Abstract
The yeast mitochondrial chaperonin Hsp60 has previously been implicated in mitochondrial DNA (mtDNA) transactions: it is found in mtDNA nucleoids associated with single-stranded DNA; it binds preferentially to the template strand of active mtDNA ori sequences in vitro; and wild-type (ρ+) mtDNA is unstable in hsp60 temperature-sensitive (ts) mutants grown at the permissive temperature. Here we show that the mtDNA instability is caused by a defect in mtDNA transmission to daughter cells. Using high resolution, fluorescence deconvolution microscopy, we observe a striking alteration in the morphology of mtDNA nucleoids in ρ+ cells of an hsp60-t s mutant that suggests a defect in nucleoid division. We show that ρ− petite mtDNA consisting of active ori repeats is uniquely unstable in the hsp60-t s mutant. This instability of ori ρ− mtDNA requires transcription from the canonical promoter within the ori element. Our data suggest that the nucleoid dynamics underlying mtDNA transmission are regulated by the interaction between Hsp60 and mtDNA ori sequences.
Keywords: yeast; mitochondria; mitochondrial DNA, nucleoids; Hsp60
Introduction
Mitochondrial DNA (mtDNA) nucleoids are protein–DNA complexes that are the heritable units of mtDNA and presumably contain many of the proteins necessary for mtDNA transactions. In the budding yeast Saccharomyces cerevisiae, a number of nucleoid proteins have been identified that were expected, such as the DNA packaging protein Abf2p and the single-stranded (ss) DNA binding protein Rim1p (Kaufman et al., 2000). It was surprising, however, to find other abundant proteins, such as Aco1p, some polypeptides of the α-ketoglutarate dehydrogenase complex, and the chaperonin Hsp60, whose functions are ostensibly unrelated to mtDNA inheritance.
Hsp60 is an essential mitochondrial chaperonin required for the proper folding of certain proteins imported into mitochondria (Cheng et al., 1989). It was found to associate with ssDNA regions and to bind in vitro to ssDNA of active ori sequences in a strand-specific manner (Kaufman et al., 2000). In an earlier study, Hsp60 was identified as an ssDNA binding protein that stimulated nuclear DNA replication in vitro (Smiley et al., 1992). DNA binding does not appear to be a general property of chaperonins of the Hsp60 type, as the Escherichia coli homologue GroEL is unable to bind to either strand of active ori mtDNA (Kaufman, 2003). It is likely that Hsp60 bound to ssDNA is the tetradecamer complex, as only small amounts of monomer are detected in Western blots of native gels of our Hsp60 preparations (Kaufman, 2003). A function for Hsp60 in mtDNA transactions has been revealed by the finding that wild-type (ρ+) mtDNA is rapidly lost from certain hsp60 temperature-sensitive (ts) mutants when grown at the permissive temperature on medium containing a fermentable carbon source (Kaufman et al., 2000), even though the chaperonin function appears near normal (Hallberg et al., 1993).
Here we show that the severe mtDNA instability phenotype in an hsp60-t s mutant is caused by a defect in mtDNA transmission to daughter cells. We find a striking alteration in the morphology of mtDNA nucleoids in the hsp60-t s mutant that strongly suggests a defect in nucleoid division. Consistent with our previous finding that Hsp60 binds preferentially in vitro to the template strand of active ori mtDNA (Kaufman et al., 2000), we find that only mtDNAs containing active ori elements are unstable in hsp60-t s mutant cells. Moreover, this instability requires transcription from the promoter element within the active ori. These findings collectively point to an interaction between Hsp60 and transcriptionally competent, active ori elements as a potential regulatory mechanism of mtDNA inheritance through nucleoid division, a process that if compromised, would lead to a decrease in the efficiency of mtDNA transmission to daughter cells.
