Abstract
As invading viruses do not harbor functional ribosomes in their virions, successful amplification of the viral genomes requires that viral mRNAs compete with cellular mRNAs for the host cell translation apparatus. Several RNA viruses have evolved remarkable strategies to recruit the host translation initiation factors required for the first steps in translation initiation by host cell mRNAs. This review describes the ways that three families of RNA viruses effectively usurp limiting translation initiation factors from the host.
Keywords: translation; picornavirus; rotavirus; cricket paralysis–like virus; start codon selection
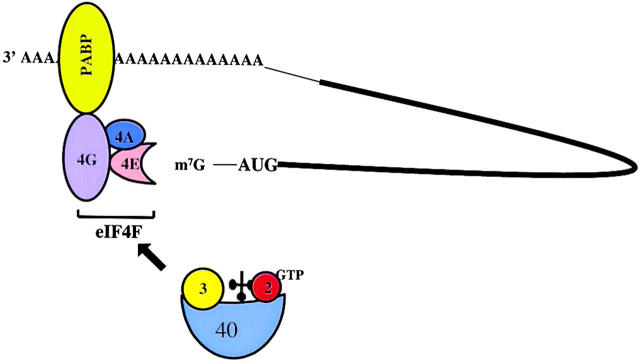
In most eukaryotic mRNAs, translation initiation commences with the recruitment of the cap binding protein complex eukaryotic initiation factor (eIF)4F, composed of factors eIF4E (cap binding protein), eIF4A, and eIF4G (Fig. 1), to the capped 5′ end (Hershey and Merrick, 2000). Subsequently, the 40S ribosomal subunit, carrying eIF3 and the ternary initiator tRNA-eIF2-GTP complex, are recruited to the 5′ end of the mRNA via interaction of eIF3 with eIF4G (Hershey and Merrick, 2000). The 40S subunits then scans the mRNA in a 5′ to 3′ direction until an appropriate start codon is encountered. At this point, the anticodon in initiator tRNA (tRNAi Met),* positioned in the ribosomal P-site, engages in base pairing with the start codon in the mRNA. The large ribosomal 60S subunit joins and protein synthesis commences (Hershey and Merrick, 2000). A variety of auxiliary proteins (Pestova et al., 2001; Dever, 2002), not covered in this review, aid in the selectivity and proccessivity of the start-site selection process. In addition, it has been shown that the polyadenosine binding protein (PABP) interacts with eIF4G (Fig. 1) (Sachs and Varani, 2000). It is thought that the resulting translational enhancement is due to efficient re-loading of ribosomes which have reached the termination codon to the 5′ end of the mRNA. Alternatively, circularization of the mRNA, oligomerization of proteins, or conformational changes in eIF4F may lead to increased mRNA translation or mRNA stability (Sachs, 2000). We will provide three examples of RNA viruses which have been found to interfere with some of these key steps in translational initiation.
Figure 1.
Model depicting the major participants that are involved in translational initiation in eukaryotic mRNAs. Interactions of eukaryotic translation initiation factors eIF2 (2), eIF3 (3), and initiator tRNA with a 40S ribosomal subunit, eIF4E (4E), eIF4A (4A), and eIF4G (4G) with the m7G cap structure, and the polyadenosine binding protein PABP with the polyadenosine tail in an mRNA are shown.
