Abstract
EWI-2, a cell surface immunoglobulin SF protein of unknown function, associates with tetraspanins CD9 and CD81 with high stoichiometry. Overexpression of EWI-2 in A431 epidermoid carcinoma cells did not alter cell adhesion or spreading on laminin-5, and had no effect on reaggregation of cells plated on collagen I (α2β1 integrin ligand). However, on laminin-5 (α3β1 integrin ligand), A431 cell reaggregation and motility functions were markedly impaired. Immunodepletion and reexpression experiments revealed that tetraspanins CD9 and CD81 physically link EWI-2 to α3β1 integrin, but not to other integrins. CD81 also controlled EWI-2 maturation and cell surface localization. EWI-2 overexpression not only suppressed cell migration, but also redirected CD81 to cell filopodia and enhanced α3β1–CD81 complex formation. In contrast, an EWI-2 chimeric mutant failed to suppress cell migration, redirect CD81 to filopodia, or enhance α3β1–CD81 complex formation. These results show how laterally associated EWI-2 might regulate α3β1 function in disease and development, and demonstrate how tetraspanin proteins can assemble multiple nontetraspanin proteins into functional complexes.
Keywords: integrins; tetraspanins; laminin-5; CD9; CD81
Introduction
The laminins are major components of normal and pathological basement membranes. Major cellular receptors for laminins are α3β1, α6β1, α6β4, and α7β1 integrins (Colognato and Yurchenco, 2000). The α6β4 integrin anchors normal epithelial cells to laminin-5 in hemidesmosomes (Borradori and Sonnenberg, 1999), whereas α3β1 integrin is implicated in laminin-5–mediated cell motility (Nguyen et al., 2000). Laminin-binding integrins also maintain epithelial integrity, and support kidney and lung morphogenesis (Kreidberg et al., 1996; DiPersio et al., 1997). Laminin-binding integrins (α3β1, α6β1, α6β4, and α7β1) are further distinguished from other integrins by their formation of robust complexes with tetraspanins (Berditchevski, 2001; Sterk et al., 2002).
Tetraspanin proteins contain four transmembrane domains, one small and one large extracellular loop, and short, cytoplasmic NH2 and COOH termini (Berditchevski, 2001; Boucheix and Rubinstein, 2001). Tetraspanins may specifically regulate integrin-dependent cell motility and morphology, but typically do not affect static cell adhesion (Stipp and Hemler, 2000; Zhang et al., 2002). Tetraspanins associate with integrins, Ig superfamily proteins, membrane-bound growth factors, growth factor receptors, and other tetraspanins to form tetraspanin-enriched microdomains on the cell surface (Berditchevski, 2001; Boucheix and Rubinstein, 2001; Hemler, 2003).
To gain insight into tetraspanin function, we used mass spectrometry to identify novel associated proteins. Tetraspanins CD9 and CD81 were targeted because of their unique pattern of associated proteins (Stipp et al., 2001b), which includes EWI-2 (also called PGRL), as a member of a subfamily of four distinct but related IgSF proteins (Clark et al., 2001; Stipp et al., 2001a; Charrin et al., 2003a). In relatively stringent detergent conditions (1% Brij 96/97), EWI-2–CD81 and EWI-2–CD9 complexes are stable, fully soluble, limited in size (<4 million D), highly stoichiometric, and can be chemically cross-linked, indicative of direct protein–protein interactions (Claas et al., 2001; Stipp et al., 2001a,b; Charrin et al., 2003a).
EWI-2 is widely expressed, with prominent mRNA expression in the brain (Clark et al., 2001; Stipp et al., 2001a), and protein expression on peripheral blood lymphocytes and hepatocytes, where it colocalizes with CD81 (Charrin et al., 2003a). We hypothesized that, as a major tetraspanin partner, EWI-2 might regulate cell motility on laminin, given the preferential association of tetraspanins with laminin-binding integrins. Our results establish EWI-2 as a novel regulator of cell reaggregation and motility on laminin-5 and reveal CD9 and CD81 as key linkers in a physical complex of EWI-2 with α3β1 integrin, a laminin-5 receptor.
Results
Expression of EWI-2 in A431 epithelial carcinoma cells
To study EWI-2 function, we transduced A431 epidermoid carcinoma cells with a retroviral vector encoding COOH-terminally FLAG-tagged EWI-2 (A431 EWI-2 cells) or with empty vector (A431 IZ cells). We detected intact EWI-2 (and a 50-kD fragment) in lysates of surface-biotinylated A431 EWI-2 cells, but not A431 IZ control cells (Fig. 1 B, inset). The level of EWI-2 expression achieved in A431 EWI-2 cells is not excessively high, but rather, is comparable to endogenous EWI-2 expression in other cell lines (e.g., 293 embryonic kidney cells; unpublished data). Levels of laminin-5 receptors (α3β1 and α6β4) on A431 cells were unaffected by EWI-2 expression. Also, A431 EWI-2 and A431 IZ cells showed equal static cell adhesion (unpublished data) and equal spreading on laminin-5 (Fig. 1, A and B). Anti-α6 antibody failed to alter A431 cell spreading (Fig. 1, C and D), indicating that α6β4 is not required. In contrast, anti-α3 blocking antibody strongly impaired spreading of both cell types (Fig. 1, E and F). Hence, α3β1 mediates A431 cell spreading on laminin-5, as seen for other cell types (Xia et al., 1996; DiPersio et al., 1997). An α6 contribution to spreading was uncovered when anti-α3 and -α6 antibodies were used in combination (Fig. 1, compare G and H with E and F), with nearly complete inhibition by both antibodies together, ∼50% reduction with anti-α3 antibody, and minimal reduction with anti-α6 antibody (Fig. 2). Thus, α3β1 alone is sufficient for A431 cell spreading, but α6β4 can partially compensate when α3β1 function is blocked.
Figure 1.
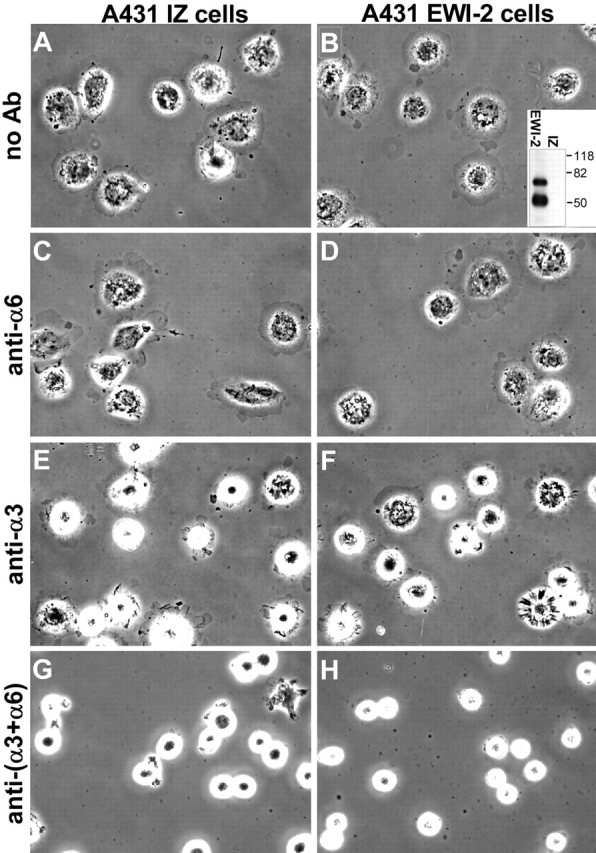
EWI-2 overexpression does not affect A431 cell spreading on laminin-5. A431 IZ control cells or A431 EWI-2 cells were plated in serum-free medium on laminin-5–coated coverslips with no antibodies (A and B), or in the presence of 5 μg/ml GoH3 anti-α6 integrin (C and D), 10 μg/ml A3-IIF5 anti-α3 integrin (E and F), or both anti-α3 and -α6 antibodies (G and H). Cells were fixed after 20 min and photographed with phase contrast. Inset in B shows anti-FLAG EWI-2 immunoprecipitate from cell surface–biotinylated A431 EWI-2 cells (EWI-2) or A431 IZ control cells (IZ) developed with HRP-ExtrAvidin®. Mol wt markers (kD) are indicated.
Figure 2.
Quantitation of A431 IZ cells and A431 EWI-2 cell spreading assays on laminin-5. The areas of 20–45 cells per coverslip were measured using Scion Image v1.62 software. Specific spreading was calculated by subtracting the mean area of cells fixed immediately after plating from the mean area of cells fixed at the end of the assay. Error bars indicate the SEM for each coverslip. Two coverslips per condition were examined; results for each coverslip are shown individually. Asterisk Indicates statistically significant difference between antibody-treated cells and the coverslips in the no antibody controls (P < 0.0005; Bonferroni post-ANOVA t test).