Results and discussion
mtDNA transmission to daughter cells is defective in hsp60-t s mutants
When certain hsp60-t s mutant strains are grown at the permissive temperature (30°C) on a fermentable carbon source such as dextrose, the Hsp60 chaperonin activity appears near normal (Hallberg et al., 1993), but these strains produce petite progeny that lack mtDNA (ρ0 petites) (Kaufman et al., 2000). The production of ρ0 petites could be caused by degradation of mtDNA, by a defect in mtDNA replication, or by deficient mtDNA transmission. Under growth conditions requiring respiration (in medium containing the nonfermentable substrate glycerol), either mtDNA degradation or a replication defect would result in a decrease in mtDNA levels to some threshold that is compatible with continued cell growth on that medium. A transmission defect, however, would result in an apparent increase in mtDNA, because progeny cells would eventually stop growing as a result of their failure to inherit enough mtDNA to sustain growth by respiration. mtDNA would therefore tend to accumulate in the growing cells in the population.
To distinguish among these possibilities, we compared the relative amounts of ρ+ mtDNA in four hsp60-t s mutant strains that differ with respect to their mtDNA instability phenotype, and in wild-type cells all grown in glycerol medium at 24°C. Among the mutant strains, there is a striking correlation between the severity of the mtDNA instability phenotype observed in dextrose medium and the mtDNA content per cell observed in glycerol medium (Fig. 1 A). The hsp60-A144V mutant, which has the strongest mtDNA instability phenotype, contains ∼3.9-fold more mtDNA than does the wild-type strain. The quantity of Abf2p, a protein whose expression level has been previously shown to influence mtDNA level (Zelenaya-Troitskaya et al., 1998), is nearly equivalent for all strains. The Hsp60 protein is present in all strains at comparable levels (Fig. 1 A, inset). These results are most consistent with a defect in mtDNA transmission in hsp60-t s mutant cells.
Figure 1.
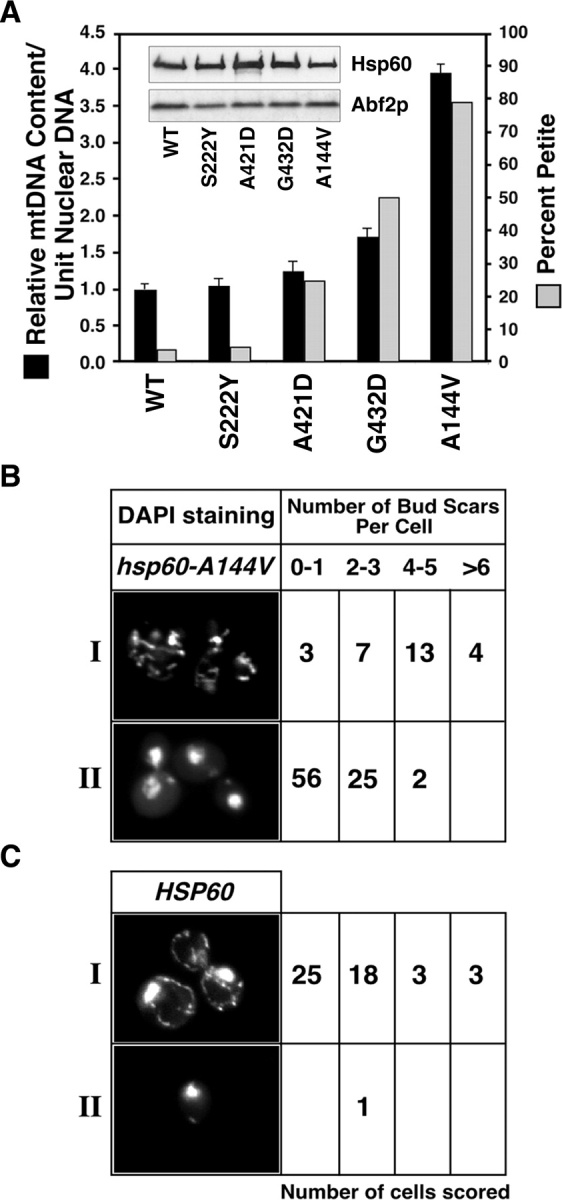
Mutations in HSP60 increase mtDNA levels and result in an mtDNA transmission defect. (A) Correlation between ρ+ mtDNA instability in hsp60-t s mutants grown in glucose medium (YPD) and mtDNA content in the same cells grown in glycerol medium (YPG). mtDNA instability data, scored as the percent petites in the population after 24 h growth on YPD at 24°C (gray bars, right Y axis), are from Kaufman et al. (2000). mtDNA levels relative to nuclear DNA (black bars, left Y axis) were determined by Southern blot analysis on whole-cell DNA. (A, inset) The elevated DNA levels in hsp60 mutants are not a result of altered Hsp60 or Abf2p protein levels. The indicated mutant strains were grown in YPG, and protein levels in whole cell lysates from equal numbers of cells were determined by Western blotting. (B and C) Older hsp60-A144V mutant cells grown in glucose medium contain more mtDNA than younger cells. Mutant (B) and wild-type (C) cells were precultured in YPG and transferred to YPD for 8–10 generations. Class I and class II cells were randomly chosen from fields to represent cells with either high (≥wild type) or low (≤two or three DAPI staining bodies) amounts of mtDNA nucleoids per cell, respectively, and then counterscored for bud scar number as described in the Materials and methods.
