Abstract
Human cytomegalovirus (HCMV) and human immunodeficiency virus type-1 (HIV-1) infect the female genital tract. A human cervical explant model was developed to study single and dual infection by these viruses in the genital compartment. An HCMV strain expressing green fluorescent protein, and two clinical HCMV strains produced peak viral DNA copies at 14 to 21 days post-infection. Peak levels of HIV-1Ba-L p24 antigen occurred at 7 days post-infection. HIV-1Ba-L appeared to enhance HCMV in co-infected tissues. Singly and dually-infected explants produced increased levels of cytokines IL-6, IL-8, and GRO-α in culture supernatants. Immunohistochemical and flow cytometric analysis showed HCMV infection of leukocytes with the phenotype CD45+/CD1a+/CD14+/HLA-DR+ but not stromal or endothelial cells. Cells expressing both GFP and HIV-1 p24 antigen were detected in co-infected tissues. The cervical explants provide an ex vivo human model for examining mechanisms of virus-virus interaction and pathogenesis in clinically relevant tissue.
Keywords: human cytomegalovirus, HIV-1, cervical explant, female genital tract
Introduction
Human cytomegalovirus (HCMV) infection is widespread as evidenced by the high HCMV seroprevalence in the adult population. Most primary infections are mild or asymptomatic, however, the virus establishes lifelong latency. Primary infection or reactivation of latent infection can cause serious disease in patients who are immunocompromised, such as congenitally-infected infants, transplant recipients, and patients co-infected with HIV-1 (Arcasoy and Kotloff, 1999; Boppana et al., 2001; Grossi et al., 1995; Jacobson, 1997; Ross et al., 2006).
HCMV may persist or reactivate in specific body compartments such as the lung and gastrointestinal tract, which can act as long-term reservoirs of infection and provide opportunities for interaction with co-infecting pathogens (Jacobson, 1998; Long et al., 1998; Nowzari et al., 2003; Paya et al., 1993; Sinzger et al., 1995; Tanaka et al., 2006). The female genital tract is a compartment where co-infection of HCMV and HIV-1 is known to occur, and several studies have examined the association between these two viruses at this site. It was reported that HIV-1-infected women shed HCMV in cervicovaginal secretions and both viruses were detected in genital secretions, even when undetectable in the blood (Clarke et al., 1996; Kovacs et al., 2001; Lurain et al., 2004; Mostad et al., 1999). Therefore, co-infection provides the potential for virus-virus interaction that may increase sexual and perinatal transmission of these viruses.
HCMV and HIV-1 are highly species specific, which limits both the cell types that support productive virus replication in vitro and the use of animal models for in vivo studies. Ex vivo human cervical tissue models were previously reported for the study of HIV-1 transmission and replication in the female genital tract (Collins et al., 2000; Greenhead et al., 2000; Gupta et al., 2002; Palacio et al., 1994). In addition, cervical tissue explants have been used to examine the efficacy of different inhibitors of HIV-1 infection (Fletcher et al., 2005; Greenhead et al., 2000; Hu et al., 2004). We report here the development of a cervical explant model to study HCMV infection and replication as well as co-infection with HIV-1.
Results
Infection of cervical explants by cell-free HCMV
The first step in developing this model was to determine the ability of the explants to support HCMV infection and HIV-1 co-infection. The lengthy course of HCMV infection requires that the tissue remain viable for at least 21 days post-infection (p.i.). To address this requirement small ectocervical explants were cut from each surgical specimen, inoculated with the cell-free HCMV strain CMVPT30-gfp, and maintained submerged in culture medium. For co-infection studies the explants were inoculated with HIV-1Ba-L 72 h after HCMV inoculation.
CMVPT39-gfp expresses green fluorescent protein (GFP), which was first detected an average of 7 days p.i., although in some tissues GFP-positive cells did not appear for 10–14 days. To follow the progression of HCMV in singly-infected and HIV-1 co-infected tissues, GFP-expressing cells were monitored daily for a total of 21 to 28 days using an inverted fluorescence microscope. The number of GFP-positive cells was scored from 1+ to 5+ (see Methods). Based on the fluorescence scores, the number of cells expressing GFP steadily increased from 1+ to 5+ over a period of 1 to 2 weeks indicating that the infection was spreading among susceptible cells in the explant. The peak number of GFP-expressing cells occurred between days 14 and 21 p.i. in both HCMV singly-infected and co-infected explants, and the average score was significantly higher at day 14 (p=0.04, Wilcoxan signed rank test) for co-infected than for explants infected with HCMV alone (Table 1).
Table 1.
Scoring of explants for HCMV-infected fluorescent cells at day 14 post-infection.
| Tissue | CMV Mean Scorea | CMV/HIV-1 Mean Score |
|---|---|---|
| A | 2.2 (nb=15) | 2.5 (n=13) |
| B | 3.1 (n=17) | 3.5 (n=16) |
| C | 1.8 (n=13) | 2.2 (n=14) |
| D | 1.5 (n=22) | 2.1 (n=16) |
| E | 2.3 (n=10) | 4.0 (n=9) |
Mean score determined from individual explants from 5 different tissue specimens scored for fluorescence 1+ to 5+ (see Materials and Methods).
n is the number of explants scored for each tissue specimen.
