Summary
Hybridization in plants and animals is more common and has more complex outcomes than previously realized. Genome-wide analyses of introgression in organisms ranging from oaks to sunflowers to fruit flies show that a substantial fraction of their genomes are permeable to alleles from related species. Hybridization can lead to rapid genomic changes, including chromosomal rearrangements, genome expansion, differential gene expression, and gene silencing, some of which are mediated by transposable elements. These genomic changes may lead to beneficial new phenotypes, and selection for fertility and ecological traits may in turn alter genome structure. Dramatic increases in the availability of genomic tools will produce a new understanding of the genetic nature of species and will resolve a century-old debate over the basis of hybrid vigour, while the natural recombinants found in hybrid zones will permit genetic mapping of species differences and reproductive barriers in non-model organisms.
In 1742, seven years after writing “nullae dantur species novae” (there are no new species) [1] Carolus Linnaeus was brought a fertile floral mutant of Linaria that he called “Peloria”. The unusual floral structure convinced Linnaeus that the plant was of hybrid origin, and the fertility of Peloria and other hybrids led Linnaeus to abandon his earlier certainty in the fixed nature of species. Instead, he proposed the radical evolutionary hypothesis that new species could arise via hybridization [2]. Despite this illustrious pedigree, hybrid speciation had little scientific support until early in 20th century when cytogenetic studies showed that hybridization may lead to speciation, especially if accompanied by chromosomal doubling (allopolyploidy). While these studies persuaded many 20th century botanists that hybridization was a common and significant force in evolution, this view was often disputed by zoologists. Now, three centuries after the birth of Linneaus, hybridization is seen as an important phenomenon in many taxa, contributing to adaptation and speciation in plants, fish, and insects.
The resurgent interest in hybridization is closely linked to the shift from genetic to genomic approaches. In this review, we take a genomic perspective on introgression and hybrid speciation. We limit ourselves to hybridization between sexually reproducing organisms, and so do not consider horizontal gene transfer in prokaryotes, between organelles and the nucleus, or interspecific transfers between organelles [reviewed in 3]. We focus on detecting hybrids and on the genomic and evolutionary consequences of introgression and hybrid speciation, while ignoring the effects of genome duplication.
Introgression and hybrid speciation
Hybridization, the production of viable offspring from interspecific matings, occurs in roughly 10% of animal species and 25% of plant species [4]. It may sometimes lead to introgression, defined here as the stable integration of genetic material from one species into another through repeated back-crossing [5]; the transient presence of alien alleles is excluded as being of little evolutionary importance. Hybridization may also lead to speciation, in which the new hybrid lineages become reproductively isolated from parental populations. While allopolyploid species have complete genomes of each parental species, homoploid hybrid species, which have not undergone chromosomal duplication, typically have unequal parental genomic contributions due to back-crossing.
Challenges in detecting introgression and hybrid speciation, and recent progress
The extent of introgression and hybrid speciation is unclear: occasional hybridization may not lead to permanent genetic exchange, so the frequency of hybridizing species may overestimate the rate of introgression. On the other hand, young species have weaker isolating barriers than older species on average, so species that do not currently hybridize may have done so in the past [4]. This uncertainty highlights both the challenges of detecting past hybridization and introgression events and the recent development of many of the essential tools.
Introgression is frequently given as an explanation when phylogenetic trees inferred from different genes are in conflict. However, processes other than introgression can produce incongruent gene trees. For example, if a large population that is polymorphic at many loci splits into several species, well-supported phylogenetic trees inferred for different genes may reflect the different histories of those genes in the parent population rather than introgression. This uncertainty can lead to conflicting interpretations of genetic data, exemplified by the recent debate over introgression in European oaks [6–8] and in the domestication of rice [9].
