Abstract
Caenhorhabditis elegans Unc104 kinesin transports synaptic vesicles at rapid velocities. Unc104 is primarily monomeric in solution, but recent motility studies suggest that it may dimerize when concentrated on membranes. Using cryo-electron microscopy, we observe two conformations of microtubule-bound Unc104: a monomeric state in which the two neck helices form an intramolecular, parallel coiled coil; and a dimeric state in which the neck helices form an intermolecular coiled coil. The intramolecular folded conformation is abolished by deletion of a flexible hinge separating the neck helices, indicating that it acts as a spacer to accommodate the parallel coiled-coil configuration. The neck hinge deletion mutation does not alter motor velocity in vitro but produces a severe uncoordinated phenotype in transgenic C. elegans, suggesting that the folded conformation plays an important role in motor regulation. We suggest that the Unc104 neck regulates motility by switching from a self-folded, repressed state to a dimerized conformation that can support fast processive movement.
Keywords: coiled coil; dimerization; microtubule; cryo-EM; C. elegans
Introduction
Kinesins are functionally diverse molecules that generate a diverse range of motile activities in the cell. Caenorhabditis elegans Unc104 kinesin is the founding member of a class of NH2-terminal kinesins (termed Unc104/KIF1) that are present in many lower and higher eukaryotic organisms (for reviews see Bloom, 2001; Vale, 2003). The Unc104/KIF1A class is characterized by a unique domain arrangement that includes a kinesin superfamily conserved catalytic core, a class-conserved neck linker and neck, and a forkhead homology associated (FHA) domain (Vale, 2003; Fig. 1). A subgroup of these motors also has a conserved pleckstrin homology (PH) domain near the COOH terminus, which specifically binds P(4,5)IP2 and appears to target the motor to membranes in vivo (Klopfenstein et al., 2002). These PH domain containing motors include C. elegans Unc104 and its mouse orthologue KIF1A, which transport synaptic vesicle precursors from the cell body to nerve terminals. Inactivating mutations or deletions of Unc104 and KIF1A lead to the accumulation of synaptic vesicle precursors in the cell body and, for KIF1A, cause neuronal cell death in mice (Hall and Hedgecock, 1991; Yonekawa et al., 1998).
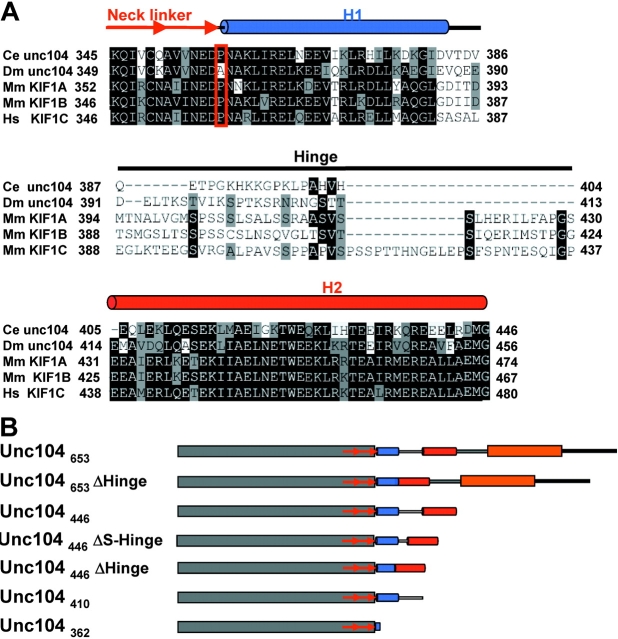
Figure 1.
Sequence comparisons and Unc104 constructs used in the paper. (A) Sequences from C. elegans (Ce) Unc104, D. Melanogaster (Dm) Unc104, M. musculus (Mm) KIF1A and KIF1B, and H. sapiens (Hs) KIF1C were compared using Clustal W (Aiyar, 2000). The Unc104 neck linker is class-conserved and ends with a conserved proline (red box). The neck consists of two predicted α-helical segments, H1 and H2, separated by a nonconserved hinge. Helical prediction was done with PHDsec (Rost and Sander, 1994). H1 consists of ∼26 residues (blue cylinder) after the neck linker. The hinge (solid line) consists of 18–49 residues. H2 consists of ∼41 residues (red cylinder). Conserved residues are shaded black, semi-conserved residues are gray. (B) Schematic representation of the Unc104 constructs used in this paper. Domains are indicated as follows: catalytic core, gray; neck linker, red arrows; H1, blue cylinder; hinge, unshaded line; H2, red cylinder; neck-FHA linker, gray line; FHA domain, orange, sequence after FHA, black line.
Unc104 and KIF1A have several unusual biochemical and biophysical properties. Although most kinesins are dimers or tetramers of heavy chains, hydrodynamic studies have suggested that Unc104 and KIF1A are primarily monomeric (Okada et al., 1995; Pierce et al., 1999). Furthermore, Unc104 and native KIF1A induce motility in vitro (Okada et al., 1995; Pierce et al., 1999) and in C. elegans and cultured mouse neurons (Zhou et al., 2001; Lee et al., 2002) at rates that are two to three times faster (1–1.6 μm/s) than those of conventional dimeric kinesin. Okada and Hirokawa (1999) demonstrated a biased diffusional motility for a single truncated KIF1A monomer, suggesting that this motor may generate processive movement by a mechanism that is distinct from the hand over hand mechanism proposed for the conventional kinesin dimer (Vale and Milligan, 2000). They suggested that this processive mechanism requires an interaction between a class-specific insertion of several lysines (the K-loop) in kinesin's primary microtubule binding site and the negatively charged, flexible COOH terminus of tubulin (Kikkawa et al., 2000; Okada and Hirokawa, 2000). Kikkawa et al. (2001) also demonstrated a ∼20° rotation of KIF1A's catalytic core (the kinesin superfamily conserved domain that includes the ATPase and microtubule binding sites) in different nucleotide states. By rotating the K-loop toward the next tubulin subunit in the plus end direction, the authors suggested that this catalytic core rotation might provide a mechanism for the processive motion of a KIF1A monomer.
The biased diffusional movement of KIF1A monomers along microtubules was much slower (0.15 μm/s; Okada and Hirokawa, 1999) than the fast movement observed in vivo (Zhou et al., 2001). However, in vitro reconstitution of Unc104 mediated vesicle motility showed that Unc104 molecules could cluster on lipid membranes and induce fast motility with velocities similar to those seen in vivo (Klopfenstein et al., 2002). Tomishige et al. (2002) subsequently showed that this fast motility was not due to the collective activities of many monomeric Unc104 motors bound to the same vesicle, but rather to the self-association of Unc104 monomers into highly processive Unc104 dimers. The mechanism of the Unc104 dimer was proposed to be similar to that envisaged for the conventional kinesin dimer. In the case of conventional kinesin, the neck linker docks onto the catalytic core upon ATP binding and, in doing so, positions the second head of the dimer so that it can bind to a new tubulin subunit toward the plus end of the microtubule (Rice et al., 1999; Vale and Milligan, 2000). The neck α-helical coiled coil tightly pairs the two catalytic cores, so that conformational changes in the neck linker can be communicated effectively to the partner head (Romberg et al., 1998; Tomishige and Vale, 2000). Unc104/KIF1A has a similar neck linker to conventional kinesin and has a predicted helix that begins at the same position as the kinesin neck coiled coil (Tomishige et al., 2002). However, the propensity for coiled-coil formation by the Unc104 neck helices is weaker than conventional kinesin, consistent with observations that dimerization and processive movement only occurs at high Unc104 concentrations that can drive the equilibrium toward dimerization.
Distinguishing between the different motility models proposed for Unc104/KIF1A would be aided by further structural and functional information. Although X-ray crystallography, cryo-EM, and mutagenesis studies have been performed on the KIF1A catalytic core (Kikkawa et al., 2000, 2001; Okada and Hirokawa, 2000), structure–function studies have not been performed on longer constructs that include the neck domain, which has been proposed to play a prominent role in recent models for motility. Here, we have performed cryo-EM, motility, and ATPase studies on Unc104 proteins containing the full neck domain or with mutations in the neck domain. Under certain experimental conditions, we show that the neck domain can form an intramolecular, parallel coiled coil that will preclude dimerization. Under other conditions, we observe neck coiled-coil–mediated dimerization of Unc104. Our results also show that the Unc104 catalytic core undergoes a much smaller nucleotide-dependent angular change than reported previously by Kikkawa et al. (2001) for KIF1A. Our functional studies show that the folded conformation of Unc104 may play a role in regulating microtubule binding affinity and in regulating transport in living C. elegans. Our work supports a model in which reorganization in the folding of the Unc104 neck may result in a transition from a self-folded monomeric conformation to a dimeric conformation that activates the fast, processive motility of Unc104.
