Abstract
We have analyzed the interactions between the signal recognition particle (SRP), the SRP receptor (SR), and the ribosome using GTPase assays, biosensor experiments, and ribosome binding assays. Possible mechanisms that could contribute to an enhanced affinity between the SR and the SRP–ribosome nascent chain complex to promote protein translocation under physiological ionic strength conditions have been explored. Ribosomes or 60S large ribosomal subunits activate the GTPase cycle of SRP54 and SRα by providing a platform for assembly of the SRP–SR complex. Biosensor experiments revealed high-affinity, saturable binding of ribosomes or large ribosomal subunits to the SR. Remarkably, the SR has a 100-fold higher affinity for the ribosome than for SRP. Proteoliposomes that contain the SR bind nontranslating ribosomes with an affinity comparable to that shown by the Sec61 complex. An NH2-terminal 319-residue segment of SRα is necessary and sufficient for binding of SR to the ribosome. We propose that the ribosome–SR interaction accelerates targeting of the ribosome nascent chain complex to the RER, while the SRP–SR interaction is crucial for maintaining the fidelity of the targeting reaction.
Keywords: protein translocation; signal recognition particle; ribosome; endoplasmic reticulum; biosensor
Introduction
Ribosomes synthesizing proteins with RER-specific signal sequences are selectively attached to protein translocation channels on the cytoplasmic surface of the RER by the combined action of the signal recognition particle (SRP) and the SRP receptor (SR) (for review see Walter and Johnson, 1994). High-affinity binding of the SRP54 subunit of SRP to the signal sequence as it emerges from the polypeptide exit site on the large ribosomal subunit is the initial sorting step that ultimately partitions ribosome nascent chain complexes (RNCs) between RER-bound and cytosolic polysome populations. Contact between the SRP–RNC complex and the SR initiates a cooperative GTPase cycle that is catalyzed by SRP54 and the SRα subunit of the SR (Connolly and Gilmore, 1993; Miller et al., 1993; Rapiejko and Gilmore, 1997). Prior to complex formation, the GTP binding sites in SRP54 and SRα exist in an “empty site” conformation that is characterized by a low nucleotide affinity with rapid and reversible binding of GTP, GDP, or Gpp(NH)p (Rapiejko and Gilmore, 1994, 1997). Cooperative binding of GTP to SRP54 and SRα is followed by the transfer of the RNC to an unoccupied protein translocation channel (Song et al., 2000). Eukaryotic translocation channels are oligomeric assemblies of Sec61 heterotrimers (Görlich and Rapoport, 1993; Beckmann et al., 2001) that incorporate additional accessory proteins (Menetret et al., 2000). The specific binding interactions that are responsible for rapid, yet selective, targeting of an SRP–RNC to the SR are not fully understood. The binding affinity between SRP and the SR is surprisingly low (Kd ≈ 125 nM) in a physiological ionic strength buffer (Connolly and Gilmore, 1993), yet targeting of the SRP–RNC complex to the SR is efficient. It has been proposed that the ribosome acts as a classical guanine nucleotide exchange factor (GEF) for SRP54 (Bacher et al., 1996), resulting in an enhanced affinity between the SR and a GTP-bound form of the SRP–RNC complex. However, SRP–RNCs target to SR proteoliposomes in the absence of GTP (Song et al., 2000), indicating that the translocon, as well as GTP, is dispensable for the targeting reaction. Here we have analyzed the interactions between SRP, the SR, and the ribosome to determine how the RNC promotes the interaction between SRP and the SR. Our results suggest a novel model for the targeting reaction that involves simultaneous recognition of the ribosome and the SRP by the SR. We propose that the ribosome–SR interaction accelerates the rate of RNC targeting, while the SRP–SR interaction is essential for the fidelity of RNC targeting to the RER.
Results
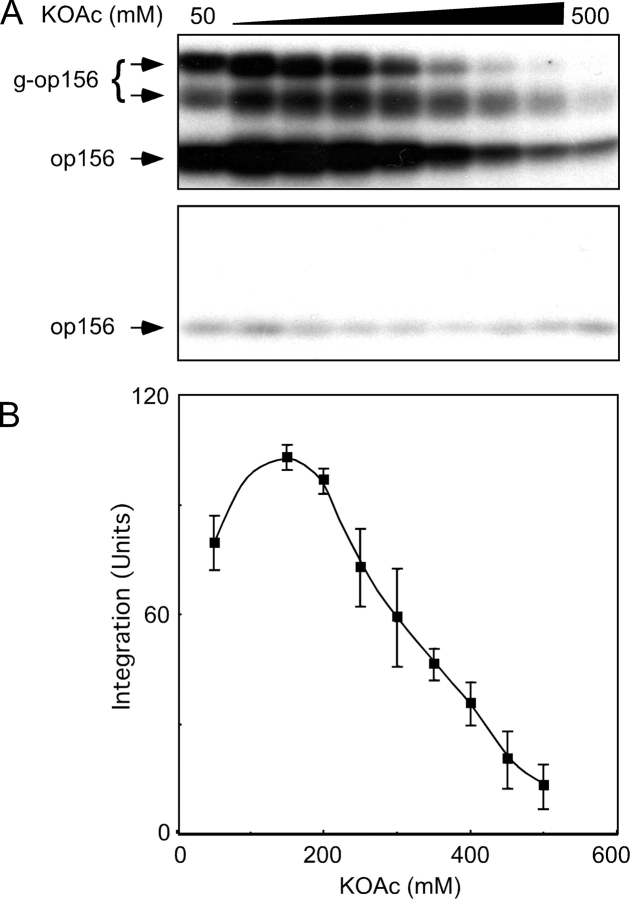
Selective binding of an RNC to the translocon is dependent upon the interaction between two ribonucleoproteins (SRP and the ribosome) and two RNP receptors (SR and the Sec61 complex). The interaction between SRP and the SR is remarkably sensitive to a physiological concentration of monovalent cations (Connolly and Gilmore, 1993). Consequently, one might predict that the targeting phase of the protein translocation reaction would be inhibited by modest increases in ionic strength. To test this prediction, SRP–RNC complexes bearing a nascent opsin polypeptide (op156) were assembled by in vitro translation of a truncated opsin mRNA transcript lacking a termination codon. The SRP–RNC complexes were incubated with microsomes in buffers containing 50 mM stepwise increases in KOAc concentration (Fig. 1 A). Integration of op156 into the membranes was detected by the decreased gel mobility that accompanies transfer of N-linked glycans onto one or both of the glycosylation sites that precede the first TM span of opsin to yield glycosylated op156 (g-op156; Fig. 1 A, top). In the absence of GTP (not depicted, but see Rapiejko and Gilmore, 1992), or when membranes lack intact SRα (C1PK-RM), we observe greatly reduced quantities of membrane-integrated op156 and no detectable g-op156 (Fig. 1 A, bottom). SRP-dependent integration of g-op156 is surprisingly insensitive to increased ionic strength, with ∼50% inhibition observed at 300 mM KOAc (Fig. 1 B).
Figure 1.
Inhibition of translocation activity by high ionic strength. (A) SRP–ribosome–op156 complexes were adjusted to final KOAc concentrations ranging between 50 and 500 mM and incubated for 40 min at 25°C with 1.2 eq (as defined in Walter et al., 1981) of PK-RM (top) or 1.2 eq of protease-digested PK-RM (C1PK-RM) that lack intact SRα (bottom). After membrane-integrated forms of op156 were separated from nonintegrated op156 on alkaline sucrose gradients, the membrane pellets were analyzed by PAGE in SDS to resolve glycosylated (g-op156) and nonglycosylated (op156) polypeptides. (B) Membrane-integrated g-op156 was quantified with a phosphorimager and is expressed in units, where 100 U corresponds to the average of the 150 mM and 200 mM KOAc data points. The average and standard deviation were calculated using data from six experiments, one of which is shown in A.