An alternative approach to detecting introgression uses comparisons of sequence data from multiple individuals per species. If species do not hybridize, then interspecific differences in sequence should accrue over time, while introgression will lead to decreased levels of interspecific divergence. The patterns of polymorphism and fixed differences depend on the migration rates among species, as well as the population size of ancestral and daughter populations—larger ancestral populations contain higher levels of sequence polymorphism and larger daughter populations retain polymorphisms longer. The statistical models required to estimate this set of parameters are complex and computationally intensive, and estimates of population sizes and migration rates often have wide confidence intervals. However, this approach has been successfully applied to detect introgression in chimps, fish, and butterflies [10–12]. Recent work has introduced an additional complication: patterns of introgression vary across the genome [13*]. Alleles at some loci do not introgress while others do so freely, presumably due to variation in the fitnesses of foreign alleles in different genetic or ecological backgrounds. These results suggest that coalescence-based approaches have considerable power, but larger genome-scale analyses will be required to understand whether rampant genetic exchange is the rule for hybridizing species or whether much of the genome is resistant to introgression.
Genomic studies allow robust detection of interspecific genetic exchange, estimation of genome-wide patterns of introgression, and determination of the sizes of parental chromosomal blocks in introgressed populations and hybrid species [14,15]. From a diagnostic standpoint, associations among alleles of one species in the genetic background of a close relative provide compelling evidence of recent introgression. This interspecific linkage disequlibrium also makes it possible to distinguish between ancestral polymorphism and introgression as explanations for phylogenetic incongruence among genes. If a group of species has a history of introgression, two classes of gene trees should be generated, representing the two species involved in the hybridization event. Also, closely linked loci should share gene trees [16].
What have genomic studies of introgression revealed about species barriers?
Genome-wide analyses of introgression in many species, including cottonwood trees [17], mice [18,19], oaks [8], fruit flies [20], mosquitoes [21], and sunflowers [22*] consistently find that different portions of the genome vary in their permeability to foreign alleles (Figure 1). There also is tremendous variation in the overall porosity of these genomes; most of the markers studied in cottonwoods did not introgress [17], while only a small fraction of the genome was differentiated in mosquitoes [21].