Results
Contributions of class-specific Unc104/KIF1 structural elements to motor ATPase and velocity of movement
The Unc104/KIF1 class of motors share several unique structural features in their neck region (domains adjacent to the superfamily conserved catalytic core; Fig. 1). A characteristic feature of this class is the presence of a FHA domain located COOH-terminal to the motor domain. The FHA domain (a 10–11 β-strand fold) mediates protein–protein interactions in a variety of proteins by binding to a phosphothreonine motif (pTxxX; Durocher and Jackson, 2002), but it is unclear if the FHA domain in Unc104/KIF1-type kinesins possesses such activity. Between the catalytic core and FHA domains are two predicted helices capable of weak intermolecular coiled-coil interactions (termed here neck helices H1 and H2; Okada et al., 1995; Pierce et al., 1999). The two helices are separated by an unstructured region (20–50 residues) that may serve as a flexible hinge (termed the neck hinge; Fig. 1 A). This sequential arrangement of the catalytic core, neck linker, helix–hinge–helix, and the FHA domain is characteristic of Unc104/KIF1-type motors from unicellular organisms to man (Vale, 2003).
To examine the functional roles of the FHA and neck domains, we prepared several constructs with deletions or mutations of these elements (Fig. 1 B). The parent “wild-type” construct used for this study was the first 653 residues of the Unc104 motor (corresponding to ∼60 residues beyond the conserved FHA domain) fused to the GFP and a 6X Histidine tag (termed Unc104653); the GFP COOH-terminal tag was necessary for anchoring the motor via anti-GFP antibodies to glass surfaces for motility assays (Pierce et al., 1999; Tomishige et al., 2002). Next, we prepared constructs in which the FHA domain (Unc104446), the neck helix 2 (Unc104410), neck helix 1 and 2 (Unc104362), or the neck hinge (Unc104653ΔHinge and Unc104446ΔHinge) were deleted (Fig. 1 B). When assayed for microtubule gliding velocity (a multiple motor assay in which glass-adsorbed motors move microtubules across the surface), all of the mutations had relatively little effect (Table I). The one exception was the virtually complete deletion of the neck by truncation at residue 362 (Unc104362), which produced a ∼5-fold decrease in velocity, as described previously (Tomishige et al., 2002). This decrease in velocity may be due to the inability of this protein to dimerize due to the truncation of the helical coiled-coil regions (Tomishige et al., 2002).
Table I. Motor activity of Unc104 constructs.
| MT-stimulated ATPase
|
Basal ATPase
|
||||
|---|---|---|---|---|---|
| Constructs
|
MT gliding velocity
|
n
|
kcat
|
KmMT
|
kcat
|
| μm/min | ATP/s | μM tubulin | ATP/s | ||
| Unc104653 | 145 ± 17 | 342 | 28 ± 6.1 | 3.0 ± 1.6 | 0.07 ± 0.01 |
| Unc104653ΔHinge | 114 ± 13 | 186 | 53 ± 9.0 | 0.9 ± 0.3 | 0.27 ± 0.05 |
| Unc104446 | 131 ± 15 | 216 | 30 ± 4.0 | 0.7 ± 0.3 | 0.14 ± 0.01 |
| Unc104446ΔHinge | 98 ± 8 | 146 | 53 ± 12 | 0.7 ± 0.1 | 0.18 ± 0.02 |
| Unc104446ΔS-Hinge | 120 ± 8 | 94 | 27 ± 1 | 1.1 ± 0.1 | 0.15 ± 0.02 |
| Unc104410 | 90 ± 4 | 187 | 43 ± 4.0 | 0.5 ± 0.2 | 0.14 ± 0.01 |
| Unc104362 | 29 ± 1 | 195 | 74 ± 39 | 4.5 ± 2.7 | 0.21 ± 0.03 |
The microtubule gliding velocity, microtubule-stimulated, and basal ATPase activities were measured on GFP-containing constructs as described in Materials and methods. n is the number of microtubules scored for each construct. The data (mean and SD) are from at least two separate protein preparations.
We also assayed the constructs for basal and microtubule-stimulated ATPase activity (Table I). The basal ATPase rates for all constructs were 200–400-fold lower than the microtubule-stimulated ATPase rates. The microtubule-stimulated ATPase rate of the parent construct Unc104653 was ∼30 ATP/s higher than reported previously by Pierce et al. (1999). Three of the constructs showed significantly higher rates (Unc104362, Unc104446ΔHinge, and Unc104653ΔHinge). The high ATPase rate and the low microtubule velocity of the monomer Unc104362 is similar to results obtained for monomeric kinesin (Jiang and Hackney, 1997), and may suggest a shorter step per ATP and/or nonproductive cycling.
Unexpectedly, four constructs with deletions or truncations outside of the catalytic domain caused a three- to sixfold increase in microtubule affinity in the ATPase assay (decrease in KmMT; Table I). Three of these constructs share in common a deletion of the FHA domain (Unc104410, Unc104446 and Unc104446ΔHinge). The fourth construct retained the FHA domain but had a deletion of the unstructured hinge region between the two neck helices preceding the FHA domain (Unc104653ΔHinge). These results indicate that the FHA domain decreases microtubule affinity and this effect is prevented by deletion of the neck hinge region. The electron microscopic examination of the Unc104 neck, described in the next sections, may provide an explanation for these results (see Discussion).
Cryo-EM visualization of the Unc104 neck in the 5′-adenylylimidodiphosphate (AMPPNP) state
To investigate the conformation of Unc104, we used cryo-EM and helical image analysis to calculate three-dimensional (3D) maps of Unc104653, Unc104446, and Unc104362 bound to microtubules. These proteins were examined initially without a COOH-terminal GFP, so that the motor alone would be imaged. The proteins were bound to microtubules in the presence of AMPPNP, resulting in a strong binding state in which the neck linker is docked onto the catalytic core (Rice et al., 1999; Kikkawa et al., 2001), as well as in the nucleotide-free and ADP states in which the neck linker detaches.
The Unc104362 3D map (Fig. 2 A) reveals that the overall shape of microtubule-bound Unc104 catalytic core is very similar to that of KIF1A, conventional kinesin and Ncd (for reviews see Sosa et al., 1997; Hoenger et al., 2000; Kikkawa et al., 2000). As described for KIF1A (Kikkawa et al., 2000), the K-loop extension of L12 appears as a protuberance in the middle of the catalytic core (Fig. 2 A, arrow).
Figure 2.
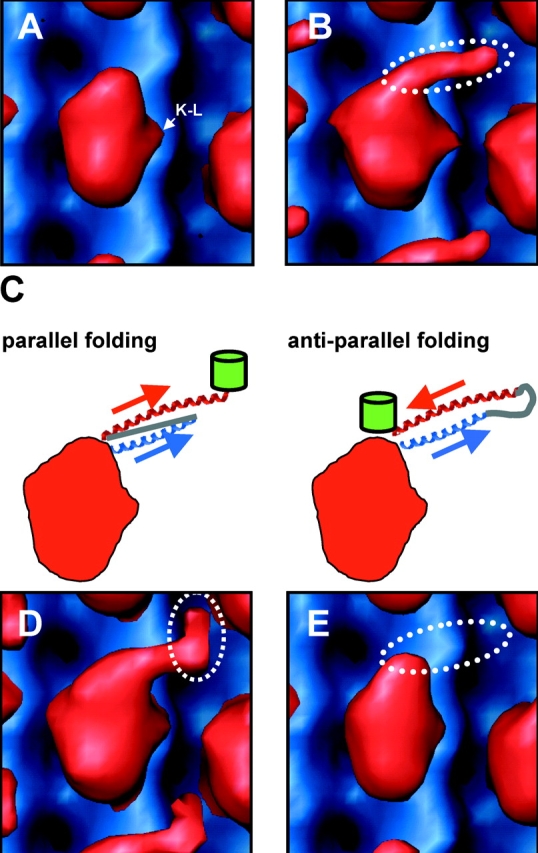
Structure of the Unc104 neck. Front view (A) of molecular surface representation the Unc104362-AMPPNP map (red) aligned and displayed with an undecorated microtubule map (blue). Density attributable to the K-loop is indicated (K–L). (B) The same view of the Unc104446-AMPPNP map. Additional density projecting from the tip of the catalytic core is attributable to the Unc104 neck (dotted ellipse). Possible self-folding models for the Unc104 neck are shown in C. In a parallel folding model, the H1 helix (blue) extends away from the catalytic core and the hinge (gray line) backtracks to allow H2 to lie parallel to H1. In the antiparallel folding model, the hinge lies distally and H2 backtracks in antiparallel fashion along H1. The green barrels represent the expected location of the GFP in the two models. The Unc104446-GFP map is shown in D. Additional density is located at the distal end of the neck density, consistent with the parallel folding model. The Unc104446-ΔHinge map is shown in E. When the hinge is absent, the neck density is not visualized, again consistent with the parallel folding model.
