Abstract
We have analyzed the abundance of SUMO-conjugated species during the cell cycle in Xenopus egg extracts. The predominant SUMO conjugation products associated with mitotic chromosomes arose from SUMO conjugation of topoisomerase II. Topoisomerase II was modified exclusively by SUMO-2/3 during mitosis under normal circumstances, although we observed conjugation of topoisomerase II to SUMO-1 in extracts with exogenous SUMO-1 protein. Inhibition of SUMO modification by a dominant-negative mutant of the SUMO-conjugating enzyme Ubc9 (dnUbc9) did not detectably alter topoisomerase II activity, but it did increase the amount of unmodified topoisomerase II retained on mitotic chromosomes after high salt washing. dnUbc9 did not disrupt the assembly of condensed mitotic chromosomes or block progression of extracts through mitosis, but it did block the dissociation of sister chromatids at the metaphase–anaphase transition. Together, our results suggest that SUMO conjugation is important for chromosome segregation in metazoan systems, and that mobilization of topoisomerase II from mitotic chromatin may be a key target of this modification.
Keywords: topoisomerase II; SUMO-1; SUMO-2; mitosis; chromosome segregation
Introduction
SUMO family proteins are small ubiquitin-related proteins that become conjugated to cellular substrates in a manner similar to ubiquitin. In mammals, there are three SUMO isoforms. Two of these isoforms, SUMO-2 and -3, are very similar in sequence (96% identity between the processed forms). Throughout this report, we will refer to SUMO-2 and SUMO-3 collectively as SUMO-2/3 whenever these isoforms cannot be unambiguously distinguished. SUMO-1 is more distinct, being only ∼45% identical with either of the other two isoforms. The SUMO conjugation pathway is similar to the ubiquitin conjugation pathway (Hochstrasser, 2000); SUMO proteins must be post-translationally processed to yield a COOH-terminal diglycine motif. After processing to generate the conjugatable forms, the first step in the SUMO conjugation pathway is the ATP-dependent formation of a thioester bond between SUMO proteins and their activating (E1) enzyme, the Aos1/Uba2 heterodimer. The second step is the formation of a thioester bond between SUMO proteins and their conjugating (E2) enzyme, Ubc9. In the last step, an isopeptide bond is formed between SUMO proteins and substrates through the cooperative action of Ubc9 and protein ligases (E3). A growing number of SUMO conjugation substrates have been reported, particularly for SUMO-1 (Melchior, 2000). SUMO-1 conjugation has been demonstrated to increase stability, to promote subcellular relocalization or to alter protein–protein interactions for various substrates (Seeler and Dejean, 2001). The cellular roles of SUMO-2 and -3 are not yet understood, nor are the functional distinctions between SUMO family members well defined.
Fission and budding yeast each contain a single SUMO family protein, pmt3 and Smt3p, respectively. These proteins have been implicated in the regulation of the cell cycle in both organisms. In fission yeast, pmt3 − mutants undergo aberrant mitosis and display high frequency loss of minichromosomes (Tanaka et al., 1999). pmt3 − mutants are also sensitive to increased temperature, UV light, DNA-damaging agents, and replication inhibitors. Mutations in the E1 and E2 proteins for pmt3 cause aberrant cell morphology, condensed and/or fragmented chromosomal DNA, mini-chromosome instability, and cells with a “cut” morphology even in the absence of radiation treatment (al-Khodairy et al., 1995; Shayeghi et al., 1997). In budding yeast, inhibition of Ubc9p synthesis blocks cell cycle progression at G2/M phase, stabilizing cyclin proteins and causing the accumulation of large budded cells with a short spindle, a single nucleus, and fully replicated DNA (Seufert et al., 1995). Moreover, budding yeast have two Smt3p isopeptidases (Ulp1p and Ulp2p/Smt4p), and disruption of either of these proteins causes defective cell cycle control. Temperature-sensitive ulp1-ts mutants arrest in the G2/M phase of the cell cycle at the restrictive temperature (Li and Hochstrasser, 1999). With prolonged incubation at elevated temperatures, ulp1-ts mutants eventually pass through mitosis and show aberrant chromosome structures, consistent with severe chromosome damage or missegregation (Li and Hochstrasser, 1999). Ulp2p/Smt4p mutants display decreased plasmid and chromosome stability, as well as failure to recover from checkpoint arrest after treatment with DNA-damaging agents, DNA replication inhibitors, or microtubule poisons (Li and Hochstrasser, 2000).
One underlying cause of the cell cycle phenotypes in budding yeast is likely to be a requirement for modification of topoisomerase II (Top2p) by Smt3p in order to release centromeric cohesion at anaphase. SMT3 was found among a number of genes whose mutants showed inability to correctly segregate chromosomes at the metaphase–anaphase transition (Biggins et al., 2001), and ULP2/SMT4 was reported as an overexpression suppressor of mutations in condensin subunits required for mitotic chromosome condensation (Strunnikov et al., 2001). More recently, Bachant et al. (2002) examined the recovery of budding yeast cells from DNA damage arrest in smt4-Δ mutants and discovered that the cells were delayed in metaphase, with their chromosomes abnormally stretched along the spindle axis. Examination of sister chromatid separation at different chromosomal positions demonstrated that centromeric cohesion was specifically decreased in smt4-Δ mutants. Top2p was suggested to be one of the substrates responsible for this phenotype, because top2 mutants lacking Smt3p modification sites could significantly suppress the smt4 centromeric cohesion defect. Human topoisomerase IIα and β have been reported to be substrates for conjugation with SUMO-1, and topoisomerase II inhibitors stimulate this modification (Mao et al., 2000). However, there has not been any report suggesting cell cycle–regulated SUMO-1 conjugation of vertebrate topoisomerase II. The mechanisms whereby Smt3p or SUMO-1 regulate topoisomerase II have not been reported in any organism.
Genetic evidence suggests that topoisomerase II plays crucial roles in both chromosome condensation and segregation during mitosis (Uemura et al., 1987). Moreover, a number of observations have shown that topoisomerase II is directly required for the assembly of condensed chromosomes in mitotic Xenopus egg extracts; topoisomerase II depletion from egg extracts blocks condensation of chromosomes from chicken erythrocyte nuclei (Adachi et al., 1991), and chemical inhibition of topoisomerase II prevents remodeling and condensation of sperm nuclei chromosomes (Hirano and Mitchison, 1993). The requirement for topoisomerase II in sister chromatid segregation can be distinguished from its role in mitotic chromosome assembly in egg extracts because chemical inhibition of topoisomerase II by VP-16 at the metaphase–anaphase transition blocks sister chromatid separation despite the assembly of intact chromosomes before VP-16 addition (Shamu and Murray, 1992). The behavior of topoisomerase II in metazoan cells during mitosis has been somewhat controversial. Early experiments indicated that topoisomerase II is tightly associated with the scaffold fraction of mitotic chromosomes (Gasser et al., 1986), and that it is distributed along with chromatid axis during metaphase (Earnshaw and Heck, 1985). From these results, it was suggested that topoisomerase II is a major structural component of mitotic chromosomes. On the other hand, the bulk of topoisomerase II can be eluted under mild, low salt conditions from mitotic chromosomes formed in Xenopus egg extracts, arguing against the notion that it is an integral component of a chromosomal scaffold (Hirano and Mitchison, 1993). Recent live-imaging experiments have shown that topoisomerase II is highly dynamic on chromosomes during mitosis (Christensen et al., 2002; Null et al., 2002; Tavormina et al., 2002). The mechanisms controlling the dynamic association of topoisomerase II to chromosomes have not been clarified.