Proteolysis of eIF4G and PABP, and sequestration of eIF4E: picornaviruses
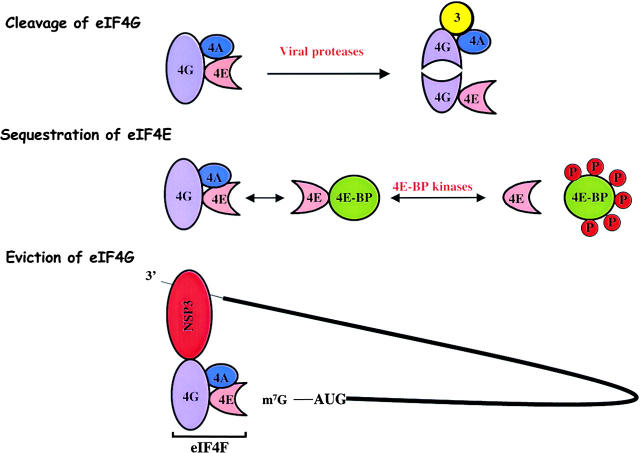
It has been known for a long time that infection of cultured cells with poliovirus, a member of the Picornaviridae, results in the translational inhibition of host but not viral mRNAs (Holland and Peterson, 1964). Subsequently, it was shown that the positive-stranded viral mRNA is translated by an internal ribosome entry mechanism that does not require an intact eIF4F complex (Pelletier and Sonenberg, 1988). Specifically, virus-encoded protease 2A cleaves the eIF4G component of eIF4F at a specific site within the 176-kD protein (Etchison et al., 1982; Lamphear et al., 1993). The NH2-terminal cleavage product contains the binding site for eIF4E, PABP, and the COOH-terminal product harbors the binding sites for eIF3 and eIF4A (Fig. 2). From these results, it was hypothesized that the NH2-terminal eIF4G fragment was not sufficient to recruit ribosomes to the complexed 5′ ends of host cell mRNAs. Viral mRNA translation was shown to be mediated by an internal ribosome entry site (IRES), an ∼500 nucleotide RNA sequence in the 5′ noncoding region of the viral RNA, which was postulated to recruit ribosomes without the need for an intact eIF4F. However, it was noted that cleavage of eIF4G precedes inhibition of host protein synthesis, and conditions could be found in which eIF4G was efficiently cleaved in infected cells in the absence of host translational shutoff (Bonneau and Sonenberg, 1987; Irurzun et al., 1995). These findings suggested that the cleavage of eIF4G was not sufficient for host protein synthesis shutoff.
Figure 2.
Alterations of the cap binding protein complex eIF4F in infected cells. (Top) Cleavage of eIF4G by picornaviral proteases. (Middle) Sequestration of eIF4E by 4E binding proteins (4E-BP) due to dephosphorylation of 4E-BPs in picornavirus-infected cells. (Bottom) Eviction of PABP by viral NSP3 from the cap binding protein complex eIF4F in rotavirus- infected cells. Interactions of eukaryotic initiation factors eIF4G (4G), eIF4A (4A), eIF4E (4E), and eIF3 (3) are indicated.
Recent discoveries showed that cleavage of eIF4G, now called eIF4GI, is only one of several eIFs that can be cleaved in picornavirus-infected cells. First, Gradi et al. (1998) discovered a functional homologue of eIF4GI, termed eIF4GII, whose cleavage correlated better with the temporal inhibition of translation in both poliovirus- and rhinovirus-infected cells (Gradi et al., 1998; Svitkin et al., 1999). Secondly, Joachims et al. (1999) and Kerekatte et al. (1999) noted that polyadenosine binding protein (PABP) is proteolyzed during both coxsackievirus and poliovirus infection by viral 2A and 3C proteases (Joachims et al., 1999; Kerekatte et al., 1999). Specifically, PABP molecules that are associated with both ribosomes and polyadenosine sequences are preferentially cleaved during poliovirus infection at four distinct sites (Kuyumcu-Martinez et al., 2002). These cleavages remove the COOH-terminal fragment of PABP, which contains bindings sites for other PABP molecules and for the 60S ribosomal subunit. Therefore, it appears that picornaviruses have evolved to cleave eIF4GI, eIF4GII and PABP to destroy both the eIF4F complex and end-to-end communication in cellular mRNAs. The hypothesis was suggested that the viral IRES could recruit the COOH-terminal fragment of eIF4GI and eIF4GII, which contain binding sites for eIF4A and eIF3 (Fig. 2), to facilitate internal initiation. Recent data from Ali et al. (2001b) have shown that the cleaved COOH-terminal fragment of eIF4GI; however, stimulates translation of both capped and IRES-containing mRNAs (Ali et al., 2001b). However, higher concentrations of the eIF4GI COOH-terminal fragment were required for optimal translation of capped mRNAs than for IRES-containing mRNAs, suggesting that the viral RNA can efficiently compete with cellular mRNAs for the limiting amounts of truncated eIF4GI and eIF4GII present in infected cells.