The 3-D map of Unc104446 in the presence of AMPPNP reveals an additional, well-defined density that is not present in Unc104362 (Fig. 2 B). This density appears as an elongated “finger” originating near the tip of the catalytic core and extending ∼45 Å in a direction roughly tangential to the microtubule surface. In the AMPPNP state, Kikkawa et al. (2001) showed that the neck linker for KIF1A, the mouse orthologue of Unc104, is docked along the catalytic core with its COOH-terminal end near the tip of the motor core, as was also described for kinesin (Rice et al., 1999; Sindelar et al., 2002). Thus, the COOH-terminal end of the neck linker coincides with the point of origin of the finger.
The 3D-map of microtubule bound Unc104653 is very similar to that of Unc104446. The well-defined finger density is again evident. In addition, there is a weak density peak lying ∼20 Å beyond the end of the finger (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb200308020/DC1), which must be due to the FHA domain.
The finger density cannot be attributed to a poorly localized, detached motor domain (e.g., the second head of an Unc104 dimer), as there is no evidence of any additional large mass beyond the attached motor domain in the mean radial density distribution (MRDD) of the map (see Fig. 4 C, blue line). Although the finger density must then be due to the neck sequences, it cannot be produced by a single α helix (e.g., H1), as such a structural element would not be visualized at the resolution of our EM paper. Therefore, two molecular conformations could explain this result: the H1 and H2 helices in the monomer form a coiled-coil interaction in either a parallel or antiparallel orientation (Fig. 2 C).
Figure 4.
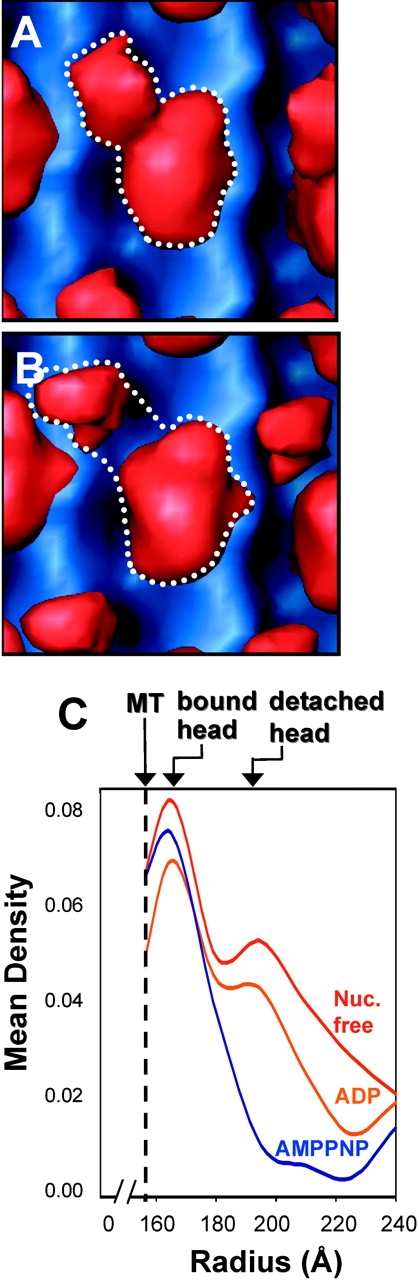
Unc104446 dimerizes in ADP and nucleotide-free states. In the(A) ADP and (B) nucleotide-free states, additional density is seen near the clearly recognizable bound head. The density is too large to be the neck and somewhat smaller than expected for a second rigidly attached catalytic core. The appearance is consistent with there being a second catalytic core that is loosely associated with the microtubule-bound catalytic core. Note that the finger density seen in Fig. 2 B is absent, and the detached catalytic core seems to occupy a slightly different position in the two states. (A and B) The dotted lines outline density attributable to the microtubule-bound Unc104 dimer. The MRDDs in the 3D-maps (C) clearly show a peak at a radius of ∼200 Å that is attributable to a second, microtubule-detached, catalytic core in the ADP (orange) and nucleotide-free (red) states. No evidence for a second catalytic core is seen in AMPPNP (blue).
To distinguish between these possibilities, we examined the Unc104446 construct with a COOH-terminal GFP, the additional mass providing a marker for the end of the H2 helix in a 3D map. In the map of the Unc104446GFP protein, additional density appeared at the distal end of the finger (Fig. 2 D), consistent with the parallel folding model. The density is smaller than would be expected for GFP, presumably because flexibility in the junction between the Unc104 and the GFP allows diffusional motion of the GFP and diminishes electron density. In the antiparallel folding model, the GFP density would be located at the opposite end of the finger, near the tip of the catalytic core (Fig. 2 C). The appearance of GFP density at the end of the finger also adds additional evidence arguing against the dimer model; were it a dimer, the GFP would be located either a considerable distance from the motor due to the long intermolecular coiled coil, or would not be visualized at all because of flexibility of the long coiled coil. Together, the results provide considerable support the intramolecular, parallel folding model.
To further test the validity of this model, we also examined the corresponding construct in which the neck hinge was completely deleted (Unc104446ΔHinge). This deletion should prevent parallel coiled-coil formation as the hinge must act as a spacer that allows H1 and H2 to be oriented with the same polarity (Fig. 2 C, parallel folding). Fig. 2 E shows that the neck hinge deletion abolished the finger density in the 3D map, indicating that it is essential for the folded conformation.
Next, we modeled a possible atomic structure of the Unc104 neck into the 3D maps (Fig. 3) to gain more insight into the folded conformation. It should be borne in mind that at the resolution of our maps, we cannot assign exact positions or orientations of secondary structure elements, but can approximate their relative locations. The H1 and H2 helices are predicted to be comprised of ∼25 (∼40 Å) and ∼42 (∼65 Å) residues, respectively. The length of the finger (45 Å) approximately matches the length of H1. Therefore, these dimensions suggest that the full length of H1 (Fig. 3, blue) projects out from the end of the neck linker, which is docked onto the motor in the AMPPNP state (Kikkawa et al., 2001). Our 3D map of Unc104446GFP identifies the COOH terminus of H2 as lying ∼40 Å from the tip of the catalytic core, indicating that the COOH-terminal ends of H1 and H2 lie together. However, H2 is predicted to be 25 Å longer than H1, implying that either the NH2-terminal 25 Å of H2 lies over the motor core (but is not resolved in the maps) or that the NH2-terminal end of H2 might become partially disordered in this state. If H2 is intact and spans 65 Å (Fig. 3), then the 18 residues of the hinge must be in a highly extended conformation in order to connect the COOH terminus of H1 to the NH2 terminus of H2. Alternatively, perhaps some disordering of the NH2-terminal region of H2 occurs to accommodate the folded conformation. These modeling experiments also suggested that a much smaller deletion of the neck hinge should disrupt intramolecular coiled-coil formation. Therefore, we prepared a protein in which only 11 aa (392–402) had been deleted from the hinge region (Unc104446ΔS-Hinge). In the AMPPNP state, this protein also failed to show a finger density, indicating that the intramolecular coiled coil could not form (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200308020/DC1).
Figure 3.