We used Xenopus egg extracts to examine cell cycle–dependent changes in SUMO-conjugated proteins. We found a set of high mol wt, chromatin-dependent mitotic SUMO-containing species, which protein sequencing revealed to be SUMO-conjugated topoisomerase II. Topoisomerase II is modified exclusively by SUMO-2/3 during mitosis under normal circumstances, although we could observe SUMO-1 conjugation of topoisomerase II in extracts with exogenous SUMO-1 protein. In cycling extracts, SUMO-2/3 modification of topoisomerase II was maximal in metaphase, followed by rapid deconjugation during anaphase. Blocking SUMO modification using a dominant-negative mutant of Ubc9 (dnUbc9 = Ubc9-C93S, L97S; Banerjee et al., 1995) did not detectably alter the activity of topoisomerase II on chromatin assembled in egg extracts. However, dnUbc9 increased the amount of unmodified topoisomerase II retained on mitotic chromosomes after high salt washing. dnUbc9 did not arrest the progression of cycling extracts through mitosis, nor did it alter the assembly of condensed chromosomes in mitotically-arrested extracts. By contrast, dnUbc9 clearly disrupted the dissociation of sister chromatids at the metaphase-anaphase transition. Together, these findings indicate that SUMO-2/3 conjugation does not drastically alter topoisomerase II enzymatic activity, but this modification is important for remodeling of topoisomerase II on mitotic chromosomes at the metaphase-anaphase transition. Failure of such remodeling could clearly play an important part in chromosome missegregation when SUMO conjugation is blocked.
Results
Regulation of SUMO-1 conjugation during the cell cycle
We sought to use Xenopus egg extract as a model system to study the relationship between SUMO conjugation and cell cycle progression. We examined the profile of SUMO-1–conjugated proteins through the cell cycle. To increase the overall level of SUMO-1 conjugation in this experiment, we added recombinant His6-tagged SUMO-1 (His-SUMO-1) to meiotically arrested cytostatic factor (CSF) extracts. The CSF extracts were then released into interphase by the addition of CaCl2 and allowed to spontaneously oscillate between interphase and mitosis (Fig. 1; Kornbluth et al., 2001). We examined the spectrum of SUMO-1–conjugated proteins at different points by Western blot analysis using affinity-purified anti-SUMO-1 antibodies.
Figure 1.
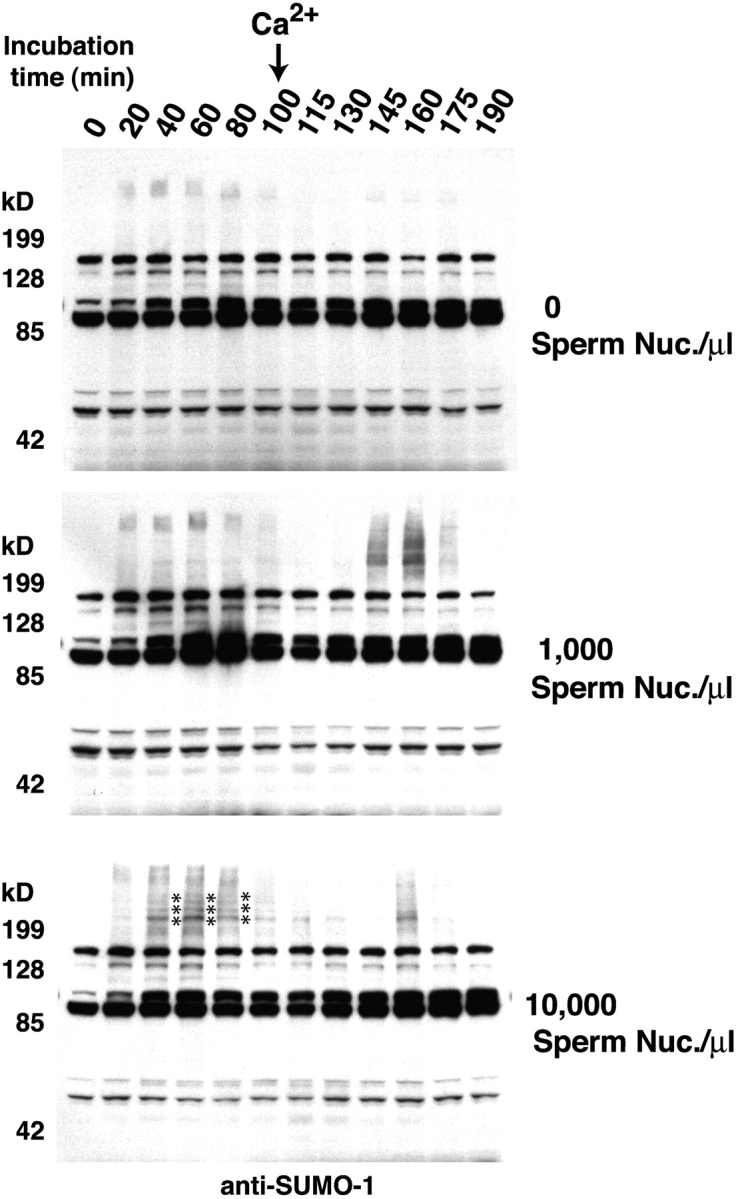
Chromatin-dependent SUMO conjugation of mitotic substrates. CSF extracts were supplemented with histidine-tagged SUMO-1 (final concentration 30 ng/μl) and with the indicated number of sperm nuclei. After formation of condensed metaphase chromosomes (90 min after the start of the reaction), CSF arrest was released into interphase by addition of 0.6 mM CaCl2. The SUMO-1–conjugated protein profile throughout the reaction was analyzed by Western blotting with affinity-purified anti-SUMO-1 antibodies. There were a series of strong M-phase–specific SUMO-1 conjugates observed in the reaction containing 10,000 sperm nuclei/μl (bottom, indicated by an asterisk). These bands could also be weakly observed in the reaction containing 1,000 sperm nuclei/μl, but were absent in the reaction without nuclei. These bands were lost after the extracts entered interphase. These characteristics indicated that the bands resulted from modification of mitotic substrates in a chromatin-dependent manner.
We observed a series of high mol wt–conjugated species before release of CSF extracts from meiotic arrest (Fig. 1, bottom; indicated with an asterisk). These conjugated species migrated with a mobility >200 kD, and they disappeared rapidly after the extracts were driven into interphase by addition of CaCl2. We could immediately confirm that these bands arose from SUMO-1 conjugation because they could be abolished by the addition of a dominant-negative form of the SUMO-1 E2 protein, Ubc9 (dnUbc9 = Ubc9-C93S, L97S; Banerjee et al., 1995). The behavior of these conjugation products was particularly remarkable because their appearance was entirely dependent on the presence of chromatin in the reaction (Fig. 1, compare top panels with bottom panels). For this reason, we chose to investigate their identity and regulation further.
Topoisomerase II is modified by conjugation to SUMO-2/3
To identify the origin of the ∼200-kD conjugated species found in M phase, we prepared a large-scale CSF extract reaction with a high concentration of added sperm nuclei and His-SUMO-1. After a 60-min incubation, we made a crude separation of chromatin-associated proteins by centrifugation. We observed by Western blotting that the desired species fractionated with the DNA (unpublished data). The His-SUMO-1–conjugated proteins from the chromatin fraction were solubilized in a buffer containing 8 M urea, and the recovered fraction was subjected to further purification using metal affinity chromatography. Upon silver staining, the resultant fraction showed a clear set of bands in the appropriate mol wt range >200 kD that were strongly recognized on Western blots by anti-SUMO-1 antibodies. These bands were isolated and subjected to protein sequencing at the Harvard Microchemistry Facility (Cambridge, MA). Protein sequence analysis showed that these conjugated species arose from His-SUMO-1 conjugation of topoisomerase II.
To confirm that topoisomerase II is modified by SUMO-1 conjugation in the absence of exogenous His-SUMO-1, we analyzed CSF egg extracts with or without exogenous SUMO-1 or dnUbc9 by Western blotting with anti-topoisomerase II antibodies (Fig. 2 A; Luke and Bogenhagen, 1989). Antibodies directed against topoisomerase II recognized a ladder of bands in the presence or absence of exogenous SUMO-1 protein. Consistent with the idea that these bands arose from conjugation to SUMO-1, they were abolished by the addition of dnUbc9 (Fig. 2 A, left). However, we were surprised to find that antibodies against SUMO-1 did not recognize any bands corresponding to conjugated topoisomerase II in the absence of exogenous SUMO-1 protein (Fig. 2 A, middle).
