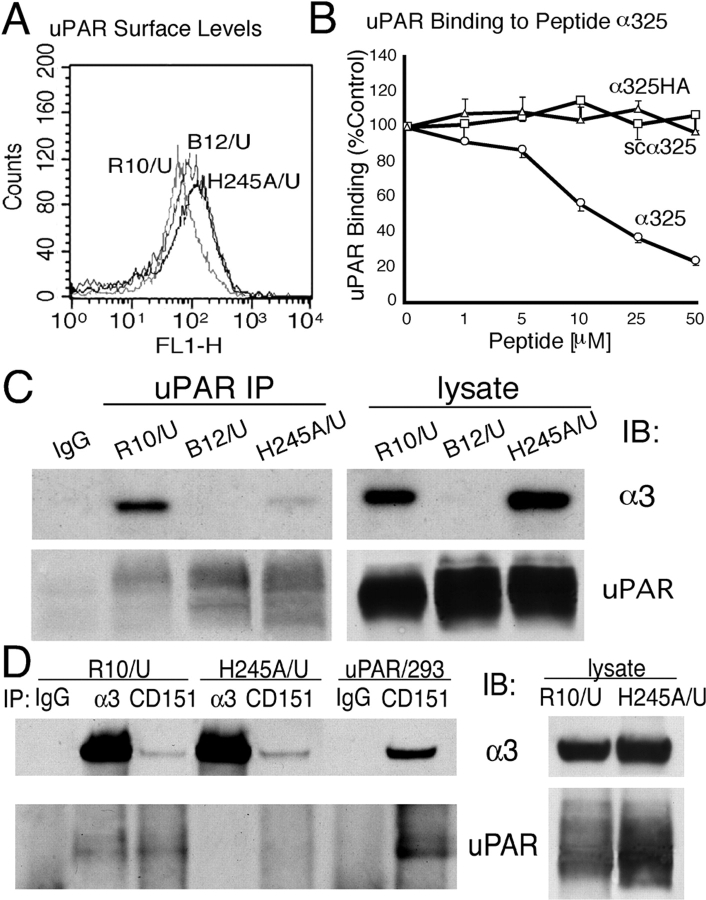
We have previously reported that recombinant soluble uPAR binds to immobilized peptides containing the exposed blade 4 BC loop (residues 242–246) of α3 (Wei et al., 2001). This assay was used to test whether the single H245A mutation was likely to affect the direct interaction of uPAR with α3. As indicated in Fig. 4 B, soluble uPAR binding to immobilized α3 residues 241–257 (α325) was progressively blocked by the addition of soluble α325 (1–50 μM), whereas addition of either scrambled α325 (scα325) or α325 containing the H245A mutation (α325HA) did not compete at any concentration tested, indicating that the H245A mutation could effect direct uPAR–α3 interaction. Although not shown, peptide 241–257 containing the R244A mutation also failed to compete at up to 100 μM. The capacity of uPAR to complex with native integrin was assessed first by coprecipitation of wt and H245A mutant α3 integrins with uPAR antibodies. Wt α3 was easily found in the uPAR immunoprecipitates (Fig. 4 C). In contrast to wt α3 coprecipitation with uPAR, little or no H245A α3 was seen in parallel immunoprecipitations of uPAR. This pattern was consistently found among two different uPAR/mutant integrin clones and in four separate immunoprecipitations. In these experiments, both the integrins and uPAR were comparably expressed as judged by FACS® (Fig. 4 A) and immunoblotting of cell lysates (Fig. 4 C). In parallel experiments, the capacity of either the R244A mutant or the G163A mutant to coprecipitate with uPAR was also examined. Results were similar to those shown for H245A; neither mutant integrin coprecipitated well with uPAR (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200304065/DC1).
Figure 4.
The H245A mutation blocks α3 coimmunoprecipitation with uPAR. (A) FACS® analysis of uPAR transfectants. uPAR-transfected α3-null (B12/U) and wt α3– and H245A mutant α3–expressing cells (R10/U and H245A/U) were stained with uPAR antibody R2. (B) Binding of biotin-suPAR to immobilized peptide α325. Biotinylated suPAR (50 nM) preincubated without or with peptide α325 (○), scα325(□), or α325HA (▵) (1–50 μM) was added to microtiter wells coated with α325 (100 μg/ml). Binding of suPAR in the presence of peptides is expressed as percentage of suPAR binding in the absence of peptides (% control). Values represent averages of triplicate determinations of three separate experiments. (C) Coimmunoprecipitation of uPAR and integrin α3. Human uPAR stably transfected into α3-null cells (B12) or cells coexpressing wt (R10) or the H245A mutant were lysed in 1% Triton X-100 lysis buffer, and the lysates were immunoprecipitated with uPAR antibody (R2). The cell lysates used and the immunoprecipitates were then separated by SDS-PAGE and blotted for integrin α3 using a polyclonal antibody (D23). The same membrane was stripped and reblotted for uPAR (R2). Data shown are representative of four independent experiments. (D) R10/U or H245A/U cells were lysed in 1% Triton X-100 buffer and immunoprecipitated with antibodies to α3 (P1B5) or CD151 (5C11). The lysates and immunoprecipitates were then blotted for α3 (D23) and uPAR (R2). Data shown is a representative of three separate experiments with similar results. UPAR/293 cells were used as a control.
Coprecipitation of uPAR and α3β1 was also observed when antibodies to α3 were used (Fig. 4 D). Again, no uPAR was seen when α3 antibodies were used to precipitate the H245A mutant, even though equivalent amounts of α3 were recovered. To probe further the complex formation between uPAR and α3β1, antibodies to CD151 were also used to precipitate α3, and then those precipitates were examined for the presence of uPAR. CD151 is known to constitutively associate with α3β1, and its site of interaction has been recently localized to the “stalk” region (residues 538–673) of the integrin α3β1 (Yauch et al., 2000). As uPAR interacts with the integrin β-propeller, both proteins could conceivably be found in the CD151 immunoprecipitations. Indeed, uPAR was observed coprecipitating with CD151 (Fig. 4 D), but this was critically dependent on the integrin sequence because no uPAR was seen in the CD151 immunoprecipitation of H245A α3 cells even though comparable amounts of α3 were recovered. The amounts of α3 coprecipitating with CD151 were markedly less than that precipitated with α3 antibodies, possibly because the CD151 antibody (5C11) was originally raised in mice against human CD151. This difference could also reflect relatively high levels of α3 in excess over CD151 in these reconstituted cells. Because no blotting antibody for murine CD151 exists to resolve this uncertainty, we also examined human cells expressing CD151, α3, and uPAR (Fig. 4 D, uPAR/293 cells). Again, precipitation of CD151 coprecipitated both α3 and uPAR. These data confirm that CD151, uPAR, and α3β1 form a multimeric complex. Collectively, these experiments provide a clear biochemical correlate to the morphological response to uPAR expression (Fig. 3); only cells bearing an α3 capable of a strong physical association with uPAR respond to uPAR with loss of stable cell–cell contacts and enhanced motility.