Ribosomes stimulate the GTPase activity of the SRP–SR complex
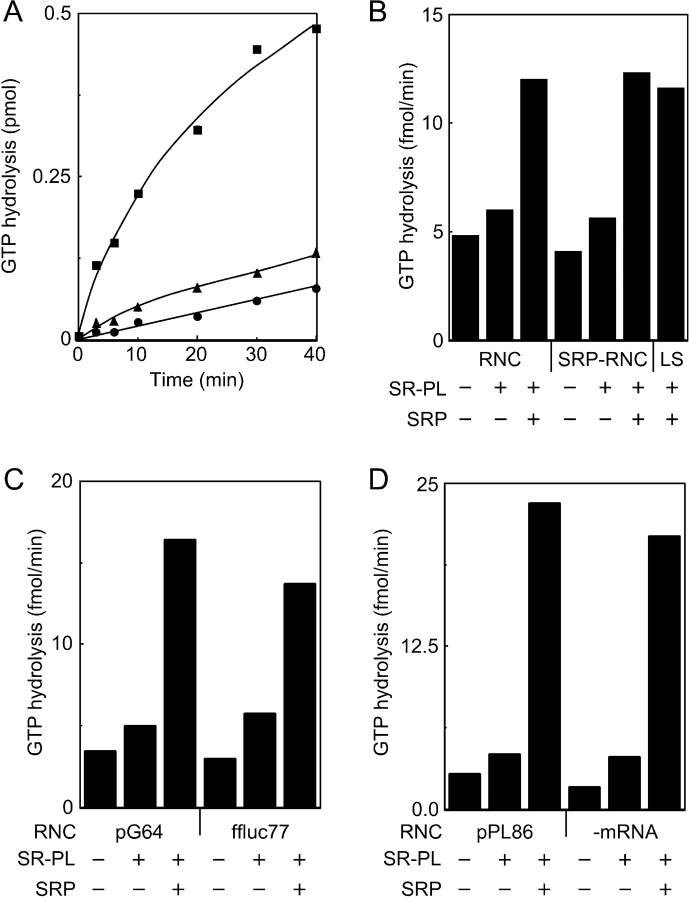
We used a previously described GTPase assay (Bacher et al., 1996, 1999) to test whether RNCs promote the interaction between SRP and the SR. The GTPase activity of the SRP–SR complex is readily detected when equimolar amounts of purified SRP, SR proteoliposomes, and in vitro–assembled RNCs are combined in an assay buffer that contains a physiological concentration (150 mM K+) of monovalent cations. RNCs that were assembled by translating a truncated mRNA that encodes the NH2-terminal 86 residues of preprolactin (pPL86) were separated from cytosolic GTPases by centrifugation through a high salt–sucrose step gradient. In agreement with previous reports (Bacher et al., 1996, 1999), the GTPase activity of the pPL86 RNCs was increased slightly by the addition of SR proteoliposomes and markedly by the addition of SRP plus the SR proteoliposomes (Fig. 2 A). Assays that lacked the pPL86 RNCs confirmed the low basal GTPase activity of the SRP–SR complex in a physiological ionic strength buffer (not depicted in Fig. 2 A, but see Fig. 3 A).
Figure 2.
Stimulation of the GTPase activity of the SRP–SR complex by RNC preparations. (A) GTPase assays were conducted in a physiological ionic strength buffer (buffer C, 150 mM K+) and contained pPL86 RNCs (all symbols) and were supplemented with 50 nM SR proteoliposomes (triangles and squares) and 50 nM SRP (squares). (B) RNCs and SRP–RNCs were assembled by translation of pPL86 mRNA in the absence or presence of SRP. The GTPase assays in buffer C (RNCs and SRP–RNCs) were supplemented with 25 nM SR proteoliposomes and 50 nM SRP as indicated. The assay designated LS lacked RNCs and was conducted in a low ionic strength buffer (buffer E, 50 mM K+). (C and D) GTPase assays in buffer C contained pG64 RNCs (C), ffluc77 RNCs (C), pPL86 RNCs (D), or mock-assembled RNCs (D) and were supplemented with 50 nM SR proteoliposomes and 50 nM SRP as indicated. (B–D) The GTPase activity was calculated using 0-, 3-, 6-, and 10-min time points.
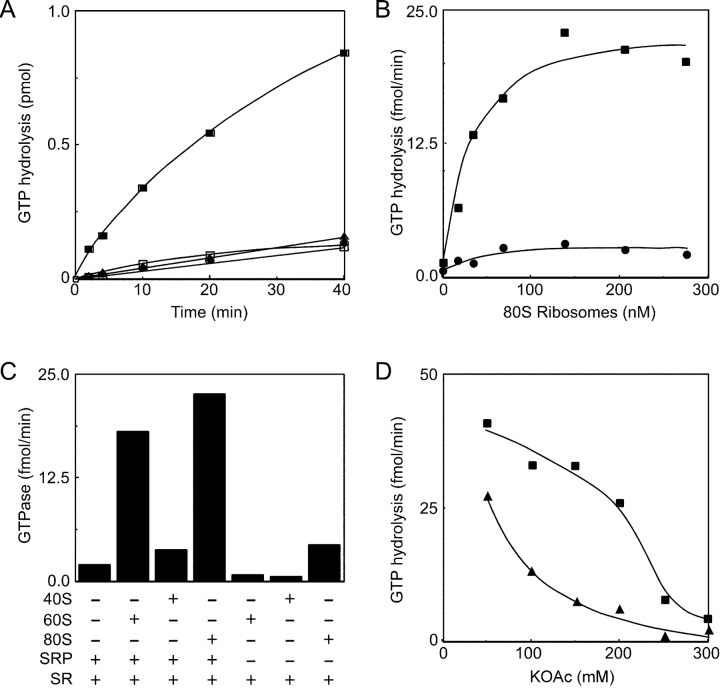
Figure 3.
80S ribosomes and 60S subunits stimulate the GTPase activity of the SRP–SR complex. (A–C) GTPase assays in buffer C (150 mM K+) were adjusted to 0.1% Nikkol. (A) GTPase assays either lacked (open squares) or contained pG64 RNCs (filled symbols) and were further supplemented with 50 nM SR (triangles and squares) and 50 nM SRP (squares). (B) GTPase assays contained 50 nM SR (circles) or 50 nM SR plus 50 nM SRP (squares), and the indicated concentration of 80S ribosomes. (C) GTPase assays contained 50 nM SR and were supplemented with 50 nM SRP, 160 nM 80S ribosomes, 120 nM 60S ribosomal subunits, or 220 nM 40S ribosomal subunits as indicated. (D) The KOAc concentration in buffer C (adjusted to 0.1% Nikkol) varied between 50 and 300 mM. The GTPase assays of 50 nM SRP and 50 nM SR were conducted in the presence (squares) or absence (triangles) of 160 nM 80S ribosomes. (B–D) The initial GTPase rates are based on 0-, 3-, 6-, and 10-min time points. GTPase rates for assays that contained an identical concentration of ribosomes or ribosomal subunits, but lacked both SRP and SR, were subtracted as background for each data point.
The robust GTPase activity observed in Fig. 2 A appears to contradict our previous conclusion that GTP hydrolysis by the SRP–RNC–SR complex is blocked in the absence of the Sec61 complex (Song et al., 2000). To address this discrepancy, we assembled pPL86 RNCs in the absence or presence of SRP. Because SRP does not dissociate from the signal sequence of RNC complexes that are isolated by centrifugation through high salt–sucrose gradients (Powers and Walter, 1996; Raden and Gilmore, 1998), we can test whether these preassembled SRP–RNC complexes obviate the requirement for additional SRP in the GTPase assay. Very similar GTPase rates were obtained when the two RNC preparations were assayed in the presence of the SR proteoliposomes, demonstrating that the prebound SRP does not satisfy the SRP requirement for GTP hydrolysis (Fig. 2 B). The addition of free SRP plus the SR proteoliposomes to both RNC preparations resulted in a GTPase rate that was equivalent to that observed when the SR proteoliposomes were incubated with SRP in a hypotonic buffer that promotes formation of the SRP–SR complex (Fig. 2 B).
The preceding experiment suggested that RNC-bound SRP was not the active component in the GTPase assay. To address this possibility, RNCs were assembled by translation of truncated mRNAs that encode the NH2-terminal 64 residues of the G protein of vesicular stomatitis virus (pG64) and the NH2-terminal 77 residues of firefly luciferase (ffluc77). Unlike pPL86 or pG64, ffluc77 lacks a signal sequence for protein translocation across the endoplasmic reticulum; hence, SRP does not bind to ffluc77 RNCs. The GTPase activity of the SRP–SR complex was stimulated by both RNC preparations (Fig. 2 C). Less than 20% of the ribosomes in a wheat germ translation reaction are assembled into RNC complexes, so the RNC preparations obtained by centrifugation contain a mixture of RNCs and nontranslating ribosomes. The GTPase assays were conducted using mock RNCs that were prepared from a translation reaction that lacked mRNA (Fig. 2 D). The GTPase cycle of SRP–SR complex was stimulated by mock RNCs, suggesting that nontranslating ribosomes and free SRP are the active components in the GTPase assay.
A significant fraction of the SR in a proteoliposome faces the liposome interior and is inaccessible to the SRP and the RNCs. As observed previously (Connolly and Gilmore, 1993), the GTPase activity of the SRP and SR is low when assayed in a physiological ionic strength buffer containing detergent micelles (Fig. 3 A, open squares). The GTPase activity was stimulated roughly eightfold by the addition of pG64 RNCs (Fig. 3 A, filled squares). Deletion of SRP (Fig. 3 A, triangles) or the SR (not depicted) reduced the GTPase activity to that shown by the RNC preparation alone (circles). The GTPase activity in assays containing detergent micelles (Fig. 3 A) was higher than in assays containing the SR proteoliposomes (Fig. 2 A). Subsequent experiments used the detergent micelle assay because the GTPase activity was not influenced by experimental variations in the efficiency of proteoliposome formation or in the asymmetry of SR reconstitution.