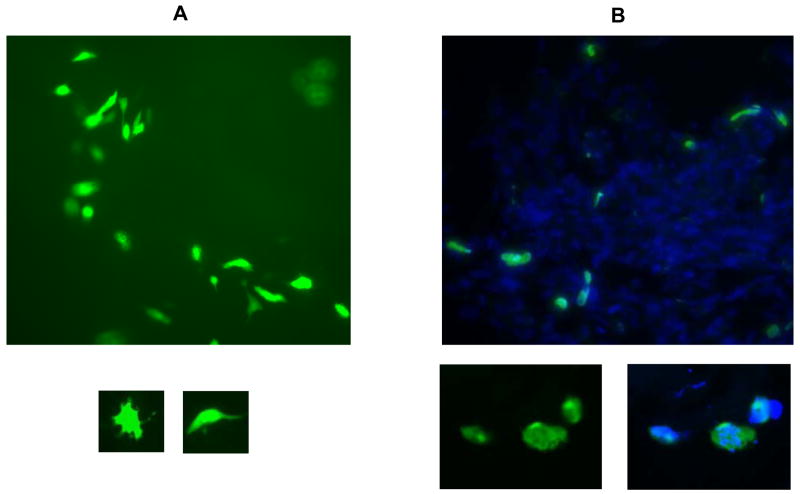
In live tissue GFP-expression appeared in individual cells distributed throughout the explant tissue (Figure 1A) rather than plaque-like clusters of adjacent cells as seen in cell culture monolayers. This suggested that infection was not occurring in epithelial, endothelial, or stromal cells. The infected cells were large with multiple extensions protruding from the main cell body (Figure 1A small images). There was some evidence of cell death after day 14 as shown by fragmentation of fluorescent cells, although the majority of GFP-expressing cells remained morphologically intact. The overall progression of CMVPT30-gfp infection appeared to be slower than that observed in cell culture with the number of fluorescent cells increasing up to at least 14 days p.i.
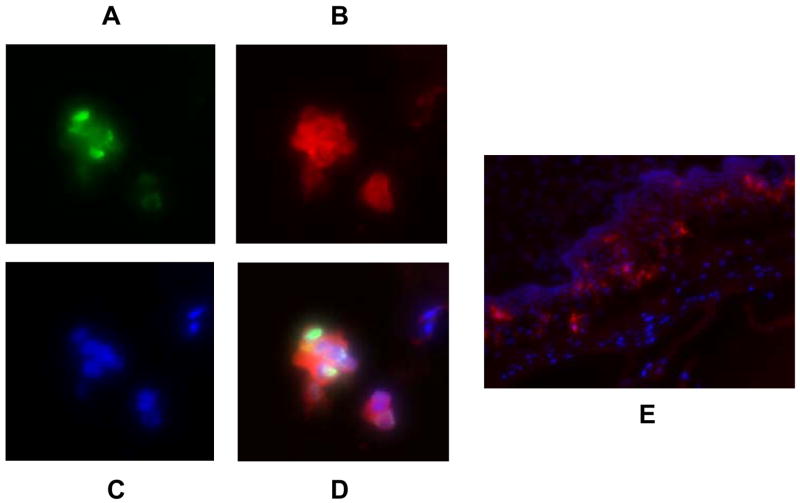
Figure 1.
Detection of HMCV-infected cells. A: Large image shows CMVPT30-gfp-infected cells in live tissue (100X). Smaller images show individual cells infected with CMVPT30-gfp (200X). B: NW23-3 infection in frozen tissue sections. Large image shows green fluorescence from FITC-conjugated anti-HCMV p52 antibody; blue fluorescence is DAPI staining of nuclei (100X). Smaller image on left below shows individual infected cells stained with FITC-conjugated anti-HCMV p52 antibody at higher magnification (200X). Smaller image on right shows same cells with overlay of DAPI staining of nuclei. All images taken with SPOT RT KE 7.3 Three Shot Color digital camera using Metamorph version 6.0 software.
To monitor tissue viability, the MTT assay was performed at the time of inoculation and at selected time points p.i.. The absorbance per mg of tissue tended to decrease after 14 days indicating a decrease in the number of viable cells, although the absorbance remained higher than the background level of control wells that did not contain tissue for as long as 28 days p.i. (data not shown).
Infection of cervical explants by HCMV cell-associated clinical strains
The production of cell-free virions in cell culture by the recombinant CMVPT30-gfp strain differs phenotypically from low-passage clinical HCMV strains, which produce only cell-associated virus in cell culture. Therefore, we tested three clinical strains, NW-23–2, CH-14, and CH-22, to confirm that they could replicate in the cervical explants. Because there was no visual means to follow infection, explants infected with the clinical strains were harvested and frozen at 14 days p.i. based on the timing of peak GFP expression observed with CMVPT30-gfp. The explants were then sectioned and stained directly for the delayed early non-structural protein, p52 (UL44), using a FITC-conjugated mouse monoclonal antibody (mAb) (Dako Glostrup, Denmark). Cells positive for p52 were detected throughout the tissue sections (Figure 1B) demonstrating the ability of these clinical strains to replicate in explant tissue. The staining properties of the anti-p52 antibody in individual cells differed from GFP expression (Figure 1B smaller images) in being more punctuate and less bright.
Quantitation of CMVPT30-gfp and HCMV clinical strains in cervical explants
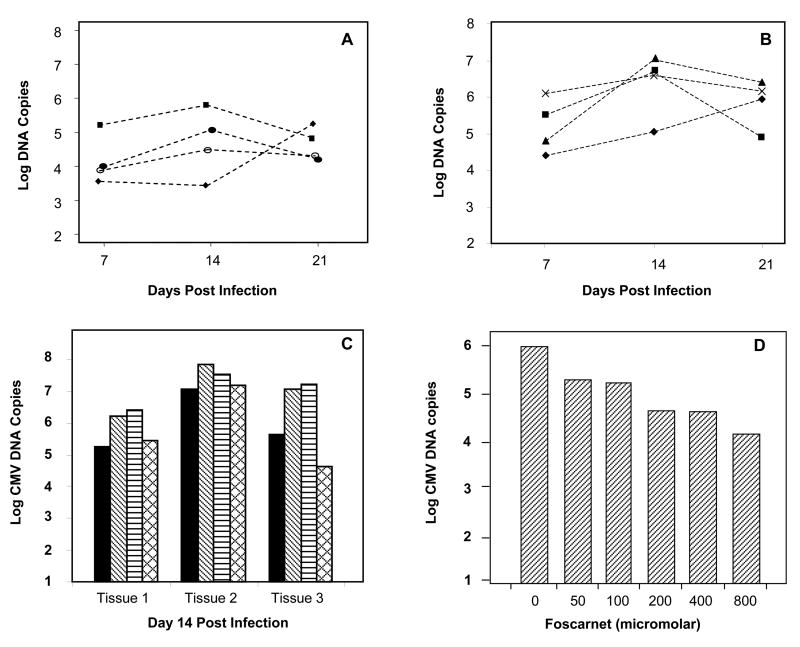
To provide a quantitative measure of HCMV replication in explant tissue, genomic copy numbers of all HCMV strains were determined by real-time PCR and normalized by tissue weight. HCMV DNA copy numbers in singly-infected explants varied, typically within the range of 103 to 106 copies per mg of tissue. Peak copy numbers of HCMVPT30-gfp occurred on days 14–21 p.i. (Figure 2A). Variability in HCMV DNA copy number occurred within the same tissue and among tissues from different hosts.