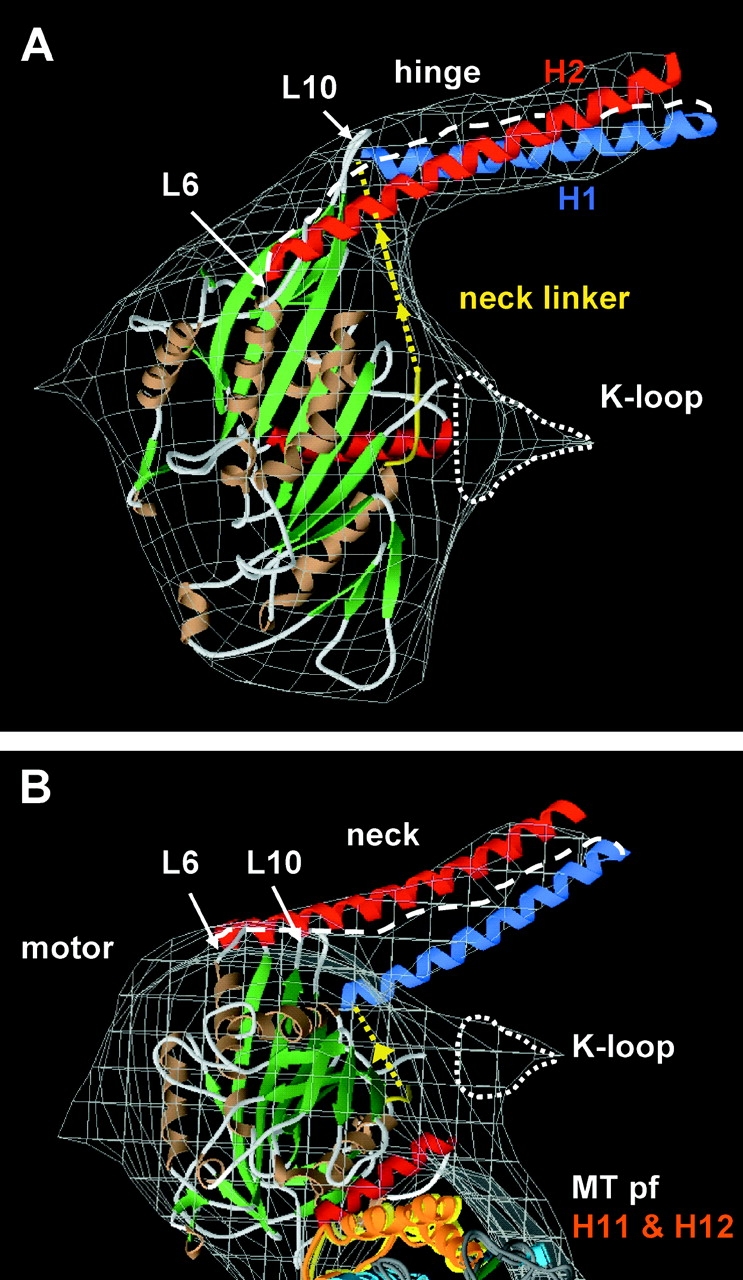
Modeling of the self-folded Unc104446 neck in the AMPPNP state. (A) Front and (B) top views (from the minus end) of a model in which the KIF1A-AMPPCP and tubulin structures were manually docked into the Unc104446 AMPPNP map (gray wire frame). A model for the ∼26 residue H1 (blue) and the ∼41 residue H2 (red) was constructed from the parallel coiled-coil cortexillin structure (Burkhard et al., 2000). A docked neck linker (yellow) joins the α-helical neck with the catalytic core. The α helices and β sheets in the motor and microtubule are shown in brown and green, respectively. There is additional density (enclosed by a dotted line) near L12 of the KIF1A crystal structure that accounts for the position of the K-loop, as seen previously in KIF1A (Kikkawa et al., 2000). The 45-Å long neck density fits the length of the ∼26-residue H1 α helix. The hinge (white dashed line) connects H1 to H2. As H2 is ∼25 Å longer than H1, and a COOH-terminal GFP is found at the end of the neck density, the NH2-terminal part of H2 must lie over the catalytic core, possibly interacting with loops 6 and 10 (L6 and L10) or associated secondary structure elements at the tip of the catalytic core. (B) In the top view, parts of the two tubulin COOH-terminal helices, H11 and H12 (orange) are closely associated with the catalytic core.
Because the entire length of the H1 helix is visualized is the 3D maps, the neck must be rigidly attached to the catalytic core, as a tenuous association would allow Brownian motion of the neck and result in a greatly attenuated density in the 3D map. From our atomic modeling (Fig. 3), loops L10 and L6 and associated secondary structure elements of the catalytic core are the most likely sites of interactions with the neck helices or neck hinge.
Unc104 dimerizes in the ADP and nucleotide-free states
Having determined the conformation of the Unc104 motor and neck domains in an ATP-like state, we next examined Unc104446 microtubule-bound conformations in the ADP and nucleotide-free states and calculated 3D maps of these complexes. In these maps (Fig. 4, A and B), we observed a large globular density lying above the attached motor at a radius of ∼200 Å from the microtubule axis. This density is unlikely to represent the neck for several reasons. First, we expect that the Unc104 neck linker is not docked onto the core in these nucleotide states (Kikkawa et al., 2001), as is true of conventional kinesin (Rice et al., 1999). Therefore, domains beyond the neck linker are expected to be disordered and not visualized in these maps. Second, the observed density is too large for the neck and is reminiscent of a second weakly attached catalytic core, as has been observed for conventional kinesin dimers (Hirose et al., 1999; Hoenger et al., 2000). Support for the idea that this density is a second head comes from comparing the MRDDs of the maps of the three nucleotide states (Fig. 4 C). In the ADP and nucleotide-free states, there is a large peak at R = ∼200 Å, but no corresponding peak in the AMPPNP map. Together, these data suggest that in the ADP and nucleotide-free states, Unc104446 is bound to microtubules as a dimer.
Rotation of the catalytic core in different nucleotide states
In work on KIF1A, a close orthologue of Unc104, Kikkawa et al. (2001) observed a ∼20° rotation of the catalytic core between the AMPPNP and ADP states and incorporated this result into a model for movement. When we compare the catalytic cores of Unc104446 in these two states (Fig. 5 A), we find no evidence for such a large rotation. The long axis of the Unc104 catalytic core in the AMPPNP state is parallel to the protofilament axis, whereas the long axis in the ADP state is only slightly rotated (Fig. 5 A, clockwise). The rotation of the catalytic core is at most 5°.
Figure 5.
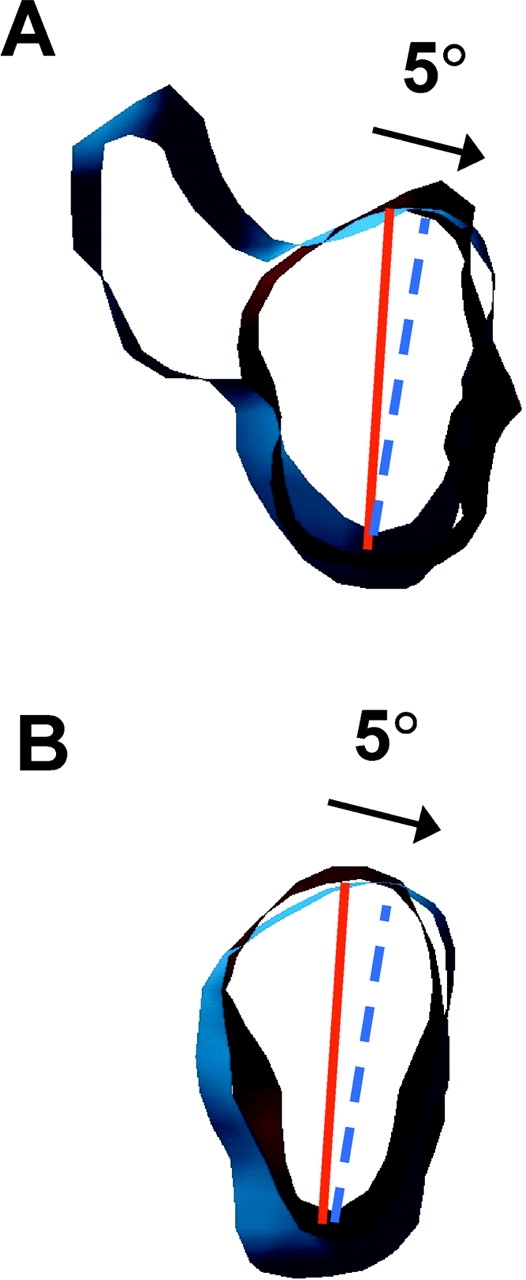
The Unc104 catalytic core rotates by ∼5° during the ATPase cycle. Thin slices of the catalytic cores of (A) Unc104446 and (B) Unc104362 in the AMPPNP (red) and ADP (blue) maps superimposed to show the small ∼5° rotation between these two nucleotide states. Solid red and dotted blue lines indicate the approximate orientation of the long axes of the catalytic core in the two states. The arrows indicate the rotational relationship between the Unc104 catalytic cores in the two nucleotide states.
The Unc104446 construct contains the neck region and in this respect differs from the KIF1A construct used by Kikkawa et al. (2001) where the neck was lacking. Therefore, to determine if the neck may account for the different results, we compared Unc104362 bound to microtubules in the AMPPNP and ADP states (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200308020/DC1). Again, we observed the same small ∼5° rotation of the catalytic core (Fig. 5 B; modeling experiments to investigate rotation of the catalytic core are shown in Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200308020/DC1). Thus, the lack of a large rotation in the Unc104 catalytic core between the AMPPNP and ADP states argues against such a rotation being a general mechanism that contributes to movement by kinesins, as was suggested by Kikkawa et al. (2001).
Role of the FHA and neck hinge regions for Unc104 function in living C. elegans
Our structural studies indicated that the neck hinge was essential for the folded state of the motor. Next, we asked whether the neck hinge and the adjacent FHA domains were important for the physiological function of the Unc104 motor in living worms. A null Unc104 mutation results in paralyzed animals due to a failure to transport synaptic vesicles to the terminals of motor neurons (Hall and Hedgecock, 1991). This paralyzed state is fully rescued by injecting the worms with transgenic Unc104. We introduced genes encoding full-length wild-type Unc104, Unc104 lacking the neck hinge, or Unc104 lacking the FHA domain into the germ line of unc104 null (e1265) worms. Expression of these genes from the endogenous promoter was identified in transgenic animals by fluorescence from GFP added to the COOH terminus, a strategy that has previously been shown not to affect the in vivo function of the motor (Zhou et al., 2001). The GFP expression pattern and levels were comparable between wild-type Unc104-GFP, Unc104-ΔHinge-GFP (deletion of amino acids 377–410), and Unc104-ΔFHA-GFP (unpublished data), indicating the differences in phenotype cannot be attributed to expression. Transgenic worms were analyzed for velocity and coordination of movement on bacterial agar lawns by video microscopy.
