Abstract
Mammalian myosin V motors transport cargo processively along actin filaments. Recent biophysical and structural studies have led to a detailed understanding of the mechanism of myosin V, making it perhaps the best understood cytoskeletal motor. In addition to describing the mechanism, this review will illustrate how “dynamic” single molecule measurements can synergize with “static” protein structural studies to produce amazingly clear information on the workings of a nanometer-scale machine.
The latest motor protein poster child is myosin V, which has experienced a meteoric rise to stardom. This fascinating group of motors transports a variety of intracellular cargo along actin (Reck-Peterson et al., 2000; Vale, 2003). In yeast, myosin V motors transport membranous organelles and mRNA from the mother cell to the bud. In vertebrates, myosin V transport pigment-containing melanosomes in melanocytes and other organelles, such as the endoplasmic reticulum, in neurons. Mutations in myosin V give rise to pigmentation and neurological defects in mice and humans. The interactions of myosin V motors with certain cargo are among the best understood examples in the motor protein field. In S. cerevisiae, an adaptor protein (She3p) links one of the two yeast myosin V motors (Myo4p) to mRNAs (Kwon and Schnapp, 2001), and a vacuolar receptor for the other myosin V motor (Myo2p) has been uncovered (Ishikawa et al., 2003). In mammals, the docking of myosin Va onto melanosomes has been shown to involve an adaptor protein (melanophilin) that interacts with GTP-loaded Rab27a on the melanosome membrane (for review see Langford, 2002).
An understanding of the mechanism of myosin V–based motility also has advanced at an incredible pace. In 1999, Spudich and colleagues (Mehta et al., 1999) showed by optical trap studies that mammalian myosin V (a dimer of two identical polypeptides joined together by a coiled coil [Fig. 1]) is processive, meaning that a single motor protein can take many consecutive steps along actin without dissociating. Processive motion was further demonstrated by observation of single fluorescently labeled myosin V molecules moving for several microns along an actin filament (Sakamoto et al., 2000). Processive motion also is a property of kinesin, a cargo-carrying microtubule motor. However, muscle myosin (as well as other members of the myosin family) takes a single “stroke” and then detaches from actin, and thus is nonprocessive. Because of several experimental advantages of myosin V revolving around its unusually large mechanical element (a long lever arm extending from its catalytic domain) and it processivity, several laboratories have shifted their attention toward this motor. As a result, myosin V has quickly leap-frogged ahead of kinesin and the much longer studied muscle myosin to become the best understood cytoskeletal motor protein. In this review, I will present the most widely accepted view of how myosin V works. I then will describe the puzzle pieces, assembled by many laboratories using single molecule biophysics, kinetics, spectroscopy, crystallography, and EM, that gave rise to this consensus mechanism. In many ways, the journey of arriving at this mechanism has been as instructive as the destination itself, illustrating the power of combining single molecule studies with information on protein structure.
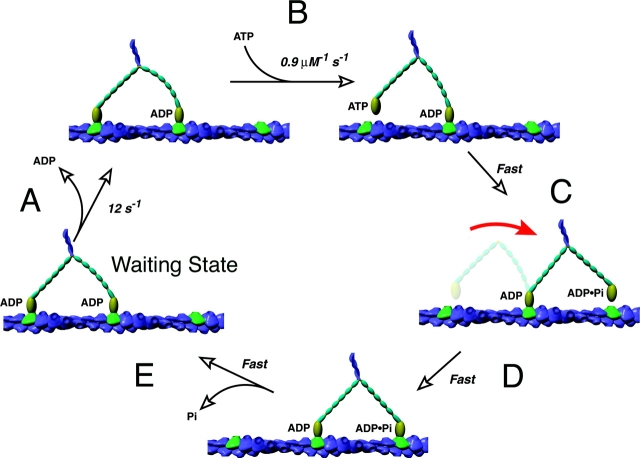
Figure 1.
A model for the processive motion of mammalian myosin V. See text for details (An emerging consensus mechanism for myosin V). This figure is a modified version of a figure provided by Drs. T. Purcell and J. Spudich.
Myosin V: why so much progress?
