Abstract
The spindle checkpoint delays anaphase onset until all chromosomes have attached properly to the mitotic spindle. Checkpoint signal is generated at kinetochores that are not bound with spindle microtubules or not under tension. Unattached kinetochores associate with several checkpoint proteins, including BubR1, Bub1, Bub3, Mad1, Mad2, and CENP-E. I herein show that BubR1 is important for the spindle checkpoint in Xenopus egg extracts. The protein accumulates and becomes hyperphosphorylated at unattached kinetochores. Immunodepletion of BubR1 greatly reduces kinetochore binding of Bub1, Bub3, Mad1, Mad2, and CENP-E. Loss of BubR1 also impairs the interaction between Mad2, Bub3, and Cdc20, an anaphase activator. These defects are rescued by wild-type, kinase-dead, or a truncated BubR1 that lacks its kinase domain, indicating that the kinase activity of BubR1 is not essential for the spindle checkpoint in egg extracts. Furthermore, localization and hyperphosphorylation of BubR1 at kinetochores are dependent on Bub1 and Mad1, but not Mad2. This paper demonstrates that BubR1 plays an important role in kinetochore association of other spindle checkpoint proteins and that Mad1 facilitates BubR1 hyperphosphorylation at kinetochores.
Keywords: spindle checkpoint; Xenopus; BubR1; Mad1; Bub1
Introduction
The metaphase to anaphase transition is delayed by the spindle checkpoint until all chromosomes have established bipolar attachment to the mitotic spindle, thus ensuring accurate sister chromatid segregation during each cell division (for review see Amon, 1999). The checkpoint signal is generated from kinetochores that fail to bind spindle microtubules or are not under tension that normally comes from bipolar attachment to the spindle (Li and Nicklas, 1995; Rieder et al., 1995). Several spindle checkpoint proteins localize to kinetochores, including Mad1, Mad2, Bub1, Bub3, BubR1, Mps1, and CENP-E (for review see Shah and Cleveland, 2000). Kinetochore binding likely allows these proteins to gain their ability to relay the checkpoint signal.
The downstream effector of the spindle checkpoint is Cdc20, an activator for the anaphase promoting complex (APC),* which is a ubiquitin protein ligase that targets key mitotic regulators for degradation. Upon binding to activators Cdc20 and Cdh1, APC recognizes the substrates Pds1 and cyclin B, whose degradation leads to sister chromatid separation and exit from mitosis, respectively (for review see Page and Hieter, 1999). It has been shown that checkpoint proteins Mad2 and BubR1 bind and inhibit Cdc20, thus preventing anaphase onset (Fang et al., 1998; Hwang et al., 1998; Kim et al., 1998; Tang et al., 2001). Purification from HeLa cell lysates also yields a protein complex that inhibits APC. This mitotic checkpoint complex contains Mad2, BubR1, Bub3, and Cdc20 (Sudakin et al., 2001).
The association of Cdc20 with spindle checkpoint proteins is increased during mitosis relative to that in interphase, partly due to accumulation of Cdc20 (Hardwick et al., 2000; Fraschini et al., 2001; Chung and Chen, 2002). Mad2–Cdc20 interaction is further enhanced when mitotic spindle is disrupted, compared with that at metaphase (Zhang and Lees, 2001; Chung and Chen, 2002). The latter result indicates that misaligned chromosomes may facilitate checkpoint protein complex formation, likely through recruiting spindle checkpoint proteins to kinetochores. Mad1 and Mad2 specifically enrich at unattached kinetochores and dissociate from kinetochores upon microtubule attachment (Chen et al., 1996, 1998; Li and Benezra, 1996; Waters et al., 1998). Mad1 is required for Mad2 to bind unattached kinetochores through direct interaction between these two proteins (Chen et al., 1998), whereas Mad1 can bind kinetochores without Mad2 (Chung and Chen, 2002). Unlike Mad1 and Mad2, some Bub1 and Bub3 proteins remain at kinetochores until early anaphase (Jablonski et al., 1998; Basu et al., 1999; Sharp-Baker and Chen, 2001). The interaction between Bub1 and Bub3 is important for Bub1 to associate with kinetochores (Taylor et al., 1998). Bub1 is also necessary for Mad1 and Mad2 to bind kinetochores independently of its protein kinase activity (Sharp-Baker and Chen, 2001). In addition to Bub1, kinetochore localization of Mad1 and Mad2 requires the protein kinase activity of Mps1, also a kinetochore-associated spindle checkpoint protein (Abrieu et al., 2001). Mps1 in budding yeast is thought to be the kinase responsible for Mad1 phosphorylation during mitosis (Hardwick et al., 1996; Weiss and Winey, 1996). It remains to be determined whether Mps1 in higher eukaryotes is the physiological kinase for Mad1.
Unattached kinetochores establish and maintain the checkpoint signal. It has been shown that Mad1 and Mad2 need to be at kinetochores continuously in order to maintain the spindle checkpoint, because blocking their binding with a dominant negative Mad1 protein abolishes the checkpoint (Chung and Chen, 2002). Kinetochores are probably the sites where checkpoint proteins Mad2 and BubR1 gain ability to interact with Cdc20. Alternatively, kinetochores may bring the checkpoint proteins to close proximity, allowing their interaction. Upon activation or protein complex assembly at kinetochores, the checkpoint proteins may then leave kinetochores to inhibit APC in the cytosol. FRAP analysis has revealed a transient interaction of Mad2 with kinetochores with a turnover rate of 24–28 s (Howell et al., 2000).
BubR1 is a central checkpoint protein on the basis of its ability to bind kinetochores (Chan et al., 1998; Taylor et al., 1998; Chan et al., 1999) and to inhibit Cdc20 in vitro (Tang et al., 2001). Human BubR1 was first identified as an interactor for kinetochore motor CENP-E (Chan et al., 1998) and also as a protein that contains homologous regions with yeast Mad3 and Bub1 (Cahill et al., 1998; Taylor et al., 1998). The protein shares similarity with Bub1 at both the amino-terminal region and the carboxy-terminal protein kinase domain, thus it was named BubR1 (for Bub1 related). Part of the homologous amino-terminal region of Bub1 and BubR1 is necessary for binding to Bub3 (Taylor et al., 1998). BubR1 is essential for the spindle checkpoint, because microinjection of antibodies against human BubR1 abolishes mitotic arrest induced by nocodazole (Chan et al., 1999). In addition, certain mutations in BubR1 have been found in some colorectal cancers that display chromosome instability (Cahill et al., 1998).
Similar to Bub1, the amino-terminal region of BubR1 is homologous to budding yeast Mad3 that lacks a kinase domain. Thus, BubR1 is thought to be the homologue of Mad3 in higher eukaryotes. Yeast Mad3 has also been shown to interact with Cdc20, Bub3, and Mad2 (Hardwick et al., 2000; Fraschini et al., 2001; Millband and Hardwick, 2002). Whereas its interaction with Bub3 is constant during the cell cycle, the interaction with Mad2 and Cdc20 is enhanced under the checkpoint-active condition (Hardwick et al., 2000; Fraschini et al., 2001). The position of Mad3 in the spindle checkpoint pathway has been studied in budding yeast. Using characteristic Mad1 phosphorylation during mitosis as a marker, it has been shown that Mad1 phosphorylation requires Mps1, Bub1, Bub3, and Mad2, but not Mad3 (Hardwick and Murray, 1995). Overexpression of Mps1 leads to hyperphosphorylation of Mad1 even without a functional Mad3 (Hardwick et al., 1996). Mitotic arrest induced by overexpression of Mps1 or a dominant active Bub1-5 mutant requires Mad1, Mad2, and Mad3 (Hardwick et al., 1996; Farr and Hoyt, 1998). These biochemical and genetic studies tentatively place Mad3 as a downstream component of the checkpoint pathway containing Mps1, Bub1, Bub3, Mad1, and Mad2. In human cells, an immunofluorescence study shows that kinetochore localization of BubR1 appears shortly after Bub1, but before CENP-E (Chan et al., 1998; Jablonski et al., 1998), consistent with the notion that BubR1 may be downstream of Bub1. To better understand the role and regulation of BubR1 in the spindle checkpoint, I have studied BubR1 in Xenopus egg extracts.