Figure 1.
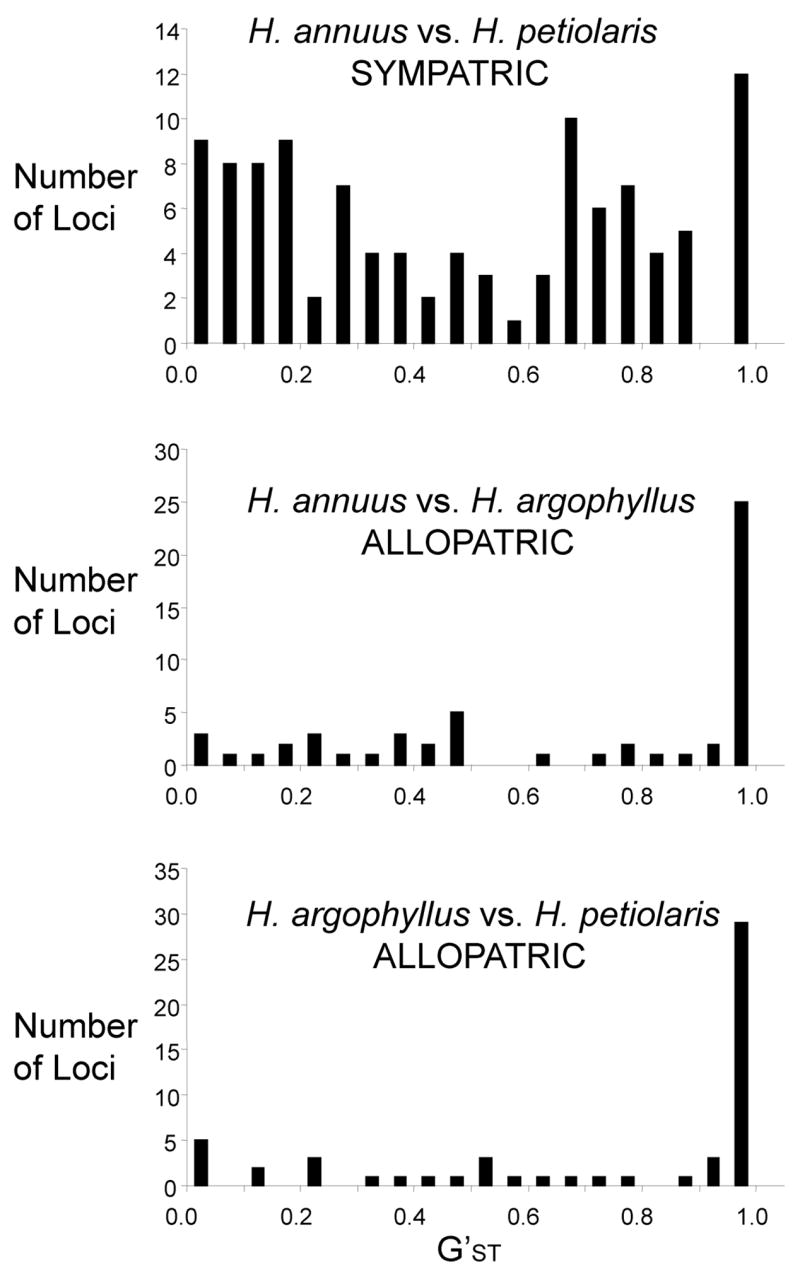
Genetic differentiation between three species of sunflower. Helianthus annuus and H. argophyllus are sister taxa that have non-overlapping ranges, while H. petiolaris is a more distantly related species that overlaps with H. annuus. Genetic differentiation varies dramatically across the genome.
How do we account for these different introgression patterns? The ease of introgression depends on the selection:recombination ratio [23], so reproductive barriers should not impede the introgression of neutral or advantageous alleles unless they are tightly linked to sites that contribute to reproductive isolation. Thus, for any given level of hybridization, less introgression is expected between species with complex, multilocus reproductive barriers. Likewise, introgression should be reduced in genomic regions with low levels of recombination. The latter prediction has been confirmed by recent work in Drosophila and Helianthus, in which introgression is diminished in low recombination regions adjacent to chromosomal breakpoints [20,22*]. In both cases, the scale of suppression was surprisingly limited: 5 centimorgans in sunflowers, 2.5 megabases in fruit flies (ca. 12.5 centimorgans).
Linking patterns of introgression to the fitness effects of different loci has been slower, but the reduced introgression of X-linked loci in mice compared to autosomal loci is consistent with the greater abundance of species incompatibilities on sex chromosomes [18,19]. These studies illustrate the potential power of genomic analyses of introgression for identifying the genetic basis of species differences, as these loci are least likely to introgress. Also, the natural recombinants found in hybrid zones or stabilized hybrid lineages permit admixture mapping of species differences and reproductive barriers [24,25]. In sunflowers, for example, chromosomal blocks associated with pollen sterility in natural hybrid zones also exhibit reduced introgression [26].
Genomic alterations in hybrids
Hybridization can result in genomic changes including alterations to gene expression, chromosomal structure, and genome size. A series of studies on synthetic allopolyploids (which combine hybridization with genome duplication) reveals that substantial changes occur immediately upon hybridization, including gene loss, gene silencing, changes in gene expression, and tissue-specific expression of copies of some loci from the two genomes [27*]. Genome duplication appears to have a weaker effect on genome change, in one case stabilizing expression patterns that were altered in the diploid hybrid [28]. Mechanistic explanations for these changes are preliminary, but are known to involve transposons in some cases [29,30].