Figure 2.
Quantitation of HCMV DNA copy numbers in cervical explants by real-time PCR. Note: all data are log scale. A) CMVPT30-gfp median copy numbers at three time points p.i. in tissues from 4 different cervical specimens. B) NW23-3 median copy numbers at three time points p.i. in tissue supernatants from 4 different cervical specimens. C) Comparison of DNA copies from infection of tissue explants from the same specimen with CMVPT30-gfp (solid bars), NW23-3 (diagonal bars), CH-22 (horizontal bars), and CH-14 (cross-hatch bars) at peak time point, Day 14 p.i. D) Inhibition of CMVPT30-gfp replication by foscarnet in explant tissue at day 14 post-infection.
Explants inoculated with cells infected with each of the clinical HCMV strains (CH-14, CH-22 or NW-23-3) produced detectable HCMV DNA copies in tissue and notably also in supernatant fluid. DNA extracted from both types of samples was quantitated by real-time PCR. Peak values occurred on days 14–21 p.i. as shown for NW23-3 (Figure 2B). When samples from the same tissue specimen were infected with CMVPT30-gfp, NW23-3, CH-14, and CH-22, the median copy numbers of two of the three clinical strains (CH-22 and NW23-3) were 0.5–1.5 log higher than CMVPT30-gfp (Figure 2C). The third clinical isolate, CH-14, produced copy numbers similar to CMVPT30-gfp. These quantitative data confirm that the cervical explants are permissive for replication of both higher passage cell-free virus strains and low passage cell-associated HCMV clinical strains.
Inhibition of CMVPT30-gfp in cervical explants
Additional evidence for HCMV replication in the explants was provided by the inhibitory activity of the HCMV antiviral agent, foscarnet. Explants were pretreated with concentrations of the drug ranging from 50 μM to 800 μM for 72 h followed by inoculation with 1 × 105 pfu of CMVPT30-gfp per well. HCMV DNA was extracted from tissue samples for quantitation at day 14 p.i. A dose-response inhibitory effect was seen in the tissue, with concentrations as low as 50 μM inhibiting viral replication by almost 80% (Figure 2D). The highest concentration of foscarnet (800 μM) inhibited HCMV replication by 98%.
Infection of cervical explants with HIV-1Ba-L
Others have shown that cervical explants are permissive for HIV-1 infection, (Collins et al., 2000; Fletcher et al., 2005; Greenhead et al., 2000). We confirmed that our stock of HIV-1Ba-L could infect and replicate in the explants. Proof of infection was determined by the qualitative detection of HIV-1 DNA at days 7, 14 and 21 p.i. HIV-1 DNA was present in all explants infected with HIV-1BaL alone, as well as those co-infected with both CMVPT30-gfp and HIV-1Ba-L (data not shown).
Quantitative analysis of HIV-1 replication was determined by p24 antigen ELISA in supernatants collected beginning at day 3 p.i., after the inoculum had been removed by repeated washings. The day 3 time point provided the residual inoculum background for the p24 antigen assay. In HIV-1-infected tissues, peak p24 antigen levels consistently occurred at day 7 (Figure 3) and then declined in supernatants collected at 14 and 21 days p.i. These data confirm that the HIV-1Ba-L strain is able to infect and replicate in the cervical explants under the experimental conditions of our protocol.
Figure 3.
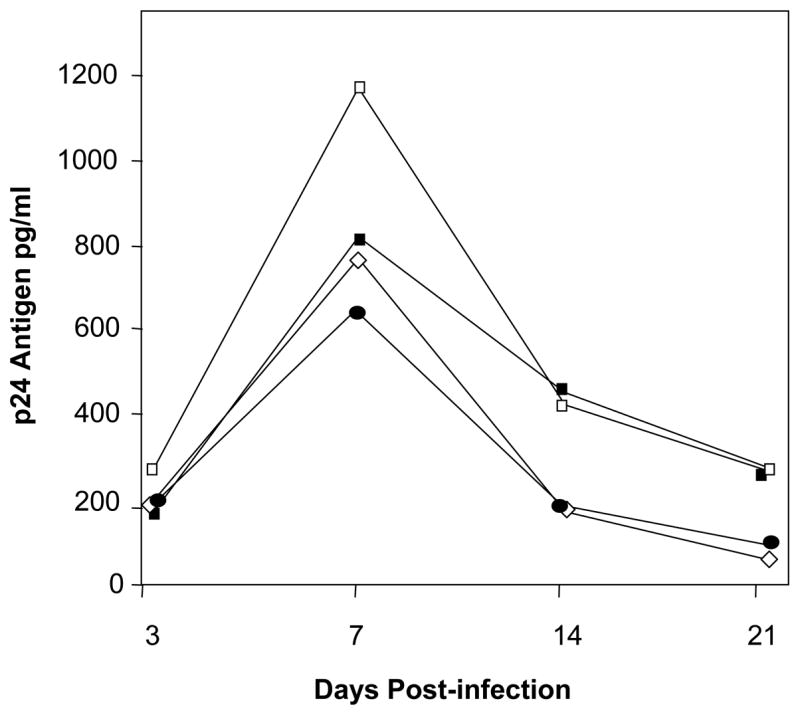
Quantitation of HIV-1Ba-L by p24 antigen ELISA. Each line shows median results at each time point p.i. for a different host tissue.