Results
BubR1 in mouse and human contains homology with the spindle checkpoint protein Bub1 and budding yeast Mad3. A sequence in the Xenopus EST database (GenBank/EMBL/DDBJ accession no. BE025630) was found to be similar to the human and mouse BubR1 and was used to isolate a full-length cDNA. This cDNA (GenBank/EMBL/DDBJ accession no. AY095442) predicts a protein of 1041 amino acids with a molecular mass of 118 kD. The protein sequence is 41.5 and 39.1% identical to the human and mouse BubR1, respectively. To gain insight into the role of BubR1 in the spindle checkpoint, anti-BubR1 antiserum was generated against amino acids 189–359, which are unique in BubR1, but not conserved in Bub1. The antibody was used to study BubR1 in Xenopus egg extracts. Mature Xenopus eggs are arrested at metaphase II by cytostatic factor (CSF). The cytoplasmic extracts prepared from eggs, termed CSF-arrested extracts, also maintain the metaphase arrest. Upon the addition of calcium, the metaphase extracts exit meiosis and enter interphase. The spindle checkpoint can be reproduced in the metaphase extract by the addition of sperm nuclei (9,000–15,000/μl extract) and nocodazole (Minshull et al., 1994). By immunoblot analysis, the anti-BubR1 antibody recognized polypeptides of ∼145 kD in interphase, metaphase, and spindle checkpoint–active extracts (Fig. 1 A, lanes 4–6). Preincubation of the antibodies with recombinant BubR1 protein abolished the signal (Fig. 1 A, lanes 1–3), showing specificity of the antibodies. These 145-kD polypeptides were BubR1, rather than Bub1, because they remained in Bub1-depleted extracts (Fig. 1 B, lane 2). Similarly, extracts depleted with anti-BubR1 antibodies still retained 150-kD polypeptides recognized by anti-Bub1 antibodies, but not by anti-BubR1 antibodies (Fig. 1 B, lane 3). Furthermore, anti-Bub1 immunoprecipitates were recognized by anti-Bub1 antibody, but not by anti-BubR1, and vice versa (Fig. 1 B, lanes 5 and 6). These results show that the antibodies against BubR1 and Bub1 are specific to corresponding proteins.
Figure 1.
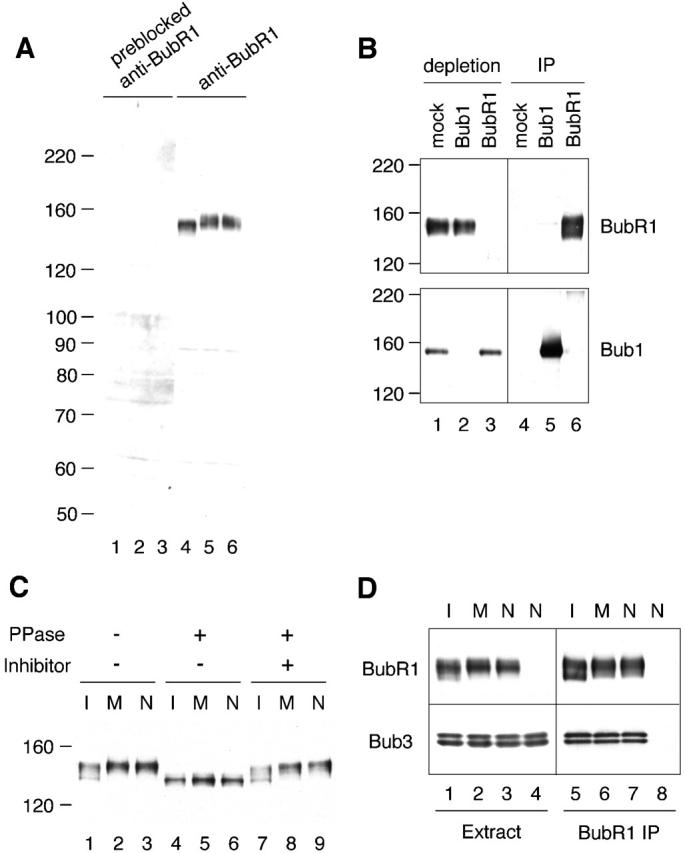
BubR1 is a phosphoprotein associated with Bub3. (A) Specificity of the anti-BubR1 antibody. Interphase (lanes 1 and 4), metaphase (lanes 2 and 5), or spindle checkpoint–active (lanes 3 and 6) extracts were immunoblotted with anti-BubR1 antibody (lanes 4–6) or the same antibody preincubated with recombinant BubR1 protein (lanes 1–3). The migration of molecular standards is indicated on the left. (B) Anti-BubR1 antibody recognizes BubR1, but not Bub1. Immunoprecipitation was performed from metaphase extracts with a control IgG (lanes 1 and 4), anti-Bub1 (lanes 2 and 5), or anti-BubR1 (lanes 3 and 6) antibody. The supernatants left after immunoprecipitation (lanes 1–3) or the immunoprecipitates (lanes 4–6) were probed with anti-BubR1 (top) or anti-Bub1 (bottom) antibody. The migration of molecular standards is indicated on the left. (C) BubR1 is a phosphoprotein. Interphase (I; lanes 1, 4, and 7), metaphase (M; lanes 2, 5, and 8), and spindle checkpoint–active (N; lanes 3, 6, and 9) extracts were treated with phosphatase buffer (lanes 1–3), LPP (lanes 4–6), or LPP and phosphatase inhibitors (lanes 7–9). The extracts were then immunoblotted with anti-BubR1 antibody. The migration of molecular standards is indicated on the left. (D) BubR1 associates with Bub3. Anti-BubR1 immunoprecipitates were prepared from interphase (lanes 1 and 5), metaphase (lanes 2 and 6), or spindle checkpoint–active (lanes 3 and 7) extracts, or from Bub1-depleted extract with the spindle checkpoint provoked (lanes 4 and 8). The extracts (lanes 1–4) or the immunoprecipitates (lanes 5–8) were immunoblotted with anti-BubR1 (top) or anti-Bub3 (bottom) antibody.
Immunoblot analysis of egg extracts shows that the electrophoretic mobility of BubR1 from metaphase and checkpoint-active extracts was slightly slower than that from interphase extracts (Fig. 1 C, lanes 1–3). The apparent size is larger than the predicted molecular weight, suggesting that the protein may become modified posttranslationally. Indeed, protein phosphatase treatment reduced the size of the protein from all three types of extracts to 135 kD (Fig. 1 C, lanes 4–6), indicating that the protein was phosphorylated.
BubR1 is required for Mad2–Cdc20 interaction
BubR1 in human cells associates with spindle checkpoint protein Bub3 (Taylor et al., 1998). Similarly, Xenopus Bub3 was coimmunoprecipitated with BubR1 from egg extracts (Fig. 1 D). Coimmunoprecipitation of Bub3 and BubR1 was specific, because Bub3 was not detectable when the immunoprecipitate was prepared from BubR1-depleted extracts (Fig. 1 D, lane 8). The level of Bub3 associated with BubR1 was constant in interphase, metaphase, or spindle checkpoint–active extracts (Fig. 1 D, lanes 5–7), showing a constitutive nature of the interaction between Bub1 and Bub3.