Figure 2.

DNA topoisomerase II is modified by SUMO-2/3 in M-phase extracts. (A) CSF extracts containing 10,000 sperm nuclei/μl were incubated at 23°C for 60 min with or without exogenous 30 ng/μl His-SUMO-1 or 150 ng/μl dnUbc9. Aliquots were analyzed by immunoblotting with anti–Xenopus topoisomerase II, anti-SUMO-1, and anti-SUMO-2/3 antibodies. (B) CSF extract containing 10,000 sperm nuclei/μl was incubated 60 min at 23°C. The reaction was split, and half was driven into interphase though the addition of 0.6 mM CaCl2. Each reaction was incubated at 23°C for an additional 30 min. Chromatin was isolated from both mitotic and interphase extracts, as described in Materials and methods. The chromatin was subjected to Western blotting with the following antibodies: (1) monoclonal anti–human topoisomerase IIα/β (DNATop2AB); (2) monoclonal anti–human topoisomerase IIα; (3) polyclonal anti– human topoisomerase IIα; (4) anti–Xenopus topoisomerase II; and (5) anti-SUMO-2/3, respectively. Note that topoisomerase IIα is essentially the sole form of topoisomerase II in egg extracts. (C) CSF extracts containing the indicated number of sperm nuclei/μl were incubated at 23°C for 60 min. Equal amounts of isolated chromatin were subjected to SDS-PAGE and analyzed by Western blotting using anti-SUMO-2/3 and anti-topoisomerase II antibodies. Asterisks indicate SUMO-conjugated forms of topoisomerase II in all panels.
Because SUMO-1 shares its E2 enzyme with the other vertebrate SUMO family members, SUMO-2 and -3 (Melchior, 2000), we examined whether they might become conjugated to topoisomerase II. SUMO-2 and -3 are closely related in their amino acid sequence (96% identical in their processed forms), whereas the identity between SUMO-1 and either SUMO-2 or -3 is more limited (∼45%). We performed Western blotting on the same samples with affinity-purified polyclonal anti-SUMO-2/3 antibodies. On Western blots of extracts without exogenous proteins, anti-SUMO-2/3 antibodies recognized bands corresponding to those recognized by anti-topoisomerase II (Fig. 2 A, right). Consistent with the idea that these bands arose from conjugation of topoisomerase II to SUMO-2/3, they were absent in Western blots of extracts containing dnUbc9. Moreover, these bands were drastically decreased in Western blots of extracts containing exogenous SUMO-1, suggesting that SUMO-1 can directly compete with SUMO-2/3 for conjugation to topoisomerase II when it is present at higher than normal physiological concentrations. Additional experiments examining the conjugation of topoisomerase II with EGFP-tagged versions of SUMO-2 and SUMO-3 in CSF extracts with sperm nuclei indicated that there may be a strong preference for SUMO-2 in this reaction (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200304088/DC1). It will obviously be of future interest to obtain reagents that can distinguish between endogenous SUMO-2 and -3 to confirm this conclusion under unperturbed conditions.
Because our earlier results demonstrated that exogenously added SUMO proteins have the capacity to distort the normal pattern of substrate modification, we wished to analyze SUMO-2/3 conjugation to topoisomerase II in unperturbed extracts, relying solely upon the endogenous proteins. First, we wished to confirm whether modification of topoisomerase II is limited to M-phase. To do this, we compared the pattern of SUMO-2/3–conjugated topoisomerase II on chromatin isolated from interphase and CSF-arrested egg extracts (Fig. 2 B). Western blotting with anti-SUMO-2/3 antibodies and four different anti-topoisomerase II antibodies showed that the conjugated forms of topoisomerase II were exclusively found in chromatin fraction from CSF extracts, leading us to conclude that this modification is truly M-phase–specific. Next, we examined the level of conjugated topoisomerase II as a function of added sperm chromatin (Fig. 2 C). The dependency of SUMO conjugation on the relative concentration of chromatin was easily observed when we purified chromosomes from reactions containing different initial concentrations of sperm chromatin by centrifugal separation through a sucrose cushion. When equal concentrations of sperm chromatin from each reaction were loaded in adjacent lanes of a Western blot, the amount of topoisomerase II–SUMO-2/3 was essentially identical, irrespective of the initial concentration of sperm chromatin in the incubations (Fig. 2 C). These data suggest that sperm chromosomes have a defined and saturable capacity to generate and/or stabilize topoisomerase II–SUMO-2/3 conjugates.
Together, our results indicate that topoisomerase II is conjugated to SUMO-2/3 in an M-phase– and chromatin-dependent manner, although SUMO-1 can compete with SUMO-2/3 in this conjugation reaction if it is present at abnormally high concentrations.
Modification of DNA topoisomerase II by endogenous SUMO-2/3
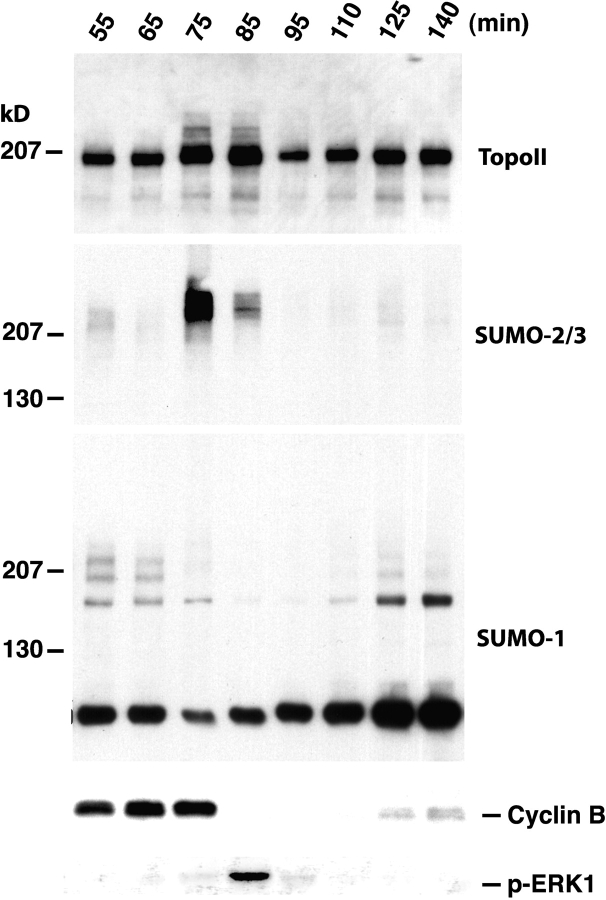
To investigate the timing of endogenous topoisomerase II modification more closely during the mitotic cell cycle, we used cycling extracts, which oscillate spontaneously and synchronously between interphase and mitosis (Kornbluth et al., 2001). We added sperm chromatin coincident with the start of the reaction and followed progression through the cell cycle by chromosome morphology (unpublished data), cyclin B abundance, and ERK1 phosphorylation (Fig. 3). Aliquots of the reaction were taken at the indicated intervals. Chromatin was prepared from these aliquots and equivalent chromatin recovery at each time point was verified by Western blotting using antibodies against the embryonic linker histone B4 (unpublished data; Dasso et al., 1994). The modification status of chromatin-associated topoisomerase II was determined by Western blotting with anti-topoisomerase II and anti-SUMO-2/3 antibodies. Examination of chromosome morphology suggested that the extract entered mitosis at ∼65 min, and that anaphase onset occurred at ∼80 min. This timing was confirmed by both the profiles of cyclin B degradation and ERK1 phosphorylation. On Western blots using anti-topoisomerase II antibodies, we observed that the maximum level of topoisomerase II conjugation occurred during metaphase at ∼75 min, close to the maximum level of cyclin B accumulation. The level of conjugated species rapidly declined as the extract passed through the metaphase–anaphase transition, and we detected no conjugated topoisomerase II in the chromosomal fraction by 95 min.
