Abstract
Dicentric chromosomes undergo a breakage–fusion–bridge cycle as a consequence of having two centromeres on the same chromatid attach to opposite spindle poles in mitosis. Suppression of dicentric chromosome breakage reflects loss of kinetochore function at the kinetochore–microtubule or the kinetochore–DNA interface. Using a conditionally functional dicentric chromosome in vivo, we demonstrate that kinetochore mutants exhibit quantitative differences in their degree of chromosome breakage. Mutations in chl4/mcm17/ctf17 segregate dicentric chromosomes through successive cell divisions without breakage, indicating that only one of the two centromeres is functional. Centromere DNA introduced into the cell is unable to promote kinetochore assembly in the absence of CHL4. In contrast, established centromeres retain their segregation capacity for greater than 25 generations after depletion of Chl4p. The persistent mitotic stability of established centromeres reveals the presence of an epigenetic component in kinetochore segregation. Furthermore, this study identifies Chl4p in the initiation and specification of a heritable chromatin state.
Keywords: yeast; centromere; kinetochore; chromatin; epigenetic
Introduction
Accurate chromosome segregation during mitosis requires the assembly of centromeric DNA and proteins to form the kinetochore, which couples chromosome movement to dynamic spindle microtubules. In Saccharomyces cerevisiae, the core centromere is defined by a distinctive nuclease-protected chromatin domain encompassing ∼160–200 bp of DNA (Bloom and Carbon, 1982; Funk et al., 1989). This protected chromatin domain is required for proper centromere function, as evidenced by the requirement of histone genes (Smith et al., 1996; Pinto and Winston, 2000) and chromatin remodeling enzymes (Tsuchiya et al., 1998), mutations in which disrupt the structure and decrease the fidelity of chromosome segregation. The CEN DNA–histone complex together with CBF3 comprises the inner kinetochore, while the outer kinetochore includes Okp1p, Ctf19p, Mcm21p, Ctf3p, and Ndc80p (Ortiz et al., 1999; Measday et al., 2002). Spindle/outer domain proteins that bind microtubules and kinetochores include components of the Dam1p–Duo1p complex (Hofmann et al., 1998; Cheeseman et al., 2001; Enquist-Newman et al., 2001; Janke et al., 2002), the Cin8p microtubule motor protein, and several nonmotor microtubule-associated proteins, such as Bik1p and Stu2p (He et al., 2001).
That higher-order chromatin structure contributes to function is evidenced by the decreased segregation fidelity of deletion derivative chromosomes (10–100 kb in size) (Newlon, 1988). Mutations that show a marked size-dependent segregation defect have been isolated (minichromosome maintenance mutants chl4/mcm17/ctf17, iml3/mcm19, and mcm21) (Roy et al., 1997). Increasing the size of minichromosomes (10 kb) with bacteriophage lambda DNA (45 kb) leads to a 17-fold increase in stabilization in chl4 and iml3 and a fourfold increase in mcm21 (Roy et al., 1997). The function of this class of mutants remains to be determined.
Whereas the presence of a single centromere on each chromosome is essential for accurate chromosome segregation, multiple centromeres on a single chromosome are deleterious. After bipolar attachment during cell division, dicentric chromosomes rearrange, giving rise to stable monocentric derivative chromosomes. Mutations in kinetochore genes that decrease the fidelity of chromosome segregation display decreased dicentric chromosome breakage as well. Three of the mutants that exhibit a size-dependent segregation phenotype, chl4, iml3, and mcm21, support the propagation of completely intact dicentric plasmids (Kouprina et al., 1993; Sanyal et al., 1998; Poddar et al., 1999; Ghosh et al., 2001). The presence of two weakened kinetochores on a dicentric chromosome may decrease the chance that both centromeres are attached to opposite spindle poles at a given time, thereby suppressing chromosome breakage (Fig. 1 B). Alternatively, dicentric chromosomes with one functional and one dysfunctional centromere might behave like a monocentric (Fig. 1 C). The complete suppression of dicentric plasmid breakage could be indicative of two functionally distinct kinetochores in these mcm mutants (Fig. 1 C).
Figure 1.
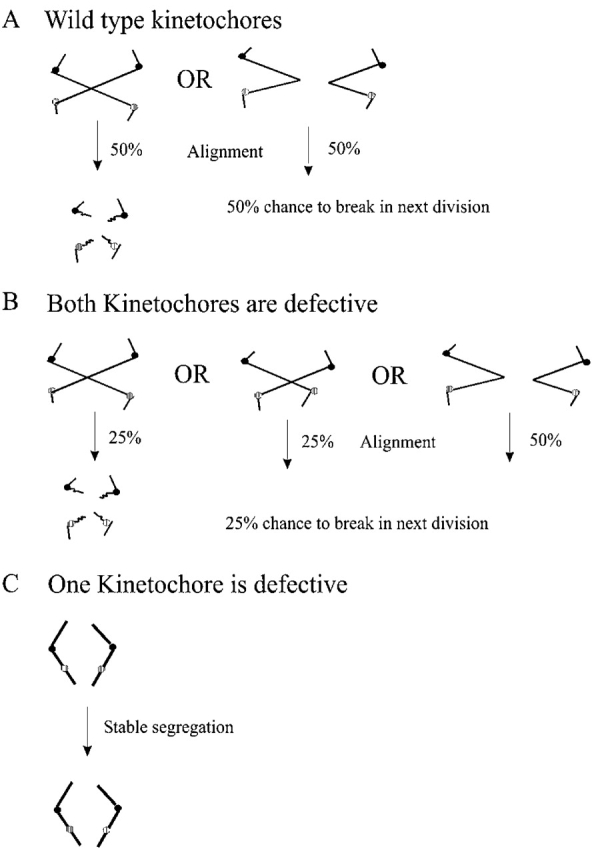
Quantitative dicentric chromosome breakage assay. (A) When two centromeres on the same sister chromatid of a dicentric chromosome form attachments to opposite spindle poles (left), progression through anaphase results in chromosome breakage followed by generation of a monocentric chromosome derivative. When both the centromeres on the same sister chromatid are oriented toward the same pole (right), dicentric chromosomes are segregated without physical rearrangement. If centromeres on the same sister chromatid have an equal chance of attaching to the same or opposite poles, then successive cell divisions will result in the accumulation of monocentric derivatives at a frequency of 50% per generation. In one division, ∼50% of dicentric chromosomes break (Brock and Bloom, 1994). (B) If either of the centromeres releases their microtubule attachment, the dicentric chromosome will segregate without physical rearrangement, resulting in partial suppression of breakage. If centromeres on the same sister chromatid have an equal chance of attaching to the same or opposite poles, then successive cell divisions will result in the accumulation of monocentric derivatives at a frequency of <50% per generation, depending on the severity of the mutation. 50% of the time, centromeres on the same sister will attach to the same pole (far right). However, not all chromosomes with centromeres attached to opposite poles will break (25% and 25%). (C) If particular kinetochore mutants differentially affect function, only one centromere attaches to the spindle pole. In this situation, the dicentric chromosome will be physically and segregationally stable.
Differential centromeric states have been reported in a number of organisms (Ault and Lyttle, 1988; Steiner and Clarke, 1994). Minichromosomes carrying only a fraction of the Schizosaccharomyces pombe centromere adopt a mitotically stable or mitotically unstable state. A mitotically unstable centromere switches to the stable state at a frequency of 0.6–0.7% (Steiner and Clarke, 1994). This was the first demonstration of the presence of two heritable states within a population in the absence of DNA structural rearrangement or chemical modification of the chromosomal DNA (Steiner and Clarke, 1994). Coexistence of different centromeric states within a cell has also been observed upon application of drugs that block histone deacetylation. Transient drug treatment induces histone hyperacetylation and increased chromosome malsegregation. After removal of the drug, histone hyperacetylation at the centromere, but not at noncentromeric sites, was propagated to subsequent generations (Ekwall et al., 1997). The mitotic stability of a modified histone state indicates an epigenetic component in the transmission of centromeric proteins in the process of kinetochore replication. Although a number of epigenetic mechanisms have been identified to promote the inheritance of a stable state, the mechanisms required to initiate or specify which heritable states are to be propagated have not been determined.
In this study, we have identified chl4/mcm17/ctf17 as a mutant that fails to assemble kinetochores on naked centromere DNA. In contrast, established centromeres are faithfully transmitted for over 25 generations in the absence of Chl4p. Thus there are distinct heritable states of centromeric chromatin in budding yeast. Chl4p is required for the specification of the mitotically stable state.