Genomic alterations seen in homoploid hybrid species may be even more dramatic. Analysis of genome structure in three synthetic and three natural hybrid species of sunflower revealed massive karyotypic change over a handful of hybrid generations [31,32]. While some of the karyotypic differences arose through the sorting of chromosomal rearrangements that distinguish the parental species, most arose de novo. The natural hybrid species also exhibit increased genome sizes – nearly 50% larger than the parental species [33]. Despite multiple independent origins [34], all species show similar increases in genome size due to the proliferation of retrotransposons [35**]. Screens of F1 to F5 synthetic hybrids and of hybrid zone plants failed to detect an increase in genome size [33], so both the trigger and timing of this retrotransposon explosion remains unknown. Lastly, microarray analyses of one of the three hybrid species, Helianthus deserticola, revealed that that approximately 2% of genes had extreme or transgressive expression in the hybrid species [36]. Although these expression differences maybe adaptively important (transport-related proteins were over-represented), it is not clear whether they arose as a consequence of hybridization.
Evolutionary consequences of introgression and hybrid speciation
First generation hybrids often exhibit an increase in size, growth rate, and yield. This hybrid vigour or heterosis has been exploited by plant and animal breeders since Darwin’s time, but its genetic basis remains controversial [37]. The dominance hypothesis, which has enjoyed long theoretical support, posits that deleterious recessive alleles are complemented in hybrids by fitter alleles from the alternate parent, generating an increase in vigour. In contrast, the overdominance hypothesis attributes hybrid vigour to the synergistic interactions of alleles at a heterozygous locus. However, it is difficult to distinguish true overdominance from the reciprocal complementation of deleterious alleles at linked loci (pseudo-overdominance), leading to scepticism about the former’s importance[38]. More recent hypotheses include the possibility that heterosis is caused by synergistic interactions among alleles at different loci (epistasis) or by non-additive changes in gene expression [39,40]. However, the latter hypothesis is best interpreted as a possible molecular mechanism underlying classical genetic models [41].
Recent gene mapping and expression studies support a pluralist explanation for heterosis. Dominance and overdominance are most frequently implicated by QTL studies, with epistasis reported less frequently [37]. Although QTL studies cannot rule out pseudo-overdominance, the preferential association of overdominant QTLs with heterotic phenotypes in tomato [42], and the demonstration of heteroic effects of the erecta mutant in Arabidopsis thaliana [43], are consistent with true overdominance. Less progress has been made toward determining the molecular mechanism(s) underlying heterosis. While non-additive gene expression is common in hybrids, it has not yet been linked to heterotic phenotypes. Also, non-additive gene expression may sometimes result from mis-expression in hybrids rather than synergistic interactions among alleles [44,45]. Lastly, studies in maize and rice suggest that small-scale duplications and deletions may contribute to heterosis via the complementation of deleted loci [46].
Although heterosis may play an important role in the establishment of asexual or allopolyploid hybrids [47], its effects are diminished in later generation hybrid segregants such as introgressive lineages or homoploid hybrid species due to increasing homozygosity. Here, success depends on the fixation of favourable new gene combinations from the two species [48]. For example, there are suggestions that new hybrid gene combinations facilitated critical ecological changes in several recently proposed hybrid species, including sculpins [49], Rhagoletis flies [50], and butterflies [51,52*]. In these cases, species of hybrid origin have colonized novel habitats, most likely through the expression of transgressive traits. Transgressive segregation, in which hybrids have more extreme trait values than either parent, can occur when parental species contain alleles with opposing effects. For example, a recent study in irises documented that although the upland Iris brevifolia cannot tolerate flooding, the introgression of its alleles into the flood-tolerant Iris fulva increases fulva’s survival [53].
The most detailed analysis of the evolutionary forces responsible for fixing favored combinations of parental alleles or chromosomal segments in hybrids focused on recombination in the homoploid hybrid sunflowers [31]. Analysis revealed that selection on ecological traits had slightly higher power than selection for fertility when predicting hybrid genomic composition, although both forms of selection were significant. Also, simulation studies suggest that while reproductive isolation between the recombinant hybrid species and the parental species arose in a few generations, the complete elimination of interspecific chromosomal polymorphism required several hundred generations [14]. The genomes of animal species of hybrid origin have received little attention as yet, so the factors determining their composition are unknown.