EWI-2 overexpression impairs A431 cell reaggregation on laminin-5
Although EWI-2 overexpression did not alter α3 or α6 integrin expression, adhesion, or spreading functions in A431 cells, dramatic effects were seen in longer-term assays. After enzymatic dissociation, a single-cell suspension of A431 cells replated on laminin-5 (Fig. 3 A) began to reaggregate. Within 5–6 h, many cells clustered into small islands (Fig. 3 B) that progressively fused to reestablish the epithelial sheet morphology of A431 cells by 18–24 h (Fig. 3 C). Like the A431 parental cell line, A431 IZ control cells reaggregated into characteristic clusters within 5–6 h (Fig. 4 A), with 20% of the cells in clusters of four cells or more and only ∼50% remaining as single cells (Fig. 4 C). By contrast, reaggregation of A431 EWI-2 cells was dramatically impaired (Fig. 4 B), as they exhibited a fivefold reduction in cluster formation and a corresponding increase in the proportion of single cells (Fig. 4 C). A431 EWI-2 cells did eventually reaggregate after overnight incubation (unpublished data). Reduced aggregation of A431 EWI-2 cells was not due to decreased viability on laminin-5. Between 1 and 24 h after plating on laminin-5, A431 IZ and A431 EWI-2 cells expanded by 124 and 113%, respectively, as measured by tetrazolium bromide (MTT) assay on quadruplicate wells. On the α2β1 integrin ligand (collagen I), both cell types reaggregated with equal efficiency (Fig. 4, D–F). Antibody inhibition experiments confirmed the α2β1 dependence of A431 attachment and spreading on collagen I (unpublished data).
Figure 3.
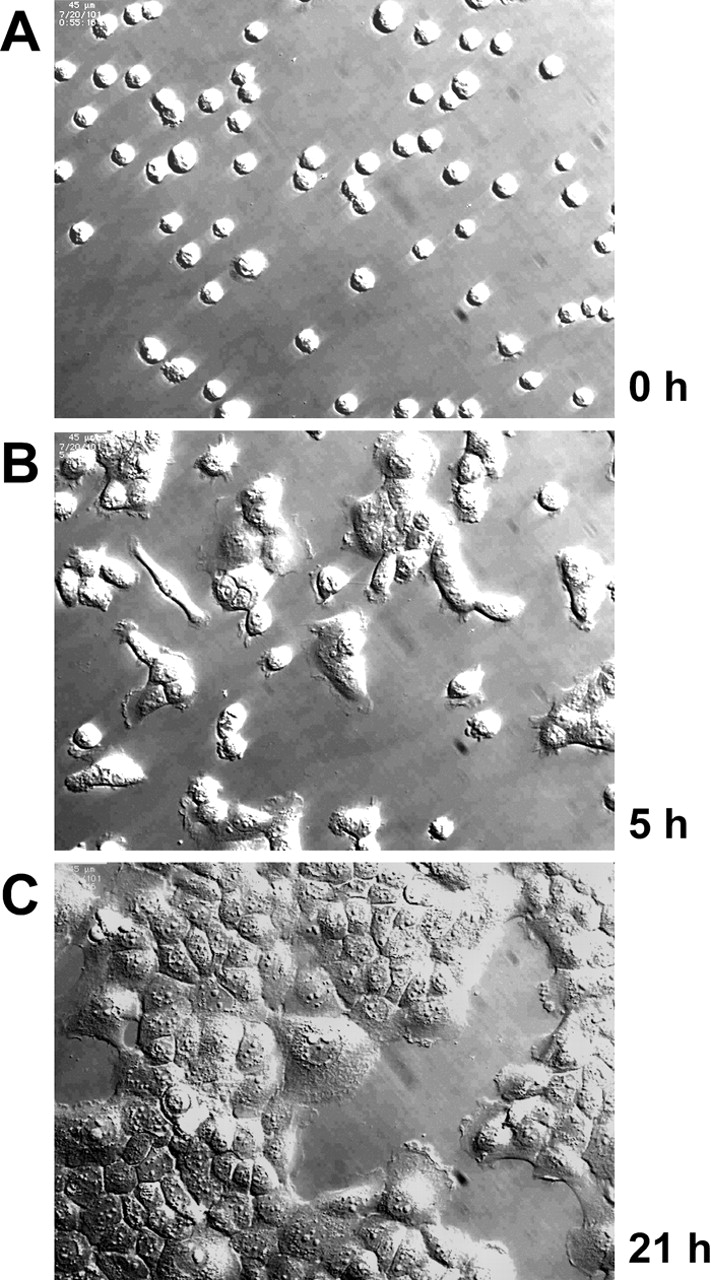
Reaggregation of dispersed A431 cell on laminin-5. A431 cells were enzymatically dissociated and plated in serum-free medium on laminin-5. The same field of cells was monitored periodically over a period of 21 h while being maintained in a microscope stage incubator. (A) Cells just after plating; (B) after 5 h; (C) after 21 h.
Figure 4.
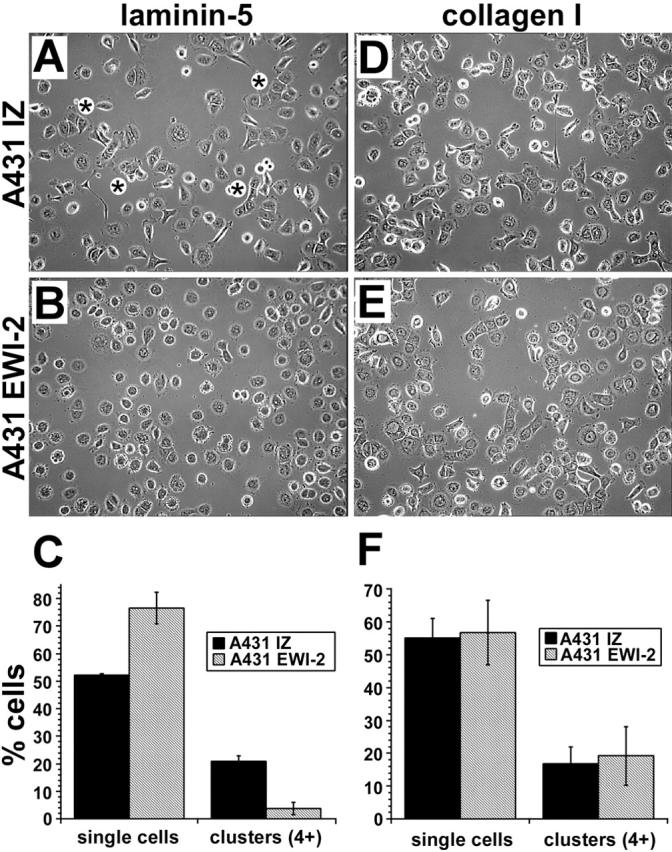
EWI-2 expression impairs A431 cell reaggregation on laminin-5. A431 IZ control or EWI-2 cells were plated, after enzymatic dissociation, in serum-free medium on coverslips coated with laminin-5 (A–C) or collagen I (D–F). After 5 h, cells were fixed and photographed in phase contrast, revealing several clusters of four or more A431 IZ cells in most fields, as indicated by asterisks in A. By contrast, few clusters of A431 EWI-2 cells were observed in B. (C) Quantitation of three separate experiments (two coverslips per condition in each experiment, five fields per coverslip) revealed a statistically significant decrease in the percentage of clustered A431 EWI-2 cells on laminin-5 (P < 0.005; t test), and a corresponding increase in the percentage of single cells (P < 0.02; t test). After 5 h on collagen I, both cell types had similar clustering properties as shown in photos (D and E) and by quantitation (F).
Decreased motility of EWI-2–overexpressing cells
The reduced reaggregation observed in Fig. 4 could result from a failure of normally migrating cells to form stable cell–cell contacts, or from decreased cell motility leading to decreased opportunity for cell–cell interactions. In a time-lapse video (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200309113/DC1), A431 IZ control cells exhibited robust random motility upon plating on laminin-5, whereas A431 EWI-2 cells attached and spread, but migrated at far slower rates, or not at all. Tracking of individual A431 EWI-2 cells revealed a 60–70% decrease in the mean migration rate (Fig. 5 A), an 84–88% decrease in the number of 10-min intervals with a mean rate of 30 μm/h or more, and a nearly twofold increase in the number of 10-min intervals with a mean migration rate of 10 μm/h or less (Fig. 5 B). A similar result was observed for cells migrating in the presence of an anti-α6 integrin-blocking antibody (unpublished data). Thus, α6β4 needn't be engaged with ligand for EWI-2 to regulate α3β1 function. To begin to define which region of EWI-2 is responsible for its anti-migratory activity, we introduced into A431 cells a chimeric EWI-2 protein, with the EWI-2 cytoplasmic tail replaced by that of CD2. This chimera, EW2xCD2c, was expressed on the surface of A431 cells and engaged a similar fraction of α3 integrin in complexes (see Fig. 9, A and B), but failed to impair migration on laminin-5 (Fig. 5 C).
Figure 5.