Not all picornaviruses inhibit host cell translation by proteolysis of eIF4GI or eIF4GII. Insights into the mechanism of inhibition of host cell translation in cells infected with encephalomyocarditis virus (EMCV) were provided with the discovery of eIF4E binding proteins (4E-BPs), a group of proteins that sequesters the eIF4E component of eIF4F (Gingras et al., 1999). Because the binding sites in eIF4E for 4E-BP and for eIF4G overlap, binding of 4E-BP to eIF4E competitively inhibits the recruitment of eIF4E into eIF4F (Fig. 2). When cell growth is stimulated by serum, growth factors or hormones, 4E-BPs are phosphorylated, reducing their affinity for eIF4E, allowing the assembly of larger amounts of functional eIF4F to ensure efficient cap-dependent translational initiation (Fig. 2). It has been shown that dephosphorylation of 4E-BPs occurs in cells infected with both EMCV and poliovirus, resulting in sequestration of eIF4E by 4E-BPs (Gingras et al., 1996). Therefore, lowering the abundance of the cap binding protein complex either by cleavage of eIF4GI and eIF4GII or by sequestration of eIF4E by 4E-BPs selectively inhibits translation of capped host cell mRNAs without inhibiting the translation of IRES-containing picornaviral mRNAs. It is now known that an eIF4GI fragment, containing the binding sites for eIF4A and eIF3, and eIF4A are sufficient to recruit ternary 40S complexes to picornavirus IRESs (Pestova et al., 1996a, 1996b). However, the IRES in hepatitis A virus is a curious exception because its activity is dependent on an intact cap binding protein complex eIF4F, and thus mediates translation initiation only poorly when eIF4E is sequestered by phosphorylated 4E-BPs (Ali et al., 2001a).
Substitution of PABP: rotaviruses
An astonishing mechanism to disrupt host cell mRNA circularization, leading to severe inhibition of host mRNA translation, has been recently discovered in cells infected with rotavirus. Rotavirus, a member of the Reoviridae, contains eleven double-stranded RNA segments. All of those are transcribed into mRNAs that possess a 5′ terminal cap structure but lack 3′ terminal poly(A) tails (Patton and Spencer, 2000). Instead, the 3′ end sequences contain a tetranucleotide motif which is conserved among different groups of rotaviruses. These features would seem to indicate that the viral mRNAs are not subjected to circularization-mediated translational enhancement. However, Piron et al. (1998) discovered that the viral NSP3 protein binds specifically to the conserved viral 3′ end sequences (Piron et al., 1998). Using two-hybrid screens in yeast, it was found that NSP3 also interacts with a binding site in eIF4G that overlaps with the binding site for PABP (Fig. 2) (Piron et al., 1998). Because eIF4G has a higher affinity for NSP3 than PABP, the interaction between PABP and eIF4G is disrupted in rotavirus-infected cells (Michel et al., 2000; Vende et al., 2000). The two consequences of NSP3 expression, then, are reduced efficiency of host mRNA translation and circularization-mediated translational enhancement of rotavirus mRNAs. Recently, the x-ray structure of NSP3 has revealed that NSP3 forms an asymmetric homodimer around the conserved sequence at the 3′ end of the viral mRNAs (Deo et al., 2002). Different amino acids in each subunit contact the viral RNA via α-helical surfaces instead of the β-sheets usually present in the RNA binding domain sequences of proteins. Because the terminal nucleotides are completely buried within NSP3 (Deo et al., 2002), it is thought that the NSP3-RNA complex confers stability to the viral mRNAs, protecting them from 3′–5′ exonucleases. This example elegantly illustrates how a virus that encodes nonpolyadenylated mRNAs can usurp the host cell translation apparatus by encoding a protein that binds to the 3′ ends of the viral mRNAs, evicting PABP from eIF4G, the key player involved in the recruitment of ribosomes to mRNAs (Figs. 1 and 2).
Bypass of initiator tRNA: cricket paralysis–like viruses
No matter by which mechanism ribosomal subunits are recruited to cellular and viral mRNAs discussed this far, subsequent positioning of the start codon in the ribosomal P-site is followed by pairing of the tRNAi Met anticodon with the mRNA start codon. Catalyzed by the GTPase activating protein eIF5, the tRNAi Met-associated factor eIF2 is then released from the 40S subunit as a binary complex with guanosine 5'-diphosphate (GDP) (Hershey and Merrick, 2000). The binary eIF2-GDP complex can not bind tRNAi Met; instead, eIF2-GDP must be recycled, forming eIF2-GTP, by the guanine nucleotide exchange factor eIF2B.