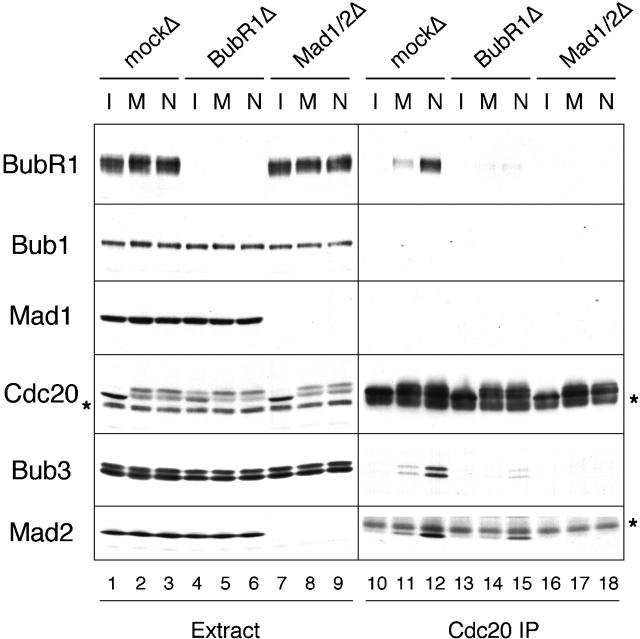
Recombinant BubR1 has been shown to bind and inhibit Cdc20 in vitro (Tang et al., 2001). I thus examined the association between BubR1 and Cdc20 in various types of egg extracts. Cdc20 was immunoprecipitated from interphase, metaphase, and spindle checkpoint–active extracts that contained the same number of chromosomes. The immunoprecipitates were then subjected to immunoblot analysis for various spindle checkpoint proteins. I did not detect BubR1, Bub3, and Mad2 in the Cdc20 immunoprecipitate prepared from interphase extract (Fig. 2, lane 10), whereas these proteins were detectable when Cdc20 was immunoprecipitated from metaphase extracts (Fig. 2, lane 11). The level of interaction was further enhanced in checkpoint-active extracts that had been incubated with nocodazole to disrupt the mitotic spindle (Fig. 2, lane 12). Bub1 and Mad1 were not detected in Cdc20 immunoprecipitates from any of the extracts (Fig. 2). These results are consistent with the model that unattached kinetochores stimulate the assembly of a protein complex containing BubR1, Bub3, Mad2, and Cdc20. I then asked the question of whether there is any dependency between BubR1, Mad2, and Bub3 in binding to Cdc20. In BubR1-depleted extracts incubated with or without nocodazole, the level of Bub3 and Mad2 present in the Cdc20 immunoprecipitates was greatly reduced (Fig. 2, lanes 14 and 15). Similarly, interaction between BubR1, Bub3 with Cdc20 was also reduced when Mad2 was depleted (Fig. 2, lanes 17 and 18). These results show that there is mutual dependency between BubR1 and Mad2 in binding to Cdc20. I cannot perform a similar experiment in Bub3-depleted extract, because our anti-Bub3 antibodies fail to efficiently deplete Bub3 from egg extracts. In addition, I could not detect Cdc20 in BubR1 immunoprecipitates (unpublished data), which may result from disruption of the protein complex by anti-BubR1 antibodies. Alternatively, Cdc20 may bind to only a small fraction of BubR1, so anti-BubR1 antibodies mask any Cdc20 signal on the blot.
Figure 2.
Mutual dependency between BubR1 and Mad2 in binding to Cdc20. CSF-arrested extracts were depleted with control IgG (mockΔ; lanes 1–3 and 10–12), anti-BubR1 (BubR1Δ; lanes 4–6 and 13–15), or anti-Mad2 (Mad1/2Δ; lanes 7–9 and 16–18) antibody. The extracts were induced to enter interphase (lanes 1, 4, 7, 10, 13, and 16), were kept at metaphase (lanes 2, 5, 8, 11, 14, and 17), or were incubated with nuclei and nocodazole to activate the spindle checkpoint (lanes 3, 6, 9, 12, 15, and 18). Anti-Cdc20 immunoprecipitates were prepared from these extracts. The extracts (lanes 1–9) or immunoprecipitates (lanes 10–18) were immunoblotted for the proteins indicated on the left. The asterisks indicate a cross-reacting protein and IgG heavy chain on the left and right panels of Cdc20 blots, respectively. The asterisk on the right of Mad2 blot indicates IgG light chain that migrates right above Mad2. Immunodepletion with anti-Mad2 antibody removes both Mad1 and Mad2.
BubR1 is important for other spindle checkpoint proteins to bind kinetochores
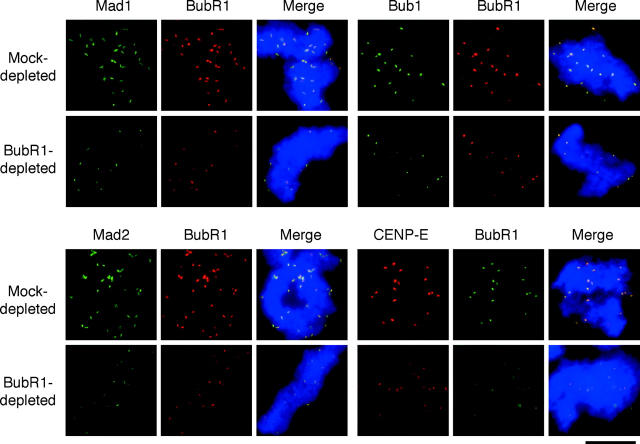
The association of Mad2 with Cdc20 is dependent on binding of Mad2 to unattached kinetochores (Chung and Chen, 2002). Thus, loss of the interaction between Cdc20 and checkpoint proteins in either BubR1- or Mad2-depleted extracts may be caused by impaired interaction of checkpoint proteins with kinetochores. To determine kinetochore localization of checkpoint proteins, mitotic chromosomes were assembled in and purified from egg extracts containing nocodazole, followed by immunofluorescence staining for various checkpoint proteins. Fig. 3 shows that anti-BubR1 antibody gave punctate staining that colocalized with the staining for Mad1, Mad2, Bub1, and CENP-E, indicating that BubR1 is associated with kinetochores. In BubR1-depleted extracts, kinetochore staining of Bub1, Mad1, Mad2, and CENP-E was greatly reduced (Fig. 3), indicating that BubR1 is important for these checkpoint proteins to localize to kinetochores. Even in extracts depleted for >98% of BubR1, some kinetochores still retained BubR1 to various levels. One possible explanation is that kinetochores may have some high-affinity binding sites for BubR1 and are able to recruit residual BubR1. Nevertheless, kinetochores that accumulated a low level of BubR1 also consistently had little other checkpoint proteins.
Figure 3.
BubR1 is required for kinetochore binding of Mad1, Mad2, Bub1, and CENP-E. Mitotic chromosomes were isolated from mock- or BubR1- depleted extracts treated with nocodazole and then incubated with rabbit antibodies against Mad1, Mad2, or Bub1, as indicated on top, followed by fluorescein-conjugated anti–rabbit antibody. BubR1 was then detected with biotinylated anti-BubR1 antibody and Texas red– conjugated streptavidin. For double staining of CENP-E and BubR1, rabbit antibody against BubR1 was used, followed by fluorescein-conjugated anti–rabbit antibody. CENP-E was then detected with biotinylated anti–CENP-E antibody and Texas red–conjugated streptavidin. Chromosomes were stained with Hoechst 33258. The merge pictures contain all three fluorochromes. All images of the same fluorochrome were taken for the same exposure time and processed in the same way. Bar, 10 μm.