uPAR overexpression leads to an integrin-dependent loss of E-cadherin at cell–cell borders
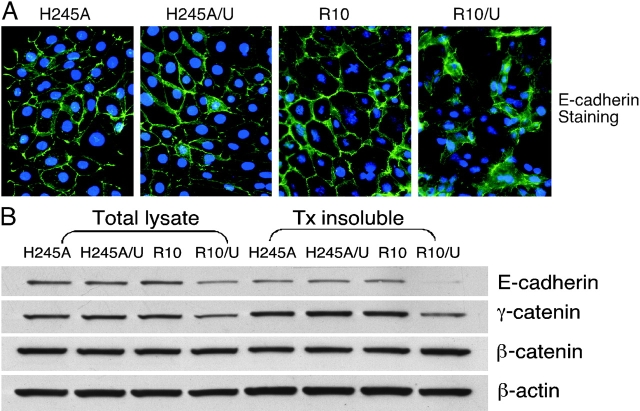
The marked dissociation in culture of epithelial cells expressing uPAR and wt α3 suggested dysfunction of cell–cell contact sites in these cells. This was assessed initially by E-cadherin immunostaining (Fig. 5 A). Expression of high levels of uPAR led to a nearly complete loss of the normal E-cadherin staining along cell–cell borders. Instead, there was less overall E-cadherin staining, and the staining was more diffuse. Strikingly, the H245A mutant showed no discernible change in E-cadherin distribution in the presence of high levels of uPAR, the cells still showing extensive E-cadherin along cell borders (Fig. 5 A).
Figure 5.
Altered E-cadherin distribution and loss of E-cadherin from the cytoskeleton in epithelial cells coexpressing α3 and uPAR. (A) Immunostaining of E-cadherin. Cells plated on chambered glass slides were fixed, permeabilized, and stained for E-cadherin as described in the text. (B) Immunoblotting of total and actin-associated E-cadherin, γ-catenin, and β-catenin. Triton-insoluble fractions and total lysates were separated by SDS-PAGE and blotted for E-cadherin, γ-catenin, and β-catenin. β-Actin (Sigma-Aldrich) was used as a loading control. The experiments above were performed at least three times with similar results.
The differences in patterns of E-cadherin immunostaining were mirrored by changes in formation of stable E-cadherin–β-actin complexes (Fig. 5 B). Immunoblots of the cytoskeletal fractions of wt and H245A α3–bearing cells showed comparable E-cadherin, γ-catenin (plakoglobin), and β-catenin in association with actin. Expression of uPAR, however, led to a marked reduction in actin-associated E-cadherin and γ-catenin in wt α3–expressing cells. No change in actin-associated β-catenin was found. In contrast, there was no reduction in actin-associated E-cadherin in H245A/U cells. Together with the coprecipitation and immunostaining data, these findings indicate that the single change of His245 to Ala, or Arg244 to Ala, within the repeat 4 BC α3 loop almost completely abrogates the physical and functional effects of uPAR on α3β1.
uPAR overexpression leads to a mesenchymal transition of kidney epithelial cells expressing α3/β1
The immunoblot illustrated in Fig. 5 B suggests that total E-cadherin and γ-catenin are lower in cells coexpressing uPAR and wt α3β1. To explore this point further, mRNA pools from triplicate cultures of cells expressing uPAR and either no α3, wt α3, or the H245A mutant were transcriptionally profiled by Affymetrix microarrays. This revealed a set of genes whose average transcript levels were significantly different only in cells coexpressing both uPAR and wt α3, no change being observed in mRNA from either α3-null or H245A-bearing cells coexpressing uPAR (not depicted). Because many of these genes are known to be associated with mesenchymal transition of epithelial cells, consistent with the observed phenotype (Fig. 3), primers for these genes were constructed for real-time PCR and transcript profiling was repeated several times (Table I). Inspection of Table I reveals down-regulation of steady-state mRNA of both E-cadherin and γ-catenin, consistent with immunoblotting (Fig. 5). In addition, there was selective up-regulation of syndecan II (fibroglycan), smooth muscle cell α actin, and the transcriptional factor SLUG. All of these proteins have been implicated in cellular migration and mesenchymal transition (Yang and Liu, 2001; Park et al., 2002; Bolos et al., 2003). SLUG is of particular interest because expression of this transcriptional factor is reported to be associated with suppression of E-cadherin and desmosomal protein mRNA and promotion of a mesenchymal transition in epithelial cells. The increase in SLUG mRNA (∼15-fold) was not seen in either the α3-negative cells or cells bearing the H245A integrin mutant and uPAR. These findings suggest that acquisition of SLUG expression in this cell system is due to uPAR overexpression, but induction requires coexpression of wt α3β1. A marked up-regulation in VEGFc (Shushanov et al., 2000), but not VEGFa or VEGFb, was also noted (Table I).
Table I. Quantitative PCR analysis of uPAR expression–induced integrin-dependent mRNA changes.
| Gene name | Copy number | Ratio | |
|---|---|---|---|
| R10/U | R10 | R10/U over R10 | |
| Syndecan II | 8918.97 | 3554.40 | 2.51 |
| VEGFc | 2198.58 | 265.29 | 8.29 |
| Smooth muscle actin α | 10634.24 | 287.40 | 37.00 |
| Fibroblast-specific protein 1 | 19440.57 | 19773.00 | 0.98 |
| SLUG | 3550.15 | 230.82 | 15.38 |
| E-cadherin | 2281.92 | 6671.68 | 0.34 |
| β-catenin | 11937.22 | 14182.87 | 0.84 |
| γ-catenin (plakoglobin) | 5812.04 | 15840.38 | 0.37 |
| GAPDH | 232273.68 | 232273.68 | 1.00 |
Total RNA from wt α3 integrin–bearing cells without uPAR (R10) or with uPAR (R10/U) was analyzed by Taqman quantitative PCR. Transcriptional levels were compared and expressed as a ratio. Data represent the average of triplicate determinations from a representative of three independent experiments with similar profile.