Quantitation of HCMV and HIV-1 co-infection
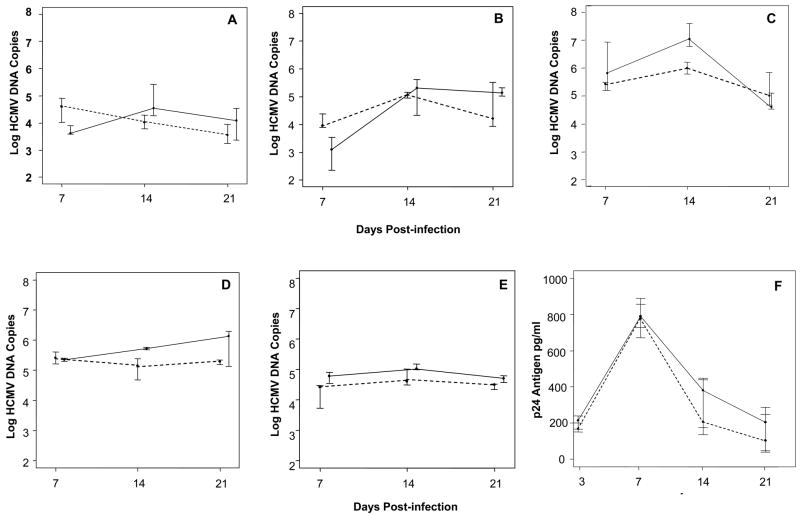
To study the potential interaction of HCMV and HIV-1 in cervical tissue, we first performed a series of co-infection studies with CMVPT30-gfp and HIV-1Ba-L. The explants were inoculated with CMVPT30-gfp for 72 h before inoculation with HIV-1Ba-L. The order and timing of exposure to the viruses was determined by preliminary experiments, which had shown that the highest HCMV loads resulted from inoculation of the tissues first with HCMV followed by inoculation with HIV-1 (data not shown). Co-infected explants were harvested for quantitation at days 7, 14 and 21 p.i. Similar to singly-infected explants, HCMV DNA copy numbers per mg of tissue in co-infected explants reached peak levels at days 14 to 21 p.i.. In four of five experiments with CMVPT30-gfp, the median peak HCMV copy values were greater in co-infected explants than in those infected with HCMV alone. Figures 4A, 4B, and 4C show the results of three of these experiments. Note that the HCMV copy numbers are log scale. For the tissue shown in Figure 4C the median values of the HCMV viral loads for HCMV/HIV-1 co-infection versus HCMV infection alone at day 14 p.i. reached statistical significance (p= 0.004). Although the median HCMV copy numbers at days 14 and 21 for the tissues shown in Figures 4A and 4B were higher for co-infection compared to single infection, the difference was not statistically significant, because the range of HCMV copy numbers at each time point was too variable.
Figure 4.
Quantitation of HCMV and HIV-1Ba-L co-infection. Graphs in Figures A-E are offset at each time point to show bars for interquartile range (25th to 75th percentile). Figures A,B,C: CMVPT30-gfp co-infection with HIV-1Ba-L. Dotted line, CMVPT30-gfp DNA copies in single infection; solid line CMVPT30-gfp DNA copies in co-infection with HIV-1. Only Day 14 difference between single and co-infection in Figure C is statistically significant (p= 0.004, Wilcoxan Rank sum test). D) CH-22 DNA copies in single infection (dotted line) and co-infection with HIV-1Ba-L (solid line); E) CH-14 single infection (dotted line) and HIV-1 co-infection (solid line). F) Results of 3 experiments showing HIV-1Ba-L p24 pg/ml determined by ELISA in singly-infected tissues (dotted line) and tissues co-infected with CMVPT30-gfp (solid line). Differences between p24 antigen pg/ml in single and dual infection at days 14 and 21 p.i. are not statistically significant by the Wilcoxan rank sum test.
We next performed similar co-infection studies using HCMV clinical strains CH-22 and CH-14. The results are shown in Figures 4D and 4E respectively. Again, the median values for HCMV genomic copy numbers were greater for co-infected explants than those singly-infected with HCMV. However, the HCMV DNA copy number differences between co-infected and singly-infected tissues did not reach statistical significance.
HIV-1 p24 antigen values for both single and dual infection peaked consistently at day 7 (Figure 4F). Although the median p24 antigen levels were always higher for co-infected explants at days 14 and 21 compared to those infected with HIV-1 alone, this difference was not statistically significant.
Based on these quantitative analyses HCMV co-infection appears to have no effect on HIV-1 replication. The quantitative co-infection data and the fluorescent cell scores (Table 1) suggest that HIV-1 may enhance HCMV replication. However, the results in most cases are not statistically significant, because of the intra-tissue variability. These experiments were conducted with only one strain of HIV-1, and the earliest time point evaluated was 7 days p.i. Earlier time points and interaction of other HIV-1 and HCMV strains remain to be examined. It should be noted, however, that similar results were obtained with both cell-associated HCMV strains and the cell-free strain CMVPT30-gfp.
Cytokine production
Infection by either HCMV or HIV-1 has been associated with the upregulation of proinflammatory cytokines (Craigen et al., 1997; Grundy et al., 1998; Humar et al., 1999; Lane et al., 2001a; Lane et al., 2001b; Lurain et al., 2004; Murayama et al., 1994; Oguariri, Brann, and Imamichi, 2007; Redman et al., 2002). Therefore, the release of cytokines from the explants into the supernatant fluid in response to HCMV and HIV-1 infection was analyzed.