Figure 3.
Topoisomerase II is modified by SUMO-2/3 during mitosis in unperturbed cycling extracts. Xenopus cycling egg extracts were supplemented with 2,000 sperm nuclei/μl and incubated at 23°C. Samples were taken from extracts at the indicated times. Aliquots of each sample were directly subjected to SDS-PAGE and Western blotting analysis to determine cyclin B abundance (fourth panel) and phosphorylated ERK1 levels (bottom). Chromatin was separately prepared from samples taken at each time point, as described in Materials and methods. The chromatin preparations were analyzed by Western blotting with antibodies against topoisomerase II (top), SUMO-2/3 (second panel), and SUMO-1 (middle). Aliquots were also removed at the indicated times and analyzed for DNA morphology after addition of Hoechst 33342 DNA dye (not depicted). We observed condensed metaphase chromosomes at 75 min, and anaphase figures at 85 min after the start of the reaction.
Western blotting with antibodies against SUMO-2/3 showed that the predominant SUMO-2/3–conjugated species in the chromosomal fraction migrated at the same mol wt as SUMO-2/3–modified topoisomerase II, and followed the same pattern of abundance, supporting the conclusion that topoisomerase II is maximally modified immediately before the onset of anaphase. By contrast, when we probed the chromosomal fractions with anti-SUMO-1 antibodies, we saw a marked down-regulation of SUMO-1 conjugation during mitosis. Instead, we observed the accumulation of a SUMO-1–conjugated species during interphase. Notably, there was never a close correspondence between the migration of a major modified topoisomerase II species and a major SUMO-1 conjugation product.
These findings further illustrate the specificity and chromatin dependence of topoisomerase II modification by SUMO-2/3 during the unperturbed egg extract cell cycle, and show that SUMO-1 is not a major modifier of topoisomerase II in cycling extracts. Moreover, this experiment documents the close coincidence between anaphase and the deconjugation of topoisomerase II under normal conditions.
SUMO-2/3 modification regulates the chromatin dynamics of topoisomerase II
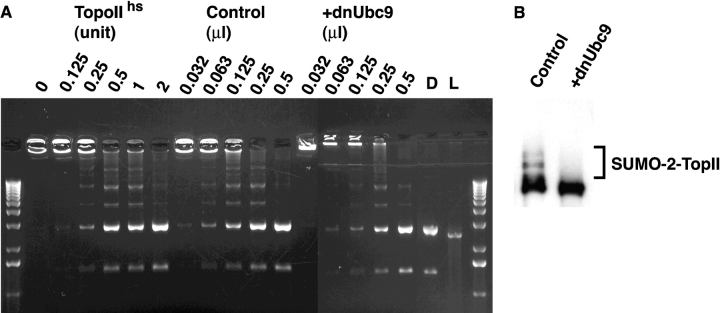
Next, we examined whether modification by SUMO-2/3 significantly alters the biochemical behavior or chromatin association of mitotic topoisomerase II. We extracted topoisomerase from mitotic chromosomes produced in egg extracts with or without dnUbc9. This allowed us to obtain a preparation of topoisomerase II that was enriched in the SUMO-conjugated form (Fig. 4 B; ∼20% modified) and a comparable preparation that did not have modified topoisomerase II. When we measured the activity of these preparations for decatenation of kinetoplast DNA, their activity levels were indistinguishable (Fig. 4 A). These results do not eliminate the possibility that SUMO-2 conjugation may inhibit topoisomerase II because this assay is not sufficiently sensitive to detect partial inhibition of the conjugated subpopulation of topoisomerase II. However, the findings in Fig. 4 would be inconsistent with dramatic stimulation of topoisomerase II activity by SUMO-2 conjugation.
Figure 4.
dnUbc9 does not alter decatenation activity of Xenopus topoisomerase II. (A) Chromosomal proteins were prepared from sperm chromatin assembled with or without 150 ng/μl dnUbc9 and assayed for decatenation activity as described in Materials and methods. Serial dilutions of chromosomal proteins were performed, in order to add the equivalent of the volumes indicated in each lane into decatenation reactions. For comparison, reactions were performed using purified human topoisomerase II, with the number of manufacturer's unit per reaction indicated. A fully decatenated marker was loaded in the lane marked D, and a linearized marker was loaded in the lane marked L. The lane marked 0 shows kinetoplast DNA from a control reaction without added chromosomal proteins. (B) Western blot of chromosomal eluates from extracts without (left) or with (right) dnUbc9 used in the decatenation reaction.
To examine chromosome association of topoisomerase II, we isolated mitotic chromosomes formed in CSF extract, using centrifugal separation through a sucrose cushion. The chromosomal pellets were sequentially extracted with buffers containing increasing concentrations of NaCl (100, 300, and 500 mM). In a previous report, it was shown that the bulk of topoisomerase II was released from chromosomes with a buffer containing 100 mM salt (Hirano and Mitchison, 1993), but we observed a slightly different extraction pattern; a significant fraction of topoisomerase II was not associated with chromosomes, and was found in the post-chromosomal supernatant (Fig. 5 A). This fraction of topoisomerase II did not show SUMO-2/3 conjugation. In our hands, undetectable amounts of chromatin-associated topoisomerase II were released by washing chromosomal preparations with buffer containing 100 mM NaCl (Fig. 5 A). The bulk of topoisomerase II was released from chromatin by washing with buffer containing 300 mM NaCl, although a small fraction of topoisomerase II was retained on the chromatin even after extraction of the chromatin with buffer containing 500 mM NaCl. SUMO-2/3–conjugated topoisomerase II was detected most abundantly in the supernatant from the 300-mM NaCl wash.
Figure 5.
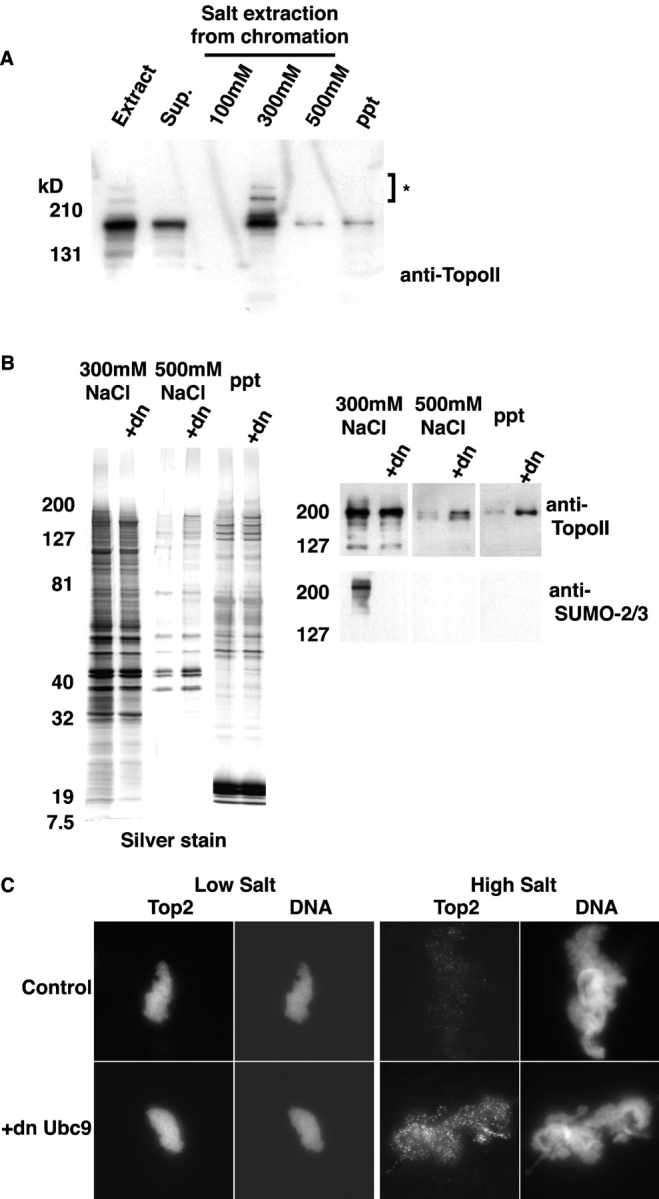
SUMO-2/3–modified DNA topo-isomerase II is associated with chromatin. (A) CSF extracts containing 10,000 sperm nuclei/μl were incubated at 23°C for 60 min, and chromatin was isolated from each reaction as described in Materials and methods. Chromatin was extracted with XB buffer supplemented with 5 mM β-glycerol phosphate plus 100, 300, or 500 mM NaCl sequentially. Equal amounts of all fractions were subjected to SDS-PAGE and Western blotting with anti-topoisomerase II antibody, as indicated. (B) CSF extracts containing 10,000 sperm nuclei/μl were incubated at 23°C for 60 min, in the absence or presence of dnUbc9 (+dn; 150 ng/μl). Chromatin was isolated from each reaction as described in Materials and methods. Chromatin was extracted as above and equal concentrations of the supernatants from the 300-mM NaCl extraction (left two lanes in each panel), 500-mM NaCl extraction (middle two lanes in each panel), and final pellet (right two lanes in each panel) were subjected to SDS-PAGE and either silver staining (left) or Western blotting (right) with anti-topoisomerase II and anti-SUMO-2/3 antibodies. (C) Mitotic chromosomes were assembled in CSF extracts at 23°C for 60 min in the presence or absence of dnUbc9. The chromosomes were low salt (CSF-XB) or high salt (CSF-XB + 200 mM NaCl) buffers and stained with monoclonal anti–human topoisomerase IIα/β antibodies, as described in Materials and methods. DNA was visualized with Hoechst 33342. All topoisomerase II immunofluorescence images were taken using the identical exposure time.