Results
De novo kinetochore function requires CHL4: dicentric chromosome breakage assay
A conditional dicentric chromosome was constructed by introducing a GAL1-regulated CEN3 cassette (GALCEN) into chromosome III. Regulation of the GAL1 promoter allows propagation of the chromosome as a monocentric on galactose and as a dicentric on glucose. Activation of the conditional centromere gives rise to monocentric derivative chromosomes in the population through breakage and rearrangement. The resulting colonies are heterogeneous in size, morphology, and chromosome III structure (Fig. 2 A) (Hill and Bloom, 1987; Koshland et al., 1987).
Figure 2.
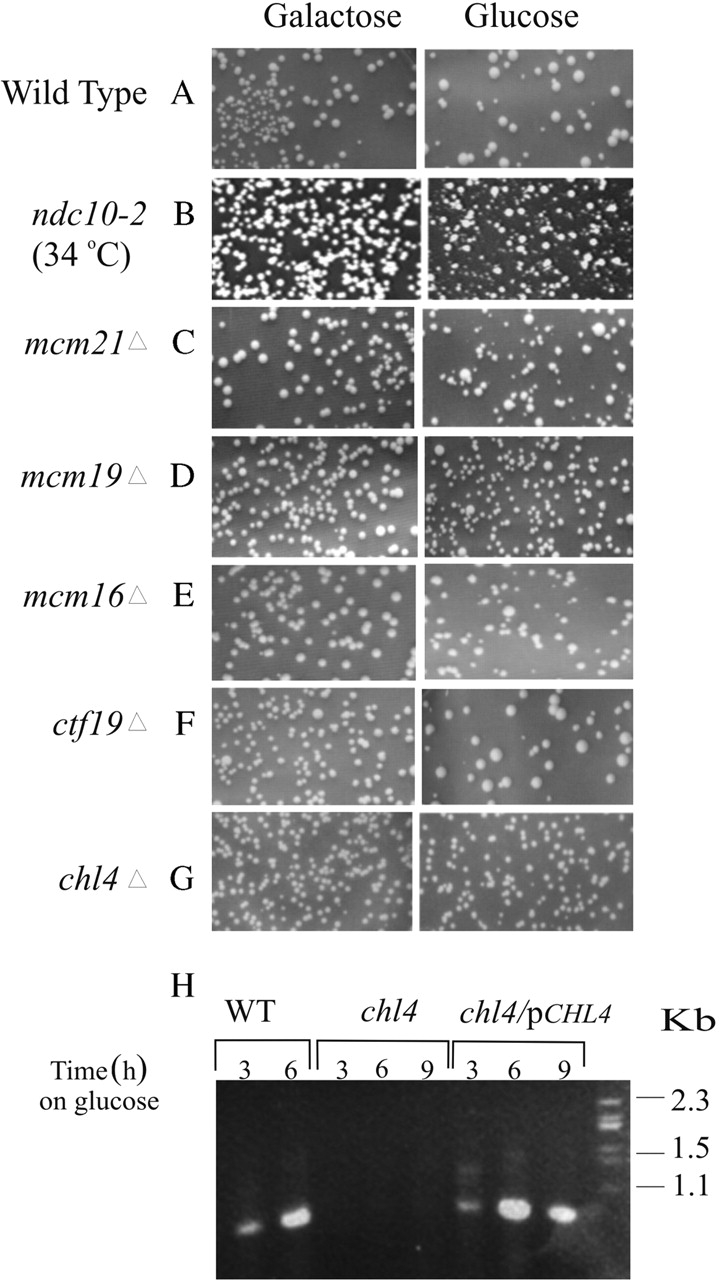
Suppression of dicentric chromosome breakage in kinetochore mutants. (A–G) Colony morphology upon dicentric chromosome activation. Wild-type (A), ndc10-2 (B), mcm21Δ (C), iml3Δ (D), mcm16Δ (E), and ctf19Δ (F) cells are uniform in size and shape on galactose (monocentric for chromosome III) and heterogeneous in size and shape on glucose (dicentric for chromosome III). In contrast, chl4Δ cells (G) appear homogenous on both galactose and glucose. (H) Physical analysis of DNA repair. DNA recovered from RAD52 dicentric chromosome–containing cells after induction on glucose, lanes 1 and 2 (3 and 6 h on glucose) show a rearrangement product of 925 bp. DNA recovered from chl4Δ dicentric chromosome–containing cells, lanes 3–5 (3, 6, 9 h on glucose) do not show diagnostic rearrangement. chl4Δ dicentric chromosome–containing cells transformed with CHL4 on a CEN plasmid show rearrangement upon transfer to glucose, lanes 6–8 (3, 6, and 9 h in glucose media).
Mutations affecting centromere function result in a suppression of dicentric chromosome breakage (Doheny et al., 1993). We examined several kinetochore mutants in the dicentric chromosome breakage assay to distinguish functional classes of mutants. Ndc10p, an essential component of the CBF3 complex, Mcm21p, Mcm16p, and Ctf19p, members of the putative outer kinetochore complex, and chl4/mcm17/ctf17 and iml3, two mutants that display a strong chromosome size-dependent segregation defect (Roy et al., 1997). Colony heterogeneity was observed upon dicentric chromosome activation in ndc10-2, mcm21Δ, iml3Δ, mcm16Δ, and ctf19Δ (Fig. 2, B–F). In contrast, chl4Δ cells containing a conditional dicentric chromosome were homogeneous and indistinguishable in size and shape from their counterparts grown on galactose (monocentric growth conditions) (Fig. 2 G).
To quantify the degree of chromosome breakage in these mutants, cell viability was determined in the absence of the major DNA repair gene RAD52. Cells lacking RAD52 and containing an active dicentric chromosome have decreased viability upon plating on glucose (2%; Table I; Brock and Bloom, 1994). In the quantitative breakage assay, each of ndc10-2, iml3Δ, mcm21Δ, mcm16Δ, ctf19Δ, and chl4Δ rad52Δ double mutants suppresses dicentric chromosome breakage. Between 40 and 50% of ndc10-2, rad52Δ, mcm21Δ, rad52Δ, and mcm16Δ rad52Δ are viable upon activation of the second centromere (Table I). 70% of iml3Δ rad52Δ, 75% of ctf19Δ rad52Δ, and 100% of chl4Δ rad52Δ were viable (Table I). The complete suppression of dicentric chromosome breakage in the chl4Δ rad52Δ mutant represents a novel kinetochore phenotype.
Table I. Viability of wild-type and mutant strains upon dicentric chromosome activation.
| Relevant genotype | RAD 52 | % Viabilitya |
|---|---|---|
| WT | + | 66.0 ± 1.0 |
| rad52Δ | − | 2.0 ± 0.2 |
| ndc10-2ts (33°C)b | + | 74.5 ± 1.0 |
| ndc10-2 ts rad52Δ (33°C)b | − | 36.0 ± 2.0 |
| mcm21Δ | + | 78.0 ± 1.0 |
| mcm21Δ rad52Δ | − | 31.0 ± 10.0 |
| mcm16 | + | 80.0 ± 2.0 |
| mcm16Δ rad52Δ | − | 43.0 ± 9.0 |
| iml3Δ | + | 81.0 ± 6.0 |
| iml3Δ rad52Δ | − | 58.0 ± 3.0 |
| ctf19Δ | + | 82.0 ± 3.0 |
| ctf19Δ rad52Δ | − | 62.0 ± 4.0 |
| ch14Δ | + | 81.0 ± 2.0 |
| ch14Δ rad52Δ | − | 80.0 ± 7.0 |
Strains containing GALCEN3 in wild type, chl4Δ, iml3Δ, mcm21Δ, and ndc10-2, in the presence or absence of RAD52 (indicated by + or −), respectively, were plated for single colonies on galactose and glucose. Dicentric viability is a measure of colony forming units on glucose/galactose. 48% of ndc10-2 rad52Δ (36% viability in rad52Δ/75% in RAD52), 40% of mcm21Δ rad52Δ (31% in rad52Δ/78% in RAD52), 55% of mcm16Δ rad52Δ (43% in rad52Δ/80% in RAD52), 70% of iml3Δ rad52Δ (58% in rad52Δ/81% in RAD52), 75% of ctf19Δ rad52Δ (62% in rad52Δ/ 82% in RAD52), and 100% of chl4Δ rad52Δ (80% in rad52Δ/81% in RAD52) cells were viable. n = 10 determinants for chl4Δ, iml3Δ, and mcm21Δ; n = 5 determinants for ctf19Δ and mcm16Δ; n = 6 determinants for ndc10-2ts; and n = 4 determinants for wild type (WT).
Mean ± SD.
The ndc10-2 ts dicentric strains were grown at 24°C and plated out at 33°C (semipermissive for growth) on glucose and galactose plates.