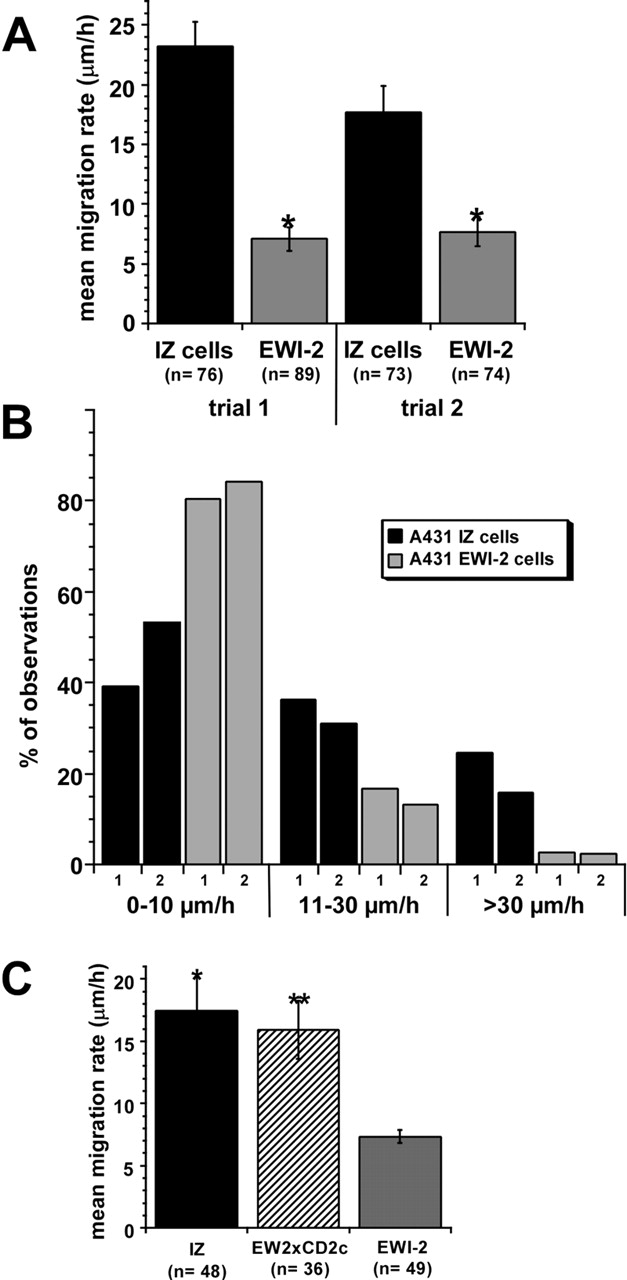
Impaired migration of A431 EWI-2 cells on laminin-5. (A) In two separate trials, XY locations of individual cells were determined once every 10 min using Scion Image v1.62 software, and the average migration rate was calculated from the series of XY measurements using KaleidaGraph v3.05 software (Abelbeck Software). A431 EWI-2 cells migration rates were 60–70% lower than those of A431 IZ control cells (*, P < 0.0001; t test). (B) The total set of migration rates calculated during each 10-min observation period was binned to show the distribution of migration rates for A431 IZ cells and A431 EWI-2 cells. On average, A431 EWI-2 cells spent 84–88% less time moving ≥30 μm/h, and 1.6–2-fold more time moving ≤10 μm/h (see Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200309113/DC1). (C) Mean migration rates of A431 IZ, A431 EW2xCD2c, and A431 EWI-2 cells were measured as in A. A431 EWI-2 cell migration rate was reduced ∼60% compared with A431 IZ or A431 EW2xCD2c (*, P < 0.001; **, P < 0.002; Bonferroni post-ANOVA t test). There was no statistically significant difference between the migration rates of A431 IZ and A431 EW2xCD2c cells (P > 0.1). For all experiments in this figure, coverslips were coated with 0.5 μg/ml rat laminin-5 and blocked with serum-free medium. Additional details regarding data acquisition and videomicroscopy appear in Zhang et al. (2002).
Figure 9.
Biochemical analysis of the EW2xCD2c chimera. (A) Wild-type and mutant EWI proteins were immunoprecipitated using anti-FLAG antibody, and then samples were analyzed by blotting using anti-α3, anti-CD81 (M38), and anti-FLAG pAbs. CD81 coprecipitating with EWI-2 is indicated with a small black arrow (middle). On the bottom, dark arrows indicate mature EWI proteins, and white arrows indicate immature forms. (B) From Brij 96 lysates of 106 cell surface–biotinylated, transduced A431 cells, EWI proteins were immunoprecipitated using anti-FLAG antibody, and then analyzed by HRP-ExtrAvidin® blotting. The locations of α3 integrin, intact EW2xCD2c, intact EWI-2, CD9, and CD81 are indicated. Two exposures of the same blot are presented to allow comparison of the relative amounts of α3, CD9, and CD81 that coprecipitate with each protein. Note that intact EW2xCD2 (∼78 kD) and intact EWI-2 (∼70 kD) are each accompanied by two smaller cleavage products. (C) 3.6 × 106 A431 cells expressing EW2xCD2c or EWI-2were lysed in Brij 96, and the indicated proteins were immunoprecipitated. Samples were then immunoblotted for FLAG (M2 anti-FLAG mAb) or CD81 as above. In all cases, small differences in total protein in the lysates were measured by amido black assay and corrected before immunoprecipitation. Lanes 1* and 2* represent shorter exposures of lanes 5 and 6, respectively. (D) α3 integrin was immunoprecipitated from Brij 96 lysates of 3.3 × 105 transduced A431 cells. Levels of CD81 and α3 within the immunoprecipitates were revealed by immunoblotting with M38 anti-CD81 mAb (top) or with anti-α3 pAb (bottom).
Altered CD81 localization in EWI-2–overexpressing cells
No obvious differences in α3β1 localization were observed as A431 IZ, A431 EW2xCD2c, and A431 EWI-2 cells all showed smooth, bright staining over the surface, with prominent labeling of filopodia at the cell perimeters (Fig. 6, A–C). In contrast, CD81 was more prominent at the cell periphery in A431 EWI-2 cells, compared with IZ or EW2xCD2c cells (Fig. 6, compare F to D and E; L to J and K). Especially striking was the enhanced punctate CD81 staining on the filopodia of A431 EWI-2 cells. In A431 IZ and EW2xCD2c cells, CD81 staining was more evenly distributed and was not as concentrated at the cell perimeter. In phase contrast (Fig. 6, G–I), A431 EWI-2 cells were typically more symmetrical, consistent with their reduced migration. This increased symmetry by itself does not explain the differences in CD81 localization because A431 IZ and EW2xCD2c cells with similar morphology still failed to show altered CD81 staining (Fig. 6, D, E, G, and H; arrows). An antibody to α4 integrin, which is not expressed by A431 cells, revealed a low level of background staining (Fig. 6 M). Thus, although EWI-2–overexpressing cells did not show altered localization of α3β1 integrin itself at the cell periphery, more CD81 became concentrated in perimeter filopodia, consistent with increased physical association with α3 (see Fig. 9 D).
Figure 6.
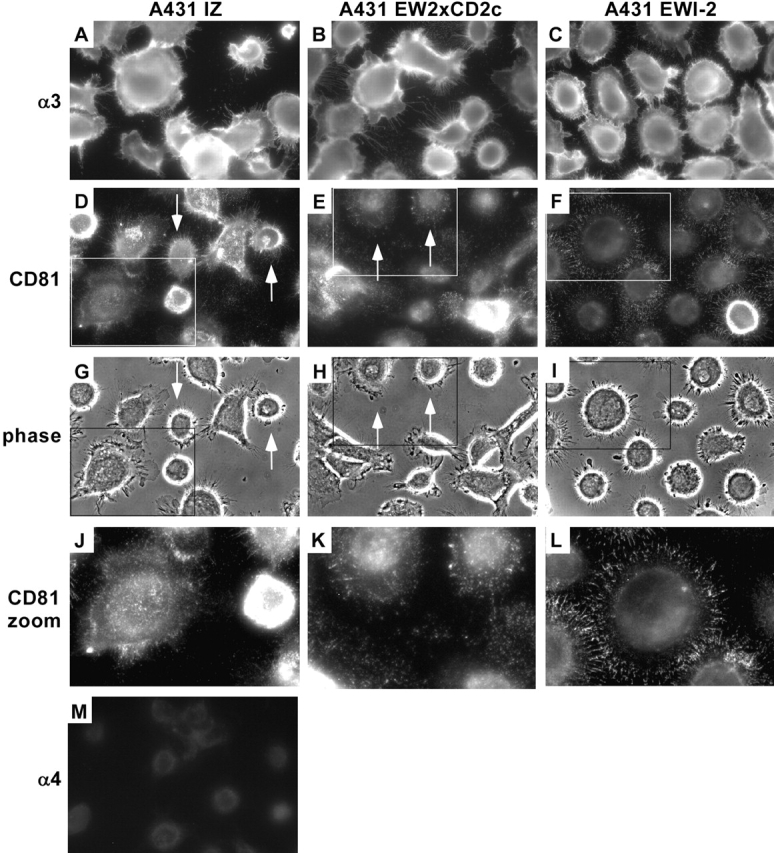
Altered CD81 localization in A431 EWI-2 cells. A431 IZ, A431 EW2xCD2c, and A431 EWI-2 cells were plated on laminin-5–coated coverslips in serum-free medium and fixed after 3.5 h. Cells were stained for α3 integrin with the A3X8 mAb (A–C), CD81 with the JS64 mAb (D–F and J–L), or α4 integrin with the B5G10 mAb (M). Staining was visualized with FITC-goat anti–mouse secondary antibody. Although α3 localization appeared similar in all three cell types, CD81 was strongly concentrated in filopodia at the perimeters of A431 EWI-2 cells. G–I shows phase-contrast images of the same fields as in D–F, to reveal cell morphology. J–L show expansion of regions boxed in D–F and G–I. Arrows in D, E, G, and H indicate a subset of cells with a round, symmetrical morphology that nevertheless failed to demonstrate the striking concentration of CD9 and CD81 at the cell perimeter that was observed in the EWI-2–overexpressing cells.