The holy grail of the motor protein field is to understand how Angstrom level changes in the nucleotide pocket (i.e., binding of ATP, nucleotide hydrolysis, phosphate release, and the release of ADP) are transmitted into nanometer-scale structural changes that elicit movement. A processive motor dimer such as myosin V possesses an additional layer of complexity, since the two motor domains must coordinate their activities. In this case, one needs to decipher the temporal sequence of kinetic and structural events for each of the two heads during the course of movement.
Why is it so hard to decipher the mechanism of a motor protein? Molecular motors are extraordinarily small and transitions in their ATPase cycle occur on the millisecond time scale. Thus, strategies for measuring motor movements and conformational changes must be compatible with these spatial and temporal parameters. Making measurements on large numbers of molecules also is problematic, because the discrete actions executed by individual molecules become “blurred” when the net output from large groups of asynchronous motors is monitored. Therefore, single molecule analyses provide the clearest window into the workings of a motor protein, particularly if combined with information on protein structure derived from high resolution EM and x-ray crystallography.
From a technical perspective, biophysical studies on myosin V began at an opportune time, since single molecule techniques were considerably advanced and atomic structures for other myosins in various nucleotide states were already solved. However, myosin V also has several experimental advantages that make it particularly well suited for exploration using the suite of available methodologies. First, myosin V possesses the granddaddy of all lever arms (24 nm), being threefold longer than the lever arm of muscle myosins and eightfold longer than kinesin's mechanical element (the neck linker). When one is attempting to visualize nanometer-scale changes, bigger is clearly better. The lever arm is easily visualized by EM (even without averaging methods), and nucleotide-dependent conformational changes can be readily discerned. In sharp contrast, the 12–amino acid neck linker of kinesin cannot be directly visualized by EM unless gold particles are attached to mark its position. Moreover, the long lever arm of myosin V gives rise to a big displacement (37 nm) per ATP hydrolyzed, and these large steps stand out clearly from Brownian noise.
A second experimental advantage of myosin V is its processivity. The ability to measure many successive steps and the intervening dwell times provides information on how chemical transitions in the ATPase site trigger mechanical events, as will be discussed later. It has been even possible to discern subtle variations in the myosin V step size from one ATPase cycle to the next, which also has provided mechanistic information. In contrast, the nonprocessive muscle myosin takes a single and much smaller (5–10 nm) step that is shrouded in a great deal of Brownian noise. Processive motility has aided the study of kinesin, but its steps are small (8 nm), which makes it difficult to clearly resolve all steps in a processive run.
A third advantage is that both heads of the myosin V dimer can bind simultaneously to actin: one head in a prestroke state and the other in a post-stroke state. Such images obtained by EM have provided clear snapshots of the before and after conformations of the myosin power stroke. In contrast, the prestroke conformation of muscle myosin has been difficult to visualize, since the motor is detached from actin in this state and may adopt multiple conformations when free in solution.
Lastly, it has been possible to express active myosin V using the baculovirus system, which has enabled investigators to vary the lever arm length and examine the effect on step size. Although this is not a unique advantage of myosin V, protein expression has been problematic for other myosins, particularly muscle myosin.
An emerging consensus mechanism for myosin V
Before describing specific investigations on myosin V motiliy, I will provide a general overview of what appears to be the emerging consensus mechanism (Fig. 1). The first optical trap study showing that myosin V takes ∼37-nm steps provided an important clue on the mechanism of processivity (Mehta et al., 1999). This step size corresponds to the crossover distance of subunits in the actin filament (also called the pseudorepeat distance), suggesting that myosin V steps along the “top” of an actin filament, rather than spirals around the helical actin filament. This interpretation was supported by EM studies, which showed that the two heads of the myosin V dimer can bind simultaneously to identically oriented actin subunits separated by 37 nm (Walker et al., 2000). Myosin V is able to span this large distance by virtue of its unusually long lever arm, an α-helix containing six IQ motifs that each bind calmodulin light chains (IQ-calmodulins are depicted as segmented domains on the lever arm in Fig. 1). However, the leading head must have its lever arm pointing backward (prestroke state), whereas the trailing head must have its lever arm pointing forward (post-stroke state). In these states, the lengths and angles of the lever arms are well-designed to allow the two heads to span the 37-nm gap without unraveling the coiled coil (Vale and Milligan, 2000; Burgess et al., 2002).