SRP binds to nontranslating 80S ribosomes (Walter et al., 1981; Powers and Walter, 1996) in addition to polysomes synthesizing secretory proteins (Walter et al., 1981). Although earlier, nonequilibrium methods indicated that SRP binds the ribosome with a relatively low affinity (Kd ≈ 50 μM; Walter et al., 1981), a recent analysis indicates that the binding affinity is substantially higher (Kd ≈ 8 nM; Flanagan et al., 2003). Purified 80S ribosomes were added to the GTPase assays of SRP and the SR to determine whether the ribosome is the active component in the RNC preparation (Fig. 3 B, squares). Notably, half-maximal stimulation of the GTPase activity was achieved when the concentration of ribosomes exceeded the concentration of the SRP and the SR. A saturable, but much lower, stimulation of GTP hydrolysis was observed in the absence of SRP (Fig. 3 B, circles). The binding site for SRP54 on the ribosome has been mapped to ribosomal proteins L23a and L35 (Pool et al., 2002), which are located in the vicinity of the polypeptide exit site on the large ribosomal subunit (Ban et al., 2000). If the ribosome stimulates the GTPase activity of the SRP–SR complex in a specific manner, one would predict that the stimulatory activity would reside on the large ribosomal subunit. Indeed, the isolated 60S subunits were almost as effective as the intact ribosome (Fig. 3 C). In contrast, 40S ribosomal subunits were comparatively ineffective even when present at a higher concentration. Neither 40S nor 60S subunits stimulated the GTPase activity of the SR in assays that were not supplemented with SRP (Fig. 3 C).
Two classes of mechanisms could explain how the ribosome could accelerate GTP hydrolysis by SRP and the SR. The addition of ribosomes could promote formation of hydrolytically active SRP–SR complexes, or the ribosome could accelerate a rate-limiting step in the hydrolysis cycle without affecting the equilibrium between SRP, the SR, and the SRP–SR complex. For example, a significant increase in the binding affinity of SRα for GTP would increase hydrolysis because the GTPase cycle is dependent upon ribonucleotide binding to both SRP54 and SRα (Powers and Walter, 1995). Of the two GTPases (SRα and SRP54), SRα has a lower affinity for GTP, hence the Km for GTP hydrolysis provides an accurate estimate of the Kd for SRα. The Km for GTP was determined in SRP–SR GTPase assays that contained or lacked 80S ribosomes. In the absence of ribosomes, the Km for GTP was 2.4 ± 1.0 μM (not depicted), which is in good agreement with a previous determination (Connolly and Gilmore, 1993). The Km for GTP was not significantly altered by the addition of ribosomes (Km = 1.4 ± 0.3 μM; not depicted), indicating that the ribosome does not stimulate the GTPase reaction by altering the nucleotide binding properties of SRα.
The interaction between the SRP and nontranslating ribosomes is reduced by increased ionic strength (Walter and Blobel, 1983; Powers and Walter, 1996). The GTPase activity of the putative ternary complex (SR–SRP–80S ribosome) was significantly less salt sensitive than the GTPase activity of the binary SRP–SR complex (Fig. 3 D), suggesting that the ribosome stabilizes the interaction between the SRP and the SR.
GTPase activities of the SR and the SR subunits
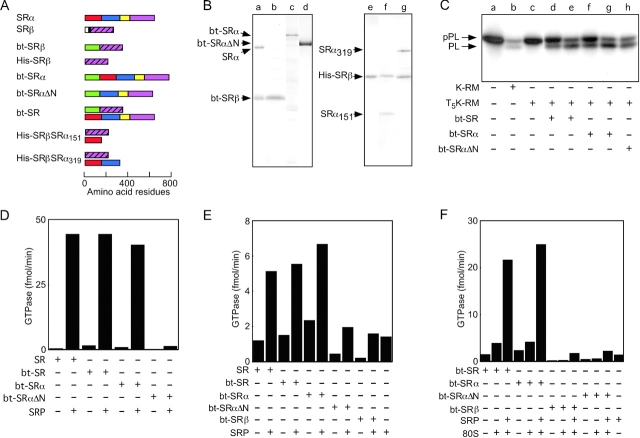
The subunits of the SR (Fig. 4 A) were expressed in Escherichia coli to investigate the mechanism of the ribosome-stimulated GTPase activity. To facilitate purification of SRβ, the lumenal and transmembrane domains of SRβ were replaced with a 13-kD domain that is biotinylated in vivo to obtain bt-SRβ (Fig. 4 B, lane b) or a hexahistidine sequence to obtain His-SRβ (Fig. 4 B, lane e). Biotinylation domain fusion constructs were also used to express bt-SRα and bt-SRαΔN (Fig. 4 B, lanes c and d). SRαΔN lacks the NH2-terminal 151 residues of SRα and corresponds to the COOH-terminal fragment of SRα that can be produced by limited digestion of microsomes with elastase (Meyer and Dobberstein, 1980; Lauffer et al., 1985). Coexpression of bt-SRβ and SRα allowed the isolation of bt-SR (Fig. 4 B, lane a). Coexpression of His-SRβ with NH2-terminal fragments of SRα (SRα151 or SRα319) allowed purification of His-SRβSRα151 (Fig. 4 B, lane f) and His-SRβSRα319 (Fig. 4 B, lane g). In vivo formation of the SR heterodimers (bt-SR, His-SRβSRα151, and His-SRβSRα319) provides evidence that SRβ and the SRX domain of SRα are correctly folded.
Figure 4.
Characterization of recombinant SR and SR subunit derivatives. (A) Diagrams of SRα, SRβ, and biotin and hexahistidine-tagged derivatives of SRα and SRβ. Segments of SRα are designated as follows: residues 1–151, red; residues 152–319, blue; N domain, yellow; G domain, magenta. Segments of SRβ are designated as follows: lumenal domain, white; TM span, black; G domain, magenta with diagonal hatching. The 13-kD biotinylation domain (bt) is designated by the green bar at the NH2 terminus of the expression constructs. (B) Coomassie blue–stained gel lanes of (a) bt-SR (bt-SRβ + SRα), (b) bt-SRβ, (c) bt-SRα, (d) bt-SRαΔN, (e) His-SRβ, (f) His-SRβSRα151, and (g) His-SRβSRα319. Individual gel lanes were aligned relative to molecular mass markers. (C) Reconstitution of protein translocation activity of protease-digested microsomes (T5K-RM) with bt-SR (d and e), bt-SRα (f and g), or bt-SRαΔN (h). The 12.5-μl reticulocyte lysate translation reactions were supplemented with either 0.5 eq of K-RM (lane b) or 0.5 eq of T5K-RM (lanes c–h). The concentrations of the E. coli–expressed SR constructs was either 24 nM (lanes d, f, and h) or 72 nM (lanes e and g). pPL and prolactin (PL) were resolved by PAGE in SDS. (D) GTPase assays (1 μM GTP) of canine SR, bt-SR, bt-SRα, and bt-SRαΔN in buffer E (50 mM K+) were supplemented with 50 nM SRP as noted in the chart below the graphs. The concentration of the SR or SR subunits was 15 nM. (E and F) GTPase assays of canine SR, bt-SR, bt-SRα, bt-SRβ, and bt-SRαΔN in buffer C (150 mM K+) were supplemented with 50 nM SRP and/or 160 nM 80S ribosomes as noted in the chart below the graphs. The concentration of the SR or SR subunits was 40 nM.
Limited proteolysis of rough microsomes (RMs) with trypsin cleaves SRα near residue 150 to liberate a COOH-terminal fragment that comigrates with SRαΔN. The translocation activity of the trypsin-inactivated microsomes can be restored by adding purified SRαΔN (Meyer and Dobberstein, 1980), in vitro–translated SRα (Andrews et al., 1989), or the recombinant SR heterodimer (Fulga et al., 2001). Translocation of pPL into the lumen of the undigested microsomes (K-RM) is accompanied by signal sequence cleavage to yield prolactin (PL; Fig. 4 C, lane b). Translocated prolactin was not observed in the absence of microsomes (Fig. 4 C, lane a) or when the protease-digested microsomes (T5K-RM) were tested (lane c). The translocation activity of the SRα-deficient microsomes could be reconstituted with bt-SR (Fig. 4 C, lanes d and e), bt-SRα (lanes f and g), or bt-SRαΔN (lane h). These results provide additional evidence that the recombinant proteins are folded and functional.
The GTPase activity of the SR purified from canine pancreas was compared with the recombinant proteins using either a hypotonic (50 mM K+) assay buffer to maximize the interaction between SRP and the SR (Fig. 4 D) or a physiological ionic strength (150 mM K+) buffer (Fig. 4 E). The E. coli–expressed proteins and the canine SR have barely detectable GTPase activities in the absence of SRP (Fig. 4, D and E). Likewise, SRP has a very low intrinsic GTPase activity (Fig. 4 E). When assayed using the low ionic strength assay buffer (50 mM K+), the bt-SR and bt-SRα form complexes with SRP that hydrolyze GTP at a rate that is comparable to the SR purified from canine pancreas (Fig. 4 D). Consistent with Fig. 3 D, the GTPase activity of complexes between SRP and the SR, or SRα, was reduced in the physiological ionic strength buffer (Fig. 4 E). SRβ did not hydrolyze GTP at a significant rate in the absence or presence of SRP (Fig. 4 E). The GTPase activities for SRP plus bt-SRαΔN showed an additive, rather than synergistic, response, indicating that active complexes were not formed between SRP and the COOH-terminal fragment of SRα (Fig. 4 E).