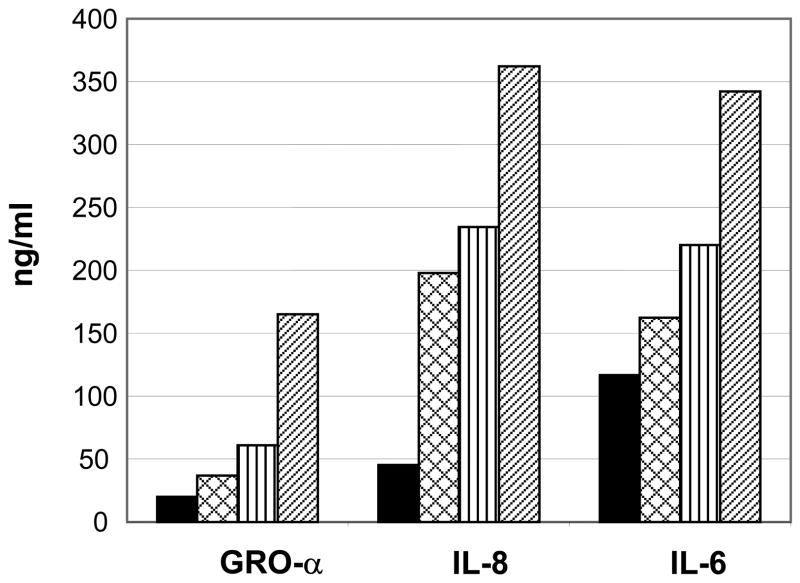
IL-6, IL-8, IL-1β, interferon (IFN)-α, IFN-β, IFN-γ and GRO-α were assayed at days 7, 14, and 21 p.i. in the supernatants from explants infected with either HCMV or HIV-1 or co-infected with HCMV and HIV-1. IFN-α, IFN-β, IFN-γ and IL-1β were not detected in any of the supernatant samples. At day 14 p.i. tissues infected with one or both viruses showed increased levels of IL-6, IL-8, and GRO-α compared to uninfected control tissues. (Figure 5). The pattern of increase under each condition (single or dual infection) was similar for each of the three cytokines.
Figure 5.
Cytokine concentrations in explant supernatant fluid from two singly-infected or co-infected tissues. Solid bar (
 ): uninfected control tissue; Cross-hatch bar (
): uninfected control tissue; Cross-hatch bar (
 ): CMVPT30-gfp; Vertical bar (
): CMVPT30-gfp; Vertical bar (
 ): HIV-1Ba-L; Diagonal bar (
): HIV-1Ba-L; Diagonal bar (
 ): CMVPT30-gfp/HIV-1Ba-L co-infection. Supernatants were collected at 14 days p.i.
): CMVPT30-gfp/HIV-1Ba-L co-infection. Supernatants were collected at 14 days p.i.
Phenotype of HCMV-infected cells
The next question we wanted to address was the identity of the specific cell type(s) in the explants that are infected with HCMV. The size and morphology of the fluorescent cells infected with CMVPT30-gfp (Figure 1A) suggested they could be macrophages, dendritic, or Langerhans cells. The first approach to identify the infected cells was to analyze frozen sections of HCMV-infected tissues. The sections were stained with mouse anti-human CD68, anti-HLA-DR, or anti-CD1a primary mAbs followed by an Alexa Fluor 594-conjugated rabbit F(ab′)2 anti-mouse IgG secondary Ab. In explants infected with the clinical strain NW23-3, there were cells that were positive for HCMV p52, and either CD68 (Figure 6 A-D) or HLA-DR (data not shown).
Figure 6.
Frozen sections of tissue infected with NW23-3 showing co-localization of anti-CMV p52 and anti-CD68 antibodies on infected cells. A: Detection of CMV with FITC-conjugated anti-CMV p52 mAb; B: Detection of anti-CD68 primary antibody with Alexa Fluor 594-conjugated F(ab′)2 rabbit anti-mouse secondary antibody; C: DAPI staining of nuclei; D: Overlay of all three fluors. E: Frozen section of uninfected control tissue showing Langerhans cells stained with anti-CD1a primary antibody and Alexa Fluor 594-conjugated F(ab′)2 rabbit anti-mouse secondary ab. Images taken with a SPOT RT KE 7.3 Three Shot Color digital camera.
The tissues in which NW23-3-specific antigens were detected had been in culture for 14–21 days. No CD1a-positive cells in either infected or uninfected control explants could be detected at this time point. However, the anti-CD1a antibody (hybridoma supernatant) detected Langerhans-like cells in uninfected tissues, which were sectioned after only 3 days in culture (Fig. 6E). Similar results were obtained for uninfected cells using an anti-CD207 (langerin) antibody (data not shown) indicating that these are Langerhans cells.
The second approach to identify the cells infected with HCMV was to dissociate tissues with collagenase and analyze the released cells using flow cytometry. The positive/negative cutoff (gate) was determined for GFP-positive cells based on analysis of uninfected control tissues (Figure 7A, 7B, and 7C) and CMVPT30-gfp-infected fibroblasts (data not shown). The same population was identified by gating on CD45-positive cells, indicating that the infected-cells are leukocytes rather than stromal or epithelial cells. CD68 was also expressed on the GFP-positive population, which is consistent with the immunohistochemical analysis described above. Anti-CD68 and anti-CD14 antibodies identified the same cell population (data not shown), therefore anti-CD14 was used in all subsequent analyses. All GFP-positive cells were analyzed for expression of CD14, CD1a, and HLA-DR. The consensus phenotype of these infected cells is CD45+/CD14+/CD1a+/HLA-DR+ (Figure 7E and F). Isotype-matched controls for all antibodies were negative (data not shown).