The two head–bound intermediate seen by EM is the “waiting state” that the motor adopts in between steps (Fig. 1). The leading head cannot swing its lever arm forward, since it is tethered by the actin-bound, trailing head. For a step to occur, ADP must be released from the trailing head (Fig. 1 A). This rate-limiting step in the ATPase cycle allows the following rapid sequence of events to occur: (Fig. 1 B) ATP binds to the empty site and causes the dissociation of the trailing head from actin, producing a transient one head–bound intermediate; (Fig. 1 C) the leading head executes a ∼20-nm swing of its lever arm, which throws the trailing head in front; and (Fig. 1 D) the “new” leading head then executes a diffusional search and rebinds preferentially to the 13th actin subunit in front of the partner head. The new leading head is believed to hydrolyze ATP, release phosphate, and enter an ADP state (Fig. 1 E); however, tension between the two heads may promote ADP release from the trailing head and slow down product release from the leading head. Through such coordination between the chemical cycles and mechanical actions of the lever arms, the myosin V dimer moves in a “hand-over-hand” manner along an actin filament. The experimental evidence for this model, as well as uncertainties, will be outlined in the next sections.
A rotating lever arm: seeing is believing
The “swinging crossbridge” hypothesis for muscle myosin–based movement was put forth by H. Huxley in 1969. However, imaging the lever arm of muscle myosin in different nucleotide states and establishing its rotation has required more than three decades of work, and this model is still not beyond controversy. In contrast, electron microscopic images of myosin V bound to actin instantly revealed a wealth of information on the conformational states of the lever arm. In the two head–bound intermediate, the forward-tilting lever arm of the trailing head matches the lever arm position in the no-nucleotide (post-stroke) muscle myosin crystal structure (Burgess et al., 2002). The lever arm position of the leading head most closely resembles a crystal structure of ADP-VO3 myosin (the prestroke state). This match is not precise due to strain on this lever arm from the trailing head, as will be discussed later.
The general conclusion from comparing EM and atomic structures is that the lever arms in the trailing and leading heads are in the post- and prestroke positions, respectively. The presumption from these static images is that the lever arm rotates between the rearward- and forward-pointing positions as myosin V moves along the filament. This hypothesis was verified in walking myosin V motors using single molecule polarization microscopy to measure angular changes in the orientation of the lever arm (Forkey et al., 2003). In this study, calmodulin was labeled with a fluorescent dye at two attachment points (to maintain a fixed orientation), and then exchanged for a single endogenous calmodulin in the myosin V dimer. The angular orientation of the lever arm was measured in single myosin V molecules using a sophisticated microscope that measures fluorescence polarization in three dimensions. Observing the motor move slowly at low ATP concentrations, the authors showed that the lever arm was oriented either at 40° or 140°, relative to the actin filament, and would undergo abrupt transitions between these two angles. The 40° state corresponds precisely to the lever arm angle of the trailing head seen by EM, whereas the 140° state is within the more variable range of angles observed for the lever arm of the leading head (Walker et al., 2000). These results conclusively show that the lever arm tilts during an ATP-driven step; the displacement of the distal end of the lever arm equals the step size observed by optical trap studies ([cos40° − cos140°] × 24 nm lever arm length = 36.8 nm). Thus, “static” visualization of the lever arm by EM and “dynamic” measurements by single molecule microscopy are in excellent agreement and provide support for a lever arm rotation mechanism.
A hand-over-hand mechanism beyond doubt
In the model shown in Fig. 1, the trailing head moves past the stationary forward head by a distance of 74 nm to reach a new actin binding site. An alternative to this hand-over-hand model is the inch worm model, which proposes that both heads advance by 37 nm during the step and that the trailing and leading heads do not exchange positions. The inch worm model has been advocated for the processive motion of kinesin by some investigators (Hua et al., 2002). The best way to distinguish between the hand-over-hand and inch worm models is to follow the displacement of one of the myosin V heads, as opposed to the center of mass, which is tracked in an optical trap experiment. This feat was accomplished by Yildiz et al. (2003) who measured the spatial position of a myosin V–bound fluorescent dye with remarkable precision using their method coined FIONA (fluorescence imaging with one-nanometer accuracy), described briefly as follows. The microscopic image of a point source of light appears as a Gaussian-shaped intensity distribution, also known as a point spread function (PSF). The ∼250-nm width of the PSF impairs resolution (distinguishing two points sources as being separate); however, if enough photons are collected by integration using a cooled ccd camera and the microscope stage is very stable, then the center position of the PSF can be accurately determined and even a 1.5-nm displacement of the PSF can be measured.