We reasoned that if hsp60-t s mutants are defective in transmission of mtDNA to daughter cells, older cells should have more mtDNA than younger cells. To test that hypothesis, we used microscopy to compare relative mtDNA levels to the number of bud scars present on cells, which reflects cell age, in cultures of wild-type and hsp60-A144V strains. Cells were precultured in glycerol medium and then grown for 8–10 generations in dextrose medium. Aliquots of cells from these cultures were fixed and stained with DAPI to visualize mtDNA and with calcofluor to visualize bud scars. Cells were visually scored as having high amounts of mtDNA (class I) or little to no mtDNA (class II) and then counter-scored for their number of bud scars (Fig. 1, B and C). Both strains showed the expected distribution of cell ages for logarithmically growing cultures. Of the 49 class I HSP60 cells observed, ∼10% had four or more bud scars (Fig. 1 C), whereas of the 27 class I hsp60-A144V cells observed, ∼60% had four or more bud scars (Fig. 1 B). Thus, hsp60-A144V cells containing high levels of mtDNA are biased toward older cells. By contrast, nearly all of the HSP60 cells scored contained mtDNA levels typical of wild-type cells, without any bias toward younger or older cells. Altogether, these data indicate that there is a defect in the transmission of mtDNA from mother to daughter cells in this hsp60-t s mutant strain.
MtDNA nucleoids in hsp60-A144V cells have aberrant morphology
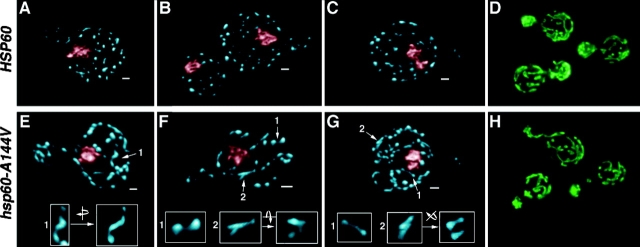
Initial inspection of the DAPI-stained mtDNA of hsp60-A144V cells (Fig. 1, B and C) suggested that the nucleoid morphology differed from that of wild-type cells. To better visualize the nucleoid architecture in these cells, we used fluorescence deconvolution microscopy to generate three-dimensional, rendered images of DAPI-stained bodies in wild-type (n = 21) and in hsp60-A144V (n = 13) cells grown in glycerol medium at 24°C. Representative images show that nucleoids in wild-type cells, which average 35 (±7) per mother cell (i.e., excluding buds), are rounded or oval structures that are well separated from each other (Fig. 2, A–C). In contrast, the nucleoids in hsp60-A144V cells (Fig. 2, E–G), which average 26 (±3) per mother cell, are significantly enlarged and elongated, sometimes forming branched (Fig. 2 F, center and right insets), serpentine (Fig. 2 E, insets), or interconnected structures resembling dumbbells (Fig. 2, F and G, left insets). As can be seen in these representative examples, the severity of the aberrant nucleoid morphology in the mutant cells varies somewhat across the nucleoid population.