This recycling step is a major translational control step in eukaryotic cells. At least four distinct cellular kinases can phosphorylate Ser-51 of the α-subunit of eIF2; phosphorylated eIF2-GDP can not be recycled to eIF2-GTP by eIF2B. Functional eIF2B is limiting in cells and, thus, accumulation of phosphorylated eIF2 results in rapid and severe inhibition of most host cell translation due to diminished amounts of 40S subunits that are competent for translational initiation by virtue of carrying ternary eIF2-GTP- tRNAi Met complexes (Kaufman, 1999). However, it is known that a few mRNAs which contain small open reading frames in their 5′ leader sequences, can be translated when phosphorylated eIF2 is abundant in cells (Dever et al., 1992; Harding et al., 2000; Novoa et al., 2001). The presence of these upstream open reading frames ensures that these mRNAs are normally poorly translated; however, accumulation of phosphorylated eIF2 causes 40S subunits to bypass translation initiation at some of these small open reading frames, allowing efficient translation initiation at the major open reading frame by a reinitiation mechanism (Hinnebusch, 1997). This mechanism ensures that translation at a certain downstream start codons can occur when ternary eIF2-GTP-tRNAi Met complexes are low; however, translation initiation remains dependent on tRNAi Met and its ternary complex. That this is not always the case is exemplified by recent discoveries in insect cricket paralysis–like viruses.
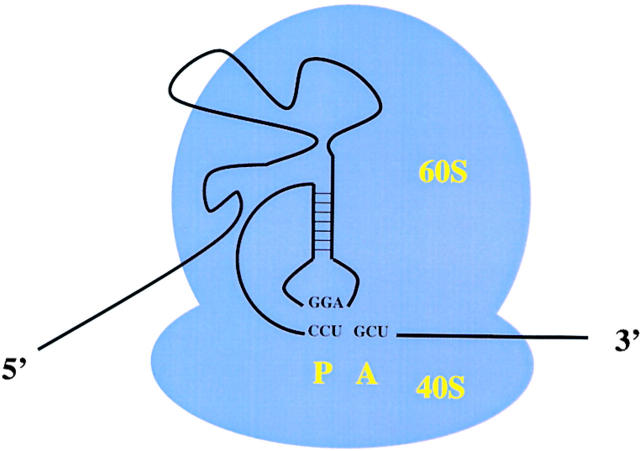
Cricket paralysis–like viruses are positive-strand RNA viruses that have a bicistronic genome organization (Liljas et al., 2002). The first large open reading which encodes the viral nonstructural proteins, is followed by a 200-nucleotide intergenic region (IGR) and an open reading frame which encodes the viral structural proteins (Hellen and Sarnow, 2001). It was known for a long time that the structural proteins are synthesized late in infection, and accumulate to higher concentrations than the nonstructural gene products (Moore et al., 1981). However, the mechanism by which the IGR mediates translation of the structural proteins from the full-length mRNA has remained an enigma until recently. Sasaki and Nakashima (1999) showed that Plautia Stali intestine virus, a member of the cricket paralysis–like virus family, contains an internal ribosome entry site (IRES) in its IGR sequence element which can mediate translation initiation at a noncognate CUU start codon without tRNAi Met (Sasaki and Nakashima, 1999, 2000). This finding was unprecedented because only cognate (i.e. AUG) or weak-cognate (i.e. CUG or GUG) codons have been known to function as start-site codons. The mechanism of this unusual start-site codon usage was unraveled by Wilson et al. (2000a)(2000b) who showed that in cricket paralysis virus the IGR-IRES sequences themselves occupy the ribosomal P-site (Fig. 3); a CCU triplet at the start site base pairs directly with upstream IGR-IRES sequences (Wilson et al., 2000a, 2000b). Biochemical analysis of the ribosome recruitment process by the IGR-IRES has revealed that the IRES can recruit both 40S and 60S subunits without any known canonical eIFs to form an 80S ribosome that can start protein synthesis from the next codon, a GCU, which is located in the ribosomal A-site (Fig. 3). Thus, the first amino acid in the protein is alanine encoded by the A-site located GCU codon (Wilson et al., 2000a, 2000b). These findings argue that the IGR-IRES element can propel the ribosome into elongation mode without prior formation of a peptide bond. Why have the cricket paralysis virus–like viruses evolved an RNA element that can initiate protein synthesis when intracellular amounts of ternary eIF2-GTP- tRNAi Met complexes are low? Recent findings have shown that eIF2 is heavily phosphorylated in cricket paralysis virus–infected cells at a time when the synthesis of viral structural proteins is at a maximum level (Bushell and Sarnow, unpublished). Efficient translation mediated by the IGR-IRES at those times suggests that the viral gene amplification has evolved to be resistant to host antiviral responses such as the activation of eIF2 kinases. Identification of the activated eIF2 kinases in infected cells should open another window through which one can view the fierce battle of viral and cellular mRNAs for the translation apparatus.