As a source of their fluorescent signal, Yildiz et al. exchanged a single fluorescently labeled calmodulin into the myosin V lever arm, as described previously. Using FIONA, the authors observed ∼74-nm displacements, a value identical to the predicted advance of the trailing head in the hand-over-hand model. After the 74-nm step, the dye-bound head should be in the leading and the next ATPase cycle should result in a ∼0-nm dye displacement. Although the 0-nm displacement could not be directly observed, tell-tale signs of these “missing” steps were apparent from the dwell times in between the 74-nm steps. Yildiz et al. showed that the dwell time distribution could be best fit by the convolution of two sequential events: one producing the 74-nm step and the other most likely reflecting the zero displacement. Other single myosin V molecules, however, did not undergo 74-nm displacements of the bound dye, but rather exhibited either alternating 52–23 nm steps or 42–33 nm steps (both adding up to ∼74 nm). These different stepping patterns could be best explained by variation in the position in which the fluorescently labeled calmodulin exchanged on the lever arm. In the majority of cases, exchange occurred on the first calmodulin-binding IQ motif closest to the head, resulting in 74–0-nm stepping motors. In other cases, labeled calmodulins exchanged into IQ positions higher up on the lever arm. For such dyes, a 37 nm − 2x nm step should occur followed by a 37 nm + 2x nm step, where x is the distance of the reporter from the joining coiled coil (the mid-point of the dimer) (Fig. 1). Indeed, the alternating 52–23-nm and 42–33-nm stepping motors are consistent with labeled calmodulins exchanged into the 5th and 6th positions, respectively; the absence of motors with other stepping behaviors suggests that exchange into positions 2, 3, and 4 rarely occurs. In summary, these data provide convincing proof for a hand-over-hand model, as an inch worm model predicts 37-nm displacements, irrespective of the dye position on the lever arm. The alternating 40° and 140° angular changes of a labeled lever arm (Forkey et al., 2003) also is best explained by the motor domains in the myosin V dimer switching between the leading and trailing positions.
When to step forward
What triggers myosin V to take a step? Clues to this question are contained within the distribution of dwell times between the steps. Remarkably, the dwell time distributions measured by optical trapping (Rief et al., 2000) or by detecting angle changes of the lever arm by polarization microscopy (Forkey et al., 2003) are virtually identical—a testimony to how accurately myosin V steps can be scored, even between different laboratories using different technologies. At high ATP concentrations, the dwell time distribution fits a single exponential, revealing that a first-order transition with a rate constant of 12 s−1 governs the waiting time between steps. Significantly, this rate constant matches the rate constant for ADP release measured by solution kinetics (De La Cruz et al., 1999). As further confirmation that ADP release triggers the step, addition of competing ADP (to drive the back reaction of ADP binding) prolongs the dwell time (Rief et al., 2000). Since the trailing head is the one that dissociates and moves during the step (Yildiz et al., 2003), ADP release from the trailing head must be rate-limiting in the myosin V cycle (Fig. 1). Collectively, these experiments illustrate how enzymatic rate constants can be deduced very accurately from single-molecule observations.
The talking heads
To enhance processivity, it seems likely that the two heads of the myosin V dimer “talk” to one another to coordinate transitions in their mechanochemical cycles. A possible mechanism for mediating head–head communication is tension generated between the two lever arms, which are constrained by the coiled coil and their connection to the actin-bound motor domains.