Figure 7.
Flow cytometric analysis of dissociated cells from CMVPT30-gfp-infected tissues. A) Histogram of dissociated cells from uninfected tissues; B) Dot plot of uninfected cells: CD14 versus HLA-DR; C) Dot plot of uninfected cells: CD14 versus CD1a; D) Histogram of GFP expression of dissociated cells from CMVPT30-gfp-infected tissues; E) Dot plot of CD14 versus HLA-DR surface expression on CMVPT30-gfp-infected cells; F) Dot plot of CD14 versus CD1a surface expression on CMVPT30-gfp-infected cells.
Co-infection of cells
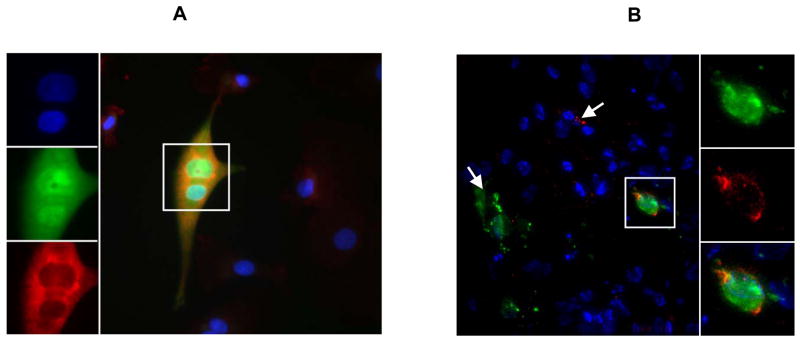
The fact that both HCMV and HIV-1 were detected at the same time in infected explants suggested that there could be co-infection of the same cell. As a first step to demonstrate that co-infection was possible, monocyte-derived macrophages, which are known to be permissive for HCMV (Bego and St Jeor, 2006) were inoculated with CMVPT30-gfp for 72 h followed by infection with HIV-1Ba-L. After an additional 48 h, approximately 10% of the cells expressed GFP and 10% of those were found to express both GFP and p24 antigen (Figure 8A).
Figure 8.
A) HCMV and HIV-1 co-infection of macrophages in cell culture. Panels at left top to bottom show DAPI stain of nuclei; HCMV GFP expression; and anti-HIV-1 p24/p17 ab binding detected by Alexa fluor 594-conjugated (Fab′)2 rabbit anti-mouse secondary ab. Large figure shows merge of all 3 fluors. Single frame of a deconvolved z-stack shown. B) HCMV and HIV-1 co-infection of cells in tissue. Cell outlined in large figure is shown enlarged at right. Top to bottom: HCMV GFP fluorescence; HIV-1 p17/p24 detected by specific primary monoclonal antibodies and Cy5-conjugated secondary ab; merge of GFP and Cy5 with DAPI stain of nuclei. Single frame of a deconvolved z-stack shown. White arrows indicate singly-infected cells: HCMV (green), HIV-1 (red).
To determine whether co-infection occurs in the cervical explants, frozen sections of tissues infected with both viruses were examined for co-localization of GFP and p17/p24 antigen. Figure 8B shows a tissue section in which there are cells singly infected with HCMV or HIV-1 as indicated by the white arrows. The cell outlined in the section is enlarged in the images to the right showing the individual fluors. Both GFP and p17/p24 antigens are expressed indicating co-infection.
Sequences of HCMV UL128-131 open reading frames (ORFs)
The immunofluorescence and flow cytometry data indicated that CMVPT30-gfp infects Langerhans-like cells in the tissue explants. Recent work has shown that the three HCMV ORFs UL128, UL130 or UL131 determine HCMV tropism, which can be altered by deletion or mutation of any one of the 3 ORFs (Hahn et al., 2004; Jarvis and Nelson, 2006; Patrone et al., 2005; Ryckman et al., 2006; Wang and Shenk, 2005). Therefore, we sequenced the stocks of CMVPT30-gfp, NW23-3, CH-14, and CH-22 for the presence of mutations affecting expression of these ORFs. No mutations were found in UL130 or UL131 in any of these stocks, and no mutations were found in the UL128 ORF from the stocks of the three clinical strains (Table 2). The supernatant and tissue extracts infected with the three clinical strains also showed no mutations in any of the three genes. However, in CMVPT30-gfp a mutation was found in the splice donor AG dinucleotide of the first intron of UL128. The specific mutation is AG → AC. By abolishing the splice junction site the predicted result would be read-through into the intron leading to an in-frame stop codon in the intron sequence, which would prematurely terminate the expressed protein four amino acids downstream of the end of the product encoded by the first exon. The same mutation in the UL128 splice donor site was found in CMVPT30-gfp isolated from infected tissue and culture supernatant fluid, but no additional mutations were found in any of the three genes. Despite the UL128 mutation, CMVPT30-gfp replicated to high copy numbers in the tissue explants, and infectious virus was released into the culture medium.
Table 2.