Although uPAR expression led to integrin-dependent changes typical of a mesenchymal transition, there was no change in vimentin mRNA and no induction of fibroblast-specific protein 1. Both of these proteins are reported to be expressed in kidney epithelial cells undergoing mesenchymal transformation in response to transforming growth factors, indicating that uPAR expression alone is not sufficient to drive frank mesenchymal transformation (Strutz et al., 2002). Because of the central role of the SLUG family of transcription factors in mesenchymal development, we focused on a signaling pathway that could plausibly connect uPAR, α3β1, and SLUG in these epithelial cells.
uPAR expression leads to Src activation: Src kinase inhibitors restore epithelial phenotype and reverse mesenchymal transition
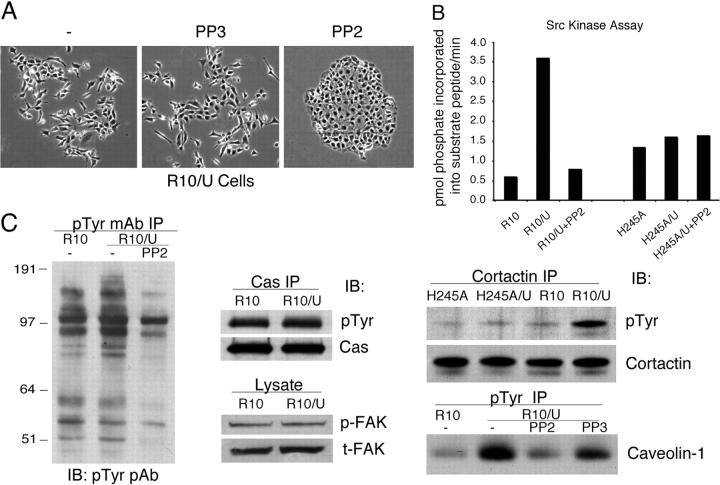
Currently, there is limited information available regarding signaling pathways in epithelial cells that regulate SLUG mRNA levels. To explore a molecular pathway by which uPAR expression could up-regulate SLUG expression and influence adherens junction stability, cells expressing uPAR and wt α3 were exposed to various inhibitors of second messenger pathways known to be activated by integrin signaling. Cells exposed to inhibitors of Rho family GTPases (exoenzyme C3, 1 ng/ml), MAP kinases (MEK [PD 98059, 10 μM], p38 kinase [SB 202190, 10 μM], c-Jun NH2-terminal kinase [JNK inhibitor, 10 μM]), PI3K (wortmanin, 100 nM), serine/threonine kinases (H7, 100 nM; bisindolylmaleimide I, 100 nM; PKC pseudosubstrate [19–31], 100 μM), or the epidermal growth factor receptor (AG1478, 1 μM) maintained a dissociated phenotype with weak or no cell–cell border formation (unpublished data). Similarly, a broad-spectrum matrix metalloproteinase (MMP) inhibitor GM6001 (100 nM) did not change the phenotype, indicating that MMP activity did not account for the loss of E-cadherin from R10/U cells (Ho et al., 2001). In contrast, a specific inhibitor of Src family kinases, PP2 (1 μM), was found to completely reverse the dissociation of these epithelial cells in culture (Fig. 6 A). Reversion to the epithelial phenotype after addition of PP2 was not immediate but instead was only apparent after 3–4 d. By 5 d, the cells were again clustered with extensive E-cadherin–rich cell–cell borders (Fig. 6 A). PP3, a structurally similar but inactive version of PP2, had no effect on cell clustering. A second structurally distinct Src kinase inhibitor, SU6656 (1 μM), also completely restored the clustered phenotype in a similar time course (not depicted).
Figure 6.
Integrin- and uPAR-dependent mesenchymal transition of epithelial cells is Src kinase dependent. (A) Src kinase inhibitor PP2 restores epithelial phenotype. R10 cells expressing uPAR (R10/U) incubated with 1 μM of PP2 or control PP3 in serum-free medium for 5 d show complete reversal of the scattering phenotype seen in Fig. 3. (B) Src kinase assay. Cells expressing wt (R10) or mutant (H245A) α3, or coexpressing uPAR (R10/U and H245A/U) with or without PP2 treatment, were lysed in RIPA buffer and immunoprecipitated using a pan-Src antibody (Src2). Src kinase activity in the precipitates was quantified by incorporation of [γ-32P]ATP into a Src substrate peptide as described in the Materials and methods. Activity was expressed as pmol phosphate incorporated into the substrate peptide/min. In four independent experiments, uPAR expression increased PP2-sensitive Src activity in cells expressing wt α3 (R10/U) but not mutant α3 (H245A/U). (C) Increased phosphorylation of Src substrates cortactin and caveolin but not p130Cas or FAK in R10/uPAR cells. P130Cas or cortactin was immunoprecipitated with a specific mAb and immunoblotted with antiphosphotyrosine pAb for phospho-p130Cas or cortactin, and the membrane was stripped and reblotted with a specific pAb for total p130Cas or cortactin. Phospho-FAK and total FAK were detected from lysates by anti–phospho-FAK and FAK mAbs, respectively. For immunoprecipitation with antiphosphotyrosine 4G10, cells were incubated with or without PP2 or PP3 (5 μM) for 2 h at 37°C before lysis. The total phosphoproteins pulled down with 4G10 were similar among each lane by Ponceau S staining of the blotting membrane. The immunoprecipitates were then blotted for phosphotyrosine using a pAb or caveolin-1. All these experiments have been repeated at least three times with similar results.