Clear visual evidence for tension acting on the lever arm of the leading head has been obtained in two EM studies. First, Burgess et al. (2002), using single-particle averaging of EM images, showed that the angle of the lever arm in the leading head is tilted further backward than the prestroke angle of the lever arm observed by x-ray crystallography. Second, earlier EM images by the same group showed individual “two-legged” myosin V motors that looked like telemark skiers, with a forward lean at the base of the leading head lever arm followed by a backward bend (Walker et al., 2000). Such images are consistent with a model in which leading head hydrolyzes ATP and releases phosphate (Fig. 1 E) and tries to undergo a power stroke. However, only the first IQ motif of the lever arm can rotate forward into a poststroke or partial poststroke conformation, whereas the distal part of the level arm is held back by the actin-bound, trailing head. This model also is supported by the FIONA data (Yildiz et al., 2003). If the first light chain was pointing straight forward and backward on the trailing and leading heads, respectively, then a stepping pattern of ∼68–6 nm should have been observed for a dye on the first calmodulin subunit. However, a myosin V with a “telemark stance” would generate the observed 74–0-nm stepping. The fluorescence polarization studies also found that some myosin Vs did not undergo alternating angular changes of the lever arm during movement (Forkey et al., 2003); this could be explained if this subset of motor had the dye-labeled calmodulin in the first IQ position and that this IQ position was tilting forward in both the trailing and leading heads.
A major question in the field is how tension mediated through the lever arms affects the chemical cycles of the two heads. It has been suspected that initiation of the power stroke by the leading head might accelerate the ADP release rate from the trailing head. Consistent with this idea, Veigel et al. (2002) showed that the mean dwell time in between steps of a processively moving myosin V motor is half of the dwell time for a single-headed myosin V interacting with actin. The authors suggested that strain generated by the leading head promotes a small (∼5 nm) rotation of the lever arm of the trailing head that might precede ADP release. In contrast, however, the ADP release rate of the rear head of a processive myosin reported by Rief et al. (2000) and Forkey et al., (2003) is identical to that of a single-headed myosin V measured by chemical kinetics (De La Cruz et al., 1999). Nevertheless, even the twofold change in ADP release kinetics observed by Veigel et al. is small. Thus, perhaps the leading head is the better candidate for tension-based regulation, which is also consistent with the visual EM images showing that its lever arm is experiencing the greatest distortion. Possible tension-mediated regulation of the leading head includes slowing down ADP dissociation (Fig. 1 A) or decreasing the subsequent rate of ATP rebinding to an empty site. Such effects would prevent the leading head from dissociating prematurely from actin and thus curtail futile cycling (ATP hydrolysis without a step).
Breaking down the 37-nm step: a power stroke followed by a diffusional search for an actin binding site
In the model shown in Fig. 1, the myosin V step is initially driven by the lever arm rotation in the leading head, and the 37-nm step is completed by the Brownian search and rebinding of the partner head to an actin subunit. Evidence for these two phases has been obtained from single-molecule measurements.
The power stroke has been most convincingly demonstrated using recombinant single-headed myosin V, which lacks partner head binding and hence is not processive. In two optical trap studies, single-headed myosin V produces displacements of ∼20 nm (Purcell et al., 2002; Veigel et al., 2002), consistent with the expected size of the lever arm rotation. Veigel et al. (2002) obtained evidence that this displacement may be broken down into 16- and 5-nm components, possibly reflecting the phosphate and ADP release steps. Steps of 20–25 nm are also evident in optical trap data from myosin V dimers operating at high loads (Mehta et al., 1999; Veigel et al., 2002; Sakamoto et al., 2003). This intermediate-sized step probably reflects the lever arm rotation of the leading head; the higher loads most likely extend the lifetime of the normally transient, one head–bound state by inhibiting the partner head from rebinding to an actin subunit.
As predicted from a lever arm rotation model, two studies show that truncation or elongation of the lever arm alters the power stroke displacement in an approximately linear manner (1xIQ, 5–7 nm; 2xIQ, 9 nm; 4xIQ, 16 nm; 8xIQ, 30 nm) (Purcell et al., 2002; Sakamoto et al., 2003). However, another study by Tanaka et al. (2002) reported processive motion and 37-nm steps by a recombinant myosin V with only a single IQ domain that was dimerized by fusion to the coiled coil domain of smooth muscle myosin. This surprising result indicates either that myosin V motion does not require a large lever arm rotation and may instead occur by the motor sliding along actin (Tanaka et al., 2002), or that the coiled coil of smooth muscle myosin can unravel and somehow permit the long step size. In either case, the construct is intriguing and warrants further investigation.