The E. coli–expressed SR and SR subunits were assayed for GTPase activity in the presence of 80S ribosomes (Fig. 4 F). The ability of 80S ribosomes to activate the GTPase activity of the SRP–SR complex was confirmed using the E. coli–expressed bt-SR and bt-SRα. Purified ribosomes did not stimulate GTP hydrolysis by bt-SRβ or bt-SRαΔN in the presence or absence of SRP. Assays of His-SRβ, His-SRβSRα151, and His-SRβSRα319 yielded results that were similar to bt-SRβ (unpublished data).
The rate of SRP-SR complex formation is not accelerated by GTP
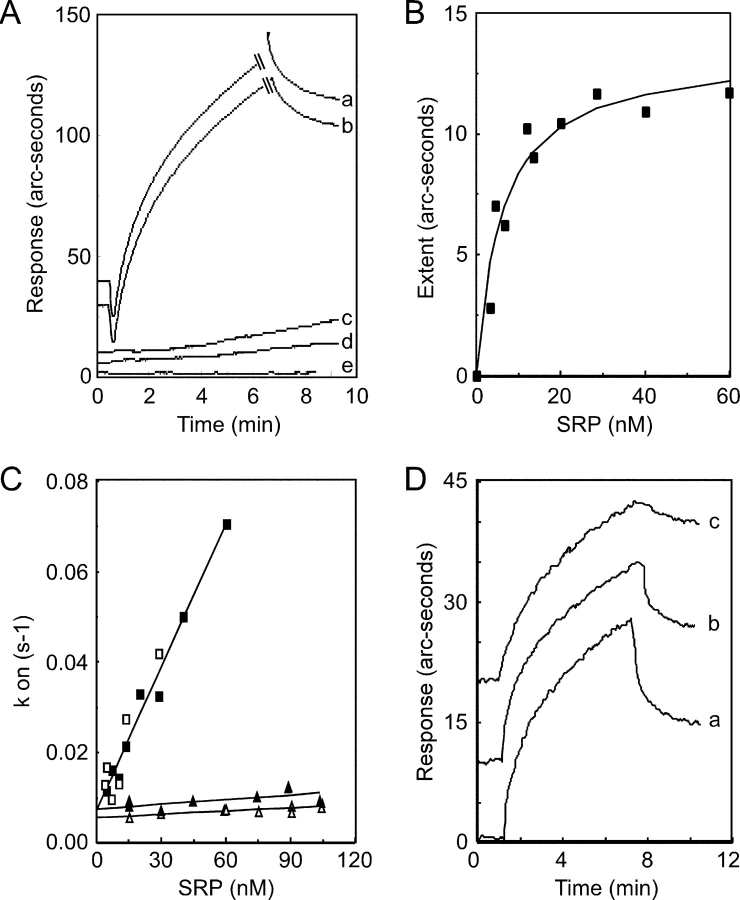
Having established that the E. coli–expressed proteins are functional by several criteria, we investigated the kinetics of SRP–SR complex formation using the IAsys optical biosensor. The immobilization strategy for the SR or the SR subunits was to coat a biotin-modified biosensor cuvette with streptavidin. After removing unbound streptavidin, the sensor surface was completed by the addition of bt-SR, bt-SRα, bt-SRβ, or bt-SRαΔN. Binding of SRP to the SR or the SR subunits was initially analyzed in a hypotonic assay buffer (50 mM K+) in the absence of GTP (Fig. 5 A). SRP binds to biosensor cuvettes containing immobilized bt-SR (Fig. 5 A, a) and bt-SRα (b). SRP did not bind to bt-SRαΔN (Fig. 5 A, c), bt-SRβ (d), or to cuvettes that contained streptavidin alone (e). The dissociation of bound SRP was monitored when applicable (Fig. 5 A, a and b). The kinetics of SRP binding to the SR was analyzed by linear regression analysis of the association curves to determine the change in refractive index caused by SRP binding (extent) and to determine the initial rate of SRP binding (kon). Hyperbolic saturation curves for binding of SRP to the SR were obtained (Fig. 5 B). The Kd value derived from the saturation curve for SRP binding to the SR in the presence of GTP is 7.6 nM, which is in reasonable agreement with the value of 15 nM that was estimated using a GTPase assay (Connolly and Gilmore, 1993). Plots of kon versus SRP concentration were linear (Fig. 5 C, filled squares). The slope and y intercept of the kon plot correspond to the rate constants for association (kass) and dissociation (kdiss), respectively, and yield a Kd value of 6.5 nM (Table I). Assay points obtained in the absence of GTP (Fig. 5 C, open squares) were adequately fit by very similar kinetic parameters (Table I). The binding kinetics of SRP to the SR was also examined in a physiological ionic strength buffer (150 mM K+) in the presence or absence of GTP (Fig. 5 C, filled and open triangles, respectively). The increase in ionic strength dramatically reduces the rate constant for complex formation, without significantly altering the rate of dissociation. The association rates for complexes formed in the absence of GTP were not significantly different from association rates obtained in the presence of GTP (Table I). The rates of dissociation were likewise not significantly influenced by the guanine nucleotide. The observation that GTP does not decrease the apparent rate constant for complex dissociation is explained by the fact that the rate of GTP hydrolysis (kcat = 3.5 × 10−2 s−1) is more rapid than kdiss for assays conducted in the absence or presence of GTP. Consequently, GTP hydrolysis is not the rate-limiting step in the dissociation reaction. The calculated Kd values for the SRP–SR complex in a physiological ionic strength buffer are not significantly different from each other (Table I) and are in reasonable agreement with the Kd value (Kd + GTP = 125 nM) estimated using the GTPase assay (Connolly and Gilmore, 1993).
Figure 5.
Kinetics of SRP binding to immobilized SR. (A) Association (a–e) and dissociation (a and b) curves for binding of SRP to the following proteins: (a) bt-SR, (b) bt-SRα, (c) bt-SRαΔN, (d) bt-SRβ, and (e) streptavidin. The association curves have been offset on the vertical axis for clarity. The biosensor cuvettes were equilibrated in buffer E (50 mM K+). Association curves were initiated at 0.5 min by the addition 22 nM SRP in the equilibration buffer, and data were collected for at least 6.5 min. Association curves for bt-SR and bt-SRα were truncated to show the dissociation curves. Dissociation curves for bt-SRα and bt-SR were initiated by replacing the SRP solution with equilibration buffer. (B) Equilibrium binding of SRP to immobilized bt-SR in buffer E containing 25 μM GTP. (C) Plots of kon for SRP binding to bt-SR in the presence (solid symbols) or absence (open symbols) of 25 μM GTP. The binding experiments were conducted in buffer D (150 mM K+, triangles) or buffer E (squares). (D) Association and dissociation curves for binding of SRP to bt-SR in the presence of buffer E adjusted to (a) 25 μM GTP, (b) 25 μM GDP, or (c) 25 μM Gpp(NH)p. The curves have been offset on the vertical axis for clarity. Association curves were initiated at 1 min by the addition 60 nM SRP in the equilibration buffer, and data were collected for at least 6 min. Dissociation curves were initiated by replacing the SRP solution with equilibration buffer containing the appropriate nucleotide.
Table I. Kinetic parameters for the SRP–SR complex and the ribosome–SR complex.
| Ligate | KOAc | GTP | kass | kdiss | Kd |
|---|---|---|---|---|---|
| mM | μM | M−1s−1 | s−1 | nM | |
| SRP | 50 | 25 | 1.1 ± 0.1 × 106 | 6.9 ± 1.8 × 10−3 | 6.5 |
| SRP | 50 | 0 | 1.2 ± 0.3 × 106 | 6.5 ± 3.5 × 10−3 | 5.3 |
| SRP | 150 | 25 | 4.8 ± 1.8 × 104 | 6.7 ± 0.5 × 10−3 | 140 |
| SRP | 150 | 0 | 2.9 ± 0.7 × 104 | 5.0 ± 0.5 × 10−3 | 175 |
| 80S | 150 | 0 | 1.4 ± 0.2 × 107 | 1.2 ± 0.2 × 10−2 | 0.9 |
| 60S | 150 | 0 | 1.9 ± 0.2 × 107 | 2.0 ± 0.5 × 10−2 | 1.1 |
The nonhydrolyzable GTP analogue Gpp(NH)p has been used to stabilize the interaction between SRP and the SR (Connolly et al., 1991; Rapiejko and Gilmore, 1992). Extensive biosensor experiments conducted in the presence of the Gpp(NH)p are not feasible because the biosensor surface cannot be completely regenerated between data points by the high salt wash procedure used to dissociate the SRP–SR complex. However, several single point experiments were conducted to compare the dissociation of SRP from the SR in the presence of GTP, GDP, or Gpp(NH)p. Dissociation of SRP from the SR was greatly reduced when GTP hydrolysis was prevented (Fig. 5 D, compare curves a and b with c).