We examined whether altering the SUMO-2/3 modification of chromosomal topoisomerase II could alter its extraction behavior. To do this, we prepared chromatin samples from CSF extracts with and without dnUbc9 and examined the salt extraction behavior of topoisomerase II from these samples (Fig. 5 B). In these experiments, it was useful to make an arbitrary distinction between subpopulations of topoisomerase II on chromatin that are loosely bound (extracted in the 300-mM NaCl wash) and subpopulations that are tightly bound (found in the supernatant and pellet from the 500-mM NaCl wash). If SUMO-2/3 conjugation does not influence the partitioning of topoisomerase II between the loosely and tightly bound fractions, addition of dnUbc9 would be predicted to cause no change in the extraction behavior of topoisomerase II. However, if SUMO-2/3 conjugation had a mechanistic role in the conversion of inextractable topoisomerase II subpopulations to extractable forms, dnUbc9 would be predicted to cause an increase in the tightly bound forms. Notably, the amount of topoisomerase II retained on the chromatin after the 300-mM NaCl extraction was dramatically increased by the presence of dnUbc9 (Fig. 5 B, right). This observation clearly favors the latter possibility.
As an alternative assay for the capacity of dnUbc9 to influence the association of topoisomerase II with chromosomes, we examined extracted chromatin by immunofluorescence using anti-topoisomerase II antibodies. Consistent with our biochemical observations, a high level of topoisomerase II remained associated with chromatin that had been extracted with CSF-XB buffer (100 mM KCl), but there was a much lower level of staining after treatment with CSF-XB buffer plus 200 mM NaCl (final salt concentration = 200 mM NaCl + 100 mM KCl; this buffer will be referred to as high salt CSF-XB; Fig. 5 C). It was noticeable that although distribution of topoisomerase II on the chromatin in the CSF-XB–washed chromatin was relatively uniform, the residual topoisomerase II retained after washing with high salt CSF-XB was punctate.
The presence of dnUbc9 throughout the chromosome assembly reaction did not change the morphology of chromosomes washed at either salt concentration. When extractions were performed with CSF-XB buffer, staining of chromosomes assembled with or without dnUbc9 showed very similar patterns, with topoisomerase II distribution closely correlating to the distribution of DNA (Fig. 5 C, left panels). By contrast, the amount of topoisomerase II associated with high salt CSF-XB–washed chromatin was dramatically increased by dnUbc9 (Fig. 5 C, right panels). The enhanced staining was not uniform, but clearly distributed into bright foci throughout the chromosomes, suggesting that dnUbc9 increases the specific retention of a particular subpopulation of topoisomerase II. Together with our biochemical data, these findings strongly indicate that SUMO-2/3 modification can alter the chromatin-binding properties of topoisomerase II, promoting conversion of tightly bound topoisomerase II to lower affinity forms.
SUMO conjugation is required for proper mitotic chromosome segregation
Our observations indicated that some tightly chromatin-associated forms of topoisomerase II are conjugated with SUMO-2/3 in order to facilitate their conversion to lower affinity forms. Such conversion could be involved in the removal of the tightly bound topoisomerase II from chromatin, or its remodeling on the chromosome. To test the biological role of mitotic SUMO modification of topoisomerase II and other potential substrates, we evaluated the effect of dnUbc9 on cycling and CSF extracts. In cycling extracts, the addition of dnUbc9 had no detectable effect on sperm chromatin decondensation, the formation of nuclei, DNA replication, or the entry of extracts into mitosis (unpublished data). Furthermore, we could not find any consistent difference between the morphology of mitotic chromosomes assembled in cycling or CSF extracts in the presence or absence of dnUbc9. These findings indicate that SUMO conjugation is not required for any of these processes. By contrast, we saw a significant difference in the behavior of chromosomes at the metaphase–anaphase transition in cycling extracts with and without dnUbc9. Although we could consistently observe segregated sister chromatids in control extracts at anaphase onset, we did not observe chromosome segregation in extracts containing dnUbc9 (unpublished data). Despite the absence of any evident sister chromatid segregation in extracts containing dnUbc9, the extracts progressed out of mitosis and formed nuclei in synchrony with control extracts.
This finding prompted us to evaluate sister chromatid segregation under conditions where we could precisely control the onset of the metaphase–anaphase transition (Fig. 6). To do this, we drove a CSF extract into interphase using CaCl2. Sperm chromatin was added to this extract, and was allowed to form nuclei and to undergo a complete round of DNA replication. The extract was then driven back into mitosis with the addition of fresh CSF extract, and dnUbc9 was added at this point (Fig. 6 A). Spindle assembly was assessed 30 min after the addition of CSF extract, and we could find no distinction between reactions with or without dnUbc9, again suggesting that dnUbc9 does not disrupt chromosome condensation, attachment to spindles, or alignment on the metaphase plate. As with unreplicated chromatin (Fig. 5 B), the bulk of topoisomerase II was extracted from replicated chromatin by washing with buffer containing 300 mM NaCl, although a small fraction of topoisomerase II was retained on the chromatin even after extraction of the chromatin with buffer containing 500 mM NaCl (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200304088/DC1). Distribution of topoisomerase II to the inextractable fraction was increased when dnUbc9 was added as mitosis was induced with fresh CSF extract.
Figure 6.