Evidence for the diffusional component of the step comes from the variability in the myosin V step size. Optical trap data shows a broader step size histogram than would be expected for a highly accurate 37-nm stepping motor (Rief et al., 2000; Purcell et al., 2002). Step size variability also is evident from the FIONA method, which shows occasional steps of 65 or 84 nm interspersed between the prevalent 74-nm steps. These data are best explained by the detached leading myosin V head undergoing a Brownian search that results in preferential binding to the 13th actin subunit, but occasionally results in rebinding to the 11th or 15th subunit (the 12th and 14th subunits are on the opposite side of the filament). Such shorter and longer spacing between the two heads also is seen in EM images (Walker et al., 2000). The 11th actin appears to be the second most favorable position for rebinding, since this explains the fact that myosin V–coated beads move in a very shallow left-hand spiral around a right-hand pitched actin filament suspended in solution (Ali et al., 2002).
The diffusional component of the mechanism also accounts for the amazing ability of myosin V motors engineered with different lever arm lengths to retain processivity. Recombinant myosin V dimers with 2xIQ, 4xIQ, and 8xIQ motifs move processively along actin filaments, although not as well as the wild-type 6xIQ motors (Purcell et al., 2002; Sakamoto et al., 2003). The 4xIQ motor, on average, takes 24-nm steps, consistent with a shorter lever arm rotation (Purcell et al., 2002). To rebind 24 nm away, the leading head must reach around the side of the filament to bind to an actin subunit. Sakamoto et al. (2003) provide suggestive evidence that this indeed takes place, indicating considerable flexibility of the heads in the myosin V dimer. Further reduction to 2xIQ motifs results in even greater difficulty in forming two-head attachments, and this motor only exhibited processive motion at low ATP (Purcell et al., 2002; Sakamoto et al., 2003). By keeping the trailing head bound longer in the ATP-free, rigor state, the detached head of the 2xIQ motor presumably has more time to dock to an actin subunit with unfavorable geometry. When reduced to 1xIQ motif, both myosin heads could no longer bind simultaneously to actin, and optical trap studies showed a single power stroke without evidence for actin binding by the partner head (Purcell et al., 2002).
New insight into actin binding by myosin V
As described earlier, studies of myosin V have benefited from a legacy of x-ray structures of muscle myosins. However, recently, the tables have turned, as a new crystal structure of the myosin V motor domain has provided insight into a long-standing question of how all myosins switch between strong and weak actin binding states (Coureux et al., 2003). Missing from the repertoire of previous myosin crystal structures has been the true rigor state, which is characterized by strong actin binding and a post-stroke lever arm position. Previous crystal structures visualized a “near-rigor” conformation with a post-stroke lever arm, but which displayed a conformation of the actin binding regions did not dock well into cryo-EM maps of the strongly bound actomyosin complex. At last, the nucleotide-free myosin V motor domain crystallized in a conformation that is likely to represent the strong-binding actin state, and this structure shows a long-suspected movement of two actin binding regions toward one another to form a more extensive polymer-binding interface. This structural interpretation agrees with recent atomic modeling of cryo-EM reconstructions of a muscle myosin–actin complex (Holmes et al., 2003). The myosin V structure also provides new insights, which are likely general to all myosins, into the coordinated conformational changes that occur between nucleotide binding pocket, the actin binding site, and the lever arm.
Evolutionary variations in the mechanism
Not all myosin Vs may work by this mechanism outlined above for mammalian myosin V. S. cerevisiae class V myosins are not processive, suggesting that either several motors are required to drive transport a membrane vesicle or that processivity is regulated in vivo (Reck-Peterson et al., 2001). An even more curious story is emerging for certain plant myosin V motors (in the literature called myosin XI, but arguably belonging to a myosin V class as they contain 6xIQ lever arms). Of particular interest is Chara (alga) myosin V, the Ferrarri of the motor protein world whose velocities of >60 μm/sec are 120-fold faster than mammalian myosin V and 10-fold faster than muscle myosin.