I also assessed kinetochore binding of checkpoint proteins by immunoblot analysis of isolated chromosomes. Fig. 4 A shows that there was very little Mad1 and Mad2 associated with chromosomes purified from metaphase extracts (lane 5). The level of Mad1 and Mad2 in the chromosomal fraction increased greatly if the mitotic spindle was disrupted with nocodazole (Fig. 4 A, compare lanes 5 and 6). Metaphase chromosomes associated with some Bub1, Bub3, and BubR1, and a higher level of these proteins was found with unattached chromosomes (Fig. 4 A, compare lanes 5 and 6). Interestingly, Bub1 and BubR1 associated with unattached chromosomes exhibited significant mobility retardation when compared with those from metaphase chromosomes (Fig. 4 A, compare lanes 5 and 6). This mobility shift was not observed with cytosolic proteins, indicating that chromosomal Bub1 and BubR1 had a higher degree of posttranslational modification than their cytosolic counterparts. This modification is due to phosphorylation, because phosphatase treatment of isolated chromosomes removed the mobility shift (Fig. 4 B). Detailed characterization of phosphorylation and its effect on the activity of Bub1 and BubR1 will be reported elsewhere. The doublet of polypeptides recognized by anti-Bub3 antibodies was not due to protein phosphorylation, because phosphatase treatment did not have any apparent effect on their electrophoretic mobility (Sharp-Baker and Chen, 2001). Consistent with immunofluorescence studies, immunoblot analysis also shows that BubR1 depletion resulted in a significant reduction of Bub1, Bub3, Mad1, and Mad2 associated with chromosomes in the presence of nocodazole (Fig. 4 A, lane 8). Without nocodazole, the levels of chromosomal Bub1 and Bub3 were also reduced in BubR1-depleted extract (Fig. 4 A, lane 7). This result indicates that BubR1 plays a role in localization of these checkpoint proteins to unattached and attached kinetochores. In addition, the remaining Bub1 was still hyperphosphorylated in the chromosomal fraction from BubR1-depleted extracts (Fig. 4 A, lane 8), suggesting that BubR1 is not involved in hyperphosphorylation of Bub1. Although immunofluorescence detected some residual BubR1 at kinetochores from BubR1-depleted extracts (Fig. 3), the protein was not detectable by immunoblot (Fig. 4, lanes 7 and 8). This discrepancy may result from a lack of linearity of the output signals or different sensitivity of the detection methods for BubR1.
Figure 4.
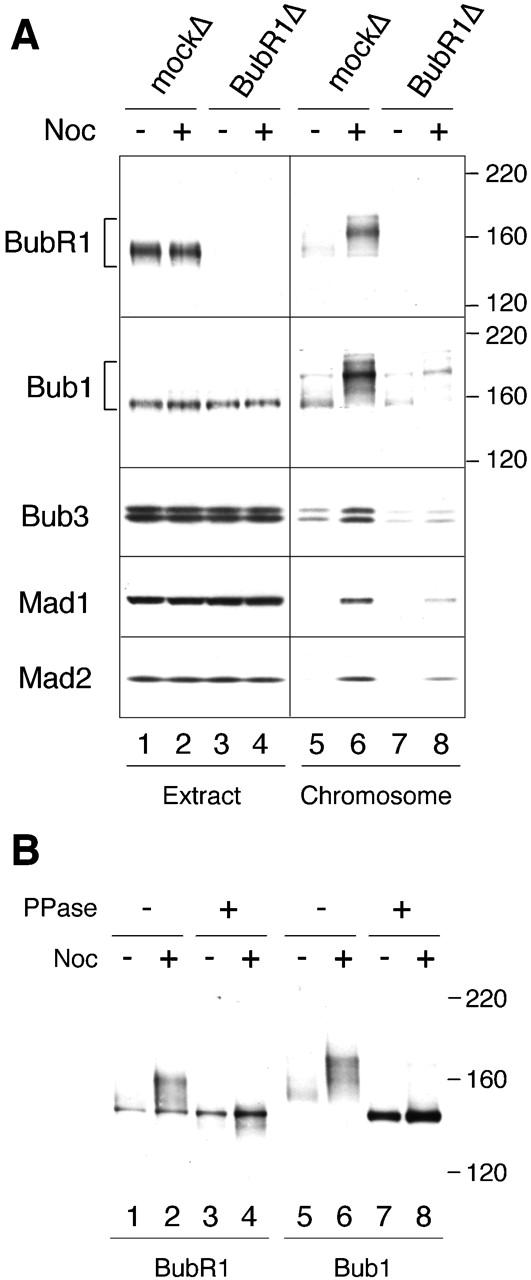
BubR1 is required for other spindle checkpoint proteins to associate with chromosomes. (A) Immunoblot analysis of various checkpoint proteins associated with chromosomes. Mitotic chromosomes were isolated from mock-depleted (lanes 1, 2, 5, and 6) or BubR1-depleted (lanes 3, 4, 7, and 8) extracts that were untreated (lanes 1, 3, 5, and 7) or treated with nocodazole (lanes 2, 4, 6, and 8). The extracts or chromosomal fractions were immunoblotted for proteins indicated on the left. The migration of molecular size standards is indicated on the right for BubR1 and Bub1 blots. (B) Phosphatase treatment removes mobility shift of chromosomal BubR1 and Bub1. Chromosomal pellets prepared from untreated extracts (lanes 1, 3, 5, and 7) or extracts treated with nocodazole (lanes 2, 4, 6, and 8) were treated with phosphatase buffer (lanes 1, 2, 5, and 6) or LPP (lanes 3, 4, 7, and 8). The samples were then immunoblotted for BubR1 (lanes 1–4) or Bub1 (lanes 5–8). The migration of molecular size standards is indicated on the right.
The effect of BubR1 depletion on kinetochore localization of other spindle checkpoint proteins suggests an essential role of BubR1 in the checkpoint. When the spindle checkpoint is provoked in CSF-arrested extracts by incubation with nuclei and nocodazole, the extracts remain arrested at metaphase even after calcium is added to inactivate the CSF activity, as determined by sustained Cdc2-associated histone H1 kinase activity and condensed chromosomes (Minshull et al., 1994). In extracts depleted for endogenous BubR1, Cdc2 activity declined by 15 min after calcium addition (Fig. 5 A) and chromosomes were decondensed by 45 min (unpublished data), indicating that BubR1 is indeed required for the spindle checkpoint.
Figure 5.
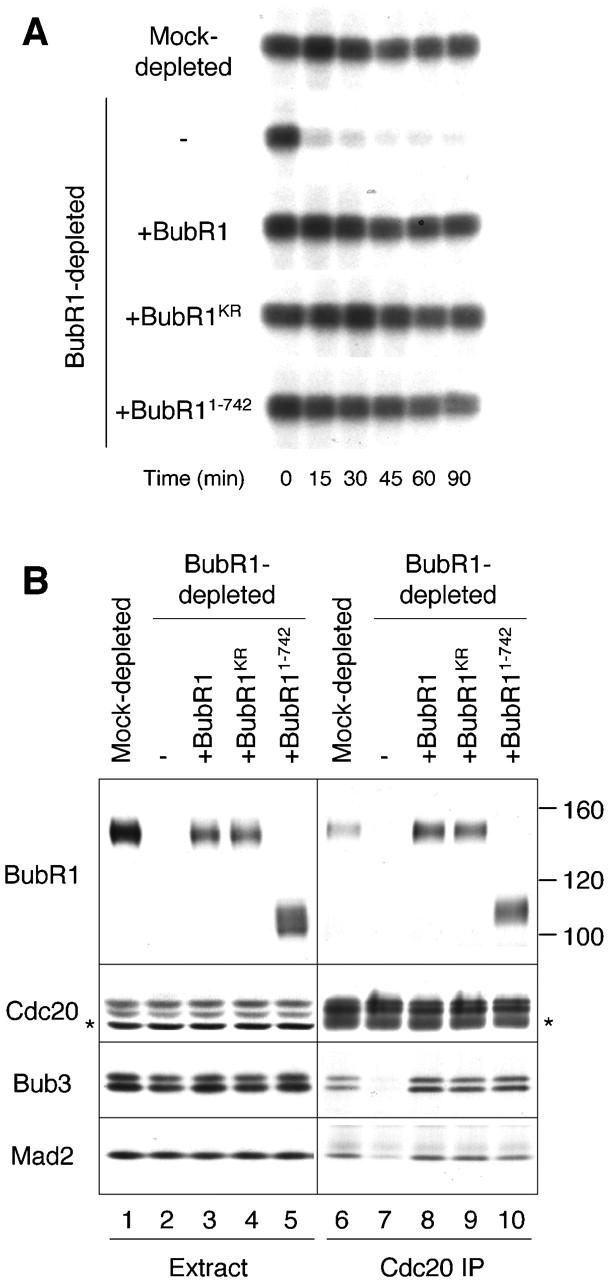
BubR1 is required for spindle checkpoint independently of its kinase activity. (A) Autoradiographs of histone H1 kinase assay. Extracts were depleted with a control IgG (mock depleted), or with anti-BubR1 antibody, and then supplemented with mock, wild-type BubR1, BubR1KR, or BubR11–742 as indicated on the left. The level of BubR1 protein in extracts was similar to that shown in B (lanes 1–5). After incubation with nuclei and nocodazole, calcium was added to the extracts to inactivate CSF activity and to trigger mitotic exit. Samples were taken immediately before calcium addition (time = 0) or every 15 min thereafter as indicated on bottom. (B) Extracts were depleted with a control IgG (lanes 1 and 6) or anti-BubR1 antibody and then supplemented with mock (lanes 2 and 7), wild-type BubR1 (lanes 3 and 8), BubR1KR (lanes 4 and 9), or BubR11–742 (lanes 5 and 10). The spindle checkpoint was induced in these extracts by incubation with nuclei and nocodazole, followed by anti-Cdc20 immunoprecipitation. The extracts (lanes 1–5) or the immunoprecipitates (lanes 6–10) were immunoblotted for proteins indicated on the left. Asterisks indicate a cross-reacting protein and IgG heavy chain on the left and right panels, respectively.