Progress is also being made toward understanding the molecular mechanisms underlying phenotypic variation in hybrids [54**]. For example, comparison of flowering time in synthetic tetraploids of Arabidopsis arenosa and A. thaliana revealed that while the autotraploids flower early, the synthetic allotetraploid plants flower late, mimicking the late flowering of the natural allotetraploid between the same two parents. The late flowering phenotype results from complementation of reciprocally down-regulated loci in the two parental species, as well as epigenetic changes, which in turn correlate with histone acetylation and methylation. These changes brought about by hybridization likely have important fitness consequences. Early flowering plants may escape drought while late flowering plants may be more reliably pollinated and may have more resources to allocate to flowering. In addition, the shift in flowering time results in an immediate reproductive barrier between allotetraploid and its parental species.
Future steps
Genomic data have already reshaped our understanding of the nature of species barriers, revealing that key morphological and ecological differences can persist even as genes move between species at many loci. So far, most of these insights derive from species that are currently hybridizing, but inexpensive sequence data make it feasible to estimate the extent and timing of introgression at all stages of species divergence. Likewise, the ability to rapidly and inexpensively estimate the size and parentage of chromosomal segments in hybrids makes it possible to determine the importance of different kinds of reproductive barriers in resisting introgression, estimate the ages of hybrid zones, calculate the speed of hybrid speciation, and identify the factors responsible for hybrid genomic composition. Finally, advances in genomics and molecular biology should rapidly reveal the genetic and molecular mechanisms underlying heterosis and other phenotypic changes in hybrids.
Acknowledgments
The authors were supported in this work by grants from the National Institutes of Health (USA), the National Science Foundation (USA), and the National Sciences and Engineering Research Council (Canada).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production procbess errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Linnaeus C. Systema naturae. Neiuwkoop: de Graaf; 1735. [Google Scholar]
- 2.Larson JL. The species concept of Linnaeus. Isis. 1968;59:291–299. [Google Scholar]
- 3.Syvanen M, Kado C, editors. Horizontal gene transfer. San Diego: Academic Press; 2002. [Google Scholar]
- 4.Mallet J. Hybridization as an invasion of the genome. Trends in Ecology & Evolution. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Rieseberg LH, Wendel JF. Introgression and its consequences in plants. In: Harrison RG, editor. Hybrid zones and the evolutionary process. Oxford University Press; 1993. pp. 70–109. [Google Scholar]
- 6.Muir G, Schlotterer C. Evidence for shared ancestral polymorphism rather than recurrent gene flow at microsatellite loci differentiating two hybridizing oaks (Quercus spp) Molecular Ecology. 2005;14:549–561. doi: 10.1111/j.1365-294X.2004.02418.x. [DOI] [PubMed] [Google Scholar]
- 7.Lexer C, Kremer A, Petit RJ. Shared alleles in sympatric oaks: recurrent gene flow is a more parsimonious explanation than ancestral polymorphism. Molecular Ecology. 2006;15:2007–2012. doi: 10.1111/j.1365-294X.2006.02896.x. [DOI] [PubMed] [Google Scholar]
- 8.Scotti-Saintagne C, Mariette S, Porth I, Goicoechea PG, Barreneche T, Bodenes K, Burg K, Kremer A. Genome scanning for interspecific differentiation between two closely related oak species [Quercus robur L. and Q petraea (Matt) Liebl.] . Genetics. 2004;168:1615–1626. doi: 10.1534/genetics.104.026849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sang T, Ge S. The puzzle of rice domestication. Journal of Integrative Plant Biology. 2007;49:760–768. [Google Scholar]
- 10.Kronforst MR, Young LG, Blume LM, Gilbert LE. Multilocus analyses of admixture and introgression among hybridizing Heliconius butterflies. Evolution. 2006;60:1254–1268. [PubMed] [Google Scholar]