How the Chara and related plant myosins V motors can move at such rapid speeds constitutes an intriguing puzzle. One potential means of creating a fast motor is to increase its ATPase rate, but the rate of ADP release from Chara myosin (10 s−1) appears to be similar to mammalian myosin V (Kimura et al., 2003). Another possibility is that the motor takes an unusually large step, but optical trap studies show that Chara myosin takes a single step of 20 nm (Kimura et al., 2003), identical to the mammalian myosin V power stroke. The third, and most likely, possibility is that Chara myosins Vs work in teams to produce fast movement. Consistent with this idea, Chara myosin V is not processive like mammalian myosin V, suggesting that it might work cooperatively like muscle myosins. As a possible mechanism, Kimura et al. (2003) provide suggestive evidence that Chara myosin's ADP release rate is accelerated by strain in the direction of motion. Thus, forward tension exerted by a “stroking” Chara myosin might accelerate the ADP release and reduce the ATPase cycle time of other myosin V heads bound to the same actin filament. This is reminiscent of the finding by Veigel et al. (2002) that forward tension generated by the leading head can accelerate ADP release in the trailing head of mammalian myosin V, but this mechanism may have become greatly exaggerated in Chara myosin V. Thus, the long lever arm in this plant motor might be used primarily as a “strain sensor” that communicates to the nucleotide site, rather than being used for processivity. If this idea proves true, then Chara and mammalian myosin Vs provide a striking example of how a similar protein structure may undergo evolutionary modifications of a kinetic mechanism to achieve distinct biological goals.
Moving onward
It has been exciting to watch the steady stream of results emerging from myosin V, and this motor has provided a wonderful example of the importance of choosing the best experimental system to study. There is clearly more work to be done on this motor. Myosin V provides an amenable single molecule system for directly visualizing chemical transitions with fluorescent nucleotides and correlating these events with force generation and stepping (Ishijima et al., 1998). Many mysteries also remain on how tension controls the ATPase cycles in the two heads. But as the “grand dame” myosin V takes its curtain calls, new starlets are waiting in the wings. Notably, the myosin VI motor has been shown to move processively with a relatively long step size (Rock et al., 2001; Nishikawa et al., 2002). However, since myosin VI has a short lever arm, biophysicists are scratching their heads, unable to explain how this motor works. Nevertheless, with the marvelous tools available to study protein machines, myosin VI will undoubtedly take center stage in the near future.
Abbreviation used in this paper: PSF, point spread function.
References
- Ali, M.Y., S. Uemura, K. Adachi, H. Itoh, K. Kinosita, Jr., and S. Ishiwata. 2002. Myosin V is a left-handed spiral motor on the right-handed actin helix. Nat. Struct. Biol. 9:464–467. [DOI] [PubMed] [Google Scholar]
- Burgess, S., M. Walker, F. Wang, J.R. Sellers, H.D. White, P.J. Knight, and J. Trinick. 2002. The prepower stroke conformation of myosin V. J. Cell Biol. 159:983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coureux, P.-D., A.L. Wells, J. Menetrey, C.M. Yengo, C.A. Morris, H.L. Sweeeney, and A. Houdusse. 2003. A structural state of the myosin V motor without bound nucleotide. Nature. 425:419–423. [DOI] [PubMed] [Google Scholar]
- De La Cruz, E.M., A.L. Wells, S.S. Rosenfeld, E.M. Ostap, and H.L. Sweeney. 1999. The kinetic mechanism of myosin V. Proc. Natl. Acad. Sci. USA. 96:13726–13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forkey, J.N., M.E. Quinlan, M.A. Shaw, J.E. Corrie, and Y.E. Goldman. 2003. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature. 422:399–404. [DOI] [PubMed] [Google Scholar]
- Holmes, K.C., I. Angert, F.J. Kull, W. Jahn, and R.R. Schroder. 2003. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 425:423–427. [DOI] [PubMed] [Google Scholar]