Figure 2.
hsp60-A144V mutant cells contain altered nucleoid structures. Wild-type (A–D) and hsp60-A144V mutant cells (E–H) were prepared as described in the Materials and methods. Nuclear DNA (A–C and E–G) was colored red after rendering. Numbered structures have been enlarged in panel insets, and some structures were rotated in three-dimensional space as indicated. The wild-type cells (and buds, A–C) contain 2.7, 2.3 (small cell not included), and 2.3 μm3 of mitochondrial DAPI staining, respectively, and hsp60-A144V cells (E–G) contain 4.3, 2.6, and 5.3 μm3, respectively. Bars, 1 μm. Unrendered, single focal plane images of GFP-labeled mitochondria in wild-type and hsp60-A144V mutant strains are shown in D and H, respectively.
Based on mtDNA volume estimates of the cells shown in Fig. 2, the mutant cell in panel F has about the same amount of mtDNA as do wild-type cells, suggesting that the overall increase in mtDNA levels in the mutant cultures is not the cause of the nucleoid morphology alterations. Moreover, the mitochondrial morphology in hsp60-A144V mutant cells (Fig. 2 H) is similar to that of wild-type cells (Fig. 2 D), suggesting that the differences in nucleoid structures do not result from any significant change in mitochondrial organization. Finally, because there does not appear to be any mtDNA replication or recombination defects in the hsp60-A144V mutant strain as detected by 2D DNA gel analysis (unpublished data), we interpret our data to mean that nucleoid division, a process obviously required for efficient transfer of mtDNA from mother to daughter cells, is defective in that strain. We have detected an occasional “aberrant” mtDNA nucleoid structure in wild-type cells (approximately one per four cells), which may be an intermediate in nucleoid division. Recently, Garrido et al. (2003) showed that mtDNA nucleoids in human cells can indeed divide and redistribute within mitochondria.
ρ− mtDNA containing active oris is uniquely unstable in hsp60-A144V cells
Our previous in vitro experiments have shown that Hsp60 binds selectively to the template strand of “active” ori sequences, which are putative origins of mtDNA replication (Kaufman et al., 2000). Although similar in organization to metazoan origins of mtDNA replication, yeast mtDNA oris are dispensable for mtDNA replication in ρ− petites (Fangman et al., 1989; Lorimer et al., 1995; MacAlpine et al., 2001). Petite mtDNA genomes consist of an excised portion of the ρ+ genome (that may or may not contain an active ori) that has usually been amplified into tandem repeats (Fig. 3 A). Active oris are characterized by the presence of conserved sequence elements, including three short GC-rich sequences and an RNA polymerase promoter (Bernardi, 1982) (Fig. 3 B). When present in ρ− petite genomes, active oris confer hypersuppressiveness, defined as the preferential transmission of that ρ− mtDNA to >95% of progeny from crosses with either ρ+ strains (Blanc and Dujon, 1980; de Zamaroczy et al., 1981) or ρ− strains that lack oris (Lorimer et al., 1995; MacAlpine et al., 2001). Inactive oris contain the sequence elements of active oris plus additional GC-rich insertions, one of which disrupts the promoter (de Zamaroczy et al., 1981), and do not confer hypersuppressiveness in crosses. It is not yet clear whether hypersuppressiveness reflects a replicative or segregational advantage of the ρ− mtDNA with active ori repeats.
Figure 3.