- 11.Won YJ, Hey J. Divergence population genetics of chimpanzees. Molecular Biology and Evolution. 2005;22:297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
- 12.Won YJ, Sivasundar A, Wang Y, Hey J. On the origin of Lake Malawi cichlid species: A population genetic analysis of divergence. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6581–6586. doi: 10.1073/pnas.0502127102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bull V, Beltran M, Jiggins CD, McMillan WO, Bermingham E, Mallet J. Polyphyly and gene flow between non-sibling Heliconius species. BMC Biology. 2006;4 doi: 10.1186/1741-7007-4-11. * Four loci were sequenced from two species of butterfly, Heliconius cydno and H. melpomene, both of which occur in Panama. Two loci were reciprocally monophyletic, but alleles of the other two were shared between the two species. Bayesian modelling suggests that this is due to introgression at one of the loci and ancestral polymorphism at the other. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buerkle CA, Rieseberg LH. The rate of genome stabilization in homoploid hybrid species. Evolution. 2007 doi: 10.1111/j.1558-5646.2007.00267.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrell PL, Williams-Coplin TD, Lattu AL, Bowers JE, Chandler JM, Patterson AH. Crop-to-weed introgression has impacted allelic composition of johnsongrass populations with and without recent exposure to cultivated sorghum. Molecular Ecology. 2005;14:2143–2154. doi: 10.1111/j.1365-294X.2005.02579.x. [DOI] [PubMed] [Google Scholar]
- 16.Linder CR, Rieseberg LH. Reconstructing patterns of reticulate evolution UN plants. American Journal of Botany. 2004;91:1700–1708. [PMC free article] [PubMed] [Google Scholar]
- 17.Martinsen GD, Whitham TG, Turek RJ, Keim P. Hybrid populations selectively filter gene introgression between species. Evolution. 2001;55:1325–1335. doi: 10.1111/j.0014-3820.2001.tb00655.x. [DOI] [PubMed] [Google Scholar]
- 18.Macholan M, Munclinger P, Sugerkova M, Dufkova P, Bimova B, Bozikova E, Zima J, Pialek J. Genetic analysis of autosomal and X-linked markers across a mouse hybrid zone. Evolution. 2007;61:746–771. doi: 10.1111/j.1558-5646.2007.00065.x. [DOI] [PubMed] [Google Scholar]
- 19.Payseur BA, Krenz JG, Nachman MW. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution. 2004;58:2064–2078. doi: 10.1111/j.0014-3820.2004.tb00490.x. [DOI] [PubMed] [Google Scholar]
- 20.Machado CA, Haselkorn TS, Noor MAF. Evaluation of the genomic extent of effects of fixed inversion differences on intraspecific variation and interspecific gene flow in Drosophila pseudoobscura and D-persimilis. Genetics. 2007;175:1289–1306. doi: 10.1534/genetics.106.064758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. Plos Biology. 2005;3:1572–1578. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yatabe Y, Kane NC, Scotti-Saintagne C, Rieseberg LH. Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and Helianthus petiolaris. Genetics. 2007;175:1883–1893. doi: 10.1534/genetics.106.064469. *This study looks at differentiation of coding and non-coding loci in three species of sunflower. Helianthus annuus and H. argophyllus are sister species with non-overlapping ranges, while H. petiolaris is less closely related but overlaps extensively with H. annuus. The sister species are more differentiated genetically than two phylogenetically more distant species that occur together and hybridize. Differentiation between H. annuus and H. petiolaris ranges from zero to complete, depending on the locus examined. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barton NH. Gene flow past a cline. Heredity. 1979;43:333–339. [Google Scholar]
- 24.Lexer C, Buerkle CA, Joseph JA, Heinze B, Fay MF. Admixture in European Populus hybrid zones makes feasible the mapping of loci that contribute to reproductive isolation and trait differences. Heredity. 2007;98:74–84. doi: 10.1038/sj.hdy.6800898. [DOI] [PubMed] [Google Scholar]