To confirm if the high viability in chl4Δ rad52Δ mutants containing a dicentric chromosome is due to a lack of chromosome breakage, we monitored the appearance of the predominant repair product after dicentric chromosome activation (Brock and Bloom, 1994). A PCR-based assay (see Materials and methods) revealed the lack of centromere rearrangement in chl4Δ RAD52 cells (Fig. 2 H, chl4), consistent with the lack of DNA breakage and repair. To determine whether Chl4p is sufficient to recover centromere function, a single copy of CHL4 was introduced into cells containing the dicentric chromosome. The 925-bp predominant repair product was evident 3 h after activation of the dicentric chromosome in the presence of Chl4p (Fig. 2 H, chl4/pCHL4). The presence of the breakage product implies that both centromeres on the dicentric chromosome are active after transfer to glucose media. Hence, Chl4p is necessary for the activation of a functional dicentric chromosome.
Chl4p is not essential for propagation of established centromeres
The presence of two weakened kinetochores on the dicentric chromosome may decrease the chance that both centromeres are attached to opposite spindle poles at a given time, thereby suppressing chromosome rearrangement (Fig. 1 B). Alternatively, if the centromeres were differentially functional, i.e., one functional and one dysfunctional centromere, the dicentric chromosome would be physically and segregationally stable (Fig. 1 C). To determine whether CHL4 is differentially required for function on new versus established centromeres, we compared the mitotic stability of centromere plasmids introduced into chl4Δ mutants (new centromeres) versus the mitotic stability of plasmids introduced into wild-type cells and subsequent removal of Chl4p (old or established centromeres). Centromere plasmids are highly unstable in chl4Δ mutants (45/45 0.08–5% stability; Table II, chl4Δ→plasmid; Maine et al., 1984; Kouprina et al., 1993). In contrast, deletion of CHL4 from centromere plasmid–bearing cells revealed a range of mitotically stable (60–90%, average of 79.2% ± 9.9) and unstable (0.08–5%, average of 0.8% ± 1.1) transformants (Table II, Wt + plasmid→chl4Δ). Centromere plasmids were faithfully segregated in 49/95 chl4Δ transformants (Table II). These assays reveal the presence of mitotically stable and unstable states upon deletion of CHL4. Thus, CHL4 is essential for de novo centromere function but not for the propagation of established centromeres.
Table II. Mitotic stability depends on the timing of Chl4p loss relative to introduction of centromere DNA.
| Plasmid | Wild type | chl4Δ→plasmid | Wt + plasmid→ chl4Δ | |||
|---|---|---|---|---|---|---|
| Stable(80–90%) | Unstable | Stable | Unstable(0.08–5%) | Stable(60–90%) | Unstable(0.08–5%) | |
| pYe (CEN3) B | 10/10 | 0/10 | 0/15 | 15/15 | 12/27 | 15/27 |
| pYe (CEN3) 30 | 10/10 | 0/10 | 0/15 | 15/15 | 20/36 | 16/36 |
| pYe (CEN3) 41 | 10/10 | 0/10 | 0/15 | 15/15 | 17/32 | 15/32 |
Centromere plasmids (pYe [CEN3] B, pYe [CEN3] 41, or pYe [CEN3] 30) were introduced into wild type and chl4Δ mutants (left and middle columns). Plasmid-bearing cells were selected, and plasmid stability was determined as described in the Materials and methods. Centromere plasmids are stably segregated (80–90%) in 30/30 wild-type cells (left column). In 45/45 chl4Δ mutants, <5% of cells contained plasmids upon nonselective growth. In the right column, cells containing the indicated centromere plasmids were transformed with chl4Δ::KANr. Mitotic stability measurements revealed heterogeneity in plasmid stability: 60–90% stability (49/95) and 0.08–5.0% stability (right column, 46/95).
Established centromere plasmids are stable in the absence of Chl4p
Although established centromere plasmids are stably segregated upon Chl4p depletion, there are indications of differentially heritable states (∼50% mitotically stable and 50% mitotically unstable; Table II). To examine the initial response after Chl4p depletion, we created an inducible degron allele of CHL4 (GAL–UBI–CHL4; see Materials and methods). 80% of cells contained the centromere plasmid pDLB2064 (CEN6/ADE3) when introduced into GAL–UBI–CHL4 on galactose (Chl4p induction) (Table III). In contrast, centromere plasmids transformed into GAL–UBI–CHL4-containing strains on glucose (Chl4p depletion) were unstable (9 ± 3%; Table III). Hence the galactose-regulated degron allele of CHL4 behaves like the chl4Δ null on glucose media and demonstrates the efficacy of the inducible degron-tagged Chl4p system. Expression of GAL–UBI–Chl4–GFP allows determination of the timing of protein loss in the population using anti-GFP antibodies for detection by Western blots and in single cells by fluorescence. No Chl4–GFP protein was detectable after 2.5 h on glucose by Western analysis (Fig. 3, inset). Similarly, no observable Chl4–GFP fluorescence was detectable in >100 cells after 3 h on glucose (unpublished data).
Table III. Mitotic stability upon depletion of Chl4p.
| Relevant genotype | Colonies (%)a | No. of colonies scoredb | n b | % Stabilityc (+URA) | ||
|---|---|---|---|---|---|---|
| Red | Red + white sectored | White | ||||
| WT | 16.0 | 67.0 | 17.0 | 980 | 5 | 89 ± 2.0 |
| chl4Δ | <0.5 | 19.0 | 81.0 | 1,397 | 5 | 10 ± 2.0 |
| GAL–UBI–CHL4 | ||||||
| Galactose | 2.0 | 75.0 | 23.0 | 415 | 3 | 80 ± 3.0 |
| Glucose | <0.5 | 13.0 | 86.0 | 2,019 | 8 | 9 ± 3.0 |
| Gal→Glu | 1.0 | 75.0 | 24.0 | 3,287 | 10 | 59 ± 14.0 |
pDLB2064 was transformed into the GAL1–UBI–CHL4 strain on glucose and galactose and selected on either Glu-uracil or Gal-uracil. Single colonies were grown on YPD/YPG overnight and plated on YPD/YPG plates. For the gal→glu transitions, single colonies from galactose transformants were grown on gal-uracil media overnight at 30°C. Cells were washed and shifted to YPD media for 8–10 h and plated onto YPD plates.
Colonies were scored as being red, white, or colonies with red and white sectors after 3 d and expressed as a percent of the total number of colonies scored.
Number of colonies scored is the sum of the number of red, white, or red and white–sectored colonies for each transformant analyzed. n, number of independent transformants assayed.
Expressed as colonies that were URA/colonies on YPD or YPG. For % stability of gal→glu, cells grown overnight in galactose were switched to YPD media for 8–10 h before plating on YPD.
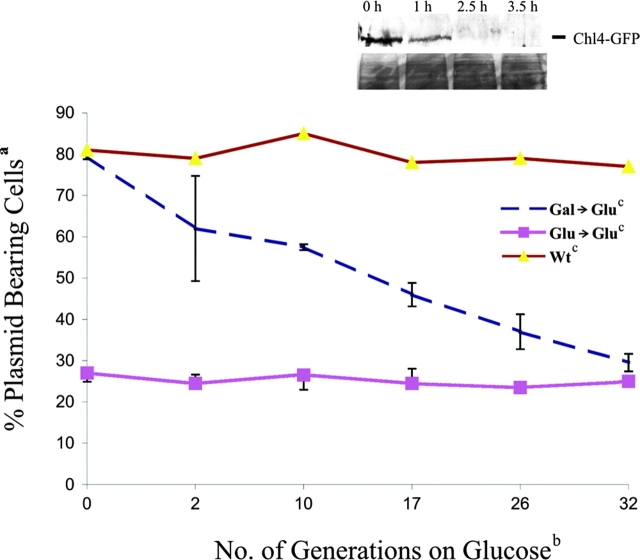
Figure 3.
Frequency of centromere switch in vivo. pDLB2064 was transformed into GAL1–UBI–CHL4 and wild-type (CDV39) cells and selected on Gal-uracil or Glu-uracil media. a,bCells were grown overnight and shifted to cGlu-ura (selective) media, and mitotic stability was determined at the generation times indicated. Colonies that were red or red and white sectored were scored as plasmid-bearing cells and are expressed as a percent of the total number of colonies plated. n = 3 transformants analyzed for Gal→Glu, 2 transformants for Glu→Glu, and 1 transformant for wild-type cells on glucose. Inset is a Western blot showing stability of the Chl4–GFP fusion protein in the degron strain. Cells from the degron strain were grown in galactose (0 h), and protein samples were prepared. Cells were washed and switched to glucose media to initiate proteolysis of the fusion protein. Protein samples were prepared at the time points indicated. The fusion protein was identified with anti-GFP antibodies in a Western blot analysis. The lower panel of the inset shows a region of the blot visualized by Ponceau S stain, indicating equal protein loading.