Mutations in HCMV ORFs UL128, UL130, UL131A
| HCMV Strain | Sample Type | Mutations UL128-131A |
|---|---|---|
| CMVPT30-gfp | Stock | Splice donor AG UL128 |
| Explant Supernatant | Splice donor AG UL128 | |
| Explant Tissue | Splice donor AG UL128 | |
| CH14/CH22/NW23-3 (Clinical Strains) | Stock | wild type |
| Explant Supernatant | wild type | |
| Explant Tissue | wild type |
Discussion
The limitations of cell culture and animal models for the study of HCMV pathogenesis prompted the development of a cervical explant model that is permissive for HCMV infection and also supports HIV-1 co-infection. Two cervical explant models have been described for HIV-1 (Collins et al., 2000; Greenhead et al., 2000). One reported by Collins, et al. (Collins et al., 2000), uses polarized 6 mm punch biopsies in Transwells surrounded by agarose with only the epithelial surface exposed to the inoculum. This model was developed for studies of virus entry and transit across the epithelial layer, however it is technically difficult to prevent leakage around the sides of the tissue. We found these punch biopsies to be permissive for HCMV infection (data not shown). However, at most 10 to 12 explants could be obtained from the same cervical specimen, and the maximum viability of the explants was approximately 7 days. This is too short a time period to adequately study HCMV infection and replication, which takes up to two weeks or longer to reach peak titers. The second model described for HIV-1 (Fletcher et al., 2005; Hu et al., 2004) uses small tissue blocks that retain an epithelial surface. The tissues are inoculated and maintained submerged in medium, which allows exposure of all surfaces to the inoculum. The advantages of this model are that the virus can access multiple cell types, and a large number of tissue samples can be obtained from the same specimen allowing comparative analyses of different experimental conditions.
The primary focus of our experiments was to examine replication of HCMV in all potentially permissive cells and to study co-infection with HIV-1. For these reasons we adapted and further developed the second explant model for HCMV infection. Importantly, the tissues remain viable up to 28 days, which allows sufficient time to achieve peak levels of HCMV replication. To our knowledge, this is the first study to use an ex vivo model for HCMV infection of tissue derived from the female genital tract, a compartment in which this virus is actively shed.
The recombinant HCMV strain CMVPT30-gfp, was chosen for the initial development of the model to provide visual confirmation of infection. The number of GFP-expressing fluorescent cells increased over the period of culture in the cervical tissue, which indicated permissive replication. A more quantitative measure of replication was obtained by real-time PCR, which showed that the peak copy numbers of HCMV DNA usually occurred at 14–21 days p.i.. However, it was evident from the comparison of results from multiple experiments using specimens from different hosts that there is considerable variability in HCMV DNA copy numbers detected in individual tissue extracts. This emphasizes the fact that comparative studies of the effects of virus replication need to be carried out in tissue derived from the same host to minimize the variability.
There is an additional caveat associated with the use of CMVPT30-gfp to study HCMV infection in cervical tissue explants, namely that CMVPT30-gfp is phenotypically different from true low-passage clinical strains. CMVPT30-gfp produces extracellular virus, while clinical strains are completely cell-associated in monolayer cell cultures. To determine the relevance of the growth characteristics of CMVPT30-gfp observed in tissue explants, cell-associated clinical isolates of HCMV were also tested for infectivity in this same model. Although there was no way visually to follow the infectivity of the clinical isolates, quantitation of viral DNA extracted from the tissues demonstrated increased DNA copy numbers, which peaked at 14–21 days post-infection. In some experiments in which both CMVPT30 and clinical isolates were compared in the same tissue, the peak copy numbers for the clinical isolates were up to 1.5 log higher than those for CMVPT30-gfp (Figure 2C). This suggests that at least some cell-associated strains might be better adapted than the cell-free strain for infection of the explants.
HCMV copy numbers were decreased by foscarnet in a dose-dependent manner. Therefore, it was possible to determine the effect of an antiviral agent with specific activity against HCMV, which further confirmed that HCMV actively replicates in the explants. In addition this suggests that the model, which has been used for HIV-1 microbicide testing (Fletcher et al., 2005; Fletcher et al., 2006; Greenhead et al., 2000; Hu et al., 2004), could be used for similar testing of potential anti-HCMV agents.
In contrast to HCMV there was much less variability in HIV-1 infectivity as measured by p24 antigen, which reached peak levels earlier (at 7 days p.i) compared to HCMV. To rule out the persistence of p24 antigen associated with the inoculum, infectivity was confirmed by the qualitative detection of HIV-1 DNA in all tissues with p24 antigen-positive supernatants. The HIV-1Ba-L strain was used in our studies, because R5 strains have been most often associated with sexual transmission, and it was shown by others to replicate well in explant tissue (Feng et al., 2003). In our model HIV-1Ba-L infection did not require PHA-stimulation. Others have reported similar findings for R5 viruses, while X4 and dual tropic HIV-1 strains did require PHA-stimulation for infection of cervical explant tissues (Greenhead et al., 2000; Hu et al., 2004). Additional evidence from other studies suggests that HIV-1 can infect multiple cell types in the female genital tract (Howell et al., 1997; Yeaman et al., 2004).
Having established the ability of both viruses to infect and replicate in the explants, we investigated the tissue response to the infection. The rationale for this approach was that the inflammatory state of the tissue at the time the specimen is taken has been shown to affect HIV-1 infectivity (Maher et al., 2005; Patterson et al., 1998; Sturm-Ramirez et al., 2000). Upregulation of the pro-inflammatory cytokines IL-6, IL-8, and GROα reportedly occurs in response to infection by HIV-1 and HCMV in cell culture (Craigen et al., 1997; Grundy et al., 1998; Humar et al., 1999; Lane et al., 2001a; Lane et al., 2001b; Murayama et al., 1994; Redman et al., 2002). At 7 and 14 days post-infection, which were the two time points tested, infected and co-infected tissues produced higher levels of IL-6, IL-8, and GROα compared to uninfected control tissues (Figure 4). The higher results obtained with co-infected tissues may reflect additive effects of replication by both viruses. Inteferons α, β, and γ were not detected, but it is possible that differential effects of single infection and co-infection on these cytokines occur before 7 days.
It was previously reported that HIV-1 infects macrophages and CD4+ T cells in cervical explants (Collins et al., 2000; Greenhead et al., 2000). We wanted to determine the target cell(s) for HCMV, which has been reported to infect a wide range of cell types in vivo including fibroblasts, epithelial cells, endothelial cells, smooth muscle cells, macrophages, and dendritic cells (Gerna et al., 2005; Hahn et al., 2004; Hertel et al., 2003; Sinzger et al., 1995). The size and morphology of the cells that expressed GFP in CMVPT30-gfp-infected tissues suggested that the target could be a macrophage, dendritic cell or Langerhans cell (Figure 1). Our results from immunohistochemical analysis of frozen sections indicated that CD68, a marker of monocyte-macrophage lineage (da Costa et al., 2005) and HLA-DR are expressed on the HCMV-infected cells, but CD1a was not detected on these same cells.