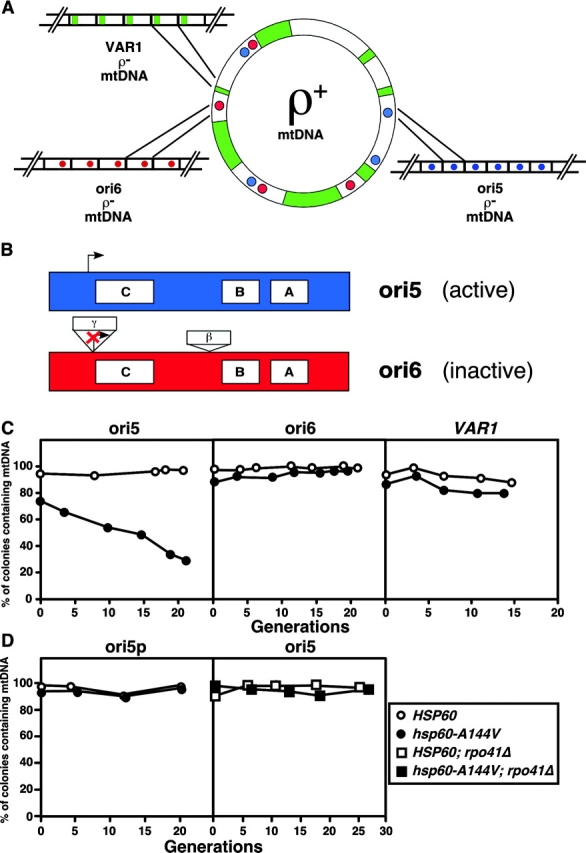
hsp60-A144V selectively destabilizes active ori-containing petite mtDNA in a promoter driven transcription–dependent manner. (A) Spontaneous ρ− petite mtDNAs are formed from random portions of the ρ+ genome that have been amplified into tandem repeats. (B) Active ori sequences contain three GC-rich sequences known as the A, B, and C boxes, as well as a nonanucleotide promoter at which transcription initiates (ori5). Inactive ori sequences have the same elements as active oris, plus two GC-rich inserts, one of which disrupts the promoter. (C) Petite strains containing different mtDNA sequences were tested for stability in HSP60 and hsp60-A144V mutant backgrounds during growth in YPD. (D) The instability of an ori5 ρ− petite genome in the hsp60-A144V background is rescued by the loss of promoter-driven transcription (left) or by inactivation of RPO41 (right). The ρ− petite strains contained an active ori (ori5) (C, left, and D, right); an inactive ori (ori6) (C, middle); no ori sequences (VAR1) (C, right); and ori5 in which the promoter was mutated (ori5p) (D, left).
Is there a relation between the observed in vitro interaction of Hsp60 with active ori DNA and the instability of ρ+ mtDNA in the hsp60-t s mutants? Experimentally, it is not yet feasible to test the stability of a ρ+ strain devoid of all active ori sequences in an hsp60-t s mutant because a ρ+ genome lacking active ori sequences is likely to be unstable in otherwise wild-type cells (Piskur, 1988). To test this relationship, we compared the stability of ρ− mtDNAs containing an active ori (ori5), containing an inactive ori (ori6), or lacking any ori sequences (VAR1) in HSP60 and hsp60-A144V strains (Fig. 3 A; see Materials and methods). As expected, all three ρ− mtDNAs were stable in HSP60 cells (Fig. 3 C). In hsp60-A144V cells, however, ori5 ρ− mtDNA was unstable (Fig. 3 C, left), whereas the ori6 and VAR1 ρ− mtDNAs were as stable as in HSP60 cells (Fig. 3 C, middle and right). These results provide genetic support for an interaction between a functional ori and Hsp60 protein, consistent with our previous finding that Hsp60 binds to active ori DNA in vitro (Kaufman et al., 2000).