To examine the stability of established centromere plasmids after depletion of the degron-tagged Chl4p, plasmid-bearing cells transformed on galactose were switched from galactose (+Chl4p) to liquid glucose media (−Chl4p) for 8–10 h (approximately three generations after loss of Chl4–GFP; Fig. 3, inset). 59 ± 14% of the cells contained centromere plasmids after colony growth on glucose in the absence of Chl4p (Table III). Established centromere plasmids were lost at a frequency of 5% per generation. In contrast, naked centromere plasmid DNA transformed into cells lacking Chl4p was lost at a frequency of 12% per generation.
Visual inspection of plasmid stability via colony color (see Materials and methods) confirms and extends the plasmid loss assay. Centromere plasmids transformed on galactose (+Chl4p) were stably segregated (77% red or red and white sectored; Table III), whereas transformation on glucose (−Chl4p) resulted in plasmid loss (86% white; Table III). Upon switch from galactose to glucose for 8–10 h followed by plating on glucose plates, 76% of the colonies were red or red and white sectored (Table III). Thus, established centromere plasmids are stably propagated in the absence of Chl4p.
Established centromeres switch to an unstable state in the absence of Chl4p
To test whether a small fraction of cells with mitotically unstable plasmids accumulates after loss of Chl4p, cells containing established centromere plasmids were grown in selective glucose media. Plasmid loss and subsequent division of plasmid-minus cells for two to three generations contribute to the fraction of plasmid-minus cells in a population (Murray and Szostack, 1985). However, in selective growth conditions, the fraction of plasmid-bearing cells is at equilibrium. The percentage of plasmid-bearing cells for stable or unstable plasmids remains constant over 30 generations of selective growth (Fig. 3). If the lower mitotic stability of centromere plasmids in GAL–UBI–CHL4 cells shifted from galactose to glucose reflects an increased minichromosome loss rate, then the percentage of plasmid-bearing cells will be lower than wild type but will remain at equilibrium over extended growth periods. Alternatively, if a small fraction of established centromeres converts to the unstable state after depletion of Chl4p, the fraction of plasmid-bearing cells upon transfer from galactose to glucose will decrease over time. Centromere plasmids transformed into GAL–UBI–CHL4 on galactose were stably maintained (80% on galactose; Table III). Cells were switched to selective glucose, and the fraction of plasmid-bearing cells was determined over time. The frequency of plasmid loss (2–3% per generation; Fig. 3, Gal→Glu) indicates that established centromeres are not in equilibrium upon depletion of Chl4p.
If the fraction lost per generation reflects a bonafide switch event from the stable to the unstable state, rather than the slow loss of Chl4p, then single colonies in a clonal population isolated after prolonged growth on glucose should be heterogeneous in their plasmid stability. A single colony with an established centromere plasmid (galactose, +Chl4p) was shifted to glucose (−Chl4p) for 8–10 h, followed by growth on glucose plates. 66 single plasmid-bearing colonies (red and white sectored) were isolated and grown on glucose media for 10 h, followed by growth on glucose plates. Of the 66 colonies, 34/66 colonies had a mitotic stability between 0 and 20%, 27/66 had a mitotic stability between 20 and 40%, and 8/66 had a mitotic stability between 40 and 60% (Table IV). In contrast, 25/25 single red and white–sectored colonies derived from a clonal population of cells transformed on glucose (−Chl4p) had a mitotic stability of between 0 and 20% after the same growth regime (Table IV, GAL–UBI–CHL4, Glu→Glu). Thus, centromere plasmids can be stably propagated for >35 generations in the absence of Chl4p.
Table IV. Persistence of the stably inherited state in the absence of Chl4p.
| Relevant genotype | % Mitotic stability | |||
|---|---|---|---|---|
| 0–20 | 20–40 | 40–60 | 60–85 | |
| Wild type (35 generations) | 14/25 (52 ± 6) | 11/25 (70 ± 7) | ||
| GAL–UBI–CHL4 | ||||
| Glu→Glu | 25/25 (9 ± 7) | |||
| Gal→Glu | 34/66 (12 ± 5) | 24/66 (27 ± 6) | 8/66 (48 ± 5) | |
Centromere plasmid transformed into either wild-type or GAL–UBI–CHL4 strains on glucose were grown in YPD for 8–10 h and plated on YPD plates. After 3–4 d, colonies were isolated and grown in selective glucose media for 10 h and plated on YPD. Colonies were counted after 3–4 d, n = 25 colonies purified. GAL–UBI–CHL4 cells transformed with centromere plasmid on galactose were washed, shifted to YPD for 8–10 h, and plated on YPD, n = 66 colonies purified. Mitotic stability was determined using the colony color assay as described in the Materials and methods.
The heterogeneous distribution of plasmid stability observed upon prolonged outgrowth on glucose is consistent with 2–3% of cells switching from the mitotically stable to unstable state. 12% of cells with stable plasmids (8/66) after 35 generations of growth on glucose is the approximate fraction expected in a population with centromeres that switch to the unstable state at a frequency of 2–3% per generation (2.5% loss/generation × 35 generations = 87.5% loss events = 12.5% stable segregants). Thus, unlike centromeres introduced into cells lacking Chl4p that are unstable and fail to segregate, established centromeres switch to the unstable state at a frequency of 2–3% per generation in the absence of Chl4p.
Differential accessibility of centromeric chromatin in chl4Δ cells
To determine whether the failure of “new” centromeres to direct chromosome segregation in chl4Δ is due to a defect in kinetochore assembly, we determined the chromatin structure of new and established centromeres in the absence of CHL4. Restriction enzyme accessibility is a quantitative assay for the degree of protection provided by centromeric protein binding (Saunders et al., 1990). Conditionally functional centromeres (GALCEN) were introduced into the HIS4 locus of chromosome III by transformation. Nuclei were incubated with increasing concentrations of DraI, which recognizes three closely apposed sites within CDEII of CEN3. The endogenous centromere was 1.5-fold more accessible in chl4Δ than in wild-type cells, consistent with the increase in chromosome loss observed in chl4Δ cells (Kouprina et al., 1993). In contrast, new GALCEN was 3.5-fold more accessible to DraI digestion. Conditional mutations in cse4, depletion of histones H2B or H4, and mutations in chromatin assembly factors cac1 and hir1 result in quantitatively similar increases in DraI accessibility (Saunders et al., 1990; Meluh et al., 1998; Sharp et al., 2002). Thus, the quantitative increase in accessibility of new centromeres indicates that Chl4p is essential for the formation of a nuclease-resistant structure at newly introduced centromeres.
Chl4p is present on functional kinetochores.
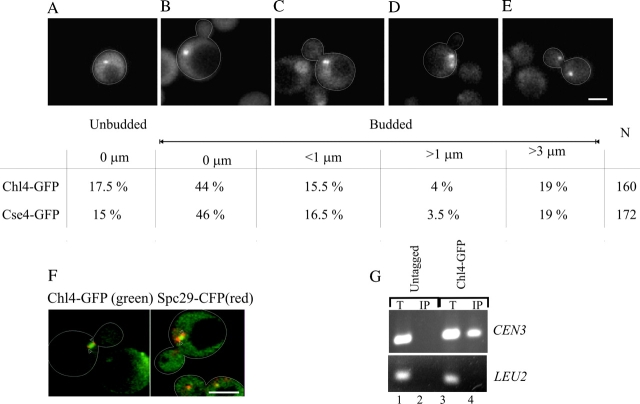
To determine whether Chl4p is associated with the centromere, we performed chromatin immunoprecipitation (ChIP)* analysis with Chl4–GFP. Centromere plasmids were stably propagated in Chl4–GFP-containing strains, indicative of wild-type Chl4–GFP function (unpublished data). Sheared chromatin was immunoprecipitated with antibodies directed against GFP and probed with PCR primers specific to CEN3 and to a noncentromeric region (LEU2). We found that Chl4p was localized to CEN3 DNA in wild-type cells (Fig. 4 G).
Figure 4.