- 25.Payseur BA, Place M. Searching the genomes of inbred mouse strains for incompatibilities that reproductively isolate their wild relatives. Journal of Heredity. 2007;98:115–122. doi: 10.1093/jhered/esl064. [DOI] [PubMed] [Google Scholar]
- 26.Rieseberg LH, Whitton J, Gardner K. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics. 1999;152:713–727. doi: 10.1093/genetics/152.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams KL, Wendel JF. Allele-specific, bidirectional silencing of an alcohol dehydrogenase gene in different organs of interspecific diploid cotton hybrids. Genetics. 2005;171:2139–2142. doi: 10.1534/genetics.105.047357. *Over the past decade, many studies have examined the genomic changes seen in synthesized polyploids, including changes in gene expression. It was not known whether these changes were due to hybridization or genome duplication. Adams and Wendel looked at gene expression of alcohol dehydrogenase in several tissues of diploid F1 cotton hybrids. They found that some crosses resulted in repeatable reciprocal tissue-specific expression of alternative alleles, and that these changes were not due to gene deletions. This study documents that repeatable changes in gene expression occur due to hybridization without chromosome doubling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hegarty MJ, Barker GL, Wilson ID, Abbott RJ, Edwards KJ, Hiscock SJ. Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Current Biology. 2006;16:1652–1659. doi: 10.1016/j.cub.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 29.Levy A, Feldman M. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biological Journal of the Linnean Society. 2004;82:607–613. [Google Scholar]
- 30.Chen ZJ, Ni ZF. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. Bioessays. 2006;28:240–252. doi: 10.1002/bies.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karrenberg S, Lexer C, Rieseberg LH. Reconstructing the history of selection during homoploid hybrid speciation. The American Naturalist. 2007;169:725–737. doi: 10.1086/516758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai Z, Nakazato T, Salmaso M, Burke JM, Tang SX, Knapp SJ, Rieseberg LH. Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics. 2005;171:291–303. doi: 10.1534/genetics.105.042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baack EJ, Whitney KD, Rieseberg LH. Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytologist. 2005;167:623–630. doi: 10.1111/j.1469-8137.2005.01433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarzbach AE, Rieseberg LH. Likely multiple origins of a diploid hybrid sunflower. Molecular Ecology. 2002;11:1703–1715. doi: 10.1046/j.1365-294x.2002.01557.x. [DOI] [PubMed] [Google Scholar]
- 35.Ungerer MC, Strakosh SC, Zhen Y. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Current Biology. 2006;16:R872–R873. doi: 10.1016/j.cub.2006.09.020. **Initial studies in Drosophila found little evidence for a role for transposons in species differences. Ungerer examined copy number of a Ty3/Gypsy-like retrotransposon in three species of sunflower that originated from hybridization and the two parental species. Quantitative PCR found 5–24 fold increases in copy number in the hybrid speices, accounting for 62–79% of the difference in genome size between the hybrid species and their parents. [DOI] [PubMed] [Google Scholar]
- 36.Lai Z, Gross BL, Zou Y, Andrews J, Rieseberg LH. Microarray analysis reveals differential gene expression in hybrid sunflower species. Molecular Ecology. 2006;15:1213–1227. doi: 10.1111/j.1365-294X.2006.02775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lippman ZB, Zamir D. Heterosis: revisiting the magic. Trends in Genetics. 2007;23:60–66. doi: 10.1016/j.tig.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 38.Cockerham CC, Zeng ZB. Design III with marker loci. Genetics. 1996;143:1437–1456. doi: 10.1093/genetics/143.3.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanson-Wagner RA, Jia Y, DeCook R, Borsuk LA, Nettleton D, Schnable PS. All possible modes of gene action are observed in a global comparison of gene expression in a maize F-1 hybrid and its inbred parents. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6805–6810. doi: 10.1073/pnas.0510430103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hua JP, Xing YZ, Wu WR, Xu CG, Sun XL, Yu SB, Zhang QF. Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2574–2579. doi: 10.1073/pnas.0437907100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Birchler JA, Auger DL, Riddle NC. In search of the molecular basis of heterosis. Plant Cell. 2003;15:2236–2239. doi: 10.1105/tpc.151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Semel Y, Nissenbaum J, Menda N, Zinder M, Krieger U, Issman N, Pleban T, Lippman Z, Gur A, Zamir D. Overdominant quantitative trait loci for yield and fitness in tomato. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12981–12986. doi: 10.1073/pnas.0604635103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Redei GP. Single locus heterosis. Z Vererbungsl. 1962;93:164–170. [Google Scholar]
- 44.Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics. 2005;171:1813–1822. doi: 10.1534/genetics.105.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michalak P, Noor MAF. Genome-wide patterns of expression in Drosophila pure species and hybrid males. Molecular Biology and Evolution. 2003;20:1070–1076. doi: 10.1093/molbev/msg119. [DOI] [PubMed] [Google Scholar]
- 46.Springer NM, Stupar RM. Allelic variation and heterosis in maize: How do two halves make more than a whole? Genome Research. 2007;17:264–275. doi: 10.1101/gr.5347007. [DOI] [PubMed] [Google Scholar]
- 47.Facon B, Jarne P, Pointier JP, David P. Hybridization and invasiveness in the freshwater snail Melanoides tuberculata: hybrid vigour is more important than increase in genetic variance. Journal of Evolutionary Biology. 2005;18:524–535. doi: 10.1111/j.1420-9101.2005.00887.x. [DOI] [PubMed] [Google Scholar]
- 48.Barton NH. The role of hybridization in evolution. Molecular Ecology. 2001;10:551–568. doi: 10.1046/j.1365-294x.2001.01216.x. [DOI] [PubMed] [Google Scholar]
- 49.Nolte AW, Freyhof J, Stemshorn KC, Tautz D. An invasive lineage of sculpins, Cottus sp (Pisces, Teleostei) in the Rhine with new habitat adaptations has originated from hybridization between old phylogeographic groups. Proceedings of the Royal Society B-Biological Sciences. 2005;272:2379–2387. doi: 10.1038/rspb.2005.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwarz D, Shoemaker KD, Botteri NL, McPheron BA. A novel preference for an invasive plant as a mechanism for animal hybrid speciation. Evolution. 2007;61:245–256. doi: 10.1111/j.1558-5646.2007.00027.x. [DOI] [PubMed] [Google Scholar]
- 51.Mavarez J, Salazar CA, Bermingham E, Salcedo C, Jiggins CD, Linares M. Speciation by hybridization in Heliconius butterflies. Nature. 2006;441:868–871. doi: 10.1038/nature04738. [DOI] [PubMed] [Google Scholar]
- 52.Gompert Z, Fordyce JA, Forister ML, Shapiro AM, Nice CC. Homoploid hybrid speciation in an extreme habitat. Science. 2006;314:1923–1925. doi: 10.1126/science.1135875. *Hybrid speciation in animals was thought to be much rarer than in plants, but recent reports are narrowing the gap. Here, genetic data document the hybrid origin of a butterfly in the genus Lycaeides. The hybrid species occupies a novel alpine habitat distinct from that of either parent. [DOI] [PubMed] [Google Scholar]
- 53.Martin NH, Bouck AC, Arnold ML. Detecting adaptive trait introgression between Iris fulva and I. brevicaulis in highly selective field conditions. Genetics. 2006;172:2481–2489. doi: 10.1534/genetics.105.053538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang JL, Tian L, Lee HS, Chen ZJ. Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis allopolyploids. Genetics. 2006;173:965–974. doi: 10.1534/genetics.106.056580. **One of the best studies connecting hybridization to changes in a phenotype with adaptive significance, including a detailed analysis of the genetic changes. The authors elucidate the factors leading an allotetraploid species to have a later flowering time than either of its diploid progenitors. [DOI] [PMC free article] [PubMed] [Google Scholar]