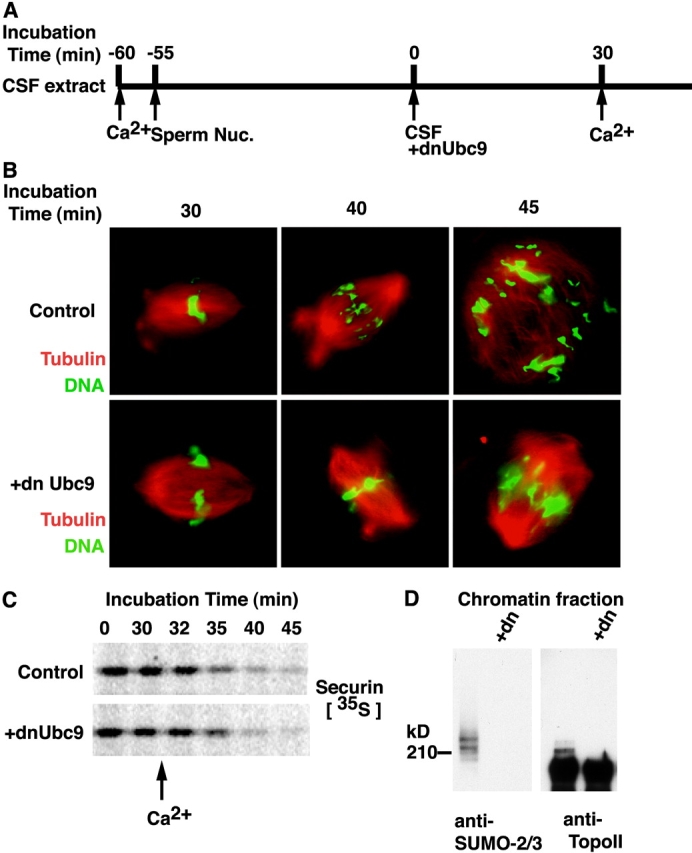
Perturbation of SUMO conjugation in mitosis causes chromosome segregation defects. (A) Schematic diagram of chromosome segregation assay. CSF extracts supplemented with rhodamine-labeled tubulin were released into interphase by CaCl2 addition. 2,000 sperm/μl were added 5 min after CaCl2, and the extract was incubated at 23°C for 55 min. Re-entry into mitosis was induced by addition of fresh CSF extract (50% of reaction volume) with or without dnUbc9 (150 ng/μl final concentration). After a 30 min incubation at 23°C, anaphase was induced with a second addition of CaCl2. (B) Aliquots were removed at the indicated times and analyzed for DNA morphology using Hoechst 33342 DNA dye (green) and for microtubule structures (red) after fixation. (C) The experiment was performed as above, with the addition of 35S-labeled securin at the time of the second aliquot of CSF extract. Samples at each time point were subjected to SDS-PAGE separation, and autoradiography using a Molecular Dynamics phosphorimager. (D) Mitotic chromatin were isolated from the reaction with or without dnUbc9 and subjected to Western blotting analysis with anti-SUMO2/3 and anti-topoisomerase II antibodies.
When a second aliquot of CaCl2 was added to reactions with or without dnUbc9, they proceeded synchronously out of mitosis, as indicated by the degradation of mitotic anaphase-promoting complex (APC) substrates securin and cyclin B (Fig. 6 C; unpublished data). This observation shows that dnUbc9 does not inhibit the APC or cause any nonspecific delay in progression out of M-phase. Observation of the control reactions revealed figures clearly indicative of sister chromatid segregation within 10 min of CaCl2 addition. Remarkably, however, the chromosome masses did not separate properly in the reactions containing dnUbc9 even after considerably longer incubations (>15 min; Fig. 6 B), clearly indicating that SUMO conjugation is essential for sister chromatid separation. This phenotype was strikingly similar to previous reports in which the topoisomerase II poison VP-16 was added to egg extracts at the metaphase–anaphase transition (Shamu and Murray, 1992).
Together, our results suggest that there is a small population of topoisomerase II that is tightly associated with mitotic chromosomes, which becomes mobilized for release from chromatin or remodeling on chromosomes through conjugation with SUMO-2/3. Sister chromatid separation is defective when SUMO conjugation is blocked by dnUbc9, indicating that this mobilization and perhaps other events regulated by SUMO conjugation are critical for correct chromosome partitioning during mitosis. Moreover, SUMO-2/3 modification of topoisomerase II is tightly regulated temporally during mitosis, suggesting that the timing of topoisomerase II mobilization is carefully controlled in coordination with other mitotic events.
Discussion
We have examined the variation of SUMO modification during the cell cycle using Xenopus egg extracts. These experiments have led to a number of remarkable findings. First, we find that topoisomerase II is the major mitotic chromatin-associated SUMO conjugation target. SUMO-2/3 modification of topoisomerase II is directly dependent on chromatin, and is tightly regulated temporally. The concentration of SUMO-2/3–conjugated topoisomerase II reaches its maximum level immediately before the onset of cyclin B degradation. Second, we find that a small population of chromatin-associated topoisomerase II is resistant to extraction from chromosomes by salt washing, indicating that it is more tightly associated with mitotic chromosomes than the bulk of topoisomerase II. The amount of extraction-resistant topoisomerase II increased with inhibition of SUMO conjugation by dnUbc9, suggesting that SUMO-2/3 modification has an important role in mobilization of topoisomerase II from the inextractable subpopulation. Finally, we find that specifically blocking SUMO modification during M-phase leads to a defect in anaphase chromosome separation. This defect is remarkably similar to defects observed when the topoisomerase II poison VP-16 is added at the metaphase–anaphase transition (Shamu and Murray, 1992).
Topoisomerase II is modified exclusively by SUMO-2/3 during mitosis under normal circumstances (Fig. 2). Such discrimination between different SUMO isoforms is noteworthy for two reasons. First, little has been elucidated about the functional differences between SUMO-1, -2 and -3. Our data show that there is strong cell cycle specificity in the use of these three SUMO isotypes, such that SUMO-1 conjugates are predominant during interphase in egg extracts, whereas SUMO-2/3 conjugates predominate during mitosis. Although we do not yet understand the molecular basis for this cell cycle preference, we have discovered that the SUMO-2/3 conjugation of topoisomerase II is tightly controlled during metaphase by mitotic kinases (unpublished data). This regulation suggests that topoisomerase II conjugation to SUMO-2/3 is closely coupled to other events occurring during mitosis. Second, exogenous SUMO-1 has the capacity to become conjugated to topoisomerase II in mitotic extracts when it is added above physiological levels, suggesting that it can override the normal controls regulating isoform selectivity during mitosis. Given that these proteins share a common set of activating and conjugation enzymes, the loss of specificity could result from direct competition between SUMO isoforms. The capacity of SUMO-1 to compete for mitotic conjugation to topoisomerase II raises the possibility that other proteins previously reported to be SUMO-1 substrates may, in fact, be more typically modified by SUMO-2 or -3.
Several findings suggest that topoisomerase II does not require SUMO-2/3 conjugation for its decatenation activity. First, only a fraction of topoisomerase II is modified by SUMO-2/3, even at the height of mitosis (Fig. 3). In most chromosomal preparations, Western blotting showed ∼20% of topoisomerase II is converted into SUMO modified forms (Fig. 2 B), although it is difficult to quantitate the fraction of modified topoisomerase II precisely because different antibodies appear to recognize SUMO-2/3–conjugated topoisomerase II with different efficiencies. More importantly, we blocked all SUMO conjugation in cycling and CSF extracts using dnUbc9, and we confirmed that dnUbc9 was highly efficient in blocking the SUMO-2/3 conjugation of topoisomerase II (Fig. 2). Chromatin preparations from these reactions were used as a source for further purification of topoisomerase II. We found that topoisomerase II from chromatin assembled with or without dnUbc9 had indistinguishable levels of activity for decatenation of kinetoplast DNA (Fig. 4). Finally, we assayed nuclear assembly, nuclear transport, DNA replication, mitotic chromosome condensation, mitotic chromosome structure, and spindle assembly using extracts with our without added dnUbc9 (Fig. 6; unpublished data). We did not find differences between extracts with or without dnUbc9 in any of these assays. Although many of these processes require topoisomerase II activity, they clearly did not require de novo SUMO conjugation of topoisomerase II or other substrates.