BubR1 contains a protein kinase domain at the carboxy-terminal region, raising the question of whether the kinase activity is required for its function. To address this question, I mutated the conserved lysine residue at position 788 in the kinase domain into arginine to generate a kinase-dead mutant (BubR1KR). I also generated a truncated BubR1 (BubR11–742) that contains amino acids 1–742 and lacks the kinase domain. I first determined if these mutant proteins were functional in the spindle checkpoint. To determine if mutant BubR1 proteins were able to support the checkpoint, RNAs corresponding to wild-type BubR1, BubR1KR, or BubR11–742 were prepared by in vitro transcription and used to produce the proteins directly in BubR1-depleted extracts as described previously for Bub1 (Sharp-Baker and Chen, 2001). Upon supplementing BubR1-depleted extracts with translations of wild-type BubR1, BubR1KR, or BubR11–742, Cdc2 activity was maintained after calcium addition (Fig. 5 A), indicating that all three proteins were able to support the spindle checkpoint. As expected from this finding, both BubR1KR and BubR11–742 coimmunoprecipitated with Cdc20, along with Mad2 and Bub3 (Fig. 5 B). The mutant proteins also restored kinetochore association of other checkpoint proteins, as determined by immunoblot analysis of isolated chromosomes (unpublished data). These results show that the kinase activity of BubR1 is not necessary for the spindle checkpoint in Xenopus egg extracts.
Mad1 is important for kinetochore localization and hyperphosphorylation of BubR1
Next, I determined if kinetochore localization of BubR1 requires other spindle checkpoint proteins. Immunofluorescence study shows that BubR1 was absent from kinetochores if chromosomes were prepared from Bub1-depleted extract in the presence of nocodazole (Fig. 6 A), indicating that kinetochore association of BubR1 is dependent on Bub1. Because Bub1 is also required for Bub3, Mad1, and Mad2 to bind kinetochores (Sharp-Baker and Chen, 2001), the effect of Bub1 depletion on BubR1 localization may be due to a lack of Bub3, Mad1, or Mad2 at kinetochores. To determine which of these checkpoint proteins is necessary for kinetochore association of BubR1, immunofluorescence staining of BubR1 was performed with chromosomes assembled in extracts depleted for Mad1 and/or Mad2. In extracts depleted for both Mad1 and Mad2, there was a slight reduction of BubR1 at kinetochores, and the remaining BubR1 was still at a higher level than that associated with metaphase kinetochores (Fig. 6 B). Adding back Mad1 alone or in combination with Mad2 to depleted extracts restored BubR1 at kinetochores to the level in mock-depleted extracts (Fig. 6 B). As shown previously (Chung and Chen, 2002), Mad1 alone without its partner Mad2 was able to bind kinetochores. On the other hand, Mad2 when added alone was unable to localize to kinetochores (Fig. 6 B; Chung and Chen, 2002) and restore BubR1 at kinetochores (Fig. 6 B). BubR1 at kinetochores was also reduced in extracts depleted for Mad1 by using anti-Mad1 antibody and was restored by the addition of Mad1 (unpublished data). These results show that Mad1 or its downstream effector, but not Mad2, is important for BubR1 to bind kinetochores.
Figure 6.
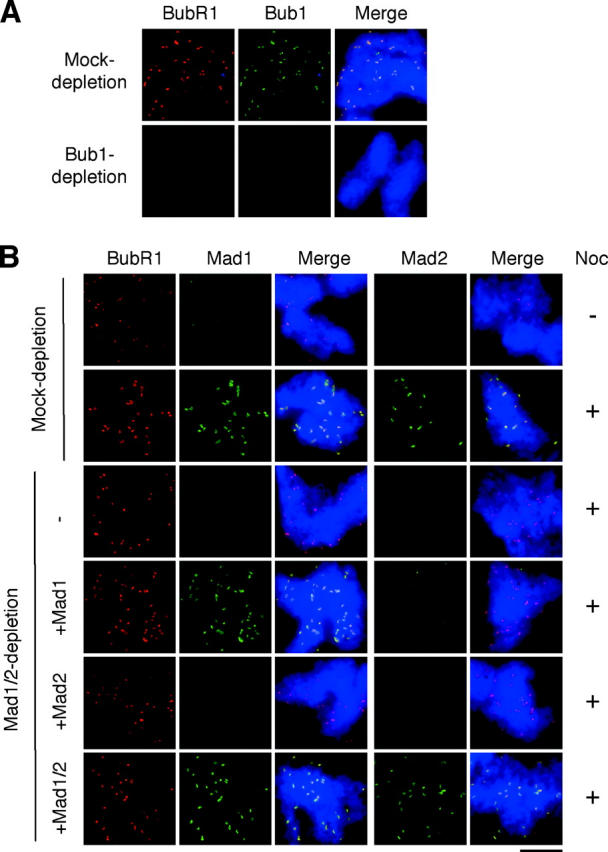
Localization of BubR1 to kinetochores is dependent on Bub1 and Mad1. (A) Bub1 is required for BubR1 to associate with kinetochores. Mitotic chromosomes were isolated from mock- or Bub1-depleted extracts in the presence of nocodazole and subjected to immunofluorescence staining for BubR1 and Bub1 as described for Fig. 3. The level of depletion is similar to that shown in Fig. 1 A. (B) Kinetochore association of BubR1 is reduced in the absence of Mad1. Extracts were mock depleted or depleted for both Mad1 and Mad2 (Mad1/2 depletion). The latter extracts were then supplemented with mock, Mad1, Mad2, or both Mad1 and Mad2 as indicated on the left. Mitotic chromosomes were assembled in these extracts lacking or containing nocodazole as indicated on the right. Chromosomes from each sample were isolated and loaded onto two coverslips. One set of the samples was stained for BubR1 and Mad1, the other for BubR1 and Mad2 as described for Fig. 3. For space conservation, BubR1 staining was shown for only one set of the samples. Bar, 10 μm. Immunoblot analysis of the extracts is similar to Fig. 7 (lanes 1–6).