Transgenic animals expressing Unc104-ΔHinge-GFP exhibited a severe movement defect compared with the transgenic animals expressing wild-type Unc104-GFP motor (Fig. 6). Their velocity of movement (1.6 ± 1.5 mm/min, n = 419) was much slower than Unc104-GFP transgenic animals (12.7 ± 3.6 mm/min, n = 454). However, a partial rescue was evident, as they displayed slightly faster movements than null animals (1.1 ± 0.8 mm/min, n = 150). The velocity of null animals appears larger than expected, because unproductive movement without directional motion is also tracked as movement. Occasional long periods of steady, directional movement were observed for the Unc104-ΔHinge-GFP expressing animals (Fig. 6, right); however, the coordination of movement was more similar to the null than wild-type Unc104-GFP worms, which display a smooth, sinusoidal wave form of movement. Transgenic worms expressing Unc104-ΔFHA-GFP also displayed a movement defect, but the decrease in velocity (2.1 ± 1.1 mm/min, n = 380) and coordination was less severe than Unc104-ΔHinge-GFP. These results demonstrate that the neck hinge and FHA domains are important for motor performance in vivo. These in vivo phenotypes cannot be due to a defect in motor velocity, because our biophysical studies described above revealed that such deletions produce minimal effect on microtubule gliding velocity.
Figure 6.
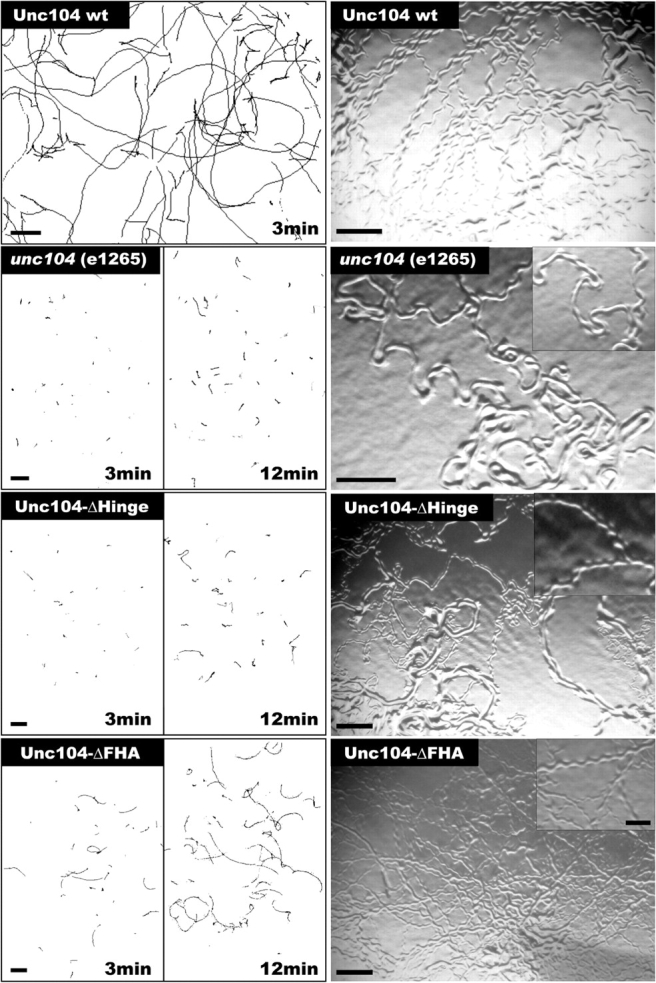
Effect of hinge and FHA domain deletions on Unc104 performance in living C. elegans . Comparison of velocities and coordinated movement of unc104 null (e1265) worms with worms expressing wild-type Unc104 wt, Unc104-ΔHinge, or Unc104-ΔFHA constructs. (Left) Worm movement was recorded over 3- or 12-min intervals and the position of the worms was tracked and plotted. Average velocities are 12.7 ± 3.6 mm/min for Unc104-wt (n = 454), 1.1 ± 0.9 mm/min for unc104 null (n = 150), 1.6 ± 1.5 mm/min for Unc104-ΔHinge (n = 419), and 2.1 ± 1.1 mm/min Unc104-ΔFHA (n = 380). The differences in velocity between the null, Unc104-ΔHinge, and Unc104-ΔFHA animals are statistically significant (P < 0.0001). Bars, 2.4 mm. (Right) Worm tracks on an agar plate reveal the degree of coordination. Although Unc104 wt animals move steadily in a sinusoidal motion in one direction over several body lengths, unc104 null worms rarely show coordinated movement. Although Unc104-ΔHinge worms move slowly, their tracks show a slight improvement of coordination compared to unc104 null (inset). Unc104-ΔFHA worms show sinusoidal tracks that are directional over several amplitudes. Bars: 0.5 mm; (inset) 0.2 mm.
Discussion
In this paper, we examined the nucleotide-dependent conformation of the microtubule-bound catalytic core of Unc104 (similar to the studies on KIF1A performed by Kikkawa et al., 2001), as well as longer Unc104 constructs that contain the neck region. We show that the catalytic core undergoes a small rotation (∼5°) between the AMPPNP and ADP states and suggest a different role for this motion than was reported previously (Kikkawa et al., 2001). We also show that the class-conserved Unc104 neck forms an ordered density that is rigidly attached to the catalytic core in the AMPPNP state; our further structural dissections suggest that this neck conformation is composed of an intramolecular, parallel coiled coil (Fig. 7 A). We also show that the neck undergoes a rearrangement in the ADP and nucleotide-free states resulting in Unc104 dimerization, a state that has been proposed on the basis of motility data (Tomishige et al., 2002) but not yet directly observed by structural methods. As will be discussed, we do not think that nucleotide state normally controls the monomer to dimer transition, but has served as an effector to shift the microtubule-bound Unc104 motor between these two states in our experimental system. However, our ATPase and transgenic worm data suggest that the folded state of the neck that we have observed is important and may regulate motor performance in vitro and in vivo. Our data, together with results from previous single molecule motility experiments (Tomishige et al., 2002), suggest that conformational changes in the Unc104 neck may regulate the monomer to dimer transition required for activating fast processive motility.
Figure 7.
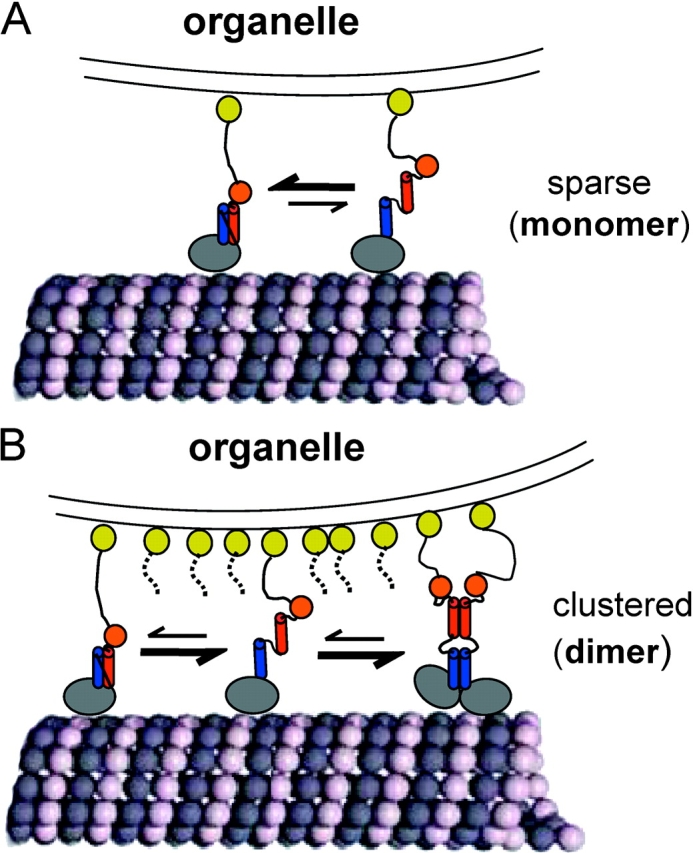
Model for regulation of monomer to dimer transition by self-folding of the neck helices. When Unc104 is sparsely distributed on an organelle membrane (A), the self-folded state is favored over the unfolded state in a dynamic equilibrium. The self-folded Unc104 monomers may move very slowly and nonprocessively, like Unc104362 (Tomishige et al., 2002). These monomers may show some plus end–directed biased diffusion, but will not generate significant force against an opposing load (Okada et al., 2003). However, when Unc104 is clustered, possibly in lipid rafts, on the organelle membrane at high local concentrations (B), unfolded monomers are recruited into dimers shifting the equilibrium away from the monomeric state. The dimerized Unc104 would then undergo fast processive motility along microtubules and generate maximal force. The monomer to dimer equilibrium shift may occur in solution, but may also be favored by the presence of microtubules. Blue and red cylinders represent H1 and H2, respectively. Orange spheres represent the FHA domains. Yellow spheres represent the membrane PH domain.
