Abstract
The social amoebas (Dictyostelia) display conditional multicellularity in a wide variety of forms. Despite widespread interest in Dictyostelium discoideum as a model system, almost no molecular data exist from the rest of the group. We have constructed the first molecular phylogeny of the Dictyostelia with parallel small subunit ribosomal RNA and α-tubulin datasets, and we found that dictyostelid taxonomy requires complete revision. A mapping of characters onto the phylogeny shows that the dominant trend in dictyostelid evolution is increased size and cell-type specialization of fruiting structures, with some complex morphologies evolving several times independently. Thus, the latter may be controlled by only a few genes, making their underlying mechanisms relatively easy to unravel.
TEXT
Multicellular animals and plants display an enormous variety of forms, but their underlying genetic diversity is small compared with the genetic diversity of microbes. Eukaryotic microbes include a broad range of unicellular life forms, with multiple independent inventions of multicellularity. One of the most intriguing challenges in biology is to understand the reason behind the repeated occurrence of this particular evolutionary stratagem.
The social amoebas, or Dictyostelia, are a group of organisms that hover on the borderline between uni- and multicellularity. Each organism starts its life as a unicellular amoeba, but they aggregate to form a multicellular fruiting body when starved. This process has been best described for the model organism Dictyostelium discoideum. The aggregate of up to 100,000 D. discoideum cells first transforms into a finger-shaped structure, the “slug.” The head region of the slug senses environmental stimuli such as temperature and light and directs the slug toward the soil’s outer surface, where spores will be readily dispersed. The slug then stands up to form the fruiting body, or sorocarp. The cells in the head region move into a prefabricated cellulose tube and differentiate into stalk cells that ultimately die. The remaining “body” cells then crawl up the stalk and encapsulate to formspores. Thus, the Dictyostelia display distinct characteristics of true multicellularity, such as cell-cell signaling, cellular specialization, coherent cell movement, programmed cell death, and altruism (1, 2).
Traditionally, social amoebas have been classified according to their most notable trait, fruiting body morphology. Based on this, three genera have been proposed: Dictyostelium, with unbranched or laterally branched fruiting bodies; Polysphondylium, whose fruiting bodies consist of repetitive whorls of regularly spaced side branches; and Acytostelium, which, unlike the other genera, forms acellular fruiting body stalks (1).
Despite the widespread use of D. discoideum as a model organism (2, 3), the Dictyostelia as a whole are poorly characterized in molecular terms; nearly all currently available data are from a single species. Nonetheless, the social amoebas provide a unique opportunity to understand the evolution of multicellularity (4-6). A primary and essential prerequisite for this is an understanding of the true phylogeny of the group. Here, we describe the phylogeny of social amoeba species and trace the acquisition of morphological and functional complexity during their evolution.
Nearly complete small subunit rRNA (SSU rDNA) gene sequences were determined from more than 100 isolates of Dictyostelia, including nearly every described species currently in culture worldwide (7). Phylogenetic analyses of these data identified four major subdivisions of the group, which we numbered 1 to 4 (Fig. 1 and fig. S1). Group 1 consists of a morphologically diverse set of Dictyostelium species. Group 2 is a mixture of species with representatives of all three traditional genera, including all pale-colored species of Polysphondylium, at least two species of Dictyostelium, and all species of Acytostelium. Group 3 is again a diverse set of purely Dictyostelium species, also including the single cannibalistic species, D. caveatum. The largest group is group 4, which consists almost entirely of Dictyostelium species but may also include a clade of two violetcolored species from two separate traditional genera, P. violaceum and D. laterosorum. With the exception of the violet-colored species, group 4 is a fairly homogeneous set of large robust species, including the model organism D. discoideum and the cosmopolitan species, D. mucoroides, which appears to be polyphyletic (8).
Figure 1.