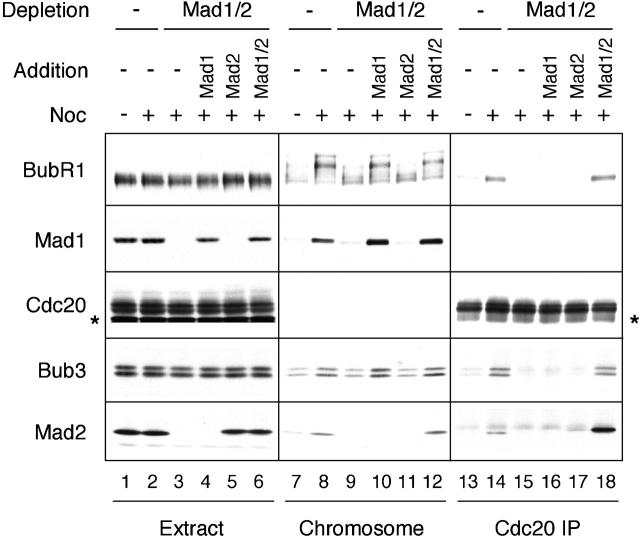
The level of BubR1 associated with kinetochores was also assessed by immunoblot analysis of purified chromosomes. In extracts depleted for both Mad1 and Mad2, there was a slight reduction of both BubR1 and Bub3 in the chromosomal fraction (Fig. 7, compare lanes 8 and 9), but both proteins were maintained at a slightly higher level than those from metaphase chromosomes (compare lanes 7 and 9). Strikingly, the remaining BubR1 protein lacks its characteristic mobility shift normally seen from unattached chromosomes (Fig. 7, compare lanes 8 and 9), indicating that the protein was not hyperphosphorylated in the absence of Mad1 and/or Mad2. Addition of Mad1 alone or both Mad1 and Mad2 together to depleted extracts restored hyperphosphorylation of BubR1 as well as the level of BubR1 and Bub3 on chromosomes (Fig. 7, lanes 10 and 12), whereas addition of Mad2 alone did not have the same effect (lane 11). These results show that hyperphosphorylation of BubR1 on unattached chromosomes requires Mad1 or its downstream effector, but not Mad2. Hyperphosphorylation of Bub1 was also dependent on Mad1, but not Mad2 (unpublished data).
Figure 7.
Hyperphosphorylation of chromosomal BubR1 requires Mad1. Mock-depleted extracts or extracts lacking Mad1 and/or Mad2 were prepared as described for Fig. 6 B (lanes 1–6). The extracts were incubated with nuclei (15,000 nuclei/μl extract) in the absence or presence of nocodazole as indicated on top. The samples were then subjected to chromosome isolation or immunoprecipitation with anti-Cdc20 antibody. The extracts (lanes 1–6), chromosomal fractions (lanes 7–12), and Cdc20 immunoprecipitates (lanes 13–18) were immunoblotted for proteins indicated on the left. The band above Mad2 in lanes 13–18 is the IgG light chain. The blots for Mad1 in Cdc20 immunoprecipitates and for Cdc20 in the chromosomal fractions are not shown, because of a lack of specific association. Exogenously added Mad1 and Mad2 tend to nonspecifically associate with Cdc20 immunoprecipitates, resulting in the high level of Mad2 in lane 18.
Extracts lacking Mad1 and/or Mad2 were also used to determine how the depletion affected coimmunoprecipitation of BubR1 and Bub3 with Cdc20. As also shown in Fig. 2, BubR1 and Bub3 were unable to interact with Cdc20 in extracts depleted for both Mad1 and Mad2 (Fig. 7, lane 15). Addition of either Mad1 or Mad2 alone to depleted extracts did not restore the interaction (Fig. 7, lanes 16 and 17), whereas addition of Mad1 and Mad2 together did (Fig. 7, lane 18). The hyperphosphorylated form of BubR1 was not detected in the Cdc20 immunoprecipitate (Figs. 2 and 7). These results show that association and hyperphosphorylation of BubR1 at kinetochores are not sufficient for BubR1 and Bub3 to interact with Cdc20 and that Mad2 is required for the interaction to occur.
Discussion
Mutual dependency between BubR1 and Mad2 in binding to Cdc20
The spindle checkpoint signal is generated from kinetochores that fail to bind spindle microtubules or are not under tension. Kinetochores recruit several spindle checkpoint proteins, including BubR1, Bub1, Bub3, Mad1, Mad2, and CENP-E (for review see Shah and Cleveland, 2000). These proteins form different complexes in the cytosol: BubR1–Bub3, Bub1–Bub3, Mad1–Mad2, and BubR1–CENP-E (for review see Shah and Cleveland, 2000). Similar to Bub1–Bub3 and Mad1–Mad2 complexes (Sharp-Baker and Chen, 2001; Chung and Chen, 2002), the interaction between BubR1 and Bub3 is also constant during the cell cycle in Xenopus egg extracts (Fig. 1 D). The bulk of Bub3 remains in the extracts depleted for BubR1 (Fig. 1 D and Fig. 2), indicating that only a small fraction of Bub3 molecules is bound with BubR1. Previous study has also shown that Bub1 associates with only a portion of Bub3 (Sharp-Baker and Chen, 2001). During mitosis, a fraction of BubR1, Bub3, and Mad2 binds to Cdc20, the activator for APC (Fig. 2). When spindle assembly is disrupted, Cdc20 associates with a higher level of BubR1, Bub3, and Mad2, compared with that at metaphase (Fig. 2). In vitro, recombinant BubR1 and Mad2 proteins are able to independently bind and inhibit Cdc20 (Fang et al., 1998; Tang et al., 2001). However, it has not been determined if BubR1 and Mad2 actually form separate complexes with Cdc20 in vivo. By immunodepletion of either BubR1 or Mad2 from extracts, I show that there is mutual dependency between BubR1 and Mad2 in binding to Cdc20 at metaphase and under the spindle checkpoint–active condition (Fig. 2).
One possible mechanism for the mutual dependency is that Cdc20 forms a complex with BubR1, Mad2, and Bub3, and that stable interaction requires both BubR1 and Mad2. Binding of BubR1 to Cdc20 may induce a conformational change in Cdc20, allowing Mad2 to interact with Cdc20, and vice versa. Alternatively, kinetochore localization of both BubR1 and Mad2 suggests that the effect of their depletion may be due to defects in regulation at kinetochores. Disruption of mitotic spindle assembly increases the level of BubR1, Bub3, and Mad2 present in Cdc20 immunoprecipitates. One possibility is that unattached chromosomes facilitate the protein complex formation by bringing these proteins into close proximity, thereby allowing their interaction. In addition, kinetochore association may convert the checkpoint proteins into a form that can readily bind Cdc20 and/or other checkpoint proteins. Upon activation or assembly of the protein complex, these checkpoint proteins may then leave kinetochores, as supported by FRAP analysis showing transient interaction of Mad2 with kinetochores (Howell et al., 2000).
In BubR1-depleted extracts, there is significant reduction of Bub1, Bub3, Mad1, Mad2, and CENP-E at unattached kinetochores (Figs. 3 and 4). Bub1 has been shown to be required for other spindle checkpoint proteins to bind kinetochores (Sharp-Baker and Chen, 2001). Thus, it is possible that the effect of BubR1 depletion is a consequence of reduced Bub1 at kinetochores. However, I cannot rule out the possibility that BubR1 may play a direct role in stabilizing Bub3, Mad1, and Mad2 at kinetochores, independently of Bub1. The reduction of kinetochore association of these checkpoint proteins in the absence of BubR1 may affect their interaction or activation, leading to an impaired assembly of Bub3 and Mad2 on Cdc20. The association between Bub3, Mad2, and Cdc20 at metaphase is also reduced without BubR1, indicating that BubR1 may also physically stabilize the complex.
Bub1 depletion abolishes kinetochore localization of BubR1 (Fig. 6 A). Bub1 is also essential for Bub3, Mad1, Mad2, and CENP-E to bind kinetochores in Xenopus egg extracts (Sharp-Baker and Chen, 2001). Bub3 has been shown to be required for BubR1 to localize to kinetochores in human cells (Taylor et al., 1998). I now further demonstrate that Mad1 also plays a role in this process. In extracts depleted with anti-Mad2 antibodies, Mad1 is removed along with Mad2. In these extracts, localization of BubR1 to kinetochores is slightly reduced (Fig. 6 B). This defect is rescued by the addition of Mad1 alone (Fig. 6 B). The lack of both Bub3 and Mad1 at kinetochores likely contributes to the dramatic effect of Bub1 depletion on kinetochore localization of BubR1.