EWI-2 physically associates with α3β1 integrin
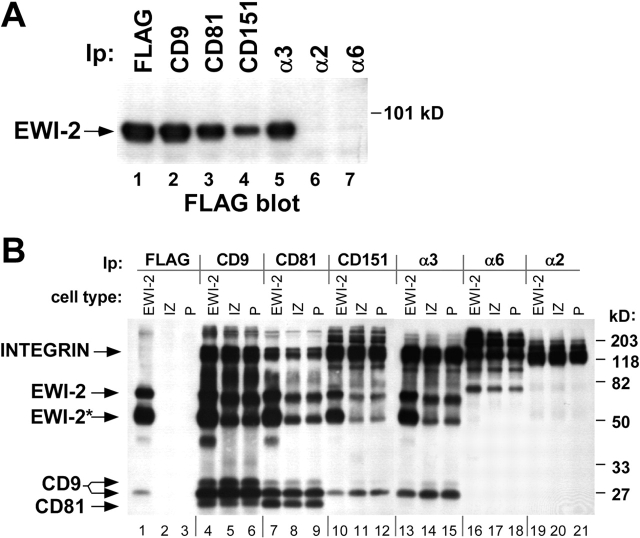
Because EWI-2 and α3 integrin can each associate with both CD9 and CD81 (Berditchevski et al., 1996; Clark et al., 2001; Stipp et al., 2001a; Charrin et al., 2003a), we hypothesized that CD9 and/or CD81 might therefore link α3β1 to EWI-2. Indeed, compared with the amount of EWI-2 recovered directly by anti-FLAG immunoprecipitation (Fig. 7 A, lane 1), a substantial fraction of EWI-2 could also be recovered in an α3β1 immunoprecipitate (Fig. 7 A, lane 5) prepared from a 1% Brij 96 lysate of A431 EWI-2 cells. In contrast, no EWI-2 was detected in association with integrins α2β1 (Fig. 7 A, lane 6) or α6β4 (Fig. 7 A, lane 7). A substantial amount of EWI-2 was also found associated with tetraspanins CD81 and CD9 (Fig. 7 A, lanes 2 and 3), as observed previously. Association of EWI-2 with tetraspanin CD151 (Fig. 7 A, lane 4) is consistent with CD151 being tightly associated with α3β1 integrin (Yauch et al., 1998). The recovery of a smaller amount of EWI-2 (Fig. 7 A, compare lane 4 with lane 5) likely occurs because the 5C11 epitope on CD151 is partly obscured in Brij 96 detergent conditions (Yang et al., 2002).
Figure 7.
EWI-2 associates with tetraspanins and α3β1 integrin. (A) A431 EWI-2 cells (∼5 × 106) expressing FLAG-tagged EWI-2 were lysed in 1% Brij 96. Equal volumes of the lysate were used for immunoprecipitations with anti-FLAG agarose or with anti-integrin or -tetraspanin antibodies. The blot was developed with biotinylated anti-FLAG antibody followed by HRP-ExtrAvidin® to detect coprecipitating EWI-2. (B) A431 EWI-2 cells (EWI-2), A431 IZ cells (IZ), and A431 parental cells (P) were labeled with sulfo-NHS biotin and lysed in 1% Brij 96. Lysates were immunoprecipitated with anti-FLAG agarose or antibodies to the indicated tetraspanins and integrins, followed by blotting with HRP-ExtrAvidin®. EWI-2* designates a 50-kD EWI-2 cleavage product, and two electrophoretic forms of CD9 are indicated. ∼7 × 105 cell equivalents were used for each immunoprecipitation. Flow cytometry results confirm that CD9, CD81, CD151, and α3β1 expression levels are similar in the different cell types. The bands of ∼210 kD in the α6 lanes (16–18) and CD151 lanes (10–12) correspond to integrin β4, a major α6 partner in A431 cells (Rabinovitz et al., 1999). α6β4 is known to associate with CD151 (Sterk et al., 2000). Densitometry indicated an approximate fourfold increase in α3-associated EWI-2 upon EWI-2 overexpression. Similar results were obtained in two independent experiments.
Immunoprecipitation of cell surface–biotinylated molecules confirmed that the EWI-2–α3β1 complex indeed occurs on the cell surface (Fig. 7 B). Using anti-FLAG immunoprecipitation, intact EWI-2 (70 kD) and the 50-kD EWI-2 cleavage product (Stipp et al., 2001a) were obtained from A431 EWI-2 cells (Fig. 7 B, lane 1), but not from mock infected (IZ), or parental (P) cells (Fig. 7 B, lanes 2 and 3). These same EWI-2 bands were present in immunoprecipitations of CD151 (Fig. 7 B, lanes 10–12), and α3 integrin (Fig. 7 B, lanes 13–15), but not α6 integrin (Fig. 7 B, lanes 16–18) or α2 integrin (Fig. 7 B, lanes 19–21). Levels of cell surface–biotinylated α3β1 integrin were similar in all three cell types (e.g., Fig. 7 B, lanes 13–15), consistent with flow cytometry results (unpublished data). As measured by densitometry, in A431 EWI-2 cells, ∼4 times more EWI-2 was associated with α3β1 integrin (Fig. 7 B, compare lane 13 with lanes 14 and 15), consistent with exogenous EWI-2 being present approximately fourfold above background levels in A431 cells.
EWI-2–α3β1 complexes: a critical role for tetraspanins
Three lines of evidence indicate that tetraspanins play a key role in linking EWI-2 to α3β1. First, tetraspanins CD9 and CD81 associate with both EWI-2 and α3β1. Immunoprecipitations of CD9 (Fig. 7 B, lanes 4–6) and CD81 (Fig. 7 B, lanes 7–9) yielded both α3β1 integrin and EWI-2, with higher levels of EWI-2 (Fig. 7 B, lanes 4 and 7) obtained from EWI-2–overexpressing cells. Integrins (α2β1, α6β4) not associating with CD9 and CD81 at the same time failed to show associated EWI-2 (Fig. 7 B, lanes 16–21). Second, removal of CD9 and CD81 resulted in diminished EWI-2–α3β1 association. After immunodepletion with anti-CD9 and -CD81 antibodies, α3 integrin immunoprecipitation yielded a level of EWI-2 (Fig. 8 A, lane 5) that was 85% reduced (as measured by densitometry) compared with that seen in control immunodepletions (Fig. 8 A, lanes 4 and 6). Removal of CD9 and CD81 was essentially complete, as seen in a subsequent CD81 immunoprecipitation (Fig. 8 A, lane 2), whereas mock immunodepletion or α2 integrin depletion failed to remove CD9 and CD81 or associated EWI-2 (Fig. 8 A, lanes 1 and 3). Conversely, α2 integrin was all removed by α2 immunodepletion (Fig. 8 A, lane 9), but not by mock or CD9/CD81 depletion (Fig. 8 A, lanes 7 and 8). Because CD9/CD81 depletion removed only a fraction (<10%) of total α3β1 integrin (Fig. 8 A, compare lane 4 with lane 5), most α3β1 appears not to be associated with CD9, CD81, or EWI-2. Third, the introduction of CD81 into CD81-deficient U937 monocytic leukemia cells (Hamaia et al., 2001) resulted in EWI-2–α3β1 association. Immunoprecipitation of α3 yielded FLAG-tagged EWI-2 when CD81 was present (Fig. 8 B, lane 6), but not when absent (Fig. 8 B, lane 5). Control CD81 immunoprecipitations from CD81+ cells yielded EWI-2 (Fig. 8 B, lane 4) and α3 integrin (Fig. 8 B, lane 8), as well as CD81 itself (Fig. 8 B, lane 12), whereas CD81 immunoprecipitations from CD81− cells did not (Fig. 8 B, lanes 3, 7, and 11). Conversely, α3 immunoprecipitation yielded CD81 only when CD81 was present (Fig. 8 B, compare lane 14 with lane 13). Immunoblotting of directly immunoprecipitated α3 confirmed that α3 levels were similar in CD81− and CD81+ cells (Fig. 8 B, lanes 9 and 10). Although smaller biosynthetic precursors of α3 were present in the total α3 blot (Fig. 8 B, lanes 9 and 10), exclusively the mature form of α3 integrin associated with CD81 (Fig. 8 B, lane 8), as previously observed (Kazarov et al., 2002).
Figure 8.

Tetraspanins support EWI-2-α3β1 integrin association. (A) ∼107 A431 EWI-2 cells were labeled with sulfo-NHS biotin and lysed in 1% Brij 96. Equal portions of lysate were depleted three times with protein G alone (mock) or with protein G plus anti- tetraspanin or α2 integrin antibodies. Depletions are indicated at the top of the figure. Depleted lysates were further divided and immunoprecipitated with anti-CD81, anti-α3 integrin, or anti-α2 integrin antibodies conjugated to agarose followed by SDS-PAGE and blotting with HRP-ExtrAvidin®. EWI-2* is a 50-kD EWI-2 cleavage fragment. Densitometry revealed that CD9 and CD81 depletion removed only ∼10% of total α3 integrin, but ∼85% of the α3-associated EWI-2. Densitometry of an independent experiment confirmed that only ∼10% of α3 is CD81-associated in A431 cells (not depicted). (B) U937 cells lacking CD9 and CD81 were transduced with CD81 and EWI-2 retroviral expression vectors and selected to obtain stable (CD81−, EWI-2+) or (CD81+, EWI-2+) cell lines. Then, both cell types were super-infected with an α3 integrin retroviral expression vector, to yield equivalent α3 expression levels, as confirmed by flow cytometry (not depicted). EWI-2 (M2 anti-FLAG mAb), CD81 (M38 mAb), or α3 integrin (A3X8 mAb) were immunoprecipitated from 1% Brij 96 lysates of CD81− or CD81+ cells. Immunoprecipitates were blotted for EWI-2 (biotinylated M2 anti-FLAG mAb), α3 integrin (D23 pAb), or CD81 (M38 mAb). Apparent mature and immature forms of EWI-2 and α3 integrin are indicated with filled and open arrows, respectively. 6 × 106 cell equivalents were analyzed in EWI-2 and CD81 immunoprecipitations, and 4.8 × 107 cell equivalents in the α3 immunoprecipitations. (C) Biotinylated CD81− or CD81+ cells were lysed in 1% Brij 96, and EWI-2 (FLAG), α3 integrin, or CD81 were immunoprecipitated as above. Cell surface–labeled proteins were revealed by HRP-ExtrAvidin® blot. 1.7 × 106 cell equivalents were analyzed in each immunoprecipitation.