Binding of ribosomes to the SR
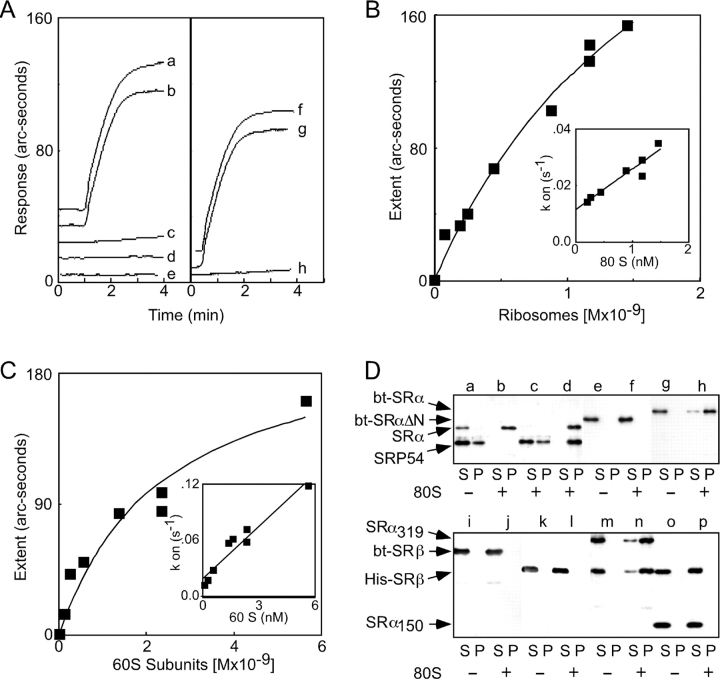
A ternary complex between the ribosome, the SRP, and the SR might involve direct contact between the ribosome and the SR. Possible interactions between the ribosome and the SR or the SR subunits were explored using the biosensor (Fig. 6 A). Ribosomes bind to immobilized bt-SR (Fig. 6 A, a) and bt-SRα (b) but not to bt-SRβ (c) or to bt-SRαΔN (e) in the physiological ionic strength buffer (150 mM K+). The addition of GTP did not increase binding of ribosomes to SRβ (Fig. 6 A, d). Large ribosomal subunits (Fig. 6 A, g), but not small subunits (h), recapitulate the binding to the SR that is observed for the intact ribosome (f). Hyperbolic binding curves for the interaction between the ribosome and immobilized SR (Fig. 6 B) yielded a Kd value of 1.9 ± 0.6 nM. The rate constants for the association (kass) and dissociation (kdiss) reactions were calculated from the kon plot (Fig. 6 B, inset) and are shown in Table I. Similar kinetic parameters were obtained for binding of the 60S subunit to bt-SR (Fig. 6 C and Table I).
Figure 6.
Kinetics of ribosome binding to immobilized SR. (A–C) Binding of ribosomes or ribosomal subunits to the SR in buffer D (150 mM K+). (A) Association curves for binding of 1 nM 80S ribosomes to the following immobilized proteins: (a) bt-SR, (b) bt-SRα, (c) bt-SRβ, (d) bt-SRβ + 25 μM GTP, or (e) bt-SRαΔN. Binding of (f) 1 nM 80S ribosomes, (g) 1.7 nM 60S subunits, or (h) 2.4 nM 40S subunits to immobilized bt-SR. Ribosomes or ribosomal subunits were added at the 1-min time point. (B and C) Equilibrium binding of ribosomes (B) or 60S ribosomal subunits (C) to immobilized bt-SR. The insets show plots of kon versus ligate concentration. (D) Cosedimentation of SR, SR subunits, and SRP with 80S ribosomes. Binding assays in buffer D were incubated for 5 min at 25°C and contained 50 nM SRP and/or 40 nM SR (or SR subunits) and 160 nM ribosomes as indicated. Individual assays contained the following proteins: (a) SRP plus bt-SR, (b) bt-SR, (c) SRP, (d) SRP plus bt-SR, (e and f) bt-SRαΔN, (g and h) bt-SRα, (i and j) bt-SRβ, (k and l) His-SRβ, (m and n) His-SRβSRα319, or (o and p) His-SRβSRα151. The assays were layered onto 50-μl cushions of 500 mM sucrose in buffer D and centrifuged for 10 min at 279,000 g av using a TLA100 rotor. After centrifugation, the samples were separated into supernatant (S) and pellet (P) fractions and resolved by PAGE in SDS, and the SRP and SR were detected using antibodies to SRP54, SRα, or SRβ. Images from sequential immunoblots using polyclonal and monoclonal antibodies were combined in the bottom panel.
Analyzing the kinetics of ternary complex formation using the biosensor was not feasible because the ligate solution (SRP plus 80S ribosomes) would consist of a mixture of ribosomes, SRP, and SRP–ribosome complexes, each of which would bind to the SR with different kinetics. Instead, complexes between the ribosome, SRP, and the SR were detected using a cosedimentation assay (Fig. 6 D, top). The concentrations of ribosomes, SRP, and the SR were identical to those used in the GTPase assays to ensure that high-affinity interactions were monitored. As detected using antibodies to SRP54 and SRα, the majority of the SRP and the SR were recovered in the supernatant (S) fraction when ribosomes were not present (Fig. 6 D, a). Binary complexes between the SR and the ribosome or SRP and the ribosome were recovered in the pellet (P) fractions (Fig. 6 D, b and c). Formation of the binary SRP–ribosome complex was less efficient than formation of the SR–ribosome complex, consistent with the 10-fold difference in binding affinities. When all three components were present, both the SRP and the SR were exclusively recovered in the pellet fraction, thereby providing strong evidence for the formation of a ternary complex between the ribosome, SRP, and the SR (Fig. 6 D, d).
The cosedimentation assay provided a facile method to map the ribosome-binding domain in the SR (Fig. 6 D). The importance of the NH2-terminal 151 residues of SRα was confirmed using the cosedimentation assay, as bt-SRα binds to ribosomes (Fig. 6 D, g and h) but bt-SRαΔN does not (e and f). Neither tagged derivative of SRβ cosedimented with 80S ribosomes (Fig. 6 D, i–l). The NH2-terminal 151 residues of SRα are not sufficient for ribosome-binding activity even when coexpressed with His-SRβ (Fig. 6 D, o and p). In contrast, His-SRβSRα319 was recovered in the pellet fraction when ribosomes were included (Fig. 6 D, m and n).
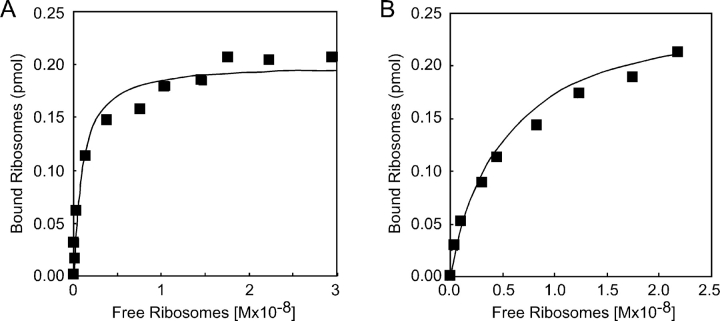
The conventional assay used to characterize the ribosome-binding activity of purified ER membrane proteins monitors binding of radiolabeled ribosomes to proteoliposomes (Kalies et al., 1994; Raden et al., 2000, and references therein). The purified canine SR was reconstituted into proteoliposomes and incubated with various quantities of 125I-labeled ribosomes in a physiological ionic strength buffer (150 mM K+). Proteoliposome-bound and unbound ribosomes were separated by gel filtration chromatography to obtain a saturation curve (Fig. 7 A). The apparent Kd value obtained in this experiment (0.87 ± 0.02 nM) is in good agreement with the Kd value obtained using the biosensor. More importantly, the stoichiometry of binding was found to be roughly 1:1 based upon the experimentally determined Bmax value and the concentration of SR in the proteoliposomes. Binding of 80S ribosomes to Sec61 proteoliposomes was analyzed as a control (Fig. 7 B), and the Kd value we obtained (5.4 ± 0.9 nM) was in good agreement with the previous literature (Kalies et al., 1994).
Figure 7.
Binding of ribosomes to SR and Sec61 proteoliposomes. Increasing amounts of 125I-labeled 80S ribosomes (0.03–0.65 pmol) were incubated with the SR proteoliposomes (A) or Sec61 proteoliposomes (B) in 50 mM TEA, 150 mM KOAc, 2.5 mM Mg(OAc)2, 1 mM DTT. Proteoliposome-bound ribosomes were separated from free ribosomes by gel filtration chromatography as described in the Materials and methods.
Discussion
Dual recognition of SRP and ribosomes by the SR
Despite the relatively low affinity between the SR and the SRP in a physiological ionic strength buffer, the SRP–RNC complex is efficiently targeted to the SR. Here, we have tested several possible mechanisms that could contribute to an enhanced affinity between the SR and an SRP–RNC that would promote the targeting reaction in isotonic or hypertonic buffers.