Rotation of the catalytic core
Kikkawa et al. (2001) reported that the microtubule-bound KIF1A catalytic core rotates by 20° between the ADP and AMPPNP states. Crystal structures of the motor in similar nucleotide states revealed a comparable rotation of the switch II (α4) helix, part of the microtubule binding interface, suggesting that rotation of the KIF1A catalytic core is generated by tight coupling to the motion of the microtubule-docked α4 helix. Based on these data, the authors proposed a mechanism for the biased processive motion of KIF1A monomers as well as a general mechanism for movement by kinesins (Kikkawa et al., 2001).
Our data on two different recombinant proteins (Unc104362 and Unc104446) showed a much smaller rotation (∼5°) of the catalytic core between the ADP and AMPPNP states. The difference between our results and those of Kikkawa et al. (2001) is due to differences in orientation of the weak binding ADP state on the microtubule; the orientation of the AMPPNP structures are very similar in the two studies. Variation in the orientation of the ADP state may be due to a species difference, although KIF1A and Unc104 are 85% homologous in their catalytic cores, or could reflect the particular construct used by Kikkawa et al. (2001), which was a chimera of the KIF1A motor core and part of the conventional kinesin neck.
Cryo-EM analyses of conventional kinesin (Rice et al., 1999) have failed to show rotation of the catalytic core in the ADP state, even though similar conformational changes of the conventional kinesin switch II helix have been observed previously (Vale and Milligan, 2000). Therefore, it seems unlikely that tight coupling of the switch II helix movement to large rotational motions of the catalytic core is a general mechanistic feature of the kinesin superfamily. However, the observed rotations in KIF1A and Unc104 (albeit different in degree) may serve some function. Despite the presence of the microtubule attracting K-loop, Unc104 and KIF1A bind much more weakly to microtubules in the ADP state than conventional kinesins (unpublished data; Tomishige et al., 2002; Online supplemental material). We speculate that catalytic core rotation may have a role in lowering Unc104's affinity for microtubules in the ADP state.
Two distinct conformations of the Unc104 neck
Under one set of experimental conditions (the AMPPNP state), the Unc104 neck forms a rigid ordered structure. The rigidity of the Unc104 neck differs from that of other kinesin neck domains, which, even though folded into stable, intermolecular coiled coils as seen in the X-ray structures (Kozielski et al., 1997; Sablin et al., 1998), are not observed in cryo-EM reconstructions of decorated microtubules due to their mobility (Sosa et al., 1997; Hoenger et al., 2000). Our data support a model in which the well-ordered neck density is due to the self-folding properties of the class-specific H1 and H2 neck helices and the intervening hinge as well as interactions of these elements with the catalytic core. By marking the COOH terminus, we showed that H1 and H2 associate in a parallel arrangement, and we confirmed that removal of all or part of the hinge precludes visualization of the neck density. A pseudo-atomic model shows that the neck density fits well the dimensions of a coiled coil comprised of helices of the same lengths as H1 and H2, and suggests that the rigidity of the Unc104 neck may be a result of contacts between H2 and loops 6 and 10 (Fig. 3).
Under other experimental conditions (ADP and nucleotide-free states), a second large density peak is seen at a higher radius. The size and location of the density peak in the 3D maps (Fig. 4, A and B) and the occurrence of a large peak in the MRDD of the maps (Fig. 4 C) suggest that this additional density is a second, weakly attached Unc104 catalytic core. Under these experimental conditions, there is no finger density in the maps, implying that the neck helices are no longer folded and rigidly attached to the catalytic core (Fig. 4, A and B). We expect that this situation allows the neck helices to participate in a dimeric intermolecular coiled coil resulting in formation of an Unc104 dimer rather similar in overall organization to conventional kinesin. Strikingly, what we observe for these experimental conditions, a detached second catalytic core and absence of neck density, is very similar to what is seen in cryo-EM maps of dimeric conventional kinesins (Hirose et al., 1999; Hoenger et al., 2000). Therefore, based on these data, we suggest that the Unc104 motor can exist in both monomeric and dimeric states.
The Unc104 FHA domain
Our results also suggest a regulatory role for the FHA domain, which is a conserved feature of all Unc104/KIF1 kinesin motors from Giardia to man (Vale, 2003). Transgenic worms expressing an Unc104 motor with an FHA domain deletion show a phenotypic defect intermediate between the severe uncoordinated movement of null and the normal movement of wild-type animals, indicating that the FHA domain is needed for proper motor function in vivo. The FHA also appears to decrease the microtubule binding affinity of the motor domain for microtubules, as its deletion causes a “gain of function” increase in microtubule affinity in the ATPase assay. To achieve this effect, the FHA domain might stabilize the intramolecular neck coiled coil and reduce the probability of forming processive Unc104 dimers (Tomishige et al., 2002). Because processive motors hydrolyze multiple ATPs per microtubule encounter, their KmMT is lower than nonprocessive motors (Hackney, 1995). Therefore, motors that lack the FHA domain might have higher affinity for microtubules (low KmMT) due to an increase in dimers in the population. Alternatively, the FHA domain may affect the neck, to which it has a defined spatial relationship (Fig. S1), in a way that alters the activity of the catalytic core. In either case, the FHA domain appears to operate coordinately with the neck to control motor activity. This may explain why the FHA domain is a universally conserved feature of all motors in the Unc104/KIF1A kinesin class.
The propensity of self-folding in the neck regulates Unc104 dimerization
Our analysis suggests that the ability of the neck to self-fold may regulate its ability to dimerize (Fig. 7). In the folded conformation, neck helices H1 and H2 would not be available to form the intermolecular coiled coil required for dimerization. However, high concentrations of Unc104 may shift the equilibrium toward dimerization (Tomishige et al., 2002).
The importance of the self-folded conformation for Unc104 function is supported by our data on Unc104 motors with full or partial deletion of the hinge, both of which prevent self-folding of the neck. We also show that transgenic animals expressing Unc104 bearing full neck hinge deletion have severe uncoordinated phenotypes similar to Unc104 null animals, underscoring its importance for biological function. An unstructured hinge at the end of the neck helix of fungal conventional kinesin also has been shown to be important for in vivo transport by this motor (Grummt et al., 1998). However, whereas deletions or substitutions in this hinge in conventional kinesin reduce motor speed and uncouple ATP turnover from gliding velocity (Grummt et al., 1998), deletions of either the neck hinge or the FHA domain in Unc104 have minimal effect on motor velocity in the microtubule gliding assay. Therefore, we believe that loss of biological activity associated with deletions of these regions in transgenic animals is related to improper motor regulation. Although the neck hinge deletions may cause other effects of which we are unaware or unable to assay, these findings suggest that the self-folded structural state described in this paper is required for normal biological function in vivo.
In our experiments, the transition between monomer and dimer conformation is affected by the nucleotide state of the motor. Normally, however, we would not expect the oligomeric state of Unc104 to be controlled by the ATPase cycle where transitions occur on a millisecond time scale. In the microtubule-bound ADP and nucleotide-free states, undocking of the neck linker in conjunction with the high concentration of motor used in these experiments (∼500 μM) might be expected to drive the equilibrium toward coiled-coil formation and dimerization (Fig. 7). This situation of high motor concentration on the microtubule might be analogous to the crowding of motors that may occur within lipid rafts on a vesicle surface. In the microtubule-bound AMPPNP state, on the other hand, the neck linker is docked and this likely favors the folded state despite the high motor density on the microtubule.
In summary, we propose that when Unc104 is sparsely distributed on vesicle membranes, the intramolecular self-folding of the neck would be favored over intermolecular coiled-coil formation (Fig. 7). Some motility and force generation by these monomeric motors may occur (Okada et al., 2003), but it would not be efficient. Upon clustering of Unc104 molecules into lipid raft domains on the membrane surface (Klopfenstein et al., 2002), the higher local motor concentration would favor intermolecular coiled-coil formation. Once dimerized, Unc104 molecules bind microtubules and generate fast processive movement (Tomishige et al., 2002). The switch between the self-folded and intermolecular coiled-coil states of the neck helices also might be influenced by posttranslational modification or association with other proteins. For example, interactions of Unc104's FHA domain with a protein partner might either promote or inhibit the folded motor conformation and thereby regulate motility.