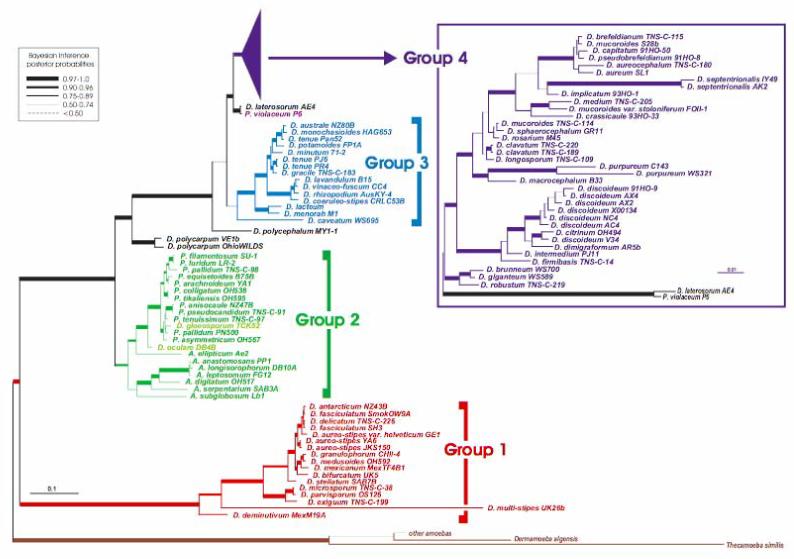
A universal phylogeny of the Dictyostelia based on SSU rDNA sequences. The tree shown was derived by Bayesian inference from 1655 aligned positions (7). The tree identifies four major taxonomic divisions (Groups 1to 4), which are indicated by separate colors and to the right of the figure beside brackets (Dictyostelium species within Group 2 are indicated in lighter green). The tree includes nearly all known and described species of Dictyostelium (D.), Polysphondylium (P.) and Acytostelium (A.). Bayesian inference posterior probabilities (biPP) are roughly indicated by line width (key at the upper left), with exact biPP and maximum likelihood bootstrap (mlBP) values given in figure S1A. Separate analyses were conducted on Group 4 sequences (7) including an additional 300 more highly divergent nucleotide positions (inset box in the upper right; fig. S1B). Branch lengths are drawn to scale (substitutions per site) as indicated by scale bars. The tree is rooted based on separate analyses (7) including closely related lobosan amoebae (fig. S1C) (10). Branches lengths for the latter were scaled up to compensate for the smaller number of alignable sites, based on the length of the first two internal branches (fig. S1C).
The four SSU rDNA groupings are confirmed by a-tubulin phylogeny (fig. S2) with two exceptions: (i) A. ellipticum is only weakly placed with group 2 in the a-tubulin tree (fig. S2), and (ii) the D. laterosorum and P. violaceum clade is grouped together with as the sister group to a weakly supported group 3 plus group 4 clade (0.64 Bayesian inference posterior probability, 51% maximum likelihood bootstrap, fig. S2). This is in contrast to its position as the exclusive sister lineage to group 4 in the SSU rDNA tree (Fig. 1). The SSU rDNA phylogeny also strongly supports group 1 as the deepest major divergence in Dictyostelia (Fig. 1 and fig. S1), as do analyses of combined SSU rDNA plus a-tubulin nucleotide sequences (fig. S3). However, an alternative root is weakly recovered in the a-tubulin amino acid phylogeny (fig. S2). Thus, the position of the dictyostelid root still requires confirmation, which will probably require multiple additional genes.
A notable feature of both phylogenies is the split of the genus Polysphondylium. The violet-colored P. violaceum is unequivocally grouped together with D. laterosorum, and these two lie together at the base of group 4 (Fig. 1) or in groups 3 and 4 (fig. S2). Meanwhile, the pale-colored polysphondylids are all found nested within group 2 (Fig. 1 and fig. S2). The dictyostelid SSU rDNA phylogeny also shows tremendous molecular depth that is roughly equivalent to that of animals and considerably greater than that of fungi (fig. S4). This suggests that Dictyostelia is a deep and complex taxon, but the true extent of this depth requires confirmation from a broader sampling of their genomes.
Social amoeba species show marked differences in the size and branching patterns of their fruiting bodies and the presence or absence and shape of support structures. They may also vary in spore characteristics, cell aggregation patterns, slug migration characteristics, and presence or absence of alternative life cycles, such as the microcyst and sexual macrocyst (1). To understand how these traits might have evolved, we mapped all well-documented dictyostelid traits onto the molecular phylogeny (Fig. 2 and fig. S5).
Figure 2.