Because of the striking effects of Src kinase inhibitors on the epithelial phenotype of uPAR/integrin-expressing cells, Src kinase activity was measured in lysates of wt and H245A mutant–expressing cells without or with concurrent uPAR expression. In three separate experiments, Src kinase activity was found to be higher in R10/uPAR as compared with R10 cells, the increased activity being completely inhibited by 1 μM PP2 (Fig. 6 B). Although there was some phosphorylation of the Src substrate peptide in H245A-expressing cells (Fig. 6 B), this kinase activity was not Src related, as it was not inhibited by PP2. This kinase activity was also unaffected by coexpression of uPAR. We next searched for Src substrates differentially phosphorylated between R10 and R10/U cells in culture. Initial analysis of tyrosine-phosphorylated (p-tyr) proteins after immunoprecipitation with 4G10 antibodies to p-tyr showed that almost all of the discernible proteins were equally phosphorylated among cells expressing wt or mutant integrins in the absence or presence of uPAR (Fig. 6 C). Whereas transient activation of FAK was apparent within minutes of plating any of the cells on laminin-5 (Fig. 2 D), there were no differences in constitutive levels of phospho-FAK among the various cells. Serum starvation led to markedly lower tyrosine phosphorylation of many proteins, but again, there were few differences between wt α3β1–bearing cells ± uPAR. Data in Fig. 6 C show comparable FAK and p130CAS phosphorylation between R10 and R10/U cells. However, two known Src kinase substrates were identified with consistently higher phosphorylation states in wt α3β1 cells expressing uPAR: cortactin and caveolin-1. Enhanced cortactin and caveolin-1 phosphorylation was only observed in wt α3β1 cells coexpressing uPAR, the H245A/U cells showing no difference for either protein. These data validate the Src kinase assays of cell lysates (Fig. 6 B) and suggest that Src is likely locally activated and to a very limited degree. Prior studies have localized uPAR with β1 integrins, cortactin, caveolin-1, and Src-family kinases, possibly providing a spatial correlate to the observed pattern of altered tyrosine phosphorylation (Wei et al., 1999).
To determine if enhanced Src activity was related to the observed increased SLUG mRNA levels (Table I), SLUG mRNA was quantified at various times after exposing cells to PP2. Increased SLUG mRNA in R10/U cells began to decrease by 24 h, and by 72 h, it was virtually at basal levels of R10 cells (Fig. 7 A). This time course corresponded to the reversion of morphological changes to that of the parent epithelial cells (Fig. 6 A). Further, E-cadherin, γ-catenin, and β-catenin mRNAs were quantified after PP2 exposure. The mRNA levels of both E-cadherin and γ-catenin also returned to baseline copy numbers of R10 cells within 4 d, whereas β-catenin changed minimally (Fig. 7 B). This corresponded to increased amounts of actin-associated E-cadherin and γ-catenin protein (not depicted), consistent with the observed reversion to epithelial organization.
Figure 7.
Src activity is required for the up-regulation of SLUG and down-regulation of E-cadherin and γ-catenin in R10/uPAR cells. (A) Time course of PP2 reversal of increased SLUG mRNA in R10/uPAR cells. R10/U cells were incubated with PP2 (1 μM) for various times (1–6 d), and SLUG mRNA was quantified by Taqman real-time PCR. (B) PP2 reversal of mesenchymal mRNAs expressed in epithelial cells by uPAR transfection. After 4 d incubation with PP2 (1 μM), mRNAs for the indicated transcripts were quantified by PCR, and relative copy numbers were expressed as percent change from baseline R10/uPAR cells. Results shown represent a mean (± SD) of three independent experiments. (C) Dominant-negative Src (DnSrc) reverses scattering phenotype of R10 cells expressing uPAR. R10/U cells were transfected with DnSrc–GFP or GFP alone, and the sorted green cells were cultured for 4 d. The data shown is a representative from three independent transfections with similar results. (D) Dominant-negative Src suppresses SLUG mRNA in R10/U cells. 1 or 4 d after transfection, cells expressing DnSrc–GFP or GFP alone were sorted by green fluorescence, and SLUG transcription was quantified by Taqman real-time PCR.
To examine the relationship between Src and SLUG with an independent method, the capacity of Src to regulate epithelial cell clustering and SLUG levels was examined by transient transfection of wt α3β1– and uPAR-expressing cells with a cDNA encoding a dominant-negative Src protein (DnSrc–GFP, 1–251). Because the efficiency of transfection is relatively low in these cells, DnSrc–GFP was expressed, and cells expressing GFP were recovered by cell sorting. These cells, or cells transfected with GFP alone, were examined immediately for SLUG mRNA by quantitative PCR or plated to observed their phenotype in culture. Plating of mock-transfected cells (GFP only) resulted in a typical R10/U phenotype (compare Fig. 7 C with Fig. 6 A). In contrast, cells expressing detectable levels of DnSrc showed striking cluster formation (Fig. 7 C). The fraction of total cells clustered after 3–4 d was at least 80%, the clustered phenotype being observed in each of three separate experiments. When the GFP-expressing cells were recovered and examined immediately for SLUG mRNA levels, the levels in DnSrc–GFP-expressing cells were found to be reduced ∼25% (1 d after transfection) and ∼80% (4 d after transfection) as compared with GFP-transfected cells (Fig. 7 D). These data extend earlier experiments with PP2 and indicate that Src kinase activity and its location are critical regulators of SLUG levels in these epithelial cells.
Discussion
Our observations are of interest for several reasons. First, the data indicate that one origin of the capacity for integrins to mediate both matrix engagement and cross-talk with other adhesion pathways arises from the integrin β-propeller itself, implying that these signaling pathways can diverge at the cell surface. Most current thinking about integrin cross-talk focuses on “spillover” between intracellular signaling cascades (Schwartz and Ginsberg, 2002). Second, the experiments elucidate a previously unrecognized, Src-dependent signaling pathway linking integrins, at least α3β1, with expression of the SLUG family of transcription factors, key elements in epithelial–mesenchymal development. And finally, the data reveal a mechanistic connection between α3β1, uPAR, and E-cadherin that may be pertinent to the observation that uPAR and α3β1 levels are commonly enhanced and E-cadherin function suppressed in invasive human carcinomas. These observations were empowered by the reported crystal structure of αvβ3, the crystal revealing the presence of a β-propeller structure at the NH2-terminal region of the α chain and its relationship to the I-like domain of the integrin β chain (Fig. 1). This allowed selection of point mutants of known location in the assembled integrin to probe for distinct functions in different regions of the α chain propeller. Our data indicate that this is the case. Matrix engagement by α3β1 leads to the expected transient wave of FAK activation, likely reflecting transient Src activation, and supports formation of stable E-cadherin connections, whereas engagement of uPAR by α3β1 leads to a low but sustained level of Src activation with eventual loss of cell–cell borders and frank mesenchymal transition as reflected by up-regulation of SLUG. Single amino acid mutations in α3β1 that abrogate effective laminin-5 or uPAR engagement, or both, block Src activation and its downstream consequences.