Mad1 depletion also abolishes binding of BubR1 to Cdc20 (Fig. 7). Interestingly, addition of Mad1 alone restores kinetochore association, but not Cdc20 binding (Fig. 6 B and Fig. 7), showing that kinetochore localization of BubR1 is not sufficient for its interaction with Cdc20 and that Mad2 is necessary for BubR1 to bind Cdc20. Several possibilities may account for this defect. First, in the absence of Mad2 at kinetochores, BubR1 may not adopt the conformational change required for its full activation, so that it cannot bind Cdc20. Second, Cdc20 may fail to localize to kinetochores in the absence of Mad2, so that it is unable to interact with BubR1. I cannot detect Cdc20 on kinetochores by either immunofluorescence staining or immunoblot analysis, possibly because of very transient interaction of Cdc20 with kinetochores. Third, BubR1, Mad2, and Bub3 may need to first assemble into a complex at kinetochores before leaving these loci along with or without Cdc20, so that BubR1 becomes immobilized at kinetochores in the absence of Mad2. The last hypothesis would predict a reduced turnover rate of BubR1 at kinetochores in Mad2-depleted extracts, which can be determined by FRAP analysis.
Kinase-deficient BubR1 supports spindle checkpoint
Human BubR1 is identified as a protein that shares homology with budding yeast Mad3 (Taylor et al., 1998). Thus, BubR1 has been thought to be the homologue of Mad3 in metazoans, even though yeast Mad3 lacks a protein kinase domain. Many aspects of the interaction between yeast Mad3 with other spindle checkpoint proteins are similar to my observation with BubR1 in egg extracts (Hardwick et al., 2000; Fraschini et al., 2001). However, unlike the Xenopus counterparts, coimmunoprecipitation of Mad2 with Cdc20 in yeast is not dependent on Mad3 and is only moderately affected by bub1 or bub3 mutations (Hardwick et al., 2000). This discrepancy suggests that budding yeast Mad3 and Xenopus BubR1 may be regulated through different mechanisms or have a different role in the spindle checkpoint.
In budding yeast, Mad3 has been placed downstream of Mad1 and Mad2 on the basis that Mad1 is still hyperphosphorylated during mitosis in mad3 mutants (Hardwick and Murray, 1995). It is not clear whether Xenopus BubR1 is also dispensable for Mad1 phosphorylation, because no mitosis-specific phosphorylation of Mad1 has been detected in egg extracts (unpublished data). Immunofluorescence staining and immunoblot analysis show that depletion of BubR1 greatly reduces kinetochore localization of Bub1, Bub3, Mad1, Mad2, and CENP-E (Figs. 3 and 4), indicating that BubR1 is a regulator of these checkpoint proteins. It remains to be determined whether yeast Mad3 is also required for other checkpoint proteins to bind kinetochores. Such comparative analysis is essential for understanding whether BubR1 and Mad3 are indeed functional homologues or if there are distinct regulatory mechanisms involving BubR1 and Mad3 in metazoans and yeast, respectively.
Extracts lacking BubR1 fail to support the spindle checkpoint (Fig. 5). This defect can be explained by the reduction of kinetochore localization of other spindle checkpoint proteins and the lack of interaction between Cdc20, Mad2, and Bub3. Interestingly, addition of wild-type BubR1, BubR1KR, or BubR11–742 to the depleted extracts restores the checkpoint and allows Cdc20 to associate with Mad2 and Bub3 (Fig. 5). These results indicate that the kinase domain of BubR1 is not necessary for the spindle checkpoint in egg extracts, consistent with a previous study showing that recombinant human BubR1 with similar mutations is able to bind and inhibit Cdc20 in vitro (Tang et al., 2001). In this regard, BubR1 appears to be similar to yeast Mad3 that lacks a kinase domain. The checkpoint function of Bub1 has also been shown to be independent of its kinase activity in egg extracts (Sharp-Baker and Chen, 2001). However, it remains a possibility that the kinase activity of BubR1 and Bub1 may enhance the efficiency of the spindle checkpoint, which is difficult to determine in egg extracts.
Regulatory mechanism of BubR1 phosphorylation
BubR1 accumulates to a higher level and becomes hyperphosphorylated at unattached kinetochores compared with that at metaphase kinetochores (Fig. 4). This phosphorylation requires Mad1 or its downstream effector, but not Mad2 (Fig. 7). It has also been shown that human BubR1 in the chromosomal fraction exhibits a higher degree of phosphorylation than the soluble counterpart (Chan et al., 1999). Hyperphosphorylation of BubR1 may be attributed to enrichment or activation of its upstream kinase at unattached kinetochores. Mad1 may recruit a kinase to unattached kinetochores, allowing efficient phosphorylation of BubR1 by bringing these molecules into close proximity. Upon microtubule binding, the kinase may dissociate from or become inactivated at kinetochores. The candidate kinase for BubR1 includes Mps1, Bub1, and MAP kinase, all of which are kinetochore associated and important for the spindle checkpoint. Alternatively, a protein phosphatase may be regulated at kinetochores by microtubules. Mad1 may prevent the phosphatase from gaining access to BubR1. Upon microtubule attachment to kinetochores, Mad1 dissociates from kinetochores, leading to dissociation of the kinase for BubR1 and/or allowing the phosphatase to target BubR1. Hyperphosphorylation of BubR1 may allow its stable interaction with kinetochores, reflecting a higher level of the protein on unattached kinetochores. Phosphorylation may also be involved in the recruitment of other spindle checkpoint proteins to kinetochores.
To date, the known function of Mad1 is to recruit Mad2 to kinetochores (Chen et al., 1998). My finding that Mad1 is also necessary for BubR1 hyperphosphorylation reveals a second function for Mad1. In addition, the dependence of BubR1 hyperphosphorylation and kinetochore association on Mad1 suggests that Mad1 is a regulator for BubR1. Together, this study demonstrates that the spindle checkpoint involves a network of regulatory mechanisms, rather than a simple, linear pathway.
Materials and methods
Isolation of Xenopus BubR1 cDNA
A Xenopus sequence in the EST database (BE025630) shows homology with the known human and mouse BubR1. To isolate the full-length cDNA, oligonucleotides were generated according to the sequences at the ends of the EST sequence. These oligonucleotides were used as primers to obtain additional sequences both 5′ and 3′ to the EST sequence using SMART™ RACE cDNA Amplification Kit (CLONTECH Laboratories Inc.). The template for the reaction was generated by reverse transcription from Xenopus egg total mRNA with oligo-dT as a primer.
Preparation of recombinant BubR1 and generation of anti-BubR1 antibodies
To generate recombinant BubR1 protein, the sequence encoding amino acids 189–359 was amplified by PCR and subcloned into pQE9 (QIAGEN) at a BamH1 site. The protein was expressed in Escherichia coli with a hexahistidine tag at the NH2 terminus and purified as previously described for Xenopus Mad2 protein (Chen et al., 1996). The purified protein was used to raise antibodies in rabbits (Covance) and to affinity purify the antibodies as previously described (Chen et al., 1996).
Preparation of Xenopus egg extracts, spindle checkpoint assay, immunofluorescence, immunoblot, and immunodepletion
CSF-arrested extracts were prepared from unfertilized Xenopus eggs that were arrested at metaphase of the second meiotic division by CSF. The extract and demembranated sperm nuclei were prepared as previously described (Murray, 1991). To prepare interphase extracts, the CSF-arrested extracts were driven into interphase by incubation with 0.5 mM calcium chloride for 30 min at 23°C, and another 30-min incubation in the presence of 100 ng/μl cycloheximide to prevent synthesis of cyclin B and mitotic entry. The checkpoint extracts were prepared in CSF-arrested extract by incubation with 15,000 sperm nuclei per μl extract at 23°C for 10 min and another 20-min incubation with 10 ng/μl nocodazole. Once the checkpoint is activated, addition of calcium is unable to induce the mitotic exit, as determined by condensed chromosomes and a sustained Cdc2-associated histone H1 kinase activity. Spindle checkpoint assays, immunofluorescence, immunoblot, and immunodepletion were performed as previously described (Chen et al., 1998; Sharp-Baker and Chen, 2001). Immunoprecipitation of Cdc20 was performed as previously described (Chung and Chen, 2002). Anti-BubR1 antibodies were biotinylated as previously described (Sharp-Baker and Chen, 2001).