Previous studies using canine microsomes or SR proteoliposomes have indicated that GTP is not required for targeting of SRP–RNCs to the SR (Rapiejko and Gilmore, 1994; Song et al., 2000), and that the targeting step precedes cooperative, stable binding of GTP to SRP54 and SRα (Rapiejko and Gilmore, 1997). Nonetheless these previous studies do not eliminate the formal possibility that the affinity of the SRP–RNC for the SR could be enhanced by rapidly reversible low-affinity binding of GTP to the empty site forms of SRP54 and SRα. As shown here, biosensor experiments designed to monitor the binding affinity between SRP and the SR demonstrated that the addition of GTP did not significantly increase the rate constant for formation of the SRP–SR complex in either isotonic or hypotonic buffers. As reported previously (Connolly et al., 1991), the nonhydrolyzable GTP analogue Gpp(NH)p stabilizes the SRP–SR complex by reducing the dissociation rate.
Could signal sequence–specific binding of SRP to an RNC cause a conformational change in SRP that enhances the affinity between SRP and the SR? This hypothesis was based upon the report that RNCs assembled in an in vitro translation system activate the GTPase activity of the SRP–SR complex (Bacher et al., 1996, 1999). Our analysis of this experimental system disclosed the remarkable finding that nontranslating ribosomes activate the GTPase activity of the SRP–SR complex. Furthermore, bona fide SRP–RNC complexes assembled using a secretory mRNA did not hydrolyze GTP when targeted to the SR proteoliposomes, consistent with our previous observation that both GTP and Gpp(NH)p stabilize the SR–SRP–RNC complex (Song et al., 2000).
The third hypothesis we considered was that the ribosome forms a platform for assembly of the SRP–SR complex. Purified 60S ribosomal subunits, but not 40S ribosomal subunits, stimulate the GTPase activity of the SRP–SR complex, consistent with the evidence that SRP54 binds to the L23a and L35 proteins in the large ribosomal subunit (Pool et al., 2002). Here, we obtained evidence that the SR has a high binding affinity for purified ribosomes or 60S ribosomal subunits. Notably, the rate constant for association of a ribosome–SR complex is 300-fold faster than the rate constant for formation of the SRP–SR complex. Consequently, the kinetics of targeting of the SRP–RNC complex to the SR should be dominated by the SR–ribosome interaction. The SR–ribosome interaction is also characterized by a relatively fast dissociation rate (kdiss ≈ 1.2 × 10−2 s-1). The rate of dissociation of SRP from the SR is less rapid, and this rate should decrease for the SRP–RNC complex. Binding of GTP to SRP54 and SRα substantially increases the stability of the SRP–RNC–SR complex because GTP hydrolysis by SRP54 and SRα is delayed until a vacant Sec61 complex is identified as an acceptor for the RNC complex (Song et al., 2000). We propose that the SR, by dual recognition of the ribosome and the SRP, will reject ribosomes that lack bound SRP.
The affinity between the SR and the ribosome appears to be conserved between eukaryotic and prokaryotic organisms. Depletion of either the translocon subunit SecE or the bacterial SRP (Ffh) leads to the in vivo accumulation of membrane-bound ribosome–FtsY complexes (Herskovits et al., 2002). The NH2-terminal acidic (A) domain of FtsY, which is involved in membrane binding (de Leeuw et al., 1997), is not homologous to the NH2-terminal 319 residues of SRα. Further work will be required to define the structural basis for the evolutionarily conserved interaction between the ribosome and the SR.
Roles for the SR subunits
The SRβ subunit of the SR was dispensable for the GTPase activity of the SRP–SR complex. Complexes formed between FtsY, the prokaryotic equivalent of SRα, and Ffh–4.5S RNA, the prokaryotic equivalent of SRP54 and the 7S RNA, hydrolyze GTP in a cooperative manner that has been investigated as a paradigm for the SRP–SR complex (Powers and Walter, 1995; Jagath et al., 2000; Peluso et al., 2000), hence it was not surprising that SRβ was dispensable for the GTPase cycles of SRP54 and SRα.
The observation that SRβ does not hydrolyze GTP when assayed alone was not unexpected, as most GTPases have very low hydrolysis rates in the absence of GEFs and GTPase-activating proteins (GAPs) (Bourne et al., 1991). SRβ does not hydrolyze GTP in the presence of 80S ribosomes, indicating that the ribosome cannot fulfill both the GEF and GAP functions for SRβ. Although photolabeling experiments had suggested that the ribosome acts as a GEF to stabilize a nucleotide-free form of SRβ (Bacher et al., 1999), a more recent report does not support this conclusion (Legate and Andrews, 2003). Our GTPase assays do not address which step, or steps, in the SRβ GTPase cycle occurs in the presence of the ribosome.
The SRX domain of SRα (residues 1–178) is necessary and sufficient for GTP-dependent heterodimerization with SRβ (Young et al., 1995; Ogg et al., 1998; Legate et al., 2000; Schwartz and Blobel, 2003). The GTP-bound, but not GDP-bound, form of SRβ forms stable heterodimers with SRX (Schwartz and Blobel, 2003). In the absence of a currently unidentified SRβ GAP, the SRβ GTPase is thought to be catalytically inert when bound to SRX (Schwartz and Blobel, 2003). An alternative model for the SRβ GTPase cycle proposes that GTP binding to SRβ regulates the release of the signal sequence from SRP54 (Fulga et al., 2001).
SRβ can be cross-linked to a 21-kD protein in the large ribosomal subunit (Fulga et al., 2001). Here, we observe an SRβ-independent, high-affinity interaction between SRα and the 60S subunit, suggesting that SRα positions SRβ adjacent to the 21-kD protein. As bt-SRα and bt-SR have similar affinities for the ribosome, we conclude that SRβ does not occlude the ribosome-binding site on SRα nor does it enhance the affinity of the SR for the ribosome. Previous studies that analyzed the ribosome–SRβ interaction have used either the recombinant SR heterodimer (Fulga et al., 2001) or trypsin-digested SR heterodimers that retain the NH2-terminal fragment of SRα (Bacher et al., 1999). The discrepancy between our results and these previous studies concerning the ribosome-binding and GTPase activities of SRβ might be explained by these structural differences in the reagents.
The NH2-terminal domain of SRα that is sufficient for ribosome-binding activity is polar (64% charged or polar residues) and basic (pI = 9.16). GTPase assays and biosensor experiments showed that SRP does not bind to bt-SRαΔN, despite the evidence that bt-SRαΔN is properly folded. It is unlikely that the NH2-terminal 151 residues of SRα are sufficient for the interaction of the SR with SRP, as the GTPase cycle of the SRP–SR complex almost certainly requires direct contact between the N and G domains of SRP54 and SRα. A model for the Ffh–FtsY complex (Montoya et al., 2000) predicts important interactions between the G domains of the two GTPases.
Regulation of the SR–ribosome interaction
Within the cell, a futile GTPase cycle catalyzed by the SRP–SR complex is not favored due to the low affinity between the SR and free SRP. However, the discovery that the ribosome can promote assembly of the SRP–SR complex in isotonic buffers raises new questions about the in vivo regulation of the SRP–SR GTPase cycle. A futile cycle involving SRP, the SR, and a ribosome would be restricted to the RER surface and would depend upon the presence of SRP and SR that are not engaged in bona fide targeting reactions. The cellular concentration of SRP and the SR may be regulated to ensure that the GTPases are substoichiometric relative to membrane-bound ribosomes.
Nontranslating ribosomes do not compete with SRP–RNCs for targeting to the Sec61 complex (Raden and Gilmore, 1998). This observation strongly suggests that there must be a mechanism to prevent the SR from being saturated by 60S ribosomal subunits, or ribosomes that are not engaged in the synthesis of secretory proteins. Although the relatively rapid dissociation rate for the ribosome–SR complex may contribute to such a mechanism, we speculate that there are additional factors that destabilize the SR–ribosome complex by selectively accelerating the dissociation rate. Ribosomes bearing nascent polypeptides that are synthesized on cytoplasmic polysomes recruit ribosome-associated chaperones, including the nascent chain–associated complex and members of the Hsc70 family (Wang et al., 1995; Bukau et al., 2000). A role for the nascent chain–associated complex in preventing signal sequence–independent binding of RNCs to the translocation channel has been proposed (Lauring et al., 1995; Moller et al., 1998), but the mechanism remains a matter of controversy (Neuhof et al., 1998; Raden and Gilmore, 1998). Future experiments will address the possibility that cytosolic chaperones regulate the binding affinity between the SR and the ribosome.
Materials and methods
Purification of the SRP, the SR, and the Sec61 complex
RMs, KOAc-washed RMs (K-RM), trypsin-digested K-RM (T5K-RM), SRP, and the SR were isolated from canine pancreas as previously described (Walter et al., 1981; Rapiejko and Gilmore, 1992; Connolly and Gilmore, 1993). Puromycin-high salt–extracted microsomes (PK-RM) and chymotrypsin-digested PK-RM (C1PK-RM) were characterized previously (Song et al., 2000). The canine Sec61 complex was purified from PK-RM by a modification of the method of Görlich and Rapoport (1993) using glycerol gradient centrifugation and anion and cation exchange chromatography.