We have previously shown that mutation of the promoter of active ori5 ρ− mtDNA (ori5p) renders it inactive (MacAlpine et al., 2001). This effect was not due to an inability of ori5p mtDNA to be transcribed, because promoter-independent transcription of ori5p mtDNA was observed. These observations allowed us to compare the stability of nearly identical active (ori5) and inactive (ori5p) sequences in the hsp60-A144V strain. We found that ori5p ρ− mtDNA was stable in hsp60-A144V cells, whereas ori5 mtDNA was unstable (Fig. 3 D, left). The promoter region of ori5 DNA does not appear to contribute to the specificity of Hsp60 binding (Kaufman, 2003; unpublished data), suggesting that additional events, such as transcription from the promoter, may influence the relation between Hsp60 and the inheritance of mtDNAs that contain an active ori. To test that notion, we inactivated the RPO41 gene, which encodes the only known mitochondrial RNA polymerase activity (Greenleaf et al., 1986), in HSP60 and in hsp60-A144V cells containing ori5 ρ− mtDNA (Fig. 3 D, right). In the rpo41Δ background, ori5 ρ− mtDNA instability in hsp60-A144V mutant cells is completely rescued. In general, ρ− mtDNAs are known to be stable in otherwise wild-type rpo41Δ cells (Fangman et al., 1990; Lorimer et al., 1995). Collectively, these data suggest that transcription from the ori5 promoter is important for mutant Hsp60 to influence the stability of the active ori-containing mtDNA.
The precise function of yeast mtDNA ori sequences is presently unresolved. Although active oris show the hallmarks of RNA-primed, leading strand replication initiating at the promoter (Graves et al., 1998; MacAlpine et al., 2001), mutation of that promoter, or elimination of mitochondrial transcription altogether in ρ− petites containing active oris (Lorimer et al., 1995; MacAlpine et al., 2001), does not destabilize ρ− mtDNA. Despite the clear evidence that active oris are dispensable for the replication and transmission of ρ− mtDNAs in otherwise wild-type cells, the current work shows that mtDNAs with active oris that can be transcribed from their promoter are unstable in cells expressing mutant Hsp60 protein. Evidently, this transcription-dependent relationship between active oris and Hsp60 for the maintenance of active ori-containing mtDNA cannot default to a strictly ori-independent mode of transmission in cells expressing mutant Hsp60 protein.
A role for transcription as a driving force for Hsp60-dependent mtDNA nucleoid division could be envisaged by a mechanism similar to that proposed by Dworkin and Losick (2002) for the separation of newly replicated bacterial chromosomes, which are attached to the bacterial membrane through the origin of replication (Draper and Gober, 2002). Hsp60, through its chaperonin activity, may also function at active ori sequences in the assembly/disassembly of components that are important to ensure proper division of mtDNA nucleoids and, hence, mtDNA segregation. The mtDNA instability in the hsp60-t s mutants might then arise because these ori-dependent segregation complexes fail to assemble or disassemble correctly due to some compromised Hsp60 function at oris, be it aberrant DNA binding or chaperone activity. Altogether, these considerations raise the interesting proposition that, rather than functioning primarily in mtDNA replication, yeast mtDNA oris together with Hsp60 may function in nucleoid division, which is likely dependent on membrane association of mtDNA (Albring et al., 1977).
Materials and methods
Strains and media
For microscopy and ρ+ mtDNA level analysis, the W303-1B ρ+ hsp60Δ::HIS3 strain carrying HSP60 or hsp60-A144V on a CEN plasmid was used. For ρ− petite mtDNA stability, strain a161-U7 ρ0 hsp60Δ::KAN carrying HSP60 or hsp60-A144V on a CEN plasmid was used. RPO41 was disrupted by the insertion of URA3. The following ρ− petite genomes were placed in the above ρ0 strain by cytoduction: A2-16/7 (2.9-kb repeat unit containing the inactive ori6; Lamb et al., 1983); sORI5w-476 (476-bp repeat unit containing ori5; MacAlpine et al., 2001); sORI5p-476 (sORI5w-476 with a mutated promoter; MacAlpine et al., 2001); and VAR1 (2.1-kb repeat unit containing the VAR1 gene; MacAlpine et al., 2001). Cells for all experiments were grown at room temperature (24°C). YP medium contains 1% yeast extract, 2% peptone, and either 3% glycerol (YPG) or 2% dextrose (YPD).