Generation of kinase-deficient BubR1 and translation of proteins in egg extracts
The kinase-dead BubR1 was generated using QuikChange (Stratagene) to change the conserved lysine 788 in the kinase domain of BubR1 to arginine. To make truncated BubR1 lacking the kinase domain, the region encoding amino acids 1–742 was amplified by PCR. To produce proteins directly in egg extracts, the coding region of wild-type or mutant BubR1, Mad1, or Mad2 was cloned into a modified pGEM transcription vector. In vitro transcription and translation in egg extracts were performed as previously described (Sharp-Baker and Chen, 2001). BubR1 was translated in extracts depleted for BubR1. Mad1 and Mad2 were separately translated in extracts depleted for endogenous Mad1 and Mad2. Different batches of translation gave consistent results.
Immunoblot of chromosomal proteins
Sperm nuclei (15,000 nuclei/μl extract) were incubated at 23°C for 10 min in 40 μl of CSF-arrested extracts, followed by incubation in the presence or absence of nocodazole (10 μg/ml) for another 20 min. At the end of incubation, samples were diluted with 360 μl ice-cold lysis buffer (10 mM K2HPO4, pH 7.2, 50 mM β-glycerophosphate, 1 mM EDTA, 5 mM EGTA, 1 mM MgCl2, 1 mM sodium vanadate, 0.1% Triton X-100, and 10 μg/ml each of leupeptin, pepstatin, and chymostatin, and 200 nM microcystin-LR), and mixed gently by inverting the tubes a few times. They were then layered over 1 ml of lysis buffer containing 30% sucrose and spun in an HB-6 rotor at 10,000 rpm for 10 min. After centrifugation, the supernatant was removed and the chromosome pellets were washed with 0.5 ml of lysis buffer containing 30% sucrose, and spun again in an HB-6 rotor at 10,000 rpm for 5 min. After removing the supernatant, the pellet was solubilized in SDS-PAGE sample buffer and analyzed by immunoblot. The major proteins obtained with these procedures were histones and XCAPs (Chung and Chen, 2002). Ponceau S staining of the blots also showed consistent levels of these proteins among samples.
Lambda protein phosphatase (LPP) treatment
Extracts (1 μl) or chromosome pellets were incubated for 30 min at 30°C with 40 U of lambda phosphatase (New England Biolabs, Inc.) in 30 μl of lambda protein phosphatase (LPP) reaction buffer. Control reactions were performed under the same conditions in 30 μl LPP reaction buffer with 2 mM ZnCl2, 1 mM sodium vanadate, and 50 mM sodium fluoride as phosphatase inhibitors.
Acknowledgments
I thank Julie Heck (Cornell University) for isolating the cDNA for Xenopus BubR1, and Arian Abrieu (University of California, San Diego, San Diego, CA) for providing biotinylated anti–CENP-E antibody.
This work was supported by grants from the National Institutes of Health and the David and Lucille Packard Foundation.
Footnotes
Abbreviations used in this paper: APC, anaphase promoting complex; CSF, cytostatic factor; LPP, lambda protein phosphatase.
References
- Abrieu, A., L. Magnaghi-Jaulin, J.A. Kahana, M. Peter, A. Castro, S. Vigneron, T. Lorca, D.W. Cleveland, and J. Labbe. 2001. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 106:83–93. [DOI] [PubMed] [Google Scholar]
- Amon, A. 1999. The spindle checkpoint. Curr. Opin. Genet. Dev. 9:69–75. [DOI] [PubMed] [Google Scholar]
- Basu, J., H. Bousbaa, E. Logarinho, Z. Li, B.C. Williams, C. Lopes, C.E. Sunkel, and M.L. Goldberg. 1999. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol. 146:13–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill, D.P., C. Lengauer, J. Yu, G.J. Riggins, J.K. Willson, S.D. Markowitz, K.W. Kinzler, and B. Vogelstein. 1998. Mutations of mitotic checkpoint genes in human cancers. Nature. 392:300–303. [DOI] [PubMed] [Google Scholar]
- Chan, G.K., B.T. Schaar, and T.J. Yen. 1998. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J. Cell Biol. 143:49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, G.K., S.A. Jablonski, V. Sudakin, J.C. Hittle, and T.J. Yen. 1999. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J. Cell Biol. 146:941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R.-H., J.C. Waters, E.D. Salmon, and A.W. Murray. 1996. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 274:242–246. [DOI] [PubMed] [Google Scholar]
- Chen, R.H., A. Shevchenko, M. Mann, and A.W. Murray. 1998. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143:283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, E., and R.-H. Chen. 2002. The spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol. Biol. Cell. 13:1501–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, G., H. Yu, and M.W. Kirschner. 1998. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 12:1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr, K.A., and M.A. Hoyt. 1998. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol. Cell. Biol. 18:2738–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini, R., A. Beretta, L. Sironi, A. Musacchio, G. Lucchini, and S. Piatti. 2001. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 20:6648–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick, K., and A.W. Murray. 1995. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 131:709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick, K.G., E. Weiss, F.C. Luca, M. Winey, and A.W. Murray. 1996. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 273:953–956. [DOI] [PubMed] [Google Scholar]
- Hardwick, K.G., R.C. Johnston, D.L. Smith, and A.W. Murray. 2000. MAD3 encodes a novel component of the spindle checkpoint that interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 148:871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B.J., D.B. Hoffman, G. Fang, A.W. Murray, and E.D. Salmon. 2000. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J. Cell Biol. 150:1233–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, L.H., L.F. Lau, D.L. Smith, C.A. Mistrot, K.G. Hardwick, E.S. Hwang, A. Amon, and A.W. Murray. 1998. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 279:1041–1044. [DOI] [PubMed] [Google Scholar]
- Jablonski, S.A., G.K. Chan, C.A. Cooke, W.C. Earnshaw, and T.J. Yen. 1998. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 107:386–396. [DOI] [PubMed] [Google Scholar]
- Kim, S.H., D.P. Lin, S. Matsumoto, A. Kitazono, and T. Matsumoto. 1998. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 279:1045–1047. [DOI] [PubMed] [Google Scholar]
- Li, X., and R.B. Nicklas. 1995. Mitotic forces control a cell cycle checkpoint. Nature. 373:630–632. [DOI] [PubMed] [Google Scholar]
- Li, Y., and R. Benezra. 1996. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 274:246–248. [DOI] [PubMed] [Google Scholar]
- Millband, D.N., and K.G. Hardwick. 2002. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol. Cell. Biol. 22:2728–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull, J., H. Sun, N.K. Tonks, and A.W. Murray. 1994. MAP-kinase dependent mitotic feedback arrest in Xenopus egg extracts. Cell. 79:475–486. [DOI] [PubMed] [Google Scholar]
- Murray, A.W. 1991. Cell cycle extracts. Methods Cell Biol. 36:581–605. [PubMed] [Google Scholar]
- Page, A.M., and P. Hieter. 1999. The anaphase-promoting complex: new subunits and regulators. Annu. Rev. Biochem. 68:583–609. [DOI] [PubMed] [Google Scholar]
- Rieder, C.L., R.W. Cole, A. Khodjakov, and G. Sluder. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130:941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J.V., and D.W. Cleveland. 2000. Waiting for anaphase: Mad2 and the spindle assembly checkpoint. Cell. 103:997–1000. [DOI] [PubMed] [Google Scholar]
- Sharp-Baker, H., and R.H. Chen. 2001. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 153:1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin, V., G.K. Chan, and T.J. Yen. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154:925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z., R. Bharadwaj, B. Li, and H. Yu. 2001. Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 1:227–237. [DOI] [PubMed] [Google Scholar]
- Taylor, S.S., E. Ha, and F. McKeon. 1998. The human homologue of bub3 is required for kinetochore localization of bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, J.C., R.H. Chen, A.W. Murray, and E.D. Salmon. 1998. Localization of mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 141:1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, E., and M. Winey. 1996. The S. cerevisiae SPB duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132:111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and E. Lees. 2001. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol. Cell. Biol. 21:5190–5199. [DOI] [PMC free article] [PubMed] [Google Scholar]