- Hua, W., J. Chung, and J. Gelles. 2002. Distinguishing inchworm and hand-over-hand processive kinesin movement by neck rotation measurements. Science. 295:844–848. [DOI] [PubMed] [Google Scholar]
- Ishijima, A., H. Kojima, T. Funastsu, M. Tokunaga, H. Higuchi, H. Tanaka, and T. Yanagida. 1998. Simulataneous observation of individual ATPase and mechanical events by a single myosin molecule during interaction with actin. Cell. 92:161–171. [DOI] [PubMed] [Google Scholar]
- Ishikawa, K., N.L. Catlett, J.L. Novak, F. Tang, J.J. Nau, and L.S. Weisman. 2003. Identification of an organelle-specific myosin V receptor. J. Cell Biol. 160:887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, Y., N. Toyoshima, N. Hirakawa, K. Okamoto, and A. Ishijima. 2003. A kinetic mechanism for the fast movement of Chara myosin. J. Mol. Biol. 328:939–950. [DOI] [PubMed] [Google Scholar]
- Kwon, S., and B.J. Schnapp. 2001. RNA localization: SHEdding light on the RNA-motor linkage. Curr. Biol. 11:R166–R168. [DOI] [PubMed] [Google Scholar]
- Langford, G.M. 2002. Myosin-V, a versatile motor for short-range vesicle transport. Traffic. 3:859–865. [DOI] [PubMed] [Google Scholar]
- Mehta, A.D., R.S. Rock, M. Rief, J.A. Spudich, M.S. Mooseker, and R.E. Cheney. 1999. Myosin-V is a processive actin-based motor. Nature. 400:590–593. [DOI] [PubMed] [Google Scholar]
- Nishikawa, S., K. Homma, Y. Komori, M. Iwaki, T. Wazawa, A. Hikikoshi Iwane, J. Saito, R. Ikebe, E. Katayama, T. Yanagida, and M. Ikebe. 2002. Class VI myosin moves processively along actin filaments backward with large steps. Biochem. Biophys. Res. Commun. 290:311–317. [DOI] [PubMed] [Google Scholar]
- Purcell, T.J., C. Morris, J.A. Spudich, and H.L. Sweeney. 2002. Role of the lever arm in the processive stepping of myosin V. Proc. Natl. Acad. Sci. USA. 99:14159–14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson, S.L., D.W. Provance, Jr., M.S. Mooseker, and J.A. Mercer. 2000. Class V myosins. Biochim. Biophys. Acta. 1496:36–51. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson, S.L., M.J. Tyska, P.J. Novick, and M.S. Mooseker. 2001. The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin-based motors. J. Cell Biol. 153:1121–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rief, M., R.S. Rock, A.D. Mehta, M.S. Mooseker, R.E. Cheney, and J.A. Spudich. 2000. Myosin-V stepping kinetics: a molecular model for processivity. Proc. Natl. Acad. Sci. USA. 97:9482–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock, R.S., S.E. Rice, A.L. Wells, T.J. Purcell, J.A. Spudich, and H.L. Sweeney. 2001. Myosin VI is a processive motor with a large step size. Proc. Natl. Acad. Sci. USA. 98:13655–13659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T., I. Amitani, E. Yokota, and T. Ando. 2000. Direct observation of processive movement by individual myosin V molecules. Biochem. Biophys. Res. Commun. 272:586–590. [DOI] [PubMed] [Google Scholar]
- Sakamoto, T., F. Wang, S. Schmitz, Y. Xu, Q. Xu, J.E. Molloy, C. Veigel, and J.R. Sellers. 2003. Neck length and processivity of myosin V. J. Biol. Chem. 278:29201–29207. [DOI] [PubMed] [Google Scholar]
- Tanaka, H., K. Homma, A.H. Iwane, E. Katayama, R. Ikebe, J. Saito, T. Yanagida, and M. Ikebe. 2002. The motor domain determines the large step of myosin-V. Nature. 415:192–195. [DOI] [PubMed] [Google Scholar]
- Vale, R.D. 2003. The molecular motor toolbox for intracellular transport. Cell. 112:467–480. [DOI] [PubMed] [Google Scholar]
- Vale, R.D., and R.M. Milligan. 2000. The way things move: looking under the hood of molecular motor proteins. Science. 288:88–95. [DOI] [PubMed] [Google Scholar]
- Veigel, C., F. Wang, M.L. Bartoo, J.R. Sellers, and J.E. Molloy. 2002. The gated gait of the processive molecular motor, myosin V. Nat. Cell Biol. 4:59–65. [DOI] [PubMed] [Google Scholar]
- Walker, M.L., S.A. Burgess, J.R. Sellers, F. Wang, J.A. Hammer, III, J. Trinick, and P.J. Knight. 2000. Two-headed binding of a processive myosin to F-actin. Nature. 405:804–807. [DOI] [PubMed] [Google Scholar]
- Yildiz, A., J.N. Forkey, S.A. McKinney, T. Ha, Y.E. Goldman, and P.R. Selvin. 2003. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 300:2061–2065. [DOI] [PubMed] [Google Scholar]