DNA hybridization
Southern blot analysis of total DNA isolated from cells grown in YPG to mid-log phase was performed in triplicate using standard methods. Membranes were hybridized with probes corresponding to ACT1 and COXI to assess relative amounts of nuclear and mtDNA, respectively; signals were quantified by phosphorimager analysis. The presence of mtDNA in petite strains was determined by colony hybridization using oligonucleotide probes specific to each petite tested.
Age determination of wild-type and mutant cells
Wild-type and hsp60-A144V mutant cells were precultured in YPG and then outgrown in YPD for 8 and 10 generations, respectively. Aliquots of the cultures were incubated with 3.7% formaldehyde and 1 μg/ml DAPI for 30 min, after which the cells were pelleted, stained for 15 min with 0.1 μg/ml Calcofluor, washed, and resuspended in ProLong (Molecular Probes).
Microscopy
To visualize mtDNA nucleoids, wild-type and hsp60-A144V mutant cells were grown to mid-log phase in YPG, fixed with formaldehyde and stained with DAPI as above, washed, resuspended in ProLong, and diluted 1:1 in 1% low melting point agarose. Images were captured by automation using Openlab (Improvision) on a Carl Zeiss MicroImaging, Inc. Axiovert 100M microscope at X100 (1.4NA PlanApo), and then deconvolved, measured, and rendered using Volocity (Improvision). The cellular content of mtDNA was estimated from total DAPI fluorescence after subtracting nuclear DNA fluorescence.
To visualize mitochondrial profiles, wild-type and hsp60-A144V strains carrying pRS416-CLG (containing the CIT1 promoter and leader sequence in frame with GFP; Okamoto et al., 2001) were grown to mid-log phase in YPG, fixed with formaldehyde, and resuspended in water. Single plane epifluorescence images were processed using Adobe Photoshop®.
Stability of petite mtDNAs
Ori5, ori5p, VAR1, and ori6 petite genomes were maintained in kar1 cells and transferred by cytoduction into wild-type and hsp60-A144V ρ0 strains to initiate each experiment. Colonies containing the ρ− mtDNA were grown in YPD, sampled at intervals, plated for single cells, and scored for mtDNA content by DNA hybridization.
Acknowledgments
We thank A. Tizenor for help with graphics and J. Waddle and C. Gilpin for assistance and advice on the microscopy. We thank our colleagues for helpful discussions and critically reading the manuscript.
This work was supported by grants from the National Institutes of Health (NIH) and The Robert A. Welch Foundation. J.E. Kolesar is an NIH predoctoral trainee (T32-HL07360).
B.A. Kaufman's present address is Montreal Neurological Institute, 3801 University St., Montreal, Quebec H3A 2B4, Canada.
Abbreviations used in this paper: mtDNA, mitochondrial DNA; ss, single stranded; ts, temperature sensitive.