A possible caveat to these findings is that while uPAR expression in kidney epithelial cells regulates E-cadherin function in an integrin-specific manner, the levels of uPAR expression in this experimental system are high and could be judged as nonphysiological. There is only a very low level of endogenous mouse uPAR, as judged by immunoblotting, in R10 cells (unpublished data). However, TGFβ1 and other growth factors implicated in tumor progression induce expression of uPAR in these cells. We have recently also observed that TGFβ1, known to promote dissolution of cell–cell contacts and mesenchymal transition of kidney epithelial cells, promotes dissociation of R10 cells in culture and does so in an α3β1-dependent manner. Cells bearing the H245A mutation do not lose stable E-cadherin junctions in response to TGFβ1 (unpublished data). Whether the TGFβ1 response is critically dependent on uPAR interacting with α3β1 is under study. In any case, these findings indicate that the mechanism linking α3β1 and E-cadherin demonstrated here may also apply to epithelial cells responding to physiological stimulation accompanied by enhanced uPAR expression and altered integrin function.
An important unanswered question is why Src activation appears sustained upon uPAR engagement, whereas Src activation is transient after matrix attachment. Classical ligand-induced integrin signaling is thought to lead to Src activation by altered conformation of the integrin cytoplasmic tails and the clustering of integrins. The latter leads to the concentration and cross-activation of signaling molecules, including Src kinases, at the site of attachment (Schwartz and Ginsberg, 2002). Tyrosine phosphatases, such as SHP-1, rapidly accumulate at attachment sites and limit or reverse protein phosphorylation (Rock et al., 1997). Engagement of uPAR by α3β1 could be expected to be different in at least two respects. There is little evidence to indicate clustering of integrins by uPAR, at least when cells are plated on laminin-5 or fibronectin. This alone may alter the accumulation of kinases and phosphatases at the site of integrin engagement. Second, as a cis-acting integrin ligand, this glycosylphosphorylinositol (GPI)-anchored membrane protein could be expected to bring its own set of associated membrane proteins to the site of integrin interaction. Because uPAR associates with lipid rafts, containing an array of signaling molecules (Sargiacomo et al., 1993), we presume that the juxtaposition of rafts and integrins leads to the distinct signaling patterns and observed downstream consequences. This speculation is supported by the pattern of enhanced tyrosine-phosphorylated proteins observed in R10/U cells. Whereas classical downstream targets of integrin signals, such as p130CAS and FAK, did not show sustained phosphorylation, the raft-localized protein caveolin-1 did (Fig. 6 C). Tyrosine-phosphorylated caveolin-1 has been reported to bind the SH2 domain of the adaptor protein Grb7, implying that this caveolin-1 phosphorylation itself could be a mediator of integrin cross-talk (Lee et al., 2000). The fact that DnSrc–GFP, which alters association of Src with its binding partners, causes reversion to a clustered phenotype (Fig. 7 C) further highlights the importance of Src localization. Exactly how raft components might regulate Src kinase activity in an integrin-specific manner, and vice versa, remains an important goal of future studies.
The observation that uPAR expression in epithelial cells can regulate E-cadherin expression and function has not been previously recognized. However, the capacity of uPAR to influence E-cadherin function specifically through α3β1 is not a complete surprise. This integrin localizes closely with adherens junctions as well as with focal adhesion sites (Wang et al., 1999). Indeed plating epithelial cells on laminin-5 reportedly promotes E-cadherin–dependent cell–cell adhesion (Dogic et al., 1998), consistent with our observations (Fig. 3 B). The expression of uPAR on the other hand has been linked to enhanced migratory capacity of epithelial cells both in vitro and in vivo (Ossowski and Aguirre-Ghiso, 2000). Migration of epithelial cells depends on both the disruption of cell–cell contacts and reversible matrix engagement. Our observations shed further light on how this occurs. Sustained expression of uPAR led to loss of cell–cell clustering and markedly increased, integrin-dependent motility (Fig. 3 C; Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200304065/DC1). This was associated with decreases in E-cadherin and γ-catenin, the latter being a key component of both adherens junctions and desmosomes (Kofron et al., 2002), and was dependent on sustained Src-kinase activation. What is the connection between activated Src and altered expression of E-cadherin and γ-catenin? Overexpression of active Src kinases in either colon carcinoma cells or keratinocytes has been reported to phosphorylate components of the adherens junction complex, leading to its dissolution and a redistribution of E-cadherin similar to our observations (Fig. 5 A) (Owens et al., 2000; Avizienyte et al., 2002). However, in contrast to these prior transfection approaches, which lead to high levels of active Src, the level of sustained Src activation as a consequence of α3β1–uPAR engagement appears much lower with no evident change in the phosphorylation states of many of the known Src substrates (Fig. 6 C). Instead, our data point to a different mechanism for loss of cell–cell borders, up-regulation of SLUG. Expression of SLUG strictly correlated with the phenotype of the kidney epithelial cells with elevated SLUG in dispersed R10/U cells and falling SLUG levels after inhibition of either Src kinase or proper Src localization (DnSrc–GFP) in parallel with reversion to an epithelial phenotype (Fig. 6 A and Fig. 7 C). In addition, uPAR had no effect on SLUG levels in H245A mutant cells, which also failed to scatter with uPAR expression or alter cadherin or catenin levels. Although we cannot be sure that the observed changes in SLUG levels alone cause reversion to epithelial levels of E-cadherin and γ-catenin and an epithelial phenotype, these observations are consonant with prior observations in several other cell systems that SLUG regulates cadherin and catenin transcription and cell–cell contact (Vallin et al., 2001; Bolos et al., 2003). To our knowledge, these are the first data directly linking integrin function with SLUG levels and further suggest that a specific integrin, α3β1, and how it is engaged by ligands, is an important determinant of SLUG expression. How low levels of active Src ultimately lead to SLUG transcription remains to be defined.