Figure 3.
Occupation of the ribosomal P-site by the cricket paralysis virus IRES. Basepair interactions between sequences in the viral IRES positioned in the ribosomal P-site (P) are diagramed. An empty ribosomal A-site (A) that can accept the first elongator tRNA molecule is shown.
Acknowledgments
We would like to thank Karla Kirkegaard for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (AI25105 and GM55979) to P. Sarnow, and from The Wellcome Trust (063233/B/00/Z) to M. Bushell.
Footnotes
Abbreviations used in this paper: eIF, eukaryotic initiation factor; EMCV, encephalomyocarditis virus; GDP, guanosine 5′-diphosphate; IGR, intergenic region; IRES, internal ribosome entry site; tRNAiMet, initiator tRNA-methionine; PABP, polyadenosine binding protein.
References
- Ali, I.K., L. McKendrick, S.J. Morley, and R.J. Jackson. 2001. a. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 75:7854–7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, I.K., L. McKendrick, S.J. Morley, and R.J. Jackson. 2001. b. Truncated initiation factor eIF4G lacking an eIF4E binding site can support capped mRNA translation. EMBO J. 20:4233–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau, A.M., and N. Sonenberg. 1987. Proteolysis of the p220 component of the cap-binding protein complex is not sufficient for complete inhibition of host cell protein synthesis after poliovirus infection. J. Virol. 61:986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo, R.C., C.M. Groft, K.R. Rajashankar, and S.K. Burley. 2002. Recognition of the rotavirus mRNA 3′ consensus by an asymmetric NSP3 homodimer. Cell. 108:71–81. [DOI] [PubMed] [Google Scholar]
- Dever, T.E. 2002. Gene-specific regulation by general translation factors. Cell. 108:545–556. [DOI] [PubMed] [Google Scholar]
- Dever, T.E., L. Feng, R.C. Wek, A.M. Cigan, T.F. Donahue, and A.G. Hinnebusch. 1992. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 68:585–596. [DOI] [PubMed] [Google Scholar]
- Etchison, D., S.C. Milburn, I. Edery, N. Sonenberg, and J.W.B. Hershey. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000 dalton polypeotide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257:14806–14810. [PubMed] [Google Scholar]
- Gingras, A.C., Y. Svitkin, G.J. Belsham, A. Pause, and N. Sonenberg. 1996. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc. Natl. Acad. Sci. USA. 93:5578–5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913–963. [DOI] [PubMed] [Google Scholar]
- Gradi, A., Y.V. Svitkin, H. Imataka, and N. Sonenberg. 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA. 95:11089–11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, H.P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 6:1099–1108. [DOI] [PubMed] [Google Scholar]
- Hellen, C.U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593–1612. [DOI] [PubMed] [Google Scholar]
- Hershey, J.W.B., and W.C. Merrick. 2000. The pathway and mechanism of initiation of protein synthesis. Translational Control of Gene Expression. N. Sonenberg, J.W.B. Hershey, and M.B. Mathews, editors. Cold Spring Harbor Press, Cold Spring Harbor, NY. 33–88.