Although EWI-2 was present in both CD81− and CD81+ cells, EWI-2 levels were somewhat lower in the CD81− cells, and much of the EWI-2 protein was shifted to a slightly lower size (Fig. 8 B, lanes 1 and 2). Immunoprecipitation of FLAG-tagged, cell surface–biotinylated EWI-2 established that only a small amount of EWI-2 was on the surface of CD81− cells (Fig. 8 C, top; lane 1), in contrast to the abundant EWI-2 on the surface of CD81+ cells (Fig. 8 C, lane 2). Hence, in the absence of CD81, EWI-2 remains mostly in an incompletely glycosylated, immature form that does not reach the U937 cell surface. Similar amounts of α3 integrin appeared on the surface of CD81− and CD81+ cells (middle), and cell surface labeled CD81 was detected only in CD81+ cells (lane 2, bottom). Thus, CD81 associates relatively early in biosynthesis with EWI-2 and then facilitates EWI-2 trafficking to the cell surface, where it is assembled into an EWI-2–CD81–α3 integrin complex.
EWI-2 mutant (EW2xCD2c) retains α3β1 association, but shows other biochemical deficiencies
Immunoprecipitation of FLAG-tagged EW2xCD2c and EWI-2 yielded similar amounts of associated α3 integrin as seen either by immunoblotting (Fig. 9 A, top; lanes 2 and 3) or by cell surface biotin labeling (Fig. 9 B, top; lanes 2 and 3). However, the EW2xCD2c protein showed substantially diminished association with CD81. CD81 was detected by blotting in immunoprecipitates of EWI-2 (Fig. 9 A, middle; lane 3), but not EW2xCD2c (Fig. 9 A, lane 2). Consistent with CD81-dependent maturation results in Fig. 8 B, the EW2xCD2c protein (with impaired CD81 association) showed a lower ratio of mature/immature forms (Fig. 9 A, bottom; compare lane 2 with lane 3). Consistent with retarded maturation, EW2xCD2c and EWI-2 showed similar levels of total FLAG-tagged protein (Fig. 9 A, bottom), but the amount of surface-labeled EW2xCD2c was somewhat diminished compared with EWI-2 (Fig. 9 B, compare lane 2 with lane 3).
If tetraspanin CD81 association is greatly diminished, then how does EW2xCD2c maintain α3 integrin association in the absence of this critical linker protein? EW2xCD2c (compared with EWI-2) retained diminished but readily detectable association with tetraspanin CD9, whereas CD81 was barely detectable even in the longer exposure (Fig. 9 B, lanes 2 and 3). In another experiment, immunoprecipitation of CD81 yielded no EW2xCD2c (Fig. 9 C, top; lane 3) and EW2xCD2c yielded no CD81 (Fig. 9 C, bottom; lane 1), whereas immunoprecipitation of CD9 yielded clearly detectable EW2xCD2c protein, exclusively in its mature form (Fig. 9 C, compare lanes 1 and 2 with lanes 1* and 2*). Notably, the level of mature EW2xCD2c in the CD9 immunoprecipitation (Fig. 9 C, lane 2) exceeded the level of mature EW2xCD2c present in the α3 integrin immunoprecipitation (Fig. 9 C, lane 4). Hence, even in the absence of CD81 association, there is sufficient EW2xCD2c-associated CD9 to allow linkage to α3 integrin.
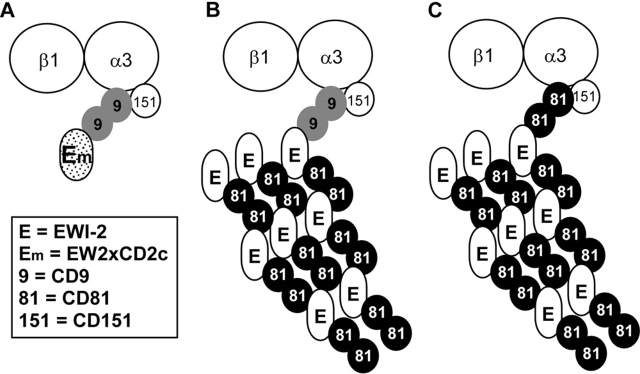
Evidence for extended EWI-2–CD81–α3β1 complexes
Several experiments suggest that additional CD81 and EWI-2 molecules may be appended to EWI-2–CD81–α3β1 and EWI-2–CD9–α3β1 “core” complexes, but not to EW2xCD2c−CD9−α3β1 core complexes. For example, immunoprecipitations of EWI-2 and EW2xCD2c yielded similar levels of associated α3 (Fig. 9, A and B), but immunoprecipitations of CD9, CD81, or α3 each yielded more EWI-2 than EW2xCD2c (Fig. 9 C, top; compare lanes 6–8 with lanes 2–4). In addition, from EWI-2–transfected A431 cells, immunoprecipitations of EWI protein, CD9, or α3 each yielded more CD81 (Fig. 9 C, bottom; compare lanes 5, 6, and 8 with lanes 1, 2, and 4). In Fig. 9 D, immunoprecipitation of α3 again yielded much more CD81 from EWI-2–expressing A431 cells (Fig. 9 D, lane 3), than from EW2xCD2–expressing cells (Fig. 9 D, lane 2). Together, these results suggest that although EW2xCD2c is adequate for assembly into core (EWI–CD9–α3β1) complexes, its loss of CD81 association coupled with diminished surface expression limits the recruitment of additional CD81 and EWI proteins (Fig. 10; see Discussion).
Figure 10.
Hypothetical model of EWI-2-α3β1 and EW2xCD2c-α3β1 complexes. (A) CD9 links EW2xCD2c to integrin α3β1-CD151 complex. Only the “core” complex is shown because EW2xCD2c is unable to associate with CD81. (B and C) Either CD9 or CD81 is capable of linking EWI-2 to α3β1 integrin in the core complex. However, overexpression of wild-type EWI-2 (but not EW2xCD2c) results in extension of the core complex to include additional EWI-2 and CD81. Hence, we can explain why immunoprecipitation of either EW2xCD2c or EWI-2 yields similar levels of associated α3β1, whereas immunoprecipitation of α3β1 yields abundant CD81 and EWI-2 (in EWI-2–transfected cells), but not much CD81 and EW2xCD2c (in EW2xCD2c-transfected cells). The model also explains why more CD81 can associate with α3β1 when EWI-2 is present. Compared with EW2xCD2c, EWI-2 expression also results in enhanced CD9–EWI-2 and CD9–CD81 complex formation (Fig. 9 C). However, these complexes are not indicated in the model because we do not know the extent to which they show association with α3β1.
Discussion
EWI-2 regulates epidermoid cell reaggregation and motility on laminin-5
Here, we establish that the IgSF protein EWI-2 associates with α3β1 integrin (a major laminin-5 receptor) and acts as a negative regulator of epithelial cell reaggregation and motility on laminin-5. Expression of EWI-2 at approximately fourfold elevated levels in A431 cells had essentially no effect on expression levels of other molecules (α3β1, other integrins, and associated tetraspanins CD9 and CD81) or on cell viability on laminin-5. Likewise, EWI-2 expression did not alter α3β1 integrin–dependent cell adhesion, the extent of cell spreading on laminin-5, α3β1 subcellular localization, or adhesion-stimulated FAK phosphorylation (unpublished data). Hence, we infer that EWI-2 did not alter α3β1 integrin affinity for ligand, or initial α3β1-dependent cell adhesion. Instead, EWI-2 exerted a highly specific effect on a select few functions (cell reaggregation and motility) that occur after cell attachment and spreading.
EWI-2 neither associated with α2β1 integrin nor affected reaggregation of A431 EWI-2 cells on collagen I, an α2β1 ligand. Although EWI-2 did reduce the mean A431 cell migration rate on collagen I (by ∼30–50%; unpublished data), this effect was not as great as that seen on laminin-5 (60–70%). This result is consistent with α3β1 functioning to some extent as a secondary receptor on a wide range of ligands to which it does not mediate initial attachment (DiPersio et al., 1995). The results also suggest a nonlinear correlation between cell migration and cell reaggregation, such that reaggregation is strongly affected by large changes in migration, but is disproportionately less affected by smaller changes in cell migration. Integrin α6β4 also did not associate with EWI-2 and, compared with α3β1, contributed much less to cell spreading on laminin-5. This is in accord with earlier analyses, in which α6β4 integrin contributed to cell attachment on laminin-5, but α3β1 was required for cell spreading (Xia et al., 1996; DiPersio et al., 1997).