High levels of SLUG repress E-cadherin promoter function, and levels of SLUG correlate inversely with E-cadherin levels in various breast cancer cell lines (Hajra et al., 2002). Although we cannot predict to what extent in human tumors uPAR expression promotes SLUG expression with its consequent negative effects on cadherin function, many carcinomas exhibit both up-regulated uPAR and SLUG and suppressed E-cadherin protein levels. Up-regulation of uPAR and down-regulation of E-cadherin are both considered risk factors for tumor progression and poor prognosis. Our data raise the possibility that one explanation for why both observations are commonly found in more invasive tumors is that they are mechanistically linked.
Materials and methods
Reagents
Full-length cDNAs for wt and G163A mutant human α3 in PBJ vector were from Y. Takada (Scripps Research Institute, La Jolla, CA). Dominant-negative Src–GFP cDNA encoding Src 1–251 was a gift from P.L. Schwartzberg (National Human Genome Research Institute, Bethesda, MD). Purified laminin-5 was a gift from J.C. Jones (Northwestern University Medical School, Chicago, IL). Anti-α3 antibody (P1B5) was purchased from Chemicon. Polyclonal anti-α3 (D23) and monoclonal anti-CD151 (5C11) were provided by M.E. Hemler (Dana Farber Cancer Institute, Boston, MA). Anti-uPAR antibody (R2) was a gift from G. Hoyer-Hansen (Finsen Lab, Copenhagen, Denmark). Inhibitor to Src family tyrosine kinases PP2 and its structurally inert isomer PP3 were from Calbiochem. Anticortactin (4F11), antiphosphotyrosine (4G10), and Src assay kit were from Upstate Biotechnology. Anti–phospho-FAK (Y397) and FAK mAbs, antiphosphotyrosine pAb, anti-p130Cas mAb, and anticaveolin pAb were from Transduction Laboratories. Polyclonal antibodies to cortactin and p130Cas were purchased from Santa Cruz Biotechnology, Inc. Anti–β-catenin, anti–γ-catenin, and anti–E-cadherin mAbs (Transduction Laboratories) were used for immunoblotting. For staining, rat anti–E-cadherin antibody (Zymed Laboratories) was used. Peptides α325 (PRHRHMGAVFLLSQEAG), scα325 (HQLPGAHRGVEARFSML), and α325HA (PRHRAMGAVFLLSQEAG) or α325RA (PRHAHMGAVFLLSQEAG) were synthesized at University of California San Francisco Biomolecular Resources Center and purified by HPLC.
Site-directed mutagenesis and cDNA cloning
PCR primers used for the H245A mutant were as follows: HP2, 5′-CGCGCCCATGGCTCGGTGCC-3′, and HP3, 5′-GGCACCGAGCCATGGGCGCG-3′. The common outside primers were P1, 5′-TACCTGCTCCTGGCTGGT-3′, and P4, 5′-ACAGGCTTCCCACTAGAAGGTCTG-3′. Purified PCR products were cut with SacI to replace the wt sequence in PBJ-α3. The mutant α3 insert was then transferred to pCDNA3.1 for expression. All constructs were confirmed by DNA sequencing before expression.
Cells and cell culture
Kidney epithelial cell line from α3 integrin–null (B12) and wild-type human α3–reconstituted B12 cells (R10) were prepared as previously described (Wang et al., 1999). B12 cells were cultured in DME supplemented with 10% FBS (Hyclone). R10 and mutant α3 (H245A, R244A, and G163A) transfected cells were cultured in DME supplemented with 50 μg/ml of zeocin (Invitrogen). B12/U was grown in DME with 100 μg/ml of hygromycin B (Invitrogen). All the uPAR-cotransfected cells were cultured in DME with both zeocin and hygromycin.
Flow cytometry
B12 cells and stable clones expressing wt or mutant α3 were stained with primary antibody to integrin α3 (P1B5) and secondary FITC-conjugated anti–mouse IgG (Sigma-Aldrich) and analyzed on a flow cytometer (FACSCaliber®; BD Biosciences). uPAR-transfected cells were stained with a primary antibody to uPAR (R2).
Adhesion assay
The cell adhesion assay was performed as previously described (Wei et al., 2001). In brief, cells were seeded on a laminin-5–coated plate and incubated for 1 h at 37°C. After washing, attached cells were fixed and stained with Giemsa. The data were quantified by measuring absorbance at a wavelength of 550 nm.
Peptide binding assay
Human recombinant suPAR, a gift of G. Deng (Berlex Laboratories, Richmond, CA), was biotinylated at 0.25 mg/ml. The binding of biotinylated suPAR to peptide α325 was performed as previously described (Simon et al., 2000). In brief, microtiter plates were coated with α325 (100 μg/ml) and incubated with biotinylated suPAR (50 nM) ± α325, scα325, or α325HA (1–50 μM). After washing, avidin peroxidase was added, and bound suPAR was quantified. Relative binding was calculated as the ratio of binding in the presence of peptide to binding in the absence of peptide.
Immunofluorescence microscopy
Cells plated on chambered slides were fixed in 3.7% paraformaldehyde, permeabilized with 0.5% Nonidet P-40, and blocked with 10% horse serum and 1% BSA in PBS. The slides were stained with primary anti–E-cadherin antibody and FITC-conjugated secondary anti–rat IgG antibody (Lab Tek). Slides were incubated with DAPI (Molecular Probes) before mounting in Prolong (Molecular Probes).
Immunoprecipitation and immunoblotting
Cells expressing uPAR were lysed in Triton lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1% Triton X-100) supplemented with protease inhibitors and 1 mM phenylmethylsulfonyl fluoride. Lysates were immunoprecipitated with antibody to uPAR (R2), and the immunoprecipitates were blotted for α3 or uPAR using rabbit polyclonal antibodies. Ultracentrifugation of lysates at 100,000 g had no effect on the amounts of subsequently recovered integrin and uPAR in the coimmunoprecipitations (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200304065/DC1). When precipitating phosphorylated proteins, RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% deoxycholate, 0.1% SDS, 1% Triton X-100) was supplemented with additional 1 mM sodium vanadate, 10 mM sodium fluoride, and 2 mM EDTA. In some cases, cells were extracted with CSK buffer (50 mM NaCl, 10 mM Pipes, pH 6.8, 3 mM MgCl2, 0.5% Triton X-100, 300 mM sucrose, supplemented with protease inhibitors and 1 mM PMSF) for 20 min at 4°C. The Triton X-100–insoluble fraction and the whole cell lysate were blotted for E-cadherin, γ-catenin, β-catenin, β-actin, phospho-FAK, or total FAK.