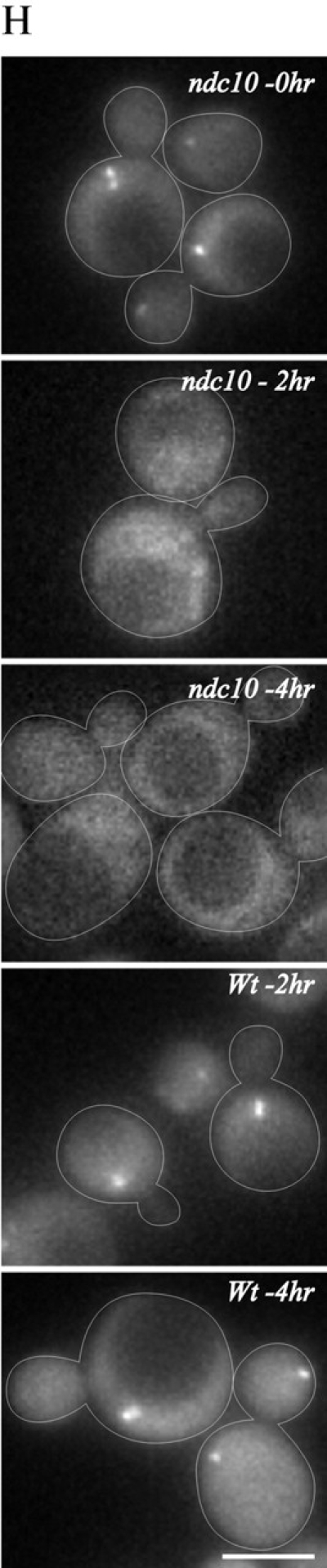
Chl4–GFP localizes to the centromere and the kinetochore and is dependent on Ndc10p. Cells containing Chl4–GFP and Cse4–GFP were examined by fluorescence microscopy. Chl4–GFP appears as one or two spots in the nucleus of the cell. The following percentages and distance between the two spots were observed, and a representative image of each is displayed: (A) single spot in unbudded cells (0 μm); (B) single spot in budded cells (0 μm); (C–E) two discrete spots in mitotic cells (<1 μm, >1 μm, and >3 μm). The fraction of cells in each morphology class with the spot to spot distance is indicated for both Chl4–GFP and Cse4–GFP. N is the total number of cells counted. Bar, 2.7 μm. (F) Cells expressing Chl4–GFP (green) and the spindle pole body component Spc29–CFP (red) were examined as described. Arrowheads indicate the position of Spc29–CFP. Bar, 6 μm. (G) ChIP assay performed by immunoprecipitation on GFP-tagged Chl4p followed by PCR analyses. Lanes 1 and 2 are samples from the untagged strain, and lanes 3 and 4 represent the strain containing the fusion protein. CEN DNA was enriched relative to the negative control locus LEU2. T, total lysate; IP, immunoprecipitated fraction. (H) Chl4–GFP localizes to the centromere in ndc10-2 ts mutant cells grown at 24°C (0 h). Upon shift of the ndc10-2 ts mutants to 37°C (restrictive temperature) for 2 or 4 h (second and third panels from the top), Chl4–GFP was no longer detectable. Upon shift of the wild-type cells to 37°C for 2 or 4 h (fourth and fifth panels from the top), Chl4–GFP spots were unperturbed. Bar, 6 μm.
In unbudded cells, a single Chl4–GFP spot was seen in the nucleus (Fig. 4 A). 44% of the population was budded cells with a single Chl4–GFP spot, 16% of cells prior to anaphase onset had two spots separated by an average of 0.6 ± 0.2 μm (Fig. 4 C), and 4% of cells had two spots separated by a mean distance of 2.0 ± 0.7 μm (Fig. 4 D). The distance and distribution of Chl4–GFP are indistinguishable from the kinetochore-specific histone-like H3 protein, Cse4–GFP (Fig. 4). The position of Chl4–GFP, with respect to the spindle apparatus, was examined by imaging Chl4–GFP together with the spindle pole body–associated protein Spc29p labeled with CFP (Fig. 4 F, Spc29–CFP). Chl4–GFP was adjacent to the spindle poles in small budded cells. In medium to large budded cells, Chl4–GFP was distributed in the region between the two poles.
The localization of several kinetochore proteins, Mtw1p, Ctf19p, and Cse4p (Hyland et al., 1999; Ortiz et al., 1999; Goshima and Yanagida, 2000; He et al., 2001), is dependent on the centromere-binding factor Cbf2p/Ndc10p. To test if Chl4–GFP localization was also dependent on functional Ndc10p, we examined Chl4–GFP in an ndc10 ts strain (ndc10-2; Kopski and Huffaker, 1997) at permissive (24°C; Fig. 4 H) and restrictive temperatures (37°C). No GFP signal was detectable after 2 or 4 h at 37°C (Fig. 4 H). In contrast, Chl4–GFP localization in wild-type cells was indistinguishable at 24°C and after 4 h at 37°C (Fig. 4 H), indicating that the fusion protein was not perturbed upon shift to 37°C. The association of Chl4p with centromeres and its in vivo distribution are indicative of its role in centromere function.
Kinetochore assembly at newly activated centromeres depends on CHL4
To determine the role of Chl4p in kinetochore assembly, we tested the association of both core and outer kinetochore components to endogenous CEN3 and GALCEN3 after its activation upon switch to glucose media. Asynchronous wild-type and chl4Δ cells harboring GALCEN3 were grown in galactose and switched to glucose medium containing nocodazole for 4 h to prevent rearrangements of the dicentric chromosome during mitosis in wild-type cells (Brock and Bloom, 1994). GFP-tagged Cse4p, Iml3p, and Ndc10p were individually immunoprecipitated from formaldehyde cross-linked extracts using anti-GFP–conjugated beads. The precipitated DNA was probed with PCR primers complementary to CEN3, GALCEN3, or LEU2 to determine the level of bound CEN3 and GALCEN3 DNA. CEN3 was quantitatively precipitated with Ndc10p, Cse4p, and Iml3p (Fig. 5, A, B, and C, CEN3) on both galactose and glucose in wild-type cells. Ndc10p, Cse4p, and Iml3p association with CEN3 was unchanged in the absence of chl4 ( Fig. 5, A, B, and C, CEN3, lanes 7–10). Association of the fusion proteins (Ndc10p, Cse4p, and Iml3p) with GALCEN3 in wild-type cells failed to occur on galactose, consistent with loss of kinetochore function on galactose (Hill and Bloom, 1987; Koshland et al., 1987). After activation of GALCEN3 upon switch to glucose for 4 h, Ndc10p, Cse4p, and Iml3p were associated with GALCEN3 (Fig. 5, A, B, and C, GALCEN3, lanes 3–6). In contrast, Ndc10p, Cse4p, and Iml3p failed to associate with GALCEN3 in chl4Δ cells after the switch to glucose (Fig. 5, A, B, and C, GALCEN3, lanes 7–10). The absence of both core and putative outer components at newly activated centromeres in the absence of chl4 demonstrates that Chl4p is essential for de novo kinetochore assembly.
Figure 5.
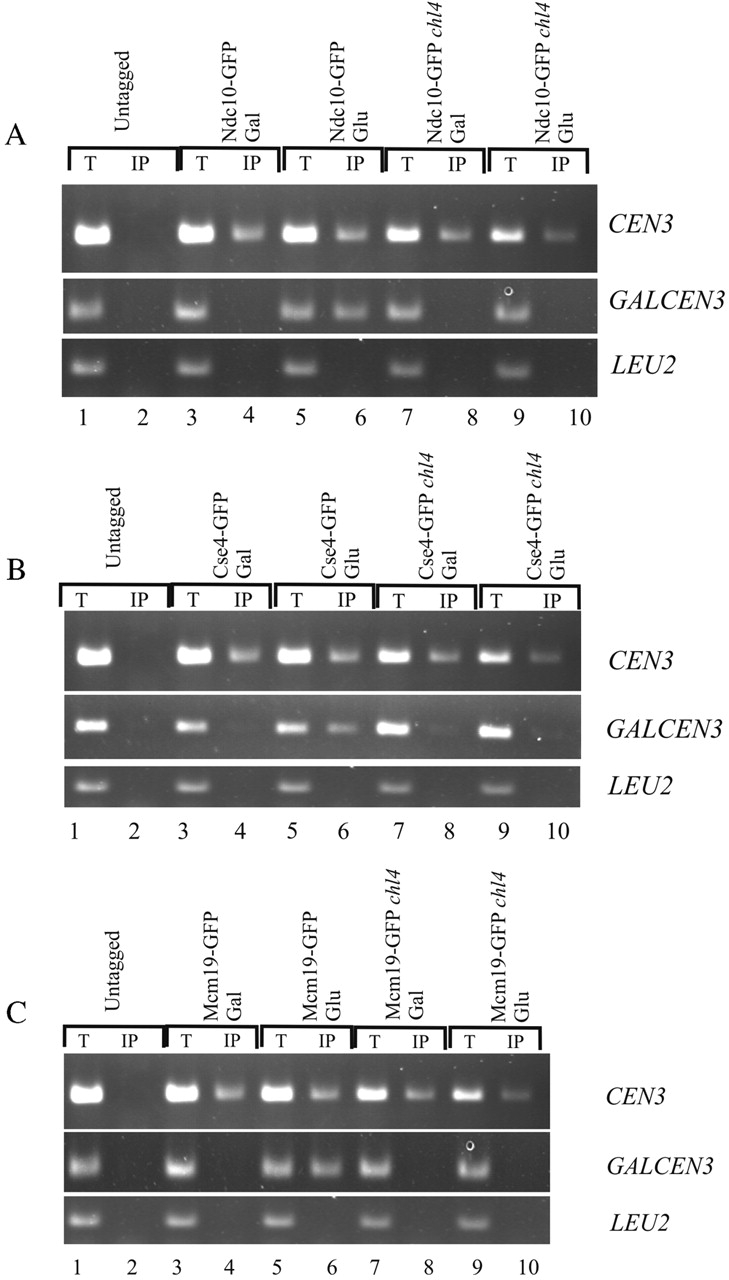
Kinetochore assembly to newly activated centromeres requires Chl4p. ChIP assay was performed by immunoprecipitation of GFP-tagged Ndc10p (A), Cse4p (B), and Iml3p (C) from wild-type and chl4Δ cells followed by PCR analysis. Cells were grown in galactose and shifted to glucose with nocodazole for 4 h. Anti-GFP immunoprecipitations were performed in tagged (lanes 3–10) and untagged strains (lanes 1 and 2) as controls. Chromatin was prepared, and PCR was performed using primers described in the Materials and methods. Chl4p is required for the interaction of Ndc10p (A), Cse4p (B), and Iml3p(C) with GALCEN3 on glucose (lanes 9 and 10). Chl4p is not required for the association of the fusion proteins with CEN3. T, total lysate; IP, immunoprecipitated fraction.