Our results yield notable clues regarding the mechanism for EWI-2 effects on α3 integrin–dependent function. First, EWI-2 caused tetraspanin CD81 protein to localize into filopodia at the cell periphery, an ideal position in which to affect cell migration. Second, EWI-2 promoted an increased biochemical association of CD81 with α3β1 integrin. A mutant form of EWI-2 (EW2xCD2c) that failed to associate with CD81, and also did not inhibit cell migration on laminin-5, in parallel failed to redirect CD81 to filopodia and failed to enhance CD81–α3β1 association. We propose that EWI-2–dependent redirection of CD81 into proximity with filopodial α3β1 should also lead to enhanced recruitment of CD81-associated cytoplasmic signaling enzymes, such as classical PKC isoforms and PI 4-kinase (Berditchevski et al., 1997; Zhang et al., 2001). These enzymes, together with other CD81-associated molecules, would then be in position to influence α3β1-mediated cell migration.
Transmembrane proteins CD98, CD36, and CD47 all associate with various β1 and/or β3 integrins and modulate adhesive and/or motility functions (Thorne et al., 2000; Brown and Frazier, 2001; Fenczik et al., 2001). However, those molecules associate with integrins in the context of detergent-insoluble light membranes (Green et al., 1999; Thorne et al., 2000; Kolesnikova et al., 2001), whereas EWI-2–tetraspanin and tetraspanin–α3β1 associations occur within detergent-soluble fractions (Claas et al., 2001; Stipp et al., 2001a). Another member of the EWI subfamily, EWI-F/CD9-P1, can also associate with α3β1 integrin (Charrin et al., 2003b), but the functional consequences have not yet been determined. In another analysis, EWI-2 overexpression impaired prostate cancer cell migration on laminin-1 and fibronectin (Zhang et al., 2003), but EWI-2 association with integrins was not studied, and the mechanism of EWI-2 function was not addressed.
EWI-2 association with α3β1 is mediated by tetraspanins CD9 and CD81
Several observations point to EWI-2 association with α3β1 being mediated by tetraspanins CD9 and CD81. First, in lysates depleted for CD9 and CD81, the majority of EWI-2 associated with α3β1 was removed, even though the majority of α3β1 itself remained undepleted. Thus, EWI-2 is linked, via tetraspanins, to a small but critical subset of α3β1. Second, more integrin was recovered in a CD9 immunoprecipitation than in EWI-2 immunoprecipitation; and more EWI-2 was recovered by CD9 (or CD81) immunoprecipitation than by anti-α3β1 (or CD151) immunoprecipitation. Again, this supports the idea that CD9 and CD81 are each more proximal to both the integrin and the EWI-2 than the integrin is to EWI-2. Third, CD9 and CD81 can associate with EWI-2 under conditions in which no integrin is present (Stipp et al., 2001a), and CD81 can associate with integrins under conditions in which no EWI-2 appears to be present (Zhang et al., 2001). Fourth, in U937 cells lacking CD9 and CD81, EWI-2 failed to associate with α3β1 unless CD81 was coexpressed. Finally, the EW2xCD2 mutant, lacking CD81 association capability, instead used CD9 for linkage to α3β1 (Fig. 10). These results strongly support a model in which either CD9 or CD81 can link EWI protein to α3β1. Differences between CD9 and CD81, with respect to EW2xCD2 association, suggest that the EWI-2 cytoplasmic tail could be more important during CD81 association.
Our works reveal a novel function for CD81 in EWI-2 trafficking. First, in the absence of CD81 (or CD9), little EWI-2 underwent full biosynthetic maturation and appeared on the surface of U937 cells. Second, the EW2xCD2c mutant, deficient in CD81 association, also showed impaired maturation and cell surface expression. In the latter case, retention of CD9 association likely accounted for a significant fraction of EW2xCD2c maturing, reaching the cell surface, and engaging α3β1 to the same extent that EWI-2 engages α3β1. The finding that only ∼10% of the α3β1 is involved in a complex with tetraspanins and EWI-2 helps to explain why EW2xCD2c and EWI-2 both engage a similar fraction of α3β1, despite the lower cell surface expression of EW2xCD2c. The 10% α3β1 estimation is consistent with previous estimates of α3β1–tetraspanin stoichiometry (Berditchevski et al., 1996). However, for the critical subset of α3β1 molecules engaged with ligand in filopodia at the cell periphery, EWI-2–tetraspanin–α3β1 association may be much greater than 10%.
Expanded EWI-2–CD81–α3β1 complexes include additional CD81 and EWI-2 molecules
Both EWI-2 and the EW2xCD2c mutant were similarly linked via tetraspanins (CD81 and/or CD9) to the α3β1 integrin. However, EWI-2 suppressed cell migration and enhanced the recruitment of CD81 to critical filopodial sites, whereas the EW2xCD2c mutant had neither of these activities. To explain these key functional differences, we look beyond the core EWI–tetraspanin–α3β1 complexes and consider the role of expanded complex formation. As illustrated in Fig. 10, both mutant and wild-type EWI-2 are linked to a similar extent to α3β1, with EW2xCD2c using CD9, and EWI-2 using CD9 or CD81. However, with CD81 association leading to more EWI-2 cell surface expression, there is capability to assemble additional CD81 and EWI-2 molecules into an expanded complex. Hence, CD81 not only links EWI-2 to α3β1, but EWI-2 also helps to recruit more CD81 to α3β1. The tendency of CD81 and CD9 to exist as homodimers (Kovalenko et al., 2003) should facilitate extension of the complex. Lacking an ability to associate with CD81, the EW2xCD2c mutant cannot support expanded complex formation.
Tetraspanin-enriched microdomains contain cholesterol, gangliosides, tetraspanins, and other proteins. These microdomains, held together at least partly due to tetraspanin palmitoylation, are physically and functionally distinct from lipid rafts (Berditchevski, 2001; Boucheix and Rubinstein, 2001; Hemler, 2003). Our EWI-2–tetraspanin–α3β1 results extend the “tetraspanin microdomain” idea by demonstrating that multiple nontetraspanin proteins (an integrin and EWI-2) can be physically and functionally linked through specific tetraspanin proteins. The association of EWI-F/CD9-P1 with α3β1 integrin provides another potential example of tetraspanins linking two nontetraspanin proteins, although the tetraspanins involved were not formally defined (Charrin et al., 2003b). Other examples of multiprotein complexes potentially linked by tetraspanins, such as α4β1 and α6β1 integrin with HLA-DR, (Rubinstein et al., 1996) and HB-EGF with β1 integrin (Nakamura et al., 1995), relied on mild CHAPS detergent conditions in which tetraspanin complexes can have sizes in excess of 20 million D (Skubitz et al., 2000). Also, the linker function of tetraspanins was not directly tested in these experiments, and the functional relevance of the complexes was not established. Complexes of CD19 with CD21 (Horvath et al., 1998), and CD36 with α3β1 or α6β1 integrin (Thorne et al., 2000) are stable in Brij96 and may be functionally relevant, but the role of tetraspanins as linkers in these complexes remains untested. In another work, we found that EWI-2 can interact with another CD81-associated integrin (α4β1) in the MOLT-4 T cell line to cause formation of expanded EWI-2–CD81–α4β1 complexes, leading to alterations in α4β1-dependent cell morphology (unpublished data).
Possible relevance in vivo
In zones of activated basement membrane underlying injury sites, cell migration on laminin-5 is mediated primarily by α3β1 integrin (Nguyen et al., 2000). Laminin-5 can also trigger α3β1-dependent scattering of squamous carcinoma cells (Kawano et al., 2001), suggesting a potential role for α3β1 in metastasis. On the other hand, α3β1 overexpression in the suprabasal layer of the epidermis inhibited malignant conversion of papillomas (Owens and Watt, 2001), suggesting a potential role for α3β1 in suppressing tumor progression in some circumstances. In all of these cases, EWI-2 could play a key regulatory role, given its relatively widespread distribution (Clark et al., 2001; Stipp et al., 2001a). In addition, EWI-2 could participate in several key developmental processes. The defects of α3-null mice in kidney, lung, and skin morphogenesis point to a critical role for α3β1 in the assembly or organization of laminin-5 in the basement membranes of these tissues (Kreidberg et al., 1996; DiPersio et al., 1997). EWI-2 may participate in these functions by regulating the outcome of α3β1 engagement of laminin-5.
In conclusion, we have established a novel structural and functional link, via CD9 and CD81, between EWI-2 and the α3β1 integrin. These results contribute to the emerging importance of tetraspanin-containing protein microdomains, and also establish a novel means of regulating integrin function through lateral interactions among transmembrane proteins.
Materials and methods
Antibodies
mAbs to integrin α2 (A2-IIE10), α3 (A3-X8 and A3-IIF5), α6 (A6-ELE), and to tetraspanins CD81 (M38 and JS64) and CD151 (5C11) were referenced previously (Kazarov et al., 2002; Yang et al., 2002). Other mAbs were anti-integrin α6 (GoH3; BD Biosciences), anti-CD9 (ALB6; CHEMICON International), and anti-CD151 (8C3; a gift from Dr. Kiyotoshi Sekiguchi, Osaka University, Japan). Anti-α3 antibody A3-X8, anti-CD81 antibody M38, and anti-CD151 antibody 5C11 were used in all cases, except where specified. The M2 anti-FLAG epitope mAb, either biotinylated or conjugated to agarose, and a rabbit anti-FLAG pAb were purchased from Sigma-Aldrich. HRP-conjugated goat-anti–mouse pAb was purchased from Transduction Laboratories, and HRP-ExtrAvidin® was purchased from Sigma-Aldrich. M38 anti-CD81, A2-IIE10 anti-α2, and A3-X8 anti-α3 antibodies were conjugated to agarose Affigel-10 (Pierce Chemical Co.) according to manufacturer's instructions.