Src kinase activity
Src family kinases were immunoprecipitated with anti–pan-Src antibody (Src2), and the activity was quantified by incorporation of [γ-32P]ATP (10 μCi; PerkinElmer) into a Src substrate peptide (Upstate Biotechnology) according to the manufacturer's instructions. Src kinase activity was expressed as pmol phosphate incorporated into the substrate peptide/min.
Taqman real-time PCR
Verification of transcript quantity in several selected cDNAs was performed using Taqman real-time PCR. The primer pairs and probe for each cDNA were designed using Primer Express software (Applied Biosystems). The quantification was performed using the standard protocol of ABI PRISM 7700 (Applied Biosystems).
Time-lapse microscopy
Cells were maintained in a heated chamber with 5% CO2 at 37°C on the microscope stage. Cell motility analyses were performed using low (10×) magnification time-lapse microscopy of random fields of cells over 18 h (one frame/10 min). Cells were filmed over the same times after plating to eliminate nonspecific differences. Cell tracking of each cell line was analyzed using SimplePCI software (Compix, Inc.).
Online supplemental material
The supplemental material (Videos 1 and 2 and Figs. S1–S3) is available at http://www.jcb.org/cgi/content/full/jcb.200304065/DC1. Videos 1 and 2 show morphology and motility of R10/U and H245A/U cells over an 18-h period by time-lapse microscopy. Frames were captured every 10 min as indicated in the figure legends. Fig. S1 shows uPAR and integrin α3 coimmunoprecipitates after ultracentrifugation. R244A and G163A mutations disrupt coimmunoprecipitation of uPAR and integrin α3 (Fig. S2) and do not change epithelial phenotype by uPAR expression (Fig. S3).
Acknowledgments
The authors thank Yoshi Takada for the wt α3 cDNA clone, Jian-Ping Xiong for the α3β1 modeling, Phuong Lan Chau, Liliane Robillard, and Rufeng Xie for technical assistance, Greg Dolganov and Samantha Donnelly for help with real-time PCR, and Carmela Bacani for help with preparation of the manuscript.
This work was supported by National Institutes of Health HL44712 (to H.A. Chapman) and a research grant from the Chiron Corporation.
The online version of this article includes supplemental material.
Abbreviations used in this paper: uPAR, urokinase receptor; wt, wild type.
References
- Avizienyte, E., A.W. Wyke, R.J. Jones, G.W. McLean, M.A. Westhoff, V.G. Brunton, and M.C. Frame. 2002. Src-induced de-regulation of E-cadherin in colon cancer cells requires integrin signalling. Nat. Cell Biol. 4:632–638. [DOI] [PubMed] [Google Scholar]
- Berditchevski, F., E. Gilbert, M.R. Griffiths, S. Fitter, L. Ashman, and S.J. Jenner. 2001. Analysis of the CD151-alpha3beta1 integrin and CD151-tetraspanin interactions by mutagenesis. J. Biol. Chem. 276:41165–41174. [DOI] [PubMed] [Google Scholar]
- Blasi, F., and P. Carmeliet. 2002. uPAR: a versatile signalling orchestrator. Nat. Rev. Mol. Cell Biol. 3:932–943. [DOI] [PubMed] [Google Scholar]
- Bolos, V., H. Peinado, M.A. Perez-Moreno, M.F. Fraga, M. Esteller, and A. Cano. 2003. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J. Cell Sci. 116:499–511. [DOI] [PubMed] [Google Scholar]
- Carriero, M.V., S. Del Vecchio, M. Capozzoli, P. Franco, L. Fontana, A. Zannetti, G. Botti, G. D'Aiuto, M. Salvatore, and M.P. Stoppelli. 1999. Urokinase receptor interacts with alpha(v)beta5 vitronectin receptor, promoting urokinase-dependent cell migration in breast cancer. Cancer Res. 59:5307–5314. [PubMed] [Google Scholar]
- Dogic, D., P. Rousselle, and M. Aumailley. 1998. Cell adhesion to laminin 1 or 5 induces isoform-specific clustering of integrins and other focal adhesion components. J. Cell Sci. 111:793–802. [DOI] [PubMed] [Google Scholar]
- Eble, J.A., K.W. Wucherpfennig, L. Gauthier, P. Dersch, E. Krukonis, R.R. Isberg, and M.E. Hemler. 1998. Recombinant soluble human alpha 3 beta 1 integrin: purification, processing, regulation, and specific binding to laminin-5 and invasin in a mutually exclusive manner. Biochemistry. 37:10945–10955. [DOI] [PubMed] [Google Scholar]
- Hajra, K.M., D.Y. Chen, and E.R. Fearon. 2002. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 62:1613–1618. [PubMed] [Google Scholar]
- Ho, A.T., E.B. Voura, P.D. Soloway, K.L. Watson, and R. Khokha. 2001. MMP inhibitors augment fibroblast adhesion through stabilization of focal adhesion contacts and up-regulation of cadherin function. J. Biol. Chem. 276:40215–40224. [DOI] [PubMed] [Google Scholar]
- Kofron, M., J. Heasman, S.A. Lang, and C.C. Wylie. 2002. Plakoglobin is required for maintenance of the cortical actin skeleton in early Xenopus embryos and for cdc42-mediated wound healing. J. Cell Biol. 158:695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H., D. Volonte, F. Galbiati, P. Iyengar, D.M. Lublin, D.B. Bregman, M.T. Wilson, R. Campos-Gonzalez, B. Bouzahzah, R.G. Pestell, et al. 2000. Constitutive and growth factor-regulated phosphorylation of caveolin-1 occurs at the same site (Tyr-14) in vivo: identification of a c-Src/Cav-1/Grb7 signaling cassette. Mol. Endocrinol. 14:1750–1775. [DOI] [PubMed] [Google Scholar]
- Morini, M., M. Mottolese, N. Ferrari, F. Ghiorzo, S. Buglioni, R. Mortarini, D.M. Noonan, P.G. Natali, and A. Albini. 2000. The alpha 3 beta 1 integrin is associated with mammary carcinoma cell metastasis, invasion, and gelatinase B (MMP-9) activity. Int. J. Cancer. 87:336–342. [PubMed] [Google Scholar]