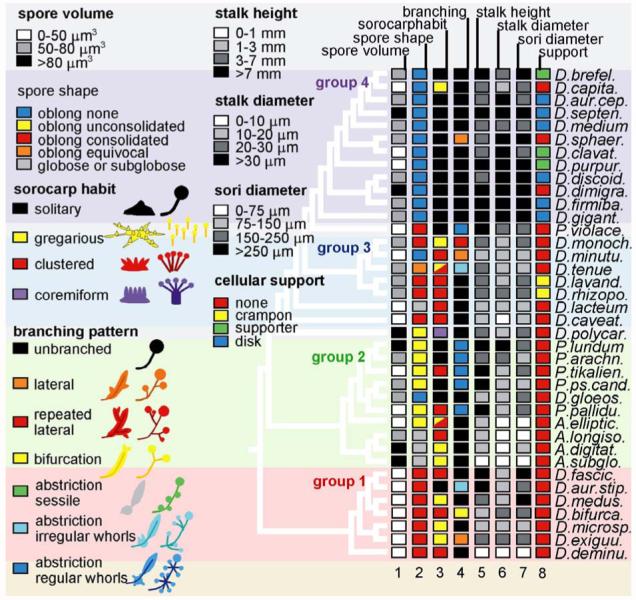
Trait mapping of dictyostelid characters. Consistently documented characters were retrieved from primary species descriptions (table S1, online material) and from Dictyostelium monographs (1, 11). Character states were numerically coded and mapped to the dictyostelid SSU rDNA phylogeny including alternate species (Fig. 1) using the MacClade 4 software package (12). For comprehensive presentation the most informative characters are combinatorially presented on a single tree with the numerical code converted into color code for qualitative traits and into greyscale for quantitative traits. The code key for the character states is shown on the left side of the figure and in table S2. A total set of 20 characters mapped to all species in the phylogeny is presented in figure S5 (7).
Few of the traditionally noted morphological characters show any clear trend across the tree, although a number show interesting within-group trends. The most globally consistent character appears to be spore shape (Fig. 2, column 2). Spores can be either round (globose) or oblong, and in the latter case they often have granules at their poles. Groups 1 and 3 are characterized by oblong spores with tightly grouped (consolidated) granules. In group 2, the granules have become loosely grouped (unconsolidated), whereas polar granules are lost entirely in group 4. Group 1 is further characterized by markedly smaller spores than the other taxa (Fig. 2, column 1).
Fruiting body (sorocarp) morphology and size are the most commonly used taxonomic characters. A primary determinant of sorocarp size is the number of cells that can be collected into one aggregate. However, most of the sorocarp size and shape variation depends on the extent and manner of subsequent aggregate subdivision (Fig. 2, columns 3 to 7; fig. S5, columns 15 to 19) (7). These characteristics are controlled by so-called organizing centers, or “tips,” the first of which appears as a small protrusion on top of a newly formed aggregate. Secondary tips may then appear during or just after aggregation, giving rise to a gregarious or clustered sorocarp habit, respectively. The rising cell masses can subdivide even further by new tips arising along their main axis, yielding lateral branches, or by groups of cells detaching themselves from the rear. The latter abstricted masses can differentiate directly into spores or form new tips, giving rise to whorls of irregular or evenly spaced branches. Species in groups 1 to 3 usually display a clustered or gregarious sorocarp habit, whereas group 4 species mainly form solitary fruiting bodies (Fig. 2, column 2). Additionally, branched forms are much more common in groups 1 to 3 than in group 4. Not surprisingly, there is an inverse relationship between a tendency for aggregates and sorogens to subdivide and the size of stalk and sorus. Thus, group 4 species also have the largest sori and the thickest and longest stalks (Fig. 2, columns 5 to 7, and fig. S5). The presence of support structures formed from stalk-like cells such as basal disks, triangular supporters, or crampons also appear to be markedly correlated with large fruiting body size (Fig. 2, column 8).
Molecular phylogenetic analyses of two independent markers show that Dictyostelia consists of four major groups, none of which correspond to traditional classifications. The molecular tree is dominated by Dictyostelium species (which appear in all four groups), Polysphondyliums are found in two very separate locations, and Acytosteliums reside in a mixed group (group 2) that also includes Dictyostelium and Polysphondylium species. Therefore, none of the four molecularly defined dictyostelid groups correspond to traditional genera, and none of the traditional genera, with the possible exception of Acytostelium, are even monophyletic. This indicates that fruiting body morphology, upon which traditional classification is based, is a very plastic trait inDictyostelia and is apparently of little use as a taxonomic determinant. This is even more evident from the scattered distribution of similar branching morphologies over the four taxon groups (fig. S5, columns 15 to 16). For instance, the rosary-type, coremiform, and laterally branched morphologies appear, respectively, two, three, and seven times independently across the tree.