References
- Albring, M., J. Griffith, and G. Attardi. 1977. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc. Natl. Acad. Sci. USA. 74:1348–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi, G. 1982. The origins of replication of the mitochondrial genome of yeast. Trends Biochem. Sci. 7:404–408. [Google Scholar]
- Blanc, H., and B. Dujon. 1980. Replicator regions of the yeast mitochondrial DNA responsible for suppressiveness. Proc. Natl. Acad. Sci. USA. 77:3942–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, M.Y., F.-U. Hartl, J. Marton, R.A. Pollock, F. Kalousek, W. Neupert, E.M. Hallberg, R. Hallberg, and A.L. Horwich. 1989. Mitochondrial heat shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature. 337:620–625. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy, M., R. Marotta, G. Faugeron-Fonty, R. Goursot, M. Mangin, G. Baldacci, and G. Bernardi. 1981. The origins of replication of the yeast mitochondrial genome and the phenomenon of suppressivity. Nature. 292:75–78. [DOI] [PubMed] [Google Scholar]
- Draper, G.C., and J.W. Gober. 2002. Bacterial chromosome segregation. Annu. Rev. Microbiol. 56:567–597. [DOI] [PubMed] [Google Scholar]
- Dworkin, J., and R. Losick. 2002. Does RNA polymerase help drive chromosome segregation in bacteria? Proc. Natl. Acad. Sci. USA. 99:14089–14094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman, W.L., J.W. Henly, G. Churchill, and B.J. Brewer. 1989. Stable maintenance of a 35-base pair yeast mitochondrial genome. Mol. Cell. Biol. 9:1917–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangman, W.L., J.W. Henly, and B.J. Brewer. 1990. RPO41-independent maintenance of ρ− mitochondrial DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido, N., L. Griparic, E. Jokitalo, J. Wartiovaara, A.M. Van Der Bliek, and J.N. Spelbrink. 2003. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 14:1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves, T., M. Dante, L. Eisenhour, and T.W. Christianson. 1998. Precise mapping and characterization of the RNA primers of DNA replication for a yeast hypersuppressive petite by in vitro capping with guanylyltransferase. Nucleic Acids Res. 26:1309–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf, A.L., J.L. Kelly, and I.R. Lehman. 1986. Yeast RPO41 gene is required for transcription and maintenance of the mitochondrial genome. Proc. Natl. Acad. Sci. USA. 83:3391–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg, E.M., Y. Shu, and R.L. Hallberg. 1993. Loss of mitochondrial hsp60 function: nonequivalent effects on matrix-targeted and intermembrane-targeted proteins. Mol. Cell. Biol. 13:3050–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, B.A. 2003. Studies on mitochondria DNA nucleoids in Saccharomyces cerevisiae: identification of bifunctional proteins. In Genetics and Development, UT Southwestern Medical Center at Dallas, Dallas, TX. 241 pp.
- Kaufman, B.A., S.M. Newman, R.L. Hallberg, C.A. Slaughter, P.S. Perlman, and R.A. Butow. 2000. In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins. Proc. Natl. Acad. Sci. USA. 97:7772–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, M.R., P.Q. Anziano, K.R. Glaus, D.K. Hanson, H.J. Klapper, P.S. Perlman, and H.R. Mahler. 1983. Functional domains in introns: RNA processing intermediates in cis- and trans-acting mutants in the penultimate intron of the mitochondrial gene for cytochrome b. J. Biol. Chem. 258:1991–1999. [PubMed] [Google Scholar]
- Lorimer, H.E., B.J. Brewer, and W.L. Fangman. 1995. A test of the transcription model for biased inheritance of yeast mitochondrial DNA. Mol. Cell. Biol. 15:4803–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine, D.M., J. Kolesar, K. Okamoto, R.A. Butow, and P.S. Perlman. 2001. Replication and preferential inheritance of hypersuppressive petite mitochondrial DNA. EMBO J. 20:1807–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, K., P.S. Perlman, and R.A. Butow. 2001. Targeting of green fluorescent protein to mitochondria. Methods Cell Biol. 65:277–283. [DOI] [PubMed] [Google Scholar]
- Piskur, J. 1988. Transmission of yeast mitochondrial loci to progeny is reduced when nearby intergenic regions containing ori/rep sequences are deleted. Mol. Gen. Genet. 214:425–432. [DOI] [PubMed] [Google Scholar]
- Smiley, J.K., W.C. Brown, and J.L. Campbell. 1992. The 66 kDa component of yeast SFI, stimulatory factor I, is hsp60. Nucleic Acids Res. 20:4913–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenaya-Troitskaya, O., S.M. Newman, K. Okamoto, P.S. Perlman, and R.A. Butow. 1998. Functions of the HMG box protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics. 148:1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]