Cell culture and retroviral transduction
A431 epithelial carcinoma cells and retroviral packaging cell lines ΦNX and PT67 were cultured in high glucose DME, and U937 pro-monocytic leukemia cells in RPMI 1640. All cultures were supplemented with 10% FBS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (all purchased from GIBCO BRL). Expression constructs were (1) a COOH-terminally FLAG-tagged EWI-2 in the pLXIZ retroviral vector (Stipp et al., 2001a); (2) also in pLXIZ, an EWI-2/CD2 chimera (EW2xCD2c) with the EWI-2 cytoplasmic tail (from Cys604) replaced by recombinant PCR with the tail of CD2 followed by the FLAG epitope; (3) human CD81 in the pLXIN retroviral vector; (4) human α3 integrin in pLXIZ; and (5) the empty pLXIZ vector. Expression vectors were transfected by calcium phosphate into ΦNX packaging cells. 48 h after transfection, ΦNX cell supernatants were passed through a 0.45-μm filter, supplemented with 4 μg/ml Polybrene® (Sigma-Aldrich), and used to infect PT67 packaging cells that had been pretreated with 200 ng/ml tunicamycin (Sigma-Aldrich) for 18 h. Stable PT67 packaging cells were obtained by selection for 2 wk in 500 μg/ml Zeocin™ (Invitrogen) or G418. The supernatant from these PT67 cells was harvested, filtered, and supplemented with Polybrene® as above, and then used to infect A431 cells or U937 cells. Stable infectants were selected and grown as uncloned populations.
Cell spreading assay
Serum-free medium (SFM) contained DME with 5 mg/ml cell culture grade BSA (#194771; ICN Biomedicals) and 20 mM Hepes, pH 7.2. A431 EWI-2 cells and A431 IZ cells were rinsed, detached by trypsin/EDTA treatment, and collected in SFM plus 0.1 mg/ml soybean trypsin inhibitor and 0.1 mg/ml DNase I (both purchased from Worthington Biochemical Corp.). After centrifugation, the supernatant was removed and cells were resuspended in SFM alone or SFM with (1) 5 μg/ml GoH3 anti-α6, (2) 10 μg/ml A3-IIF5 anti-α3, or (3) both antibodies. Cells were then plated on acid-washed glass coverslips that had been coated with 1 μg/ml rat laminin-5 (provided by Desmos, Inc.) and blocked with SFM. Cells attached and spread for 20 min before being fixed with 3.7% formaldehyde in PBS with 5% sucrose and 2 mM MgCl2 for 15 min at RT. Coverslips were rinsed and mounted on slides in FluoroSave reagent (Calbiochem). Cell images were acquired with a monochrome CCD camera (Spot RT; Diagnostic Instruments) on a microscope (Axiovert 135; Carl Zeiss MicroImaging, Inc.), and were controlled by IP Lab software (Scanalytics) running on a G4 Macintosh computer. Areas of cells (20–45 cells per coverslip) were calculated using the Scion Image v1.62 program (Scion Corp.).
Reaggregation assay
Acid-washed glass coverslips (12-mm circles) were coated overnight at 4°C with 0.5 μg/ml rat laminin-5 in PBS with 0.005% Tween 20, or with 40 μg/ml collagen I in PBS. Coverslips were then blocked for 1 h at RT with SFM. A431 EWI-2 and A431 IZ cells that had been starved overnight in SFM were harvested as for the spreading assay above, and 1.5 × 105 cells per well were plated to laminin-5– or collagen I–coated coverslips in a 24-well dish. After 5–6 h, cells were fixed, stained with DAPI HCl (Sigma-Aldrich) to label nuclei, mounted, and imaged as above. Clustered and unclustered cells were counted in five fields per coverslip (typically >500 cells per coverslip). Two coverslips per condition were examined in each experiment, and always yielded statistically similar results. DAPI staining enabled unambiguous scoring of cell numbers in large, tightly packed cell clusters. Viability of A431 EWI-2 cells and A431 IZ cells under these assay conditions was tested using thiazolyl blue tetrazolium bromide (MTT) in Cell Proliferation Kit I (Roche).
Immunofluorescent localization
A431 cells were plated on laminin-5 in SFM. After 3 h, cells were fixed for 15 min with prewarmed 4% PFA, 2 mM MgCl2, and 4% sucrose in PBS. Fixed cells were rinsed three times with 50 mM Tris-HCl, pH 7.4, and 150 mM NaCl, and were blocked for 30 min in 20% goat serum in PBS. Primary antibodies were added at 5 μg/ml in blocking buffer and allowed to bind overnight at 4°C. After rinsing in PBS, FITC-goat anti–mouse second antibody was added in blocking buffer for 1 h at RT. Rinsed coverslips were mounted in FluoroSave (Calbiochem). Fluorescent images were acquired on an upright microscope (Axioskop; Carl Zeiss MicroImaging, Inc.) using a Spot RT camera driven by IP Lab software, as described above.
Immunoprecipitation, immunoblotting, and immunodepletion
A431 cells were lysed by scraping into 20 mM Hepes, pH 7.5, 150 mM NaCl, and 5 mM MgCl2 (HBSM) supplemented with 1% Brij 96 (Fluka), 2 mM PMSF (Sigma-Aldrich), 20 μg/ml aprotinin, and 10 μg/ml leupeptin (Boehringer). In some experiments, cells were biotinylated with 0.2 mg/ml sulfo-NHS-LC biotin (Pierce Chemical Co.) in HBSM for 1 h at RT, then rinsed three times with HBSM before lysis. After a 1-h extraction at 4°C with rocking, insoluble material was removed by centrifugation, and lysates were precleared for 1 h at 4°C with protein G–Sepharose (BD Biosciences). M2 anti-FLAG agarose or specific antibodies plus protein G–Sepharose were added, and immune complexes were collected overnight at 4°C. After rinsing four times with lysis buffer, immune complexes were eluted by boiling in sample buffer, resolved by SDS-PAGE, and transferred to nitrocellulose. Blots were blocked with 5% nonfat milk in PBS with 0.1% Tween 20.
FLAG epitope immunoblots were rinsed with TBS and blotted for 30 min with biotinylated M2 anti-FLAG mAb (10 μg/ml in TBS). After 10 rinses, blots were developed with a 30-min exposure to HRP-ExtrAvidin® in TBS, followed by 10 more rinses, and chemiluminescence detection (Renaissance® reagent; NEN Life Science Products). Myc epitope and CD151 immunoblots were developed for 1 h with anti-myc 9E10 ascites or the 8C3 anti-CD151 mAb diluted in blocking buffer, followed by 1 h with an HRP-goat anti–mouse pAb and chemiluminescence.
For immunodepletion, cell surface–biotinylated cells were lysed in 1% Brij 96 as described above. The lysate was divided into four parts and precleared three times for 1 h with (1) protein G–Sepharose alone, (2) protein G–Sepharose plus a combination of M38 and ALB6, anti-CD81, and CD9 antibodies, or (3) protein G–Sepharose plus A2-IIE10 anti-α2 integrin. The depleted samples were further divided into three parts each, immunoprecipitated with agarose-conjugated anti-CD81, anti-α3, or anti-α2 antibodies, and then analyzed by SDS-PAGE and chemiluminescence as above. Semi-quantitative densitometry was performed using GeneTools software (Syngene) on digitized images captured from trans-illuminated films with a CCD camera driven by GeneSnap software (Syngene).
Online supplemental material
Video 1 shows that EWI-2 overexpression impairs carcinoma cell motility on laminin-5. A431 IZ cells (left) and A431 EWI-2 cells (right) were plated on laminin-5 in serum-free medium. Migration was monitored for 2 h at 1 frame/min, and the video play rate is 6 frames/s. Online supplemental material available at http://www.jcb.org/cgi/content/full/jcb.200309113/DC1.
Supplemental Material
Acknowledgments
This work was supported the National Institutes of Health (grants CA42368 and GM38903), the Dana-Farber Claudia Adams Barr Program in Cancer Research, a Medical Foundation Charles A King Trust Fellowship (to C.S. Stipp) and by the Leukemia Society of America (T.V. Kolesnikova).
The online version of this article includes supplemental material.