Discussion
We have demonstrated that, first, the segregation state of centromeric DNA is inherited; second, this state is dependent on the function of Chl4p; and third, naked centromere DNA fails to assemble into chromatin in the absence of Chl4p, indicating a differential requirement for de novo and template-directed propagation of centromeric states. Thus, Chl4p is required to initialize and designate a chromatin state to be heritably propagated.
The introduction of centromere plasmid DNA into a host cell via transformation requires kinetochore assembly on a naked DNA template. Using a degron-tagged Chl4p, we demonstrated that centromere plasmid stability is dependent upon the time of loss of Chl4p (Tables III and IV). 12% of cells containing centromere plasmids after 35 generations in the absence of Chl4p retained mitotically stable plasmids, whereas the remainder of cells had mitotically unstable centromere plasmids (Table IV). The heterogeneity observed in the mitotic stability of minichromosomes after loss of Chl4p suggests that preexisting kinetochores switch to the unstable state upon loss of Chl4p at a frequency of 2–3% per generation. The switch events were revealed by the presence of both mitotically stable and unstable states after deletion of CHL4 from centromere plasmid–bearing cells (Table II). The stability of a fraction of centromeres in the absence of Chl4p for many generations is indicative of an epigenetic mechanism in centromere inheritance in S. cerevisiae.
CHL4 encodes a 53-kD predicted protein that shares a low degree of homology with the family of bacterial RecA proteins (25% identical and 47% similarity over 350 amino acids; Kouprina et al., 1993). In addition, it has 29% DNA sequence homology to an uncharacterized ORF in S. pombe (Pi022p) and Neurospora crassa. The relationship to RecAp is unclear, as Chl4p lacks critical residues for ATP hydrolysis. Nevertheless, the region meets the necessary requirements for coding an α helix-turn-helix motif at position 384–406, indicative of a potential DNA binding property. Chl4p–GFP localizes to the centromere and is dependent on the presence of the centromere core component Ndc10p (Fig. 4). The localization of Chl4p is consistent with its role in centromere propagation and chromosome segregation. The foundation of the centromere–kinetochore complex is likely to be comprised of centromere-specific histones (S. cerevisiae Cse4p, Homo sapiens CENP-A, Caenorhabditis elegans HCP-3, and Drosophila melanogaster Cid). Mutations in core centromeric histones and centromere chromatin remodeling components result in increased accessibility to the restriction enzyme DraI (Saunders et al., 1990; Meluh et al., 1998; Tsuchiya et al., 1998; Sharp et al., 2002); likewise mutations in the conserved centromeric DNA core that disrupt mitotic stabilization disrupt the chromatin structure as well (Saunders et al., 1988). Endogenous centromeres were found to be 1.5–2-fold more accessible to DraI in chl4Δ cells, and new centromeres were three–fourfold more accessible to DraI in chl4Δ cells (unpublished data), indicating that Chl4p may contribute to the specification of centromeric chromatin.
Established centromeres seem to be capable of recruiting kinetochore components in the absence of Chl4p. Lack of Chl4p, however, abolishes recruitment of the same kinetochore components to newly activated centromeres (Fig. 5). The apparently normal recruitment of components to established centromeres even in the absence of Chl4p, loss of which results in increased chromosome loss, is consistent with the idea that established centromeres are propagated in an epigenetic manner in contrast to new centromeres that require de novo assembly. These data provide the first step toward understanding the mechanism that specifies kinetochore protein loading onto naked DNA.
Evidence for the hypothesis that the de novo formation of a functional kinetochore is distinct from propagation/duplication can be derived from the observation that mutations in the kinetochore proteins cse4-1, mif2-3, and ndc10-1 have a very small effect on the cohesin recruitment (Scc1p) to an endogenous centromere (CEN6); whereas the same mutations abolish recruitment of Scc1p to conditional centromeres (GALCEN) switched from galactose to glucose for 1 h (Tanaka et al., 1999). Thus, cohesin recruitment to new centromeres, and not established centromeres, like centromeric chromatin, is sensitized in kinetochore protein mutants.
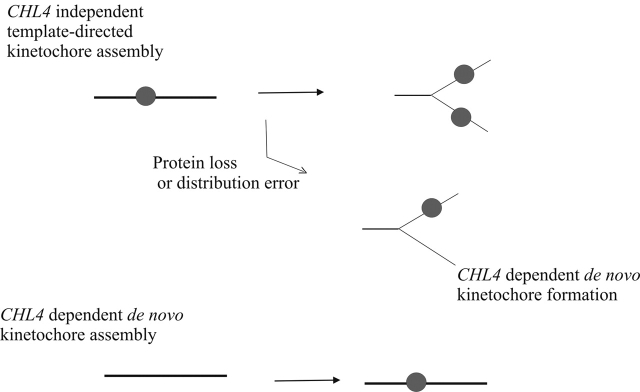
An occasional failure to distribute or segregate the kinetochore protein complex during replication, or a failure to maintain proper kinetochore conformation during mitosis, would require de novo assembly of the kinetochore complex as a “repair” function (Fig. 6). Failure to promote this dynamic de novo assembly, as revealed in chl4Δ cells, results in chromosome loss and is deleterious to cell survival. Centromere function can be epigenetically regulated in a number of organisms, including humans (for review see Karpen and Allshire, 1997). Centromeres are frequently inactivated on dicentric chromosomes, and new or neocentromeres can be formed in chromosomal regions not usually associated with centromere activity. The requirements for forming a fully functional centromere during normal centromere propagation and for neocentromere formation are likely to be different at the outset because the former can work from a preexisting template, whereas the latter has to create one. In addition, mechanisms that act at centromeres, such as hypoacetylation, poly (ADP) ribosylation, and replication timing, are features also shared to varying degrees by other chromosomal domains, such as the telomere. Hence, the primary driver for centromere formation must involve a centromere-specific factor whose activity provides a chromosomal mark to endorse the formation of a centromere. Our data demonstrating that de novo kinetochore function is distinct from the propagation of preexisting kinetochores in S. cerevisiae implicate Chl4p in driving CEN kinetochore formation during de novo assembly. Although the S. cerevisiae kinetochore may only promote a single microtubule–chromosome interaction, the structure and motility features of this point kinetochore (Pearson et al., 2001) and the heritability of different chromatin states reveal these complexes to exhibit many of the features of metazoan kinetochores.
Figure 6.
Model for the role of Chl4p in kinetochore assembly. Functional kinetochore assembly during replication requires kinetochore protein deposition at the centromere DNA. Initial dispersal of old centromere components onto nascent daughter duplexes provides a template for the recruitment of proteins to form a functional kinetochore complex. De novo centromere assembly is dependent on CHL4, as revealed by the introduction of naked centromeric DNA into chl4Δ cells. An occasional failure to distribute or segregate the template during replication would lead to a requirement for de novo kinetochore assembly in wild-type cells. The aberrant Cse4–GFP localization and the increased accessibility of CDEII in chl4Δ cells are consistent with a role for CHL4 in maintaining the fidelity of chromosome segregation.