By contrast, dnUbc9 disrupted chromosome segregation at the metaphase–anaphase transition in egg extracts (Fig. 6). It is clearly possible that the lack of conjugation for many substrates may have contributed toward this phenotype. However, several arguments suggest that the failure to conjugate topoisomerase II and the resultant failure to promote its partitioning into lower affinity fractions may be among the key factors responsible for defective chromosome segregation: First, topoisomerase II is by far the most abundant target of SUMO conjugation on mitotic chromosomes (Fig. 3). Second, topoisomerase II poisons, such as VP-16, which cause the accumulation of arrested topoisomerase II–DNA complexes, produce chromosome segregation defects that are remarkably similar to dnUbc9 (Fig. 6; Shamu and Murray, 1992). Notably, topoisomerase II inhibitors that do not cause accumulation of arrested topoisomerase II–DNA complexes, such as Merbarone (Fortune and Osheroff, 1998), do not block chromosome segregation despite the fact that they produce comparable levels of inhibition in decatenation assays (unpublished data). Finally, consistent with our conclusion that SUMO conjugation regulates chromosome segregation in egg extracts, misregulated Smt3p conjugation in smt4-Δ mutants of budding yeast can promote premature loss of centromeric cohesion (Bachant et al., 2002). top2 mutants lacking Smt3p modification sites could suppress this defect, suggesting that Top2p is one of the primary substrates whose conjugation to Smt3p can alter cohesion.
It may be appropriate to reevaluate the role of topoisomerase II on mitotic chromosomes. It was initially suggested that topoisomerase II forms part of a tightly bound chromosome scaffold (Earnshaw and Heck, 1985), but this view was overturned by the observation that the bulk of topoisomerase II can be removed from in vitro–assembled chromosomes without significant changes in chromosome structure (Hirano and Mitchison, 1993). More recent demonstrations that the majority of topoisomerase II is extremely dynamic on mitotic chromosomes (Christensen et al., 2002; Null et al., 2002; Tavormina et al., 2002) also argue against a static role for topoisomerase II as a chromosome scaffold. On the other hand, we find that there is a small population of topoisomerase II that is very tightly bound to mitotic chromatin, being retained even after extraction in a 500-mM NaCl-containing buffer. dnUbc9 enlarged this population, showing that SUMO-2/3 conjugation negatively controls the apportionment of topoisomerase II to such tightly bound forms. Our findings present the interesting idea that this small, high affinity fraction of topoisomerase II may act as part of a dynamic tether between sister chromatids, and that its removal at the metaphase–anaphase transition may be an essential event during chromosome distribution into daughter cells.
Materials and methods
Recombinant protein and antibodies
The bacterial expression and production of mutated human Ubc9 were as described previously (Saitoh et al., 1998; Azuma et al., 2001). dnUbc9 was made in analogy to a previously described mutant of cdc34 that forms a stable oxygen ester with ubiquitin and which is therefore unable to transfer ubiquitin to E3 enzymes or conjugation substrates (Banerjee et al., 1995). For production of the human form of His-SUMO-1 (processed form), the SUMO-1 coding sequence was subcloned into a pET30 vector and expressed in Escherichia coli. His-SUMO-1 was purified by chromatography on Talon (Contac) and Mono Q (Amersham Biosciences) resins. Full-length SUMO-2 (GenBank/EMBL/DDBJ accession no. NP_008868.2) and SUMO-3 (GenBank/EMBL/DDBJ accession no. NP_008867.1) were generated by PCR from constructs designed for the expression of the processed form of each protein (provided by H. Saitoh, Kumamoto University, Kumamoto, Japan; Saitoh and Hinchey, 2000), using oligonucleotides encoding the COOH-terminal extensions removed during processing. The processed and full-length forms of SUMO-2 and -3 were cloned into pGEX4T-1, fused at their NH2 termini to an EGFP fragment that was excised from a pEGFP C-1 plasmid (CLONTECH Laboratories, Inc.). The GST/EGFP-tagged SUMO-2 and -3 proteins were purified using glutathione agarose (Amersham Biosciences), according to the manufacturer's protocol. After elution by thrombin cleavage, the proteins were further purified by chromatography on a Mono Q column.
Rabbit polyclonal anti-SUMO-1 antibodies were raised against the processed form of human SUMO-1 (Research Genetics). Untagged, recombinant SUMO-1 (processed form) was coupled to CNBr-activated Sepharose (Amersham Biosciences) and used for affinity purification of the antibodies from immune sera. Rabbit pAbs against SUMO-2/3 were provided by H. Saitoh (Saitoh and Hinchey, 2000). Untagged, recombinant SUMO-2 (processed form) was coupled to CNBr-activated Sepharose (Amersham Biosciences) and used for affinity purification of the antibodies from immune sera. Mouse mAbs against Xenopus cyclin B2 were provided by T. Hunt, (Cancer Research UK, South Mimms, UK). Monoclonal anti–human topoisomerase IIα/β (Fig. 2 B, lanes 1 and 2), monoclonal anti–human topoisomerase IIα (Fig. 2 B, lanes 3 and 4), and rabbit polyclonal anti–human topoisomerase IIα (Fig. 2 B, lanes 5 and 6) were purchased from Research Diagnostics, Inc. Rabbit pAbs against Xenopus topoisomerase II α/β (Fig. 2 B, lanes 7 and 8) were as described previously (provided by Dr. P.L. Jones, University of Illinois at Urbana-Champaign, Urbana, IL); Luke and Bogenhagen, 1989). Unless otherwise indicated, Western blotting was performed using anti–Xenopus topoisomerase II α/β. Antibodies against phosphorylated ERK1/2 were purchased from Cell Signaling.
Xenopus egg extracts and cell cycle analysis
Xenopus sperm nuclei, low speed extracts of Xenopus eggs arrested by CSF, and cycling extracts were prepared as described previously (Kornbluth et al., 2001). For visualization of spindles, rhodamine-labeled tubulin (Cytoskeleton, Inc.) was added to a final concentration of 0.1 mg/ml, as indicated. In cases where reactions were supplemented with exogenous proteins, the proteins were added to the extract and incubated on ice for 5 min before the addition of sperm chromatin and the initiation of the experiment by transfer of the reaction to 23°C, unless otherwise indicated. In Fig. 6, destruction of securin was monitored as described previously (Arnaoutov and Dasso, 2003).
Isolation of chromatin from egg extract
For preparation of chromatin from CSF or cycling extracts, 100-μl aliquots of each reaction were diluted fivefold with CSF-XB buffer (10 mM Hepes-KOH, pH 7.7, 100 mM KCl, 2 mM MgCl2, 5 mM EGTA, and 50 mM sucrose) containing 0.5% Triton X-100, and were incubated for 1 min at RT. The samples were then layered onto a 40% glycerol-containing CSF-XB cushion, and centrifuged at 10,000 g for 10 min at 4°C. The pellets were resuspended in 40% glycerol-containing CSF-XB, and the centrifugation was repeated. 50 μl SDS-PAGE sample buffer was added to the resulting pellet, and the sample was heated to 100°C for 5 min.
Preparation of chromatin-bound topoisomerase II and decatenation assay
Chromatin was prepared from 500 μl of egg extract with or without 150 ng/μl dnUbc9 containing 10,000 sperm nuclei/μl as described above, except that the elution was performed with 100 μl XB buffer supplemented with 5 mM β-glycerol phosphate and 300 mM NaCl instead of sample buffer. DNA and other insoluble material was removed by centrifugation at 20,000 g for 15 min. The post-chromosomal supernatant from this elution was assayed for kinetoplast DNA decatenation using a topoisomerase II assay kit (TopoGEN, Inc.) according to the manufacturer's protocol. Unless otherwise noted, all reactions contained 10 ng/μl of kinetoplast DNA in a 20-μl reaction, and were incubated for 15 min at 23°C. Products of the decatenation assay were analyzed by electrophoresis on 1% agarose gels in TBE. For comparison, decatenation by purified human topoisomerase II is shown in Fig. 4.