References
- Berditchevski, F. 2001. Complexes of tetraspanins with integrins: more than meets the eye. J. Cell Sci. 114:4143–4151. [DOI] [PubMed] [Google Scholar]
- Berditchevski, F., M.M. Zutter, and M.E. Hemler. 1996. Characterization of novel complexes on the cell surface between integrins and proteins with 4 transmembrane domains (TM4 proteins). Mol. Biol. Cell. 7:193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berditchevski, F., K.F. Tolias, K. Wong, C.L. Carpenter, and M.E. Hemler. 1997. A novel link between integrins, transmembrane-4 superfamily proteins (CD63 and CD81), and phosphatidylinositol 4-kinase. J. Biol. Chem. 272:2595–2598. [DOI] [PubMed] [Google Scholar]
- Borradori, L., and A. Sonnenberg. 1999. Structure and function of hemidesmosomes: more than simple adhesion complexes. J. Invest. Dermatol. 112:411–418. [DOI] [PubMed] [Google Scholar]
- Boucheix, C., and E. Rubinstein. 2001. Tetraspanins. Cell. Mol. Life Sci. 58:1189–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, E.J., and W.A. Frazier. 2001. Integrin-associated protein (CD47) and its ligands. Trends Cell Biol. 11:130–135. [DOI] [PubMed] [Google Scholar]
- Charrin, S., F. Le Naour, V. Labas, M. Billard, J.P. Le Caer, J.F. Emile, M.A. Petit, C. Boucheix, and E. Rubinstein. 2003. a. EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem. J. 373:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin, S., S. Manie, M. Billard, L. Ashman, D. Gerlier, C. Boucheix, and E. Rubinstein. 2003. b. Multiple levels of interactions within the tetraspanin web. Biochem. Biophys. Res. Commun. 304:107–112. [DOI] [PubMed] [Google Scholar]
- Claas, C., C.S. Stipp, and M.E. Hemler. 2001. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. J. Biol. Chem. 276:7974–7984. [DOI] [PubMed] [Google Scholar]
- Clark, K.L., Z. Zeng, A.L. Langford, S.M. Bowen, and S.C. Todd. 2001. PGRL is a major CD81-associated protein on lymphocytes and distinguishes a new family of cell surface proteins. J. Immunol. 167:5115–5121. [DOI] [PubMed] [Google Scholar]
- Colognato, H., and P.D. Yurchenco. 2000. Form and function: the laminin family of heterotrimers. Dev. Dyn. 218:213–234. [DOI] [PubMed] [Google Scholar]
- DiPersio, C., S. Shah, and R. Hynes. 1995. α3Aβ1 integrin localizes to focal contacts in response to diverse extracellular matrix proteins. J. Cell Sci. 108:2321–2336. [DOI] [PubMed] [Google Scholar]
- DiPersio, C.M., K.M. Hodivala-Dilke, R. Jaenisch, J.A. Kreidberg, and R.O. Hynes. 1997. α3β1 Integrin is required for normal development of the epidermal basement membrane. J. Cell Biol. 137:729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenczik, C.A., R. Zent, M. Dellos, D.A. Calderwood, J. Satriano, C. Kelly, and M.H. Ginsberg. 2001. Distinct domains of CD98hc regulate integrins and amino acid transport. J. Biol. Chem. 276:8746–8752. [DOI] [PubMed] [Google Scholar]
- Green, J.M., A. Zhelesnyak, J. Chung, F.P. Lindberg, M. Sarfati, W.A. Frazier, and E.J. Brown. 1999. Role of cholesterol in formation and function of a signaling complex involving αVβ3, integrin-associated protein (CD47), and heterotrimeric G proteins. J. Cell. Biol. 146:673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaia, S., C. Li, and J.P. Allain. 2001. The dynamics of hepatitis C virus binding to platelets and 2 mononuclear cell lines. Blood. 98:2293–2300. [DOI] [PubMed] [Google Scholar]
- Hemler, M.E. 2003. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19:397–422. [DOI] [PubMed] [Google Scholar]
- Horvath, G., V. Serru, D. Clay, M. Billard, C. Boucheix, and E. Rubinstein. 1998. CD19 is linked to the integrin-associated tetraspans CD9, CD81, and CD82. J. Biol. Chem. 273:30537–30543. [DOI] [PubMed] [Google Scholar]
- Kawano, K., S.S. Kantak, M. Murai, C.C. Yao, and R.H. Kramer. 2001. Integrin α3β1 engagement disrupts intercellular adhesion. Exp. Cell. Res. 262:180–196. [DOI] [PubMed] [Google Scholar]
- Kazarov, A.R., X. Yang, C.S. Stipp, B. Sehgal, and M.E. Hemler. 2002. An extracellular site on tetraspanin CD151 determines α3 and α6 integrin-dependent cellular morphology. J. Cell Biol. 158:1299–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova, T.V., B.A. Mannion, F. Berditchevski, and M.E. Hemler. 2001. β1 integrins show specific association with CD98 protein in low density membranes. BMC Biochem. 2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko, O.V., X. Yang, T.V. Kolesnikova, and M.E. Hemler. 2003. Evidence for specific tetraspanin homodimers: inhibition of palmitoylation makes cysteines available for crosslinking. Biochem J. 10.1042/BJ20031037. [DOI] [PMC free article] [PubMed]
- Kreidberg, J.A., M.J. Donovan, S.L. Goldstein, H. Rennke, K. Shepherd, R.C. Jones, and R. Jaenisch. 1996. α3β1 integrin has a crucial role in kidney and lung organogenesis. Development. 122:3537–3547. [DOI] [PubMed] [Google Scholar]
- Nakamura, K., R. Iwamoto, and E. Mekada. 1995. Membrane-anchored heparin-binding EGF-like growth factor (HB-EGF) and diphtheria toxin receptor-associated protein (DRAP27)/CD9 form a complex with integrin α3β1 at cell–cell contact sites. J. Cell Biol. 129:1691–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, B.P., M.C. Ryan, S.G. Gil, and W.G. Carter. 2000. Deposition of laminin 5 in epidermal wounds regulates integrin signaling and adhesion. Curr. Opin. Cell Biol. 12:554–562. [DOI] [PubMed] [Google Scholar]
- Owens, D.M., and F.M. Watt. 2001. Influence of β1 integrins on epidermal squamous cell carcinoma formation in a transgenic mouse model: α3β1, but not α2β1, suppresses malignant conversion. Cancer Res. 61:5248–5254. [PubMed] [Google Scholar]
- Rabinovitz, I., A. Toker, and A.M. Mercurio. 1999. Protein kinase C-dependent mobilization of the α6β4 integrin from hemidesmosomes and its association with actin-rich cell protrusions drive the chemotactic migration of carcinoma cells. J. Cell Biol. 146:1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein, E., F. Le Naour, C. Lagaudriere-Gesbert, M. Billard, H. Conjeaud, and C. Boucheix. 1996. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA integrins. Eur. J. Immunol. 26:2657–2665. [DOI] [PubMed] [Google Scholar]
- Skubitz, K.M., K.D. Campbell, and A.P. Skubitz. 2000. CD63 associates with CD11/CD18 in large detergent-resistant complexes after translocation to the cell surface in human neutrophils. FEBS Lett. 469:52–56. [DOI] [PubMed] [Google Scholar]
- Sterk, L.M., C.A. Geuijen, L.C. Oomen, J. Calafat, H. Janssen, and A. Sonnenberg. 2000. The tetraspan molecule CD151, a novel constituent of hemidesmosomes, associates with the integrin α6β4 and may regulate the spatial organization of hemidesmosomes. J. Cell Biol. 149:969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterk, L.M., C.A. Geuijen, J.G. van den Berg, N. Claessen, J.J. Weening, and A. Sonnenberg. 2002. Association of the tetraspanin CD151 with the laminin-binding integrins α3β1, α6β1, α6β4 and α7β1 in cells in culture and in vivo. J. Cell Sci. 115:1161–1173. [DOI] [PubMed] [Google Scholar]
- Stipp, C.S., and M.E. Hemler. 2000. Transmembrane-4-superfamily proteins CD151 and CD81 associate with α3β1 integrin, and selectively contribute to α3β1-dependent neurite outgrowth. J. Cell Sci. 113:1871–1882. [DOI] [PubMed] [Google Scholar]
- Stipp, C.S., T.V. Kolesnikova, and M.E. Hemler. 2001. a. EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem. 276:40545–40554. [DOI] [PubMed] [Google Scholar]
- Stipp, C.S., D. Orlicky, and M.E. Hemler. 2001. b. FPRP, a major, highly stoichiometric, highly specific CD81- and CD9-associated protein. J. Biol. Chem. 276:4853–4862. [DOI] [PubMed] [Google Scholar]
- Thorne, R.F., J.F. Marshall, D.R. Shafren, P.G. Gibson, I.R. Hart, and G.F. Burns. 2000. The integrins α3β1 and α6β1 physically and functionally associate with CD36 in human melanoma cells. Requirement for the extracellular domain of CD36. J. Biol. Chem. 275:35264–35275. [DOI] [PubMed] [Google Scholar]
- Xia, Y., S.G. Gil, and W.G. Carter. 1996. Anchorage mediated by integrin α6β4 to laminin 5 (epiligrin) regulates tyrosine phosphorylation of a membrane-associated 80-kD protein. J. Cell Biol. 132:727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., C. Claas, S.K. Kraeft, L.B. Chen, Z. Wang, J.A. Kreidberg, and M.E. Hemler. 2002. Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol. Biol. Cell. 13:767–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch, R.L., F. Berditchevski, M.B. Harler, J. Reichner, and M.E. Hemler. 1998. Highly stoichiometric, stable, and specific association of integrin α3β1 with CD151 provides a major link to phosphatidylinositol 4-kinase, and may regulate cell migration. Mol. Biol. Cell. 9:2751–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.A., A.L. Bontrager, and M.E. Hemler. 2001. Transmembrane-4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific β1 integrins. J. Biol. Chem. 276:25005–25013. [DOI] [PubMed] [Google Scholar]
- Zhang, X.A., A.R. Kazarov, X. Yang, A.L. Bontrager, C.S. Stipp, and M.E. Hemler. 2002. Function of the tetraspanin CD151-α6β1 integrin complex during cellular morphogenesis. Mol. Biol. Cell. 13:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.A., W.S. Lane, S. Charrin, E. Rubinstein, and L. Liu. 2003. EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Cancer Res. 63:2665–2674. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.