Materials and methods
Strain construction and plasmids
The S. cerevisiae strains and plasmids used in this study are described in Table V. The S. cerevisiae dicentric strains were described previously (Brock and Bloom, 1994). chl4Δ::KAN r , iml3Δ::KAN r , and mcm21Δ::KAN r were constructed by transforming with a PCR-amplified product that was generated using the KANMX cassette (Wach et al., 1994) and 5′ and 3′ primers flanking CHL4 (5′-GTATAGCAAGTTCTAACCCAGAATCAGGTTCTTTATTATTGTCAAGACAGTGAAGCTTCGTACGC-3′ and 5′-GCTAGACAGATTATCGAAAACGGAACAATTACTTTCAAGTGCCCATCTGCATAGGCCAC-TAGTGGATCTG-3′). Transformants were selected on YPD + 300 μg/ml G418 or YPG + 150 μg/ml G418 (as the case may be) and screened using a 3′ internal primer to kanamycin, 3′ internal primers to the gene, and a 5′ primer external to the deletion cassette. CHL4, IML3, MCM21, MCM16, and CTF19 are nonessential genes, and strains were grown at 32°C. RAD52Δ::LEU2 disruptions were made as described previously (Brock and Bloom, 1994). All the COOH-terminal fusions of GFP were made as described previously (Wach et al., 1994). The fragment was integrated into wild-type strains (Table V) and KKY186 (ndc10-2). The Spc29–CFP (Pearson et al., 2001) integration was made in the wild-type and GAL–UBI–CHL4 strains containing either Chl4–GFP fusion or pCse4-GFPTRP1. Transformations were performed using the one-step gene replacement method. Strains with stable integrations were maintained in YPD media (2% glucose, 2% peptone, and 1% yeast extract).
Table V. S. cerevisiae strains and plasmids used in this study.
| Strains and plasmids | Relevant genotype | Source/reference |
|---|---|---|
| 1D | MAT a ade1 met14 ura3-52 leu2 his3 his4 | K. Bloom |
| 8a | MAT a his3 ura3 leu2 | K. Bloom |
| 9a | MAT α lys2 trp1Δ ura3 leu2 | K. Bloom |
| KKY186 | MAT a ndc10-2ts his3-200 leu2-3, 112 ura3-52 | T. Huffakera |
| CDV39 | MAT α ade2 ade3 leu2 trp1Δ ura3 lys2 | J. Pringleb |
| KBY4001B | MAT a ade1 met14 ura3-52 leu2 his3 his4::GALCEN3URA3 | K. Bloom |
| KBY4002B | MAT a ade1 met14 ura3-52 leu2 his3 his4::GALCEN3URA3 rad52Δ::LEU2 | K. Bloom |
| KBY4005 | MAT a ade1 met14 leu2 his3 his4::GALCEN3URA3 chl4Δ::KANr | This study |
| KBY4062 | MAT a ade1 met14 leu2 his his4::GALCEN3URA3 mcm21Δ::KANr | This study |
| KBY4063 | MAT a ade1 met14 leu2 his his4::GALCEN3URA3 iml3Δ::KANr | This study |
| KBY4011 | MAT a ndc10-2ts his3-200 leu2-3, 112 ura3-52 his4::GALCEN3URA3 | This study |
| KBY4006 | MAT a ade1 met14 leu2 his3 his4::GALCEN3URA3 chl4Δ::KANr rad52::LEU2 | This study |
| KBY4066 | MAT a ade1 met14 leu2 his3 his4::GALCEN3URA3 iml3Δ::KANr rad52::LEU2 | This study |
| KBY4067 | MAT a ade1 met14 leu2 his3 his4::GALCEN3URA3 mcm21Δ::KANr rad52::LEU2 | This study |
| KBY4012 | MATa ndc10-2ts his3-200 leu2-3, 112 ura3-52 his4::GALCEN3URA3 rad5Δ::LEU2 | This study |
| KBY4013 | MAT a ndc10-2ts his3-200 leu2-3, 112 ura3-52 Chl4–GFP KANr | This study |
| KBY4014 | MAT α lys2 trp1Δ ura3 leu2 Chl4–GFP KANr | This study |
| KBY4033 | MAT α ade2 ade3 leu2 trp1Δ ura3 lys2 chl4Δ::KANr | This study |
| KBY4039 | MAT α lys2 trp1Δ ura3 leu2 pCse4-GFP–TRP1 | This study |
| KBY4040 | MAT α lys2 trp1Δ ura3 leu2 Chl4–GFP HYGr Spc29–CFP KANr | This study |
| KBY4048 | MAT α ade2 ade3 leu2 trp1Δ ura3 lys2 LEU2GAL–UBI–CHL4 | This study |
| KBY4072 | MAT α ade2 ade3 leu2 trp1Δ ura3 lys2 LEU2GAL–UBI–CHL4 Spc29–CFP HYGr pKK1 | This study |
| pDLB2064 | pRS316/ADE3 | D. Lewc |
| pKK1 | pRS314/TRP1,Cse4–GFP fusion | R. Bakerd/M. Fitzgerald-Hayese |
| YipGALpUb | UBI4 LEU2 | E. Perkinsf |
| p315CHL4 | pRS315/CHL4 | V. Larionovg |
Cornell University, Ithaca, NY.
University of North Carolina at Chapel Hill.
Duke University, Durham, NC.
University of Massachusetts Medical School, Worcester, MA.
University of Massachusetts at Amherst, Amherst, MA.
University of Minnesota, Duluth, MN.
National Institutes of Health, Bethesda, MD.
Promoter exchange system for CHL4 (degron tagging)
The NH2 terminus of CHL4 was tagged with ubiquitin under the GAL1 promoter. The UBI4 gene adjacent to the GAL1 promoter was inserted into the yeast integrating plasmid Yiplac128 (YIpGALpUb). YIpGALpUb was used as a template to PCR across the plasmid with primers targeted to exchange the CHL4 promoter. The primer is designed to create an NH2-terminal arginine-tagged CHL4 with ubiquitin so that the half-life of the protein is considerably reduced. The inclusion of UBI4 constitutively targets the protein for degradation. High levels of expression on galactose maintain protein levels when grown on galactose and leads to rapid depletion when grown on glucose. The sequence of the ubiquitin tag is MQIFVKTLTGKTITLEVESSDTIDNVKSKIQDKEGIPPDQQRLIFAGKQLEDG-RTLSDYNIQKESTLHLVLRLRGG.
Analysis of dicentric chromosome breakage in chl4Δ
Genomic DNA was recovered from dicentric chromosome–containing cells (chl4Δ, wild type, and chl4Δ transformed with pRS315-CHL4) after induction of the dicentric chromosome on glucose for 3–9 h (Brock and Bloom, 1994). A PCR reaction for detecting the rearrangement product generated as a result of breakage was set up using primers to amplify the rearrangement product.
Mitotic stability assays
Cells transformed with centromere plasmids were plated on selective plates. Single colonies were grown overnight (unless otherwise indicated) in 5 ml of rich media (YPD or YPGalactose) at 30°C. 150–250 cells were plated after sonication on YPD or YPG and replica plated to media selective for the plasmid. The mitotic stability of the plasmids is expressed in percent as the number of colonies on selective plates/number of colonies on YPD or YPG × 100.
Colony color assays for mitotic stability
A colony color assay (Koshland et al., 1985) was employed to exclude selective bias for mitotically stable plasmids after the transformation experiment above. ade2 ade3 cells (CDV39) containing a plasmid bearing the ADE3 gene are red or red and white sectored. Plasmid loss is visualized as white colonies or increased white sectoring. CHL4 was deleted from cells harboring a CEN3/ADE3 minichromosome (pCH1023). Transformants were plated on rich media, without selection for the plasmid. 47.3% (98/207) of the colonies were red or red and white sectored, and 52.6%(109/207) of the colonies were white. The red chl4Δ colonies were stable (55–85%) upon outgrowth. Thus, heritable stable states persist for >20 generations after the deletion of CHL4. White colonies were found to contain unstable plasmids (>90% white colonies) or no plasmid, indicative of plasmid loss before plating.
Microscopy
Cells expressing Cse4–GFP, Chl4–GFP, Spc29–CFP, and GAL–UBI–Chl4–GFP were grown overnight to midlogarithmic growth phase in rich media. Cells were applied to microscope slides coated with 0.1% poly-l-lysine and imaged with Metamorph software (Universal Imaging Corp.) using a 100×/1.4NA Plan Apo objective on a Microphot FXA (Nikon) (Salmon et al., 1994). Images were collected with a cooled CCD camera (C4880; Hamamatsu). Five fluorescence images were acquired at 0.75-μm intervals through the cell and projected into a single reconstruction. For images at 37°C, cells were shifted from 24°C to media that was prewarmed to 37°C, allowed to grow for 2–5 h, as indicated in the text, and imaged as described.