The neck coiled coil may be a site of regulation for other kinesin motors as well. The conventional kinesin tail domain has been suggested to bind to and possibly induce a conformational change in its neck coiled-coil region (Stock et al., 1999). Also, genetic and biochemical studies indicate that the C. elegans protein CYK-4 can bind to the neck of the ZEN-4/CeMKLP1 kinesin and regulate motor function in vivo (Mishima et al., 2002). It will be interesting to examine whether these protein interactions cause alterations in neck coiled-coil stability that affect processivity of these motors.
Materials and methods
Proteins
Unc104 constructs were derived from a pET17b Unc104653-GFP plasmid that encodes amino acids 1–653 of C. elegans Unc104 (Pierce et al., 1999; GenBank/EMBL/DDBJ accession no. JN0114) with COOH-terminal GFP (S65T variant) and 6X Histidine tag. The GFP-enabled oriented motor attachment to the glass-adsorbed, anti-GFP antibodies in microtubule gliding assay. In Unc104653ΔHinge and Unc104446ΔHinge, residues 377–410 were removed from both constructs. In Unc104446ΔS-Hinge, only residues 392–402 were removed. Constructs for cryo-EM lacked GFP. All constructs were verified by sequencing.
Recombinant Unc104 was produced in a bacterial expression system and purified as described previously (Pierce and Vale, 1998). Fractions from the final column containing pure protein, were pooled and used within 1 wk. For cryo-EM, protein was concentrated to >25 mg/ml. A Sephedex-25 column was used to remove excess salt and nucleotide, and to exchange the buffer for 20 mM Pipes, pH 6.8, 20 mM NaCl, 4 mM MgCl2, 1 mM EGTA, and 1 mM DTT (binding buffer). Before use, the solution was centrifuged at 13,000 g for 5 min at 4°C.
Microtubule gliding and ATPase assays
Motors were subject to a final step of microtubule affinity purification (Case et al., 1997) before assaying activity. Microtubule-stimulated ATPase was measured in BRB80 (80 mM Pipes, pH 6.8, 2 mM MgCl2, and 1 mM EGTA) using an NADH-coupled enzyme assay (Huang et al., 1994; Pierce et al., 1999) with microtubule concentrations ranging from 0 to 15 μM. The kcat and KmMT (half maximal tubulin concentration for ATPase stimulation) were determined by fitting these curves to a hyperbolic function using Kaleidagraph. Basal ATPase (no microtubules present) was assayed using a malachite green phosphate assay (Kodama et al., 1986). Microtubule gliding assays were performed as described previously (Pierce et al., 1999).
Cryo-EM and image analysis
Microtubules were polymerized at 10 mg/ml in 80 mM Pipes, pH 6.8, 4 mM MgCl2, 1 mM EGTA, 13% DMSO 50 μM taxol, diluted to 5 mg/ml and adsorbed to freshly glow discharged Quantifoil grids (∼2 min). Grids were rinsed with 4 μl of binding buffer and 4 μl of the Unc104 protein in binding buffer was added for 2–3 min before blotting and quick freezing in liquid ethane slush. Frozen grids were stored under liquid nitrogen. To produce the nucleotide-free state, 1 μl of apyrase (100–200 U/ml) was added to the grid before freezing. For the other states, the Unc104 solution contained 5 mM Mg.ADP or 5 mM Mg.AMPPNP.
Examination and imaging of the frozen grids were performed essentially as described in Al-Bassam et al. (2002). Images of helical 15–protofilament microtubules were screened for optical quality and full decoration using laser diffraction, and digitized at spot and step sizes equivalent to 4.97 Å at the specimen. Representative images and power spectra are provided in Online supplemental material (Fig. S5, available at http://www.jcb.org/content/full/jcb.200308020/DC1). Helical image analysis was performed with PHOELIX (Whittaker et al., 1995; Carragher et al., 1996) with some modifications. In brief, near and far side layer lines were obtained from consecutive short segments (2,200-Å long) of straightened, contrast transfer function-corrected, microtubule images. Generally, for each 3D map, 31–51 short segments were used (see Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200308020/DC1). The layer line data were subjected to three cycles of fitting and averaging. The final averaged layer lines (Fig. S6, available at http://www.jcb/org/cgi/content/full/jcb.200308020/DC1) were truncated at 18-Å resolution and 3D maps were calculated by Fourier Bessel inversion and summation.
Modeling
The tubulin dimer structure (Nogales et al., 1998; PDB id 1TUB) was manually docked into the undecorated microtubule 3D map using the program O (Jones et al., 1991), using the high resolution microtubule model as a guide (Li et al., 2002). The KIF1A-AMPPCP and KIF1A-ADP structures (PDB id 1I6I and 1I5S, respectively; Kikkawa et al., 2001) were similarly docked into the Unc104 density. An α-helical coiled-coil segment from cortexillin (Burkhard et al., 2000; PDB id 1D7M) was used to model the neck helices. Models were displayed with AVS (AVS Corp.).
C. elegans transformation and phenotypic assays
The full-length wild-type Unc104-GFP construct was provided by M. Zhou and J. Scholey (University of California, Davis, Davis, CA; Zhou et al., 2001). The neck hinge (residues 377–410) and FHA deletions (residues 462–593) were introduced using standard PCR and DNA cloning techniques. The Unc104 null strain CB1265 [unc-104 (e1265) II] was used for analysis. C. elegans were grown and maintained as described previously (Brenner, 1974).
Heritable lines of transgenic worms carrying extrachromosomal arrays of the Unc104-GFP construct (Zhou et al., 2001) were created by microinjection of Unc104-GFP plasmid into hermaphrodites by methods described previously (Fire, 1986; Mello et al., 1991). After microinjecting Unc104-GFP wt, Unc104-ΔHinge-GFP, or Unc104-ΔFHA-GFP (each at 70 μg/ml) into the unc104 (e1265) animals, transgenic lines were selected on the basis of their sinusoidal, wild-type movement and/or GFP expression. Locomotion was assayed by touching the head or tail of the worm with an eyelash to stimulate backward or forward movement. Young adult hermaphrodites' movement velocities were assayed by transferring well-fed, M9-buffer–washed worms to a fresh agar plate (Brenner, 1974). After 1 h on these plates, movement was recorded on a dissecting microscope at 1–3 frames/s for 3–12 min with a CCD TV camera (Javelin) and Adobe Premiere 4.0. Movements were tracked using a custom programmed ImageJ's multitracker plug in (NIH, ImageJ version 1.29; Nico Stuurman, University of California, San Francisco, San Francisco, CA). Worm track images were taken on a dissecting microscope (Nikon) with a digital camera (model Coolpix5000; Nikon) at full resolution.
Online supplemental material
Fig. S1 is a 3D map of Unc104653 (AMPPNP); Fig. S2 is a 3D map of Unc104446 ΔS-Hinge (AMPPNP); Fig. S3 shows a rotation of the catalytic core; Fig. S4 is a pseudo-atomic model for rotation of the catalytic core; Fig. S5 shows a representative image and power spectra; and Fig. S6 is the averaged layer line data for the 3D maps. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200308020/DC1.
Supplemental Material
Acknowledgments
We thank Michio Tomishige for preparing the Unc104362-GFP construct, Nora Hom-Booher for preparing the Unc104410-GFP construct, and Cori Bargmann and her laboratory for their advice on working with C. elegans. We thank Carolyn Moores, Michio Tomishige, and Nicole Mahoney for helpful discussions and Brian Sheehan for help with computing.
Jawdat Al-Bassam was a predoctoral fellow of the American Heath Association (AHA-0010004Y), and Y. Cui is supported by an National Institutes of Health (NIH) postdoctoral fellowship (GM20453). This work was supported by grants from the NIH (GM52468 to R.A. Milligan.; RR17573 and GM61939 to B.O. Carragher).
J. Al-Bassam and Y. Cui contributed equally to this work.
The online version of this paper contains supplemental material.
Abbreviations used in this paper: 3D, three-dimensional; AMPPNP, 5′-adenylylimidodiphosphate; FHA, forkhead homology associated; MRDD, mean radial density distribution; PH, pleckstrin homology.