Immunofluorescence staining of chromatin
2,000 nuclei/μl sperm chromatin was incubated for 60 min in CSF extract. The reactions were diluted with either CSF-XB buffer (low salt) or CSF-XB buffer supplemented with an additional 200 mM NaCl (high salt), and incubated for 5 min at 23°C. The chromosomes were fixed by addition of an equal volume of the same buffer with 1.6% PFA. After a 10-min incubation at 23°C, the chromosomes were spun through a 20% glycerol cushion onto glass coverslips. Chromatin was stained with monoclonal anti-topoisomerase IIα/β antibody (TopoIIA/B; Research Diagnostics, Inc.) at 1/200 dilution for 1 h at 23°C followed by Alexa® 488–conjugated anti–mouse IgG (Molecular Probes, Inc.) for 1 h at 23°C. After DNA was counterstained by 4 μg/ml Hoechst 33342, samples were mounted in medium (Vectashield®; Vector Laboratories) and sealed. Specimens were observed by a fluorescent microscope (Axioskop; Carl Zeiss MicroImaging, Inc.) with either a Plan-neo 63×/1.25 or Iris 100×/1.4 objective (Carl Zeiss MicroImaging, Inc.). Images were taken with a CCD camera (Orca II; Hamamatsu Photonics K.K.) operated by Openlab software (Improvision).
Online supplemental material
Fig. S1 shows that SUMO-2 is preferred over SUMO-3 for mitotic conjugation to topoisomerase II. Fig. S2 shows association of SUMO-2/3–modified topoisomerase II with replicated chromatin. Online supplemental material available at http://www.jcb.org/cgi/content/full/jcb.200304088/DC1.
Supplemental Material
Acknowledgments
We would like to thank Rohinton Kamakaka for critical advice on protein purification. We also thank Peter Jones, Hisato Saitoh, and Tim Hunt for providing antibodies and clones. We would like to thank the members of the Dasso group for critical comments on the manuscript.
This work was supported in part by Human Frontiers Science Program research grant RG0229/1999-M to M. Dasso.
The online version of this article includes supplemental material.
Abbreviations used in this paper: CSF, cytostatic factor; dnUbc9, dominant-negative mutant of Ubc9; His-SUMO, His6-tagged SUMO.
References
- Adachi, Y., M. Luke, and U.K. Laemmli. 1991. Chromosome assembly in vitro: topoisomerase II is required for condensation. Cell. 64:137–148. [DOI] [PubMed] [Google Scholar]
- al-Khodairy, F., T. Enoch, I.M. Hagan, and A.M. Carr. 1995. The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J. Cell Sci. 108:475–486. [DOI] [PubMed] [Google Scholar]
- Arnaoutov, A., and M. Dasso. 2003. The Ran GTPase regulates kinetochore function. Dev. Cell. 5:99–111. [DOI] [PubMed] [Google Scholar]
- Azuma, Y., S.H. Tan, M.M. Cavenagh, A.M. Ainsztein, H. Saitoh, and M. Dasso. 2001. Expression and regulation of the mammalian SUMO-1 E1 enzyme. FASEB J. 15:1825–1827. [DOI] [PubMed] [Google Scholar]
- Bachant, J., A. Alcasabas, Y. Blat, N. Kleckner, and S.J. Elledge. 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 9:1169–1182. [DOI] [PubMed] [Google Scholar]
- Banerjee, A., R.J. Deshaies, and V. Chau. 1995. Characterization of a dominant negative mutant of the cell cycle ubiquitin-conjugating enzyme Cdc34. J. Biol. Chem. 270:26209–26215. [DOI] [PubMed] [Google Scholar]
- Biggins, S., N. Bhalla, A. Chang, D.L. Smith, and A.W. Murray. 2001. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics. 159:453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, M.O., M.K. Larsen, H.U. Barthelmes, R. Hock, C.L. Andersen, E. Kjeldsen, B.R. Knudsen, O. Westergaard, F. Boege, and C. Mielke. 2002. Dynamics of human DNA topoisomerases IIα and IIβ in living cells. J. Cell Biol. 157:31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasso, M., S. Dimitrov, and A.P. Wolffe. 1994. Nuclear assembly is independent of linker histones. Proc. Natl. Acad. Sci. USA. 91:12477–12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw, W.C., and M.M. Heck. 1985. Localization of topoisomerase II in mitotic chromosomes. J. Cell Biol. 100:1716–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune, J.M., and N. Osheroff. 1998. Merbarone inhibits the catalytic activity of human topoisomerase IIα by blocking DNA cleavage. J. Biol. Chem. 273:17643–17650. [DOI] [PubMed] [Google Scholar]
- Gasser, S.M., T. Laroche, J. Falquet, E. Boy de la Tour, and U.K. Laemmli. 1986. Metaphase chromosome structure. Involvement of topoisomerase II. J. Mol. Biol. 188:613–629. [DOI] [PubMed] [Google Scholar]
- Hirano, T., and T.J. Mitchison. 1993. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J. Cell Biol. 120:601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser, M. 2000. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2:E153–E157. [DOI] [PubMed] [Google Scholar]
- Kornbluth, S., J. Yang, and M. Powers. 2001. Analysis of the cell cycle using Xenopus egg extracts. Current Protocols in Cell Biology. J.S. Bonifacino, M. Dasso, J. Lippincott-Schwartz, J.B. Harford, and K.M. Yamada, editors. John Wiley & Sons, Inc., New York. 11.11.1–11.11.13.
- Li, S.J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature. 398:246–251. [DOI] [PubMed] [Google Scholar]
- Li, S.J., and M. Hochstrasser. 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol 20:2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke, M., and D.F. Bogenhagen. 1989. Quantitation of type II topoisomerase in oocytes and eggs of Xenopus laevis. Dev. Biol. 136:459–468. [DOI] [PubMed] [Google Scholar]
- Mao, Y., S.D. Desai, and L.F. Liu. 2000. SUMO-1 conjugation to human DNA topoisomerase II isozymes. J. Biol. Chem. 275:26066–26073. [DOI] [PubMed] [Google Scholar]
- Melchior, F. 2000. SUMO–nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591–626. [DOI] [PubMed] [Google Scholar]
- Null, A.P., J. Hudson, and G.J. Gorbsky. 2002. Both alpha and beta isoforms of mammalian DNA topoisomerase II associate with chromosomes in mitosis. Cell Growth Differ. 13:325–333. [PubMed] [Google Scholar]
- Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252–6258. [DOI] [PubMed] [Google Scholar]
- Saitoh, H., D.B. Sparrow, T. Shiomi, R.T. Pu, T. Nishimoto, T.J. Mohun, and M. Dasso. 1998. Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr. Biol. 8:121–124. [DOI] [PubMed] [Google Scholar]
- Seeler, J.S., and A. Dejean. 2001. SUMO: of branched proteins and nuclear bodies. Oncogene. 20:7243–7249. [DOI] [PubMed] [Google Scholar]
- Seufert, W., B. Futcher, and S. Jentsch. 1995. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 373:78–81. [DOI] [PubMed] [Google Scholar]
- Shamu, C.E., and A.W. Murray. 1992. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J. Cell Biol. 117:921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayeghi, M., C.L. Doe, M. Tavassoli, and F.Z. Watts. 1997. Characterisation of Schizosaccharomyces pombe rad31, a UBA-related gene required for DNA damage tolerance. Nucleic Acids Res. 25:1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov, A.V., L. Aravind, and E.V. Koonin. 2001. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics. 158:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., J. Nishide, K. Okazaki, H. Kato, O. Niwa, T. Nakagawa, H. Matsuda, M. Kawamukai, and Y. Murakami. 1999. Characterization of a fission yeast SUMO-1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol. Cell. Biol. 19:8660–8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina, P.A., M.G. Come, J.R. Hudson, Y.Y. Mo, W.T. Beck, and G.J. Gorbsky. 2002. Rapid exchange of mammalian topoisomerase II alpha at kinetochores and chromosome arms in mitosis. J. Cell Biol. 158:23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura, T., H. Ohkura, Y. Adachi, K. Morino, K. Shiozaki, and M. Yanagida. 1987. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 50:917–925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.