Isolation of ribosomes, ribosomal subunits, and RNC complexes and radioiodination of ribosomes
Ribosomes were isolated from canine RM by extraction with high salt as previously described (Collins and Gilmore, 1991). Residual SRP was separated from 80S ribosomes by two sequential centrifugations through a high salt–sucrose cushion (Collins and Gilmore, 1991) followed by centrifugation through a physiological salt–sucrose cushion and resuspension of the ribosomes in buffer A (50 mM triethanolamine-acetate [TEA], pH 7.5, 150 mM KOAc, 5 mM Mg[OA]2, 1 mM DTT). Canine 80S ribosomes were dissociated into 40S and 60S subunits by treatment with 1 mM puromycin in buffer A, after which the sample was applied to a 14-ml 10–30% sucrose gradient in 50 mM TEA, 500 mM KOAc, 5 mM Mg(OAc)2, 1 mM DTT. The ribosomal subunits were resolved by centrifugation for 4.5 h at 200,000 g av using a Beckman Coulter SW40 rotor.
Sucrose gradient–purified 80S ribosomes (Raden et al., 2000) were resuspended in DTT-free buffer A and labeled with 125I Bolton-Hunter reagent (Amersham Biosciences) as previously described (Raden et al., 2000). Proteoliposomes were prepared as previously described (Song et al., 2000) using a modification of the method of Görlich and Rapoport (1993). Binding of 125I-labeled ribosomes to proteoliposomes was assayed as previously described (Raden et al., 2000). In brief, 0.03–0.65 pmol of 125I-labeled ribosomes was incubated with aliquots of the proteoliposomes in buffer A. The 25-μl sample was applied to a 1.2-ml Sepharose CL-2B column equilibrated in buffer A to resolve proteoliposome-bound ribosomes (0.3–0.6 ml of eluate) from unbound ribosomes (0.6–1.5 ml of eluate).
Truncated mRNAs encoding the NH2-terminal 86 residues of pPL (pPL86), 64 residues of vesicular stomatitis virus glycoprotein (pG64), 77 residues of firefly luciferase (ffluc77), or 156 residues of bovine opsin (op156) were prepared as previously described (Rapiejko and Gilmore, 1994).
SRP–RNC–op156 complexes were assembled in a reticulocyte lysate reaction as previously described (Rapiejko and Gilmore, 1997) and adjusted to 375 μM cycloheximide to block further translation. Membrane integration and N-linked glycosylation of op156 were assayed as previously described (Rapiejko and Gilmore, 1997).
RNC complexes bearing pPL86, pG64, or ffluc77 were assembled by translating truncated mRNAs for 15 min in a wheat germ reaction that lacked radiolabeled amino acids and SRP, unless noted otherwise. After blocking further translation by the addition of 2 mM cycloheximide, the translation products were adjusted to 500 mM KOAc before centrifugation for 1 h at 400,000 g av at 4°C through a high salt–sucrose cushion (1 M sucrose, 25 mM Hepes-KOH, pH 7.8, 500 mM KOAc, 5 mM Mg(OAc)2, 1 mM cycloheximide, 1 mM DTT). The RNCs were resuspended in half of the volume of the translation reaction in buffer B (25 mM Hepes-KOH, pH 7.8, 5 mM Mg[OAc]2, 1 mM cycloheximide, 1 mM DTT) adjusted to 500 mM KOAc and reisolated by centrifugation as described above. Finally, the RNCs were resuspended in buffer B adjusted to 150 mM KOAc at a concentration of 1 μM ribosomes.
GTPase assays
GTPase assays were conducted at 25°C in a total volume of 5 μl and contained 25–50 nM SR (canine SR, recombinant SR, or SR subunits), 50 nM SRP, 140 nM RNCs or mock RNCs, and 0.5 μM [α-32P]GTP (410 Ci/mmol) in buffer C (50 mM TEA-OAc, 150 mM KOAc, 5 mM Mg[OAc]2, 2 mM DTT, 2 mM cycloheximide) unless noted otherwise. The detergent micelle GTPase assays contained 0.1% Nikkol. Aliquots of the GTPase assays were removed at frequent time intervals and spotted onto PEI-cellulose thin layer plates to resolve GDP from GTP (Connolly and Gilmore, 1993).
Expression and purification of SR and SR subunits
DNA encoding a canine SRβ derivative lacking the NH2-terminal 54 residues (SRβΔN) was obtained by PCR amplification of the SRβ plasmid pMAC455 (Young et al., 1995) using appropriate primers and standard PCR conditions. The SRβΔN coding sequence was inserted into the PinPoint vector (Promega) to obtain pbt-SRβΔN. The dicistronic plasmid pbt-SRβ-SRα encoding bt-SRβ and SRα was constructed by inserting the SRα coding sequence derived from plasmid pG4α (Rapiejko and Gilmore, 1992) into pbt-SRβΔN. The plasmids pbt-SRα and pbt-SRαΔN encode fusion proteins between the biotinylation domain and canine SRα or canine SRα lacking the NH2-terminal 151 residues, respectively. The SRβΔN sequence was subcloned into pET14b (Novagen) to obtain pHis-SRβ. Heterodimers consisting of His-SRβ and NH2-terminal fragments of SRα (SRα151 or SRα319) were expressed from dicistronic plasmids. All constructs were verified by DNA sequencing. The biotinylated proteins were purified from the E. coli (J109) lysates by affinity chromatography (Soft-Avidin resin; Promega) and anion and cation exchange chromatography. The His-tagged proteins were purified from E. coli (Rosetta; Novagen) lysates by Ni-NTA (QIAGEN) affinity chromatography and cation exchange chromatography.
IAsys affinity sensor experiments
Binding of SRP or ribosomes to the SR or SR subunits was assayed using an IAsys affinity sensor (Affinity Sensors). The binding surface was constructed by incubating saturating amounts of streptavidin (Promega) with a biotin-coated cuvette for 5 min. After a brief wash with buffer D (50 mM TEA, 150 mM KOAc, 2.5 mM Mg[OAc]2, 0.1% Nikkol) the bt-SR or a bt-SR subunit was added and incubated until equilibrium binding was observed (5 min). Preparation of the sensor surface was followed by a brief wash with buffer D. Binding time courses were performed at 25°C using a variety of ligates (SRP, ribosomes, or ribosomal subunits) in buffers D or E (buffer D with KOAc reduced to 50 mM) in either the absence or presence of 25 μM GTP. The ligate was preincubated for 2 min at 25°C (with GTP when appropriate) before the addition to a cuvette containing the immobilized bt-protein. Analysis of binding experiments showed that 6.5 min was sufficient to calculate equilibrium binding values. After binding, the cuvette was rapidly washed three times with buffer D or E (with or without GTP), and dissociation of ligate was monitored for 3 min. The binding surface was regenerated by dissociating residual ligate with buffer F (50 mM TEA, 500 mM KOAc, 5 mM Mg[OAc]2, 0.1% Nikkol). The cuvette was then washed and reequilibriated in buffer D or E containing GTP as indicated. The high salt wash procedure removes the ligate without damaging or detaching the bt-protein. Dissociation of Gpp(NH)p–stabilized bt-SR–SRP complexes was incomplete.
Binding of ligate to the sensor surface is measured as a response (arc seconds of change in the refractive index), which corresponds to the accumulation of mass within the optical window at the binding surface. The extent (in arc seconds) refers to the calculated maximum response (Rmax) at equilibrium for a given concentration of ligate. The rates of ligate binding (kon) and the extent (Rmax) were calculated from association curves using FASTfit software supplied with the instrument.
Acknowledgments
We thank Dan Kelleher for useful discussion during the preparation of this manuscript. We thank David Andrews (McMaster University, Hamilton, Ontario) and Chris Nicchitta (Duke University, Durham, NC) for providing antibodies to SRP54 and SRα, respectively.
This work was supported by National Institutes of Health grant PHS GM35687.
Abbreviations used in this paper: GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; pPL, preprolactin; RM, rough microsome; RNC, ribosome nascent chain complex; SR, SRP receptor; SRP, signal recognition particle.