The strongest evolutionary trend in dictyostelid fruiting body morphology appears to be related to size. Whereas the species in groups 1, 2, and 3 generally split up their aggregates into multiple sorogens, which then subdivide even further to yield branched fruiting bodies, the aggregates of group 4 species usually give rise to a solitary fruiting body that is only rarely branched. As a result, the group 4 species have more robust fruiting structures with much larger spore heads than the other groups. These large structures are typically supported at their base by basal disks or triangular supporters that are derived from a third cell type, the anterior-like cells. In at least one species, D. discoideum, this cell type diverges even further to produce two more structures, the upper and lower cup that support the spore head. This is an interesting example of the correlation between the size of an organism and its cell type diversity, which marks the evolution of many multicellular organisms (9).
The DNA-based phylogeny of the Dictyostelids indicates four high-level taxa, none of which correspond to the three traditional genera. Therefore, we sought unique descriptive names for each group. For group 1, we propose the name “Parvisporids” (parvi means small), because these species all have small spores. For group 2, we propose “Heterostelids,” signifying their wide variety of fruiting body and stalk morphologies. We propose “Rhizostelids” for group 3, which includes species with rootlike support structures for their fruiting bodies. Finally, we propose that group 4 exclusively retain the name “Dictyostelid,” because it includes the widely studied model organism D. discoideum.
Supplementary Material
References and Notes
- 1.Raper KB. The Dictyostelids. Princeton, NJ: Princeton Univ. Press; 1984. [Google Scholar]
- 2.Kessin RH. Dictyostelium: Evolution, Cell Biology and the Development of Multicellularity. Cambridge, UK: Cambridge Univ. Press; 2001. [Google Scholar]
- 3.Eichinger L, et al. Nature. 2005;435:43. [Google Scholar]
- 4.Strassmann JE, Zhu Y, Queller DC. Nature. 2000;408:965. doi: 10.1038/35050087. [DOI] [PubMed] [Google Scholar]
- 5.Queller DC, Ponte E, Bozzaro S, Strassmann JE. Science. 2003;299:105. doi: 10.1126/science.1077742. [DOI] [PubMed] [Google Scholar]
- 6.Alvarez-Curto E, et al. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6385. doi: 10.1073/pnas.0502238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Materials and methods are available as supporting material on Science Online.
- 8. D. mucoroides appears in three separate clades in group 4. This is partly because the original holotype of Brefeld (10) was lost, and subsequent researchers made different diagnoses of more recent isolates. Hagiwara’s D. mucoroides (strain number TNS-C-114) was diagnosed by Raper (1) as D. sphaerocephalum, whereas Raper’s D. mucoroides (strain number S28b) was diagnosed as D. brefeldianum by Hagiwara (11)
- 9.Bonner JT. Evolution Int. J. Org. Evolution. 2004;58:1883. doi: 10.1111/j.0014-3820.2004.tb00476.x. [DOI] [PubMed] [Google Scholar]
- 10.Brefeld O. Abh. Senckenberg. Naturforsch. Ges. 1869;7:85. [Google Scholar]
- 11.Hagiwara H. The Taxonomic Study of Japanese Dictyostelid Cellular Slime Molds. Tokyo: Natural Science Museum; 1989. pp. 131–135. [Google Scholar]
- 12.Nikolaev SI, et al. Protist. 2005;156:191. doi: 10.1016/j.protis.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Maddison WP, Maddison DR. Folia Primatol. (Basel) 1989;53:190. doi: 10.1159/000156416. [DOI] [PubMed] [Google Scholar]
- 14. We thank honors research students L. Paternoster, S. Saleem, S. Wilkinson, and C. Williams for help with sequencing and early analyses and E. Vadell and J. C. Landolt for gifts of dictyostelid cultures. T.D. and T.W. thank R. Marschalek for SSU rDNA sequencing early in the project. This research was supported by Biotechnology and Biological Sciences Research Council (BBSRC) grants COD16760 and COD16761 to P.S. and S.L.B., BBSRC grant R01362 to S.L.B., Dutch Science Foundation (NWO) grant 805.17.047 to P.S., Wellcome Trust University Award Grant 057137 to P.S., and an NSF postdoctoral fellowship in Microbiology to D.E.R. Sequences reported in this paper have been deposited in the European Molecular Biology Laboratory (EMBL) database under accession numbers AM168028 to AM168115 (SSU rDNA) and AM168453 to AM168491 (a-tubulin)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