- Hinnebusch, A.G. 1997. Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J. Biol. Chem. 272:21661–21664. [DOI] [PubMed] [Google Scholar]
- Holland, J.J., and J.A. Peterson. 1964. Nucleic acid and protein synthesis during poliovirus infection of human cells. J. Mol. Biol. 8:556–573. [DOI] [PubMed] [Google Scholar]
- Irurzun, A., S. Sanchez-Palomino, I. Novoa, and L. Carrasco. 1995. Monensin and nigericin prevent the inhibition of host translation by poliovirus, without affecting p220 cleavage. J. Virol. 69:7453–7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachims, M., P.C. Van Breugel, and R.E. Lloyd. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, R.J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211–1233. [DOI] [PubMed] [Google Scholar]
- Kerekatte, V., B.D. Keiper, C. Badorff, A. Cai, K.U. Knowlton, and R.E. Rhoads. 1999. Cleavage of Poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J. Virol. 73:709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez, N.M., M. Joachims, and R.E. Lloyd. 2002. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J. Virol. 76:2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear, B.J., R. Yan, F. Yang, D. Waters, H.D. Liebig, H. Klump, E. Kuechler, T. Skern, and R.E. Rhoads. 1993. Mapping the cleavage site in protein synthesis initiation factor eIF-4 gamma of the 2A proteases from human Coxsackievirus and rhinovirus. J. Biol. Chem. 268:19200–19203. [PubMed] [Google Scholar]
- Liljas, L., J. Tate, T. Lin, P. Christian, and J.E. Johnson. 2002. Evolutionary and taxonomic implications of conserved structural motifs between picornaviruses and insect picorna-like viruses. Arch. Virol. 147:59–84. [DOI] [PubMed] [Google Scholar]
- Michel, Y.M., D. Poncet, M. Piron, K.M. Kean, and A.M. Borman. 2000. Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 275:32268–32276. [DOI] [PubMed] [Google Scholar]
- Moore, N.F., B. Reavy, and J.S. Pullin. 1981. Processing of cricket paralysis virus induced polypeptides in Drosophila cells: production of high molecular weight polypeptides by treatment with iodoacetamide. Arch. Virol. 68:1–8. [DOI] [PubMed] [Google Scholar]
- Novoa, I., H. Zeng, H.P. Harding, and D. Ron. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153:1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, J.T., and E. Spencer. 2000. Genome replication and packaging of segmented double-stranded RNA viruses. Virology. 277:217–225. [DOI] [PubMed] [Google Scholar]
- Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 334:320–325. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., C.U. Hellen, and I.N. Shatsky. 1996. a. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., V.G. Kolupaeva, I.B. Lomakin, E.V. Pilipenko, I.N. Shatsky, V.I. Agol, and C.U. Hellen. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA. 98:7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., I.N. Shatsky, and C.U. Hellen. 1996. b. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 16:6870–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piron, M., P. Vende, J. Cohen, and D. Poncet. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 17:5811–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, A.B. 2000. Physical and functional interactions between the mRNA cap structure and the poly(A) tail. Translational Control of Gene Expression. N. Sonenberg, J.W.B. Hershey, and M.B. Mathews, editors. Cold Spring Harbor Press, Cold Spring Harbor, NY. 447–465.
- Sachs, A.B., and G. Varani. 2000. Eukaryotic translation initiation: there are (at least) two sides to every story. Nat. Struct. Biol. 7:356–361. [DOI] [PubMed] [Google Scholar]
- Sasaki, J., and N. Nakashima. 1999. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 73:1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, J., and N. Nakashima. 2000. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. USA. 97:1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin, Y.V., A. Gradi, H. Imataka, S. Morino, and N. Sonenberg. 1999. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J. Virol. 73:3467–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vende, P., M. Piron, N. Castagne, and D. Poncet. 2000. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J. Virol. 74:7064–7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.E., T.V. Pestova, C.U. Hellen, and P. Sarnow. 2000. a. Initiation of protein synthesis from the A site of the ribosome. Cell. 102:511–520. [DOI] [PubMed] [Google Scholar]
- Wilson, J.E., M.J. Powell, S.E. Hoover, and P. Sarnow. 2000. b. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 20:4990–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]