References
- Aiyar, A. 2000. The use of CLUSTAL W and CLUSTAL X for multiple sequence alignment. Methods Mol. Biol. 132:221–241. [DOI] [PubMed] [Google Scholar]
- Al-Bassam, J., R.S. Ozer, D. Safer, S. Halpain, and R.A. Milligan. 2002. MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J. Cell Biol. 157:1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom, G.S. 2001. The UNC-104/KIF1 family of kinesins. Curr. Opin. Cell Biol. 13:36–40. [DOI] [PubMed] [Google Scholar]
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard, P., R.A. Kammerer, M.O. Steinmetz, G.P. Bourenkov, and U. Aebi. 2000. The coiled coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges. Structure Fold. Des. 8:223–230. [DOI] [PubMed] [Google Scholar]
- Carragher, B., M. Whittaker, and R.A. Milligan. 1996. Helical processing using PHOELIX. J. Struct. Biol. 116:107–112. [DOI] [PubMed] [Google Scholar]
- Case, R.B., D.W. Pierce, N. Home-Booher, C.L. Hart, R.D. Vale. 1997. The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell. 90:959–966. [DOI] [PubMed] [Google Scholar]
- Durocher, D., and S.P. Jackson. 2002. The FHA domain. FEBS Lett. 513:58–66. [DOI] [PubMed] [Google Scholar]
- Fire, A. 1986. Integrative transformation of Caenorhabditis elegans. EMBO J. 5:2673–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt, M., G. Woehlke, U. Henningsen, S. Fuchs, M. Schleicher, and M. Schliwa. 1998. Importance of a flexible hinge near the motor domain in kinesin-driven motility. EMBO J. 17:5536–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney, D.D. 1995. Highly processive microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains. Nature. 377:448-450. [DOI] [PubMed] [Google Scholar]
- Hall, D.H., and E.M. Hedgecock. 1991. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 65:837–847. [DOI] [PubMed] [Google Scholar]
- Hirose, K., J. Lowe, M. Alonso, R.A. Cross, and L.A. Amos. 1999. Congruent docking of dimeric kinesin and ncd into three-dimensional electron cryomicroscopy maps of microtubule-motor ADP complexes. Mol. Biol. Cell. 10:2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger, A., M. Thormahlen, R. Diaz-Avalos, M. Doerhoefer, K.N. Goldie, J. Muller, and E. Mandelkow. 2000. A new look at the microtubule binding patterns of dimeric kinesins. J. Mol. Biol. 297:1087–1103. [DOI] [PubMed] [Google Scholar]
- Huang, T.G., J. Suhan, and D.D. Hackney. 1994. Drosophila kinesin motor domain extending to amino acid position 392 is dimeric when expressed in Escherichia coli. J. Biol. Chem. 269:16502–16507. [PubMed] [Google Scholar]
- Jiang, W., and D.D. Hackney. 1997. Monomeric kinesin head domains hydrolyze multiple ATP molecules before release from a microtubule. J. Biol. Chem. 272:5616–5621. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., J.Y. Zou, S.W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 47:110–119. [DOI] [PubMed] [Google Scholar]
- Kikkawa, M., Y. Okada, and N. Hirokawa. 2000. 15 Å resolution model of the monomeric kinesin motor, KIF1A. Cell. 100:241–252. [DOI] [PubMed] [Google Scholar]
- Kikkawa, M., E.P. Sablin, Y. Okada, H. Yajima, R.J. Fletterick, and N. Hirokawa. 2001. Switch-based mechanism of kinesin motors. Nature. 411:439–445. [DOI] [PubMed] [Google Scholar]
- Klopfenstein, D.R., M. Tomishige, N. Stuurman, and R.D. Vale. 2002. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 109:347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama, T., K. Fukui, and K. Kometani. 1986. The initial phosphate burst in ATP hydrolysis by myosin and subfragment-1 as studied by a modified malachite green method for determination of inorganic phosphate. J. Biochem. 99:1465–1472. [DOI] [PubMed] [Google Scholar]
- Kozielski, F., S. Sack, A. Marx, M. Thormahlen, E. Schonbrunn, V. Biou, A. Thompson, E.M. Mandelkow, and E. Mandelkow. 1997. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 91:985–994. [DOI] [PubMed] [Google Scholar]
- Lee, J.R., H. Shin, J. Ko, J. Choi, H. Lee, and E. Kim. 2002. Characterization of the movement of the kinesin motor KIF1A in living cultured neurons. J. Biol. Chem. 278:2624–2629. [DOI] [PubMed] [Google Scholar]
- Li, H., D.J. DeRosier, W.V. Nicholson, E. Nogales, and K.H. Downing. 2002. Microtubule structure at 8 A resolution. Structure. (Camb). 10:1317–1328. [DOI] [PubMed] [Google Scholar]
- Mello, C.C., J.M. Kramer, D. Stinchcomb, and V. Ambros. 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10:3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima, M., S. Kaitna, and M. Glotzer. 2002. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell. 2:41–54. [DOI] [PubMed] [Google Scholar]
- Nogales, E., S.G. Wolf, and K.H. Downing. 1998. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 391:199–203. [DOI] [PubMed] [Google Scholar]
- Okada, Y., and N. Hirokawa. 1999. A processive single-headed motor: kinesin superfamily protein KIF1A. Science. 283:1152–1157. [DOI] [PubMed] [Google Scholar]
- Okada, Y., and N. Hirokawa. 2000. Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc. Natl. Acad. Sci. USA. 97:640–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada, Y., H. Yamazaki, Y. Sekine-Aizawa, and N. Hirokawa. 1995. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 81:769–780. [DOI] [PubMed] [Google Scholar]
- Okada, Y., H. Higuchi, and N. Hirokawa. 2003. Processivity of the single-headed kinesin KIF1A through biased binding to tubulin. Nature. 424:574–577. [DOI] [PubMed] [Google Scholar]
- Pierce, D.W., and R.D. Vale. 1998. Assaying processive movement of kinesin by fluorescence microscopy. Methods Enzymol. 298:154–171. [DOI] [PubMed] [Google Scholar]
- Pierce, D.W., N. Hom-Booher, A.J. Otsuka, and R.D. Vale. 1999. Single-molecule behavior of monomeric and heteromeric kinesins. Biochemistry. 38:5412–5421. [DOI] [PubMed] [Google Scholar]
- Rice, S., A.W. Lin, D. Safer, C.L. Hart, N. Naber, B.O. Carragher, S.M. Cain, E. Pechatnikova, E.M. Wilson-Kubalek, M. Whittaker, et al. 1999. A structural change in the kinesin motor protein that drives motility. Nature. 402:778–784. [DOI] [PubMed] [Google Scholar]
- Romberg, L., D.W. Pierce, and R.D. Vale. 1998. Role of the kinesin neck region in processive microtubule-based motility. J. Cell Biol. 140:1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost, B., and C. Sander. 1994. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 19:55–72. [DOI] [PubMed] [Google Scholar]
- Sablin, E.P., R.B. Case, S.C. Dai, C.L. Hart, A. Ruby, R.D. Vale, and R.J. Fletterick. 1998. Direction determination in the minus-end-directed kinesin motor ncd. Nature. 395:813–816. [DOI] [PubMed] [Google Scholar]
- Sindelar, C.V., M.J. Budny, S. Rice, N. Naber, R. Fletterick, and R. Cooke. 2002. Two conformations in the human kinesin power stroke defined by X-ray crystallography and EPR spectroscopy. Nat. Struct. Biol. 9:844–848. [DOI] [PubMed] [Google Scholar]
- Sosa, H., D.P. Dias, A. Hoenger, M. Whittaker, E. Wilson-Kubalek, E. Sablin, R.J. Fletterick, R.D. Vale, and R.A. Milligan. 1997. A model for the microtubule-Ncd motor protein complex obtained by cryo-electron microscopy and image analysis. Cell. 90:217–224. [DOI] [PubMed] [Google Scholar]
- Stock, M.F., J. Guerrero, B. Cobb, C.T. Eggers, T.G. Huang, X. Li, and D.D. Hackney. 1999. Formation of the compact confomer of kinesin requires a COOH-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J. Biol. Chem. 274:14617–14623. [DOI] [PubMed] [Google Scholar]
- Tomishige, M., and R.D. Vale. 2000. Controlling kinesin by reversible disulfide cross-linking. Identifying the motility-producing conformational change. J. Cell Biol. 151:1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige, M., D.R. Klopfenstein, and R.D. Vale. 2002. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 297:2263–2267. [DOI] [PubMed] [Google Scholar]
- Vale, R.D. 2003. The molecular motor toolbox for intracellular transport. Cell. 112:467–480. [DOI] [PubMed] [Google Scholar]
- Vale, R.D., and R.A. Milligan. 2000. The way things move: looking under the hood of molecular motor proteins. Science. 288:88–95. [DOI] [PubMed] [Google Scholar]
- Whittaker, M., B.O. Carragher, and R.A. Milligan. 1995. PHOELIX: a package for semi-automated helical reconstruction. Ultramicroscopy. 58:245–259. [DOI] [PubMed] [Google Scholar]
- Yonekawa, Y., A. Harada, Y. Okada, T. Funakoshi, Y. Kanai, Y. Takei, S. Terada, T. Noda, and N. Hirokawa. 1998. Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein–deficient mice. J. Cell Biol. 141:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H.M., I. Brust-Mascher, and J.M. Scholey. 2001. Direct visualization of the movement of the monomeric axonal transport motor UNC-104 along neuronal processes in living Caenorhabditis elegans. J. Neurosci. 21:3749–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.