- Nakamura, K., R. Iwamoto, and E. Mekada. 1995. Membrane-anchored heparin-binding EGF-like growth factor (HB-EGF) and diphtheria toxin receptor-associated protein (DRAP27)/CD9 form a complex with integrin α3β1 at cell–cell contact sites. J. Cell Biol. 129:1691–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski, L., and J.A. Aguirre-Ghiso. 2000. Urokinase receptor and integrin partnership: coordination of signaling for cell adhesion, migration and growth. Curr. Opin. Cell Biol. 12:613–620. [DOI] [PubMed] [Google Scholar]
- Owens, D.W., G.W. McLean, A.W. Wyke, C. Paraskeva, E.K. Parkinson, M.C. Frame, and V.G. Brunton. 2000. The catalytic activity of the Src family kinases is required to disrupt cadherin-dependent cell-cell contacts. Mol. Biol. Cell. 11:51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H., Y. Kim, Y. Lim, I. Han, and E.S. Oh. 2002. Syndecan-2 mediates adhesion and proliferation of colon carcinoma cells. J. Biol. Chem. 277:29730–29736. [DOI] [PubMed] [Google Scholar]
- Rock, M.T., W.H. Brooks, and T.L. Roszman. 1997. Calcium-dependent signaling pathways in T cells. Potential role of calpain, protein tyrosine phosphatase 1b, and p130Cas in integrin-mediated signaling events. J. Biol. Chem. 272:33377–33383. [DOI] [PubMed] [Google Scholar]
- Sargiacomo, M., M. Sudol, Z. Tang, and M.P. Lisanti. 1993. Signal transducing molecules and glycosyl-phosphatidylinositol-linked proteins form a caveolin-rich insoluble complex in MDCK cells. J. Cell Biol. 122:789–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, M.A., and M.H. Ginsberg. 2002. Networks and crosstalk: integrin signalling spreads. Nat. Cell Biol. 4:E65–E68. [DOI] [PubMed] [Google Scholar]
- Shushanov, S., M. Bronstein, J. Adelaide, L. Jussila, T. Tchipysheva, J. Jacquemier, A. Stavrovskaya, D. Birnbaum, and A. Karamysheva. 2000. VEGFc and VEGFR3 expression in human thyroid pathologies. Int. J. Cancer. 86:47–52. [DOI] [PubMed] [Google Scholar]
- Simon, D.I., Y. Wei, L. Zhang, N.K. Rao, H. Xu, Z. Chen, Q. Liu, S. Rosenberg, and H.A. Chapman. 2000. Identification of a urokinase receptor-integrin interaction site. Promiscuous regulator of integrin function. J. Biol. Chem. 275:10228–10234. [DOI] [PubMed] [Google Scholar]
- Springer, T. 1997. Folding of the N-terminal, ligand-binding region of integrin alpha-subunits into a beta-propeller domain. Proc. Natl. Acad. Sci. USA. 94:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutz, F., M. Zeisberg, F.N. Ziyadeh, C.Q. Yang, R. Kalluri, G.A. Muller, and E.G. Neilson. 2002. Role of basic fibroblast growth factor-2 in epithelial-mesenchymal transformation. Kidney Int. 61:1714–1728. [DOI] [PubMed] [Google Scholar]
- Tarui, T., A.P. Mazar, D.B. Cines, and Y. Takada. 2001. Urokinase-type plasminogen activator receptor (CD87) is a ligand for integrins and mediates cell-cell interaction. J. Biol. Chem. 276:3983–3990. [DOI] [PubMed] [Google Scholar]
- Vallin, J., R. Thuret, E. Giacomello, M.M. Faraldo, J.P. Thiery, and F. Broders. 2001. Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/beta-catenin signaling. J. Biol. Chem. 276:30350–30358. [DOI] [PubMed] [Google Scholar]
- Wang, Z., J.M. Symons, S.L. Goldstein, A. McDonald, J.H. Miner, and J.A. Kreidberg. 1999. (Alpha)3(beta)1 integrin regulates epithelial cytoskeletal organization. J. Cell Sci. 112:2925–2935. [DOI] [PubMed] [Google Scholar]
- Wei, Y., X. Yang, Q. Liu, J.A. Wilkins, and H.A. Chapman. 1999. A role for caveolin and the urokinase receptor in integrin-mediated adhesion and signaling. J. Cell Biol. 144:1285–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Y., J.A. Eble, Z. Wang, J.A. Kreidberg, and H.A. Chapman. 2001. Urokinase receptors promote beta1 integrin function through interactions with integrin alpha3beta1. Mol. Biol. Cell. 12:2975–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, J.P., T. Stehle, B. Diefenbach, R. Zhang, R. Dunker, D.L. Scott, A. Joachimiak, S.L. Goodman, and M.A. Arnaout. 2001. Crystal structure of the extracellular segment of integrin alphaVbeta3. Science. 294:339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., and Y. Liu. 2001. Dissection of key events in tubular epithelial to myofibroblast transition and its implications in renal interstitial fibrosis. Am. J. Pathol. 159:1465–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., D. Seetoo, Y. Wang, M. Ranson, C.R. Berney, J.M. Ham, P.J. Russell, and P.J. Crowe. 2000. Urokinase-type plasminogen activator and its receptor in colorectal cancer: independent prognostic factors of metastasis and cancer-specific survival and potential therapeutic targets. Int. J. Cancer. 89:431–439. [DOI] [PubMed] [Google Scholar]
- Yauch, R., A. Kazarov, B. Desai, R. Lee, and M. Hemler. 2000. Direct extracellular contact between integrin alpha(3)beta(1) and TM4SF protein CD151. J. Biol. Chem. 275:9230–9238. [DOI] [PubMed] [Google Scholar]
- Zhang, X.P., W. Puzon-McLaughlin, A. Irie, N. Kovach, N.L. Prokopishyn, S. Laferte, K. Takeuchi, T. Tsuji, and Y. Takada. 1999. Alpha 3 beta 1 adhesion to laminin-5 and invasin: critical and differential role of integrin residues clustered at the boundary between alpha 3 N-terminal repeats 2 and 3. Biochemistry. 38:14424–14431. [DOI] [PubMed] [Google Scholar]