ChIP
Cells were grown in YPG to an OD660 of 1.0, washed, transferred to YPD or YPG containing 15 μg/ml nocodazole, and reincubated for 4 h. The cells were then formaldehyde fixed and processed for ChIP analysis as described in Hetch and Grunstein (1999). PCR of nonimmunoprecipitated DNA and immunoprecipitated DNA was performed with primers to CEN3 (5′-gatcagcgccaaacaatatgg-3′ and 5′-attctacgtcaacgagtcct-3′), which produces a 490-bp fragment, primers to GALCEN3 (5′-gatcagcgccaaacaatatgg-3′ and 5′-cacgatgcgtccggcgtaga-3′), which produces a 350-bp fragment, and primers to LEU2 (5′-cgtcgttttgcaggtgacc-3′ and 5′-catttaggaccacccacagc-3′), which produces a 220-bp fragment. PCR products were separated on 1.5% agarose gels and visualized by ethidium bromide staining.
Acknowledgments
We thank E. Yeh, L. Topper, D. Beach, P. Maddox, C. Pearson, and J. Molk for critical reading and comments on the manuscript. We thank Isabelle Pot and P. Hieter for stimulating discussions and helpful comments on the manuscript. We thank all the other members of the Bloom lab for technical help. We thank Ed Perkins for the YIpGALpUb plasmid, Danny Lew for the pDLB2064 plasmid, and Tim Huffaker for the ndc10-2 strain.
This work was supported by National Institutes of Health grant GM32238 to K.S. Bloom.
Footnotes
Abbreviation used in this paper: ChIP, chromatin immunoprecipitation.
References
- Ault, J.G., and T.W. Lyttle. 1988. A transmissible dicentric chromosome in Drosophila melanogaster Chromosoma. 97:71–79. [Google Scholar]
- Bloom, K.S., and J. Carbon. 1982. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 29:305–317. [DOI] [PubMed] [Google Scholar]
- Brock, J.A., and K. Bloom. 1994. A chromosome breakage assay to monitor mitotic forces in budding yeast. J. Cell Sci. 107:891–902. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., M. Enquist-Newman, T. Muller-Reichert, D.G. Drubin, and G. Barnes. 2001. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 152:197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny, K.F., P.K. Sorger, A.A. Hyman, S. Tugendreich, F. Spencer, and P. Hieter. 1993. Identification of essential components of the S. cerevisiae kinetochore. Cell. 73:761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall, K., T. Olsson, B.M. Turner, G. Cranston, and R.C. Allshire. 1997. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 91:1021–1032. [DOI] [PubMed] [Google Scholar]
- Enquist-Newman, M., I.M. Cheeseman, D. Van Goor, D.G. Drubin, P.B. Meluh, and G. Barnes. 2001. Dad1p, third component of the duo1p/dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell. 12:2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk, M., J.H. Hegemann, and P. Philippsen. 1989. Chromatin digestion with restriction endonucleases reveals 150-160 bp of protected DNA in the centromere of chromosome XIV in Saccharomyces cerevisiae. Mol. Gen. Genet. 219:153–160. [DOI] [PubMed] [Google Scholar]
- Ghosh, S.K., A. Poddar, S. Hajra, K. Sanyal, and P. Sinha. 2001. The IML3/MCM19 gene of Saccharomyces cerevisiae is required for a kinetochore-related process during chromosome segregation. Mol. Genet. Genomics. 265:249–257. [DOI] [PubMed] [Google Scholar]
- Goshima, G., and M. Yanagida. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 100:619–633. [DOI] [PubMed] [Google Scholar]
- He, X., D.R. Rines, C.W. Espelin, and P.K. Sorger. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 106:195–206. [DOI] [PubMed] [Google Scholar]
- Hetch, A., and M. Grunstein. 1999. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction methods. Enzymology. 304:399–414. [DOI] [PubMed] [Google Scholar]
- Hill, A., and K. Bloom. 1987. Genetic manipulation of centromere function. Mol. Cell. Biol. 7:2397–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, C., I.M. Cheeseman, B.L. Goode, K.L. McDonald, G. Barnes, and D.G. Drubin. 1998. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 143:1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland, K.M., J. Kingsbury, D. Koshland, and P. Hieter. 1999. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 145:15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., J. Ortiz, T.U. Tanaka, J. Lechner, and E. Schiebel. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen, G.H., and R.C. Allshire. 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13:489–496. [DOI] [PubMed] [Google Scholar]
- Kopski, K.M., and T.C. Huffaker. 1997. Suppressors of the ndc10-2 mutation: a role for the ubiquitin system in Saccharomyces cerevisiae kinetochore function. Genetics. 147:409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland, D., J.C. Kent, and L.H. Hartwell. 1985. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 40:393–403. [DOI] [PubMed] [Google Scholar]
- Koshland, D., L. Rutledge, M. Fitzgerald-Hayes, and L.H. Hartwell. 1987. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell. 48:801–812. [DOI] [PubMed] [Google Scholar]
- Kouprina, N., A. Kirillov, E. Kroll, M. Koryabin, B. Shestopalov, V. Bannikov, V. Zakharyev, and V. Larionov. 1993. Identification and cloning of the CHL4 gene controlling chromosome segregation in yeast. Genetics. 135:327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine, G.T., P. Sinha, and B.-K. Tye. 1984. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 106:365–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday, V., D.W. Hailey, I. Pot, S.A. Givan, K.M. Hyland, G. Cagney, S. Fields, T.N. Davis, and P. Hieter. 2002. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 16:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., P. Yang, L. Glowczewski, D. Koshland, and M.M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 94:607–613. [DOI] [PubMed] [Google Scholar]
- Murray, A.W., and J.W. Szostack. 1985. Chromosome segregation in mitosis and meiosis. Annu. Rev. Cell Biol. 1:289–315. [DOI] [PubMed] [Google Scholar]
- Newlon, C.S. 1988. Yeast chromosome replication and segregation. Microbiol. Rev. 52:568–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, J., O. Stemmann, S. Rank, and J. Lechner. 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13:1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, C.G., P.S. Maddox, E.D. Salmon, and K. Bloom. 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152:1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, I., and F. Winston. 2000. Histone H2A is required for normal centromere function in Saccharomyces cerevisiae. EMBO J. 19:1598–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poddar, A., N. Roy, and P. Sinha. 1999. MCM21 and MCM22, two novel genes of the yeast Saccharomyces cerevisiae are required for chromosome transmission. Mol. Microbiol. 31:349–360. [DOI] [PubMed] [Google Scholar]
- Roy, N., A. Poddar, A. Lohia, and P. Sinha. 1997. The mcm17 mutation of yeast shows a size-dependent segregational defect of a mini-chromosome. Curr. Genet. 32:182–189. [DOI] [PubMed] [Google Scholar]
- Salmon, E.D., T. Inoue, A. Desai, and A.W. Murray. 1994. High resolution multimode digital imaging system for mitosis studies in vivo and in vitro. Biol. Bull. 187:231–232. [DOI] [PubMed] [Google Scholar]
- Sanyal, K., S.K. Ghosh, and P. Sinha. 1998. The MCM16 gene of the yeast Saccharomyces cerevisiae is required for chromosome segregation. Mol. Gen. Genet. 260:242–250. [DOI] [PubMed] [Google Scholar]
- Saunders, M., M. Fitzgerald-Hayes, and K. Bloom. 1988. Chromatin structure of altered yeast centromeres. Proc. Natl. Acad. Sci. USA. 85:175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, M.J., E. Yeh, M. Grunstein, and K. Bloom. 1990. Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol. Cell. Biol. 10:5721–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, J.A., A.A. Franco, M.A. Osley, and P.D. Kaufman. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16:85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, M.M., P. Yang, M.S. Santisteban, P.W. Boone, A.T. Goldstein, and P.C. Megee. 1996. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol. Cell. Biol. 16:1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, N.C., and L. Clarke. 1994. A novel epigenetic effect can alter centromere function in fission yeast. Cell. 79:865–874. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., M.P. Cosma, K. Wirth, and K. Nasmyth. 1999. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 98:847–858. [DOI] [PubMed] [Google Scholar]
- Tsuchiya, E., T. Hosotani, and T. Miyakawa. 1998. A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res. 26:3286–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast . 10:1793–1808. [DOI] [PubMed] [Google Scholar]