References
- Andrews, D.W., L. Lauffer, P. Walter, and V.R. Lingappa. 1989. Evidence for a two-step mechanism involved in assembly of functional signal recognition particle receptor. J. Cell Biol. 108:797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher, G., H. Lütcke, B. Jungnickel, T.A. Rapoport, and B. Dobberstein. 1996. Regulation by the ribosome of the GTPase of the signal-recognition particle during protein targeting. Nature. 381:248–251. [DOI] [PubMed] [Google Scholar]
- Bacher, G., M. Pool, and B. Dobberstein. 1999. The ribosome regulates the GTPase of the β-subunit of the signal recognition particle receptor. J. Cell Biol. 146:723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, N., P. Nissen, J. Hansen, P.B. Moore, and T.A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 289:905–920. [DOI] [PubMed] [Google Scholar]
- Beckmann, R., C.M. Spahn, N. Eswar, J. Helmers, P.A. Penczek, A. Sali, J. Frank, and G. Blobel. 2001. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 107:361–372. [DOI] [PubMed] [Google Scholar]
- Bourne, H.R., D.A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 349:117–127. [DOI] [PubMed] [Google Scholar]
- Bukau, B., E. Deuerling, C. Pfund, and E.A. Craig. 2000. Getting newly synthesized proteins into shape. Cell. 101:119–122. [DOI] [PubMed] [Google Scholar]
- Collins, P., and R. Gilmore. 1991. Ribosome binding to the endoplasmic reticulum: A 180-kD protein identified by crosslinking to membrane-bound ribosomes is not required for ribosome binding activity. J. Cell Biol. 114:639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, T., and R. Gilmore. 1993. GTP hydrolysis by complexes of the signal recognition particle and the signal recognition particle receptor. J. Cell Biol. 123:799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, T., P.J. Rapiejko, and R. Gilmore. 1991. Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science. 252:1171–1173. [DOI] [PubMed] [Google Scholar]
- de Leeuw, E., D. Poland, O. Mol, I. Sinning, C.M. ten Hagen-Jongman, B. Oudega, and J. Luirink. 1997. Membrane association of FtsY, the E. coli SRP receptor. FEBS Lett. 416:225–229. [DOI] [PubMed] [Google Scholar]
- Flanagan, J.J., J.C. Chen, Y. Miao, Y. Shao, J. Lin, P.E. Bock, and A.E. Johnson. 2003. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the chain lengthens. J. Biol. Chem. 278:18628–18637. [DOI] [PubMed] [Google Scholar]
- Fulga, T.A., I. Sinning, B. Dobberstein, and M.R. Pool. 2001. SRβ coordinates signal sequence release from SRP with ribosome binding to the translocon. EMBO J. 20:2338–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich, D., and T.A. Rapoport. 1993. Protein translocation into proteoliposomes reconstituted from purified components of the ER membrane. Cell. 75:615–630. [DOI] [PubMed] [Google Scholar]
- Herskovits, A.A., E. Shimoni, A. Minsky, and E. Bibi. 2002. Accumulation of endoplasmic membranes and novel membrane-bound ribosome-signal recognition particle receptor complexes in Escherichia coli. J. Cell Biol. 159:403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagath, J.R., M.V. Rodnina, and W. Wintermeyer. 2000. Conformational changes in the bacterial SRP receptor FtsY upon binding of guanine nucleotides and SRP. J. Mol. Biol. 295:745–753. [DOI] [PubMed] [Google Scholar]
- Kalies, K.-U., D. Görlich, and T.A. Rapoport. 1994. Binding of ribosomes to the rough endoplasmic reticulum is mediated by the Sec61p complex. J. Cell Biol. 126:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffer, L., P.D. Garcia, R.N. Harkins, L. Coussens, A. Ullrich, and P. Walter. 1985. Topology of signal recognition particle receptor in endoplasmic reticulum membrane. Nature. 318:334–338. [DOI] [PubMed] [Google Scholar]
- Lauring, B., G. Kreibich, and M. Wiedmann. 1995. The intrinsic ability of ribosomes to bind to endoplasmic reticulum membranes is regulated by signal recognition particle and nascent-polypeptide-associated complex. Proc. Natl. Acad. Sci. USA. 92:9435–9439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate, K.R., and D.W. Andrews. 2003. The β-subunit of the SRP receptor is a novel GTP binding protein without intrinsic GTPase activity. J. Biol. Chem. 278:27712–27720. [DOI] [PubMed] [Google Scholar]
- Legate, K.R., D. Falcone, and D.W. Andrews. 2000. Nucleotide-dependent binding of the GTPase domain of the signal recognition particle receptor β-subunit to the α-subunit. J. Biol. Chem. 275:27439–27446. [DOI] [PubMed] [Google Scholar]
- Menetret, J., A. Neuhof, D.G. Morgan, K. Plath, M. Radermacher, T.A. Rapoport, and C.W. Akey. 2000. The structure of ribosome-channel complexes engaged in protein translocation. Mol. Cell. 6:1219–1232. [DOI] [PubMed] [Google Scholar]
- Meyer, D.I., and B. Dobberstein. 1980. A membrane component essential for vectorial translocation of nascent proteins across the endoplasmic reticulum: requirements for its extraction and reassociation with membranes. J. Cell Biol. 87:498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.D., H. Wilhelm, R. Gierasch, R. Gilmore, and P. Walter. 1993. GTP binding and hydrolysis by the signal recognition particle during initiation of protein translocation. Nature. 366:351–354. [DOI] [PubMed] [Google Scholar]
- Moller, I., M. Jung, B. Beatrix, R. Levy, G. Kreibich, R. Zimmermann, M. Wiedmann, and B. Lauring. 1998. A general mechanism for regulation of access to the translocon: competition for a membrane attachment site on ribosomes. Proc. Natl. Acad. Sci. USA. 95:13425–13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya, G., K. Kaat, R. Moll, G. Schafer, and I. Sinning. 2000. The crystal structure of the conserved GTPase of SRP54 from the archaeon Acidianus ambivalens and its comparison with related structures suggests a model for the SRP-SRP receptor complex. Structure Fold. Des. 8:515–525. [DOI] [PubMed] [Google Scholar]
- Neuhof, A., M.M. Rolls, B. Jungnickel, K.-U. Kalies, and T.A. Rapoport. 1998. Binding of signal recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction. Mol. Biol. Cell. 9:103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg, S.C., W.P. Barz, and P. Walter. 1998. A functional GTPase domain, but not its transmembrane domain, is required for function of the SRP receptor β-subunit. J. Cell Biol. 142:341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso, P., D. Herschlag, S. Nock, D.M. Freymann, A.E. Johnson, and P. Walter. 2000. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science. 288:1640–1643. [DOI] [PubMed] [Google Scholar]
- Pool, M.R., J. Stumm, T.A. Fulga, I. Sinning, and B. Dobberstein. 2002. Distinct modes of signal recognition particle interaction with the ribosome. Science. 297:1345–1348. [DOI] [PubMed] [Google Scholar]
- Powers, T., and P. Walter. 1995. Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science. 269:1422–1424. [DOI] [PubMed] [Google Scholar]
- Powers, T., and P. Walter. 1996. The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr. Biol. 6:331–338. [DOI] [PubMed] [Google Scholar]
- Raden, D., and R. Gilmore. 1998. Signal recognition particle-dependent targeting of ribosomes to the rough endoplasmic reticulum in the absence and presence of the nascent polypeptide associated complex. Mol. Biol. Cell. 9:117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raden, D., W. Song, and R. Gilmore. 2000. Role of the cytoplasmic segments of Sec61-α in the ribosome-binding and translocation-promoting activities of the Sec61 complex. J. Cell Biol. 150:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapiejko, P.J., and R. Gilmore. 1992. Protein translocation across the ER requires a functional GTP binding site in the α subunit of the signal recognition particle receptor. J. Cell Biol. 117:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapiejko, P.J., and R. Gilmore. 1994. Signal sequence recognition and targeting of ribosomes to the endoplasmic reticulum by the signal recognition particle do not require GTP. Mol. Biol. Cell. 5:887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapiejko, P.J., and R. Gilmore. 1997. Empty site forms of the SRP54 and SRα GTPases mediate targeting of ribosome-nascent chain complexes to the endoplasmic reticulum. Cell. 89:703–713. [DOI] [PubMed] [Google Scholar]
- Schwartz, T., and G. Blobel. 2003. Structural basis for the function of the β subunit of the eukaryotic signal recognition particle receptor. Cell. 112:793–803. [DOI] [PubMed] [Google Scholar]
- Song, W., D. Raden, E. Mandon, and R. Gilmore. 2000. Role of Sec61α in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell. 100:333–343. [DOI] [PubMed] [Google Scholar]
- Walter, P., and G. Blobel. 1983. Subcellular distribution of signal recognition particle and 7S-RNA determined with polypeptide-specific antibodies and complementary DNA probe. J. Cell Biol. 97:1693–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, P., and A.E. Johnson. 1994. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 10:87–119. [DOI] [PubMed] [Google Scholar]
- Walter, P., I. Ibrahimi, and G. Blobel. 1981. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in vitro–assembled polysomes synthesizing secretory protein. J. Cell Biol. 91:545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., H. Sakai, and M. Wiedmann. 1995. NAC covers ribosome-associated nascent chains thereby forming a protective environment for regions of nascent chains just emerging from the peptidyl transferase center. J. Cell Biol. 130:519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, J.C., J. Ursini, K.R. Legate, J.D. Miller, P. Walter, and D.W. Andrews. 1995. An amino-terminal domain containing hydrophobic and hydrophilic sequences binds the signal recognition particle receptor α subunit to the β subunit on the endoplasmic reticulum. J. Biol. Chem. 270:15650–15657. [DOI] [PubMed] [Google Scholar]