CD1a-positive cells morphologically resembling Langerhans cells were detected in uninfected tissues at 3 days p.i. but not at 14 days p.i. It is likely that uninfected Langerhans cells would have migrated from the tissues after 14 days, but cells expressing GFP do not appear to leave the tissue during the period of culture. The failure to detect CD1a expression on infected cells in these sections compared to detection by flow cytometry (see below) may reflect potential differences in CD1a expression over time and lower sensitivity of the anti-CD1a hybridoma supernatant used for detection in the sections.
HIV-1 infection of cervical explants has been reported by others to target cells that express CD68, which were described as subepithelial macrophages. No CD1a+ cells were found to express p24 antigen (Greenhead et al., 2000).
For further determination of the HCMV-infected cell phenotype in the explants, cells dissociated from tissues were analyzed by flow cytometry. There was no evidence that stromal, epithelial, or endothelial cells were infected. This was surprising, because extracellular strains of HCMV only grow in vitro in normal diploid fibroblasts, and low-passage clinical isolates grow in both fibroblasts and endothelial cells. The GFP+ cell population was also CD45+ indicating that the infected cells were leukocytes. CD68 and CD14 were both expressed by the GFP+ cell population. The consensus phenotype of the GFP+ cells was found to be CD45+/CD1a+/CD14+/HLA-DR+. Although the morphology of the cells as seen in Figure 1 resembles macrophages, dendritic, or Langerhans cells, this phenotype is not strictly consistent with any of these cell types. Macrophages would be predicted to be CD14+/CD1a−, and dendritic cells and Langerhans cells are usually CD14−/CD1a+. However, antigen presenting cells that are CD14/CD1a double positive have been reported under both normal and disease conditions such as Langerhans cell histiocytosis. The double-positive cells may represent a subset of functionally immature Langerhans cells (Angel et al., 2006; Caux et al., 1992; Geissmann et al., 2002; Geissmann et al., 2001; Paczesny et al., 2007). Regardless of the actual cell identity, CMVPT30-gfp appears to infect a migratory cell, which is not necessarily uniformly distributed throughout the tissue sample. Others have observed a wide variability in the number of dendritic and Langerhans cells in the normal uterine cervix (Poppe et al., 1998), which could partially explain the intra-tissue variability in HCMV copy numbers that we have observed.
Infection of both HCMV and HIV-1 in the same tissue allows for the possibility of co-infection of the same cell. Recent reports have described HCMV and HIV-1 interaction in monocyte-derived macrophages. King et al. (King, Baillie, and Sinclair, 2006) have shown in cultures of myeloid cells and primary macrophages that HCMV-infected cells upregulate CCR5 expression on uninfected bystander cells, which then have enhanced susceptibility to HIV-1. At the same time CCR5 expression was decreased for HCMV-infected monocyte-derived-macrophages. However, rare cells expressed both HCMV immediate-early antigens and HIV-1 Gag. Maheshwari et al. (Maheshwari et al., 2006) also reported infrequent observations of both HIV-1 and HCMV antigens in monocyte-derived macrophages in culture as well as in cells in the intestinal mucosa of co-infected patients. Our immunohistochemical analysis of macrophage cultures definitively showed expression of both GFP and the HIV-1-specific antigens p17/p24 (Figure 8A). The observation of co-infection of the same cell in culture was corroborated in frozen sections of infected explants, in which occasional cells were found to express both GFP and p17/p24 (Figures 8A and 8B). How these viruses interact within the same cell remains to be determined.
HCMV cell tropism has been linked to the HCMV ORFs UL128, UL130, or UL131A. Mutations, which can appear in any of these 3 ORFs after short passage in vitro, may produce altered cell tropism (Gerna et al., 2005; Hahn et al., 2004; Jarvis and Nelson, 2006; Patrone et al., 2005; Ryckman et al., 2006; Wang and Shenk, 2005). CMVPT30-gfp, NW23-3, CH-14, and CH-22 DNA extracted from viral stocks, infected tissue, and culture supernatants was sequenced in the region UL128 to UL131 to check for mutations. The stocks of the three clinical strains, CH-14, CH-22 and NW23-3, contain no mutations in any of the three ORFs. The CMVPT30-gfp strain was found to carry a mutation in a splice-junction dinucleotide in UL128 but no mutations in UL130 or UL131. Despite the mutation in CMVPT30-gfp, it appears to replicate to high copy numbers in infected tissues. The question remains, however, whether the mutation influences the tropism for the cell phenotype that we have observed. It also remains to be determined whether strains with mutations in the other two ORFs will have altered infectivity in the cervical explant tissue. The explants provide a human tissue model to study HCMV strains with mutations in this region, which should allow individual mutations to be linked with specific cell-tropism phenotypes.
Beyond analysis of cell tropism the model could be used for characterization of HCMV infection at the level of gene expression. Recently it was reported that the pattern of rat CMV gene expression varies in vivo in different tissues (Streblow et al., 2007). Approximately 95% of the known ORFs were expressed in vitro in different cell types, while in vivo the pattern of expression varied with the type of tissue infected and tended to be restricted to genes that encode products involved in immune evasion rather than viral replication. These findings for rat CMV demonstrate that viral gene expression in cell culture monolayers may not accurately reflect gene expression in vivo. An ex vivo tissue model could be used to determine whether HCMV infection undergoes similar restriction of gene expression.
The importance of the cervical explant model that we have developed is the ability to study the pathogenicity of human viruses, such as HCMV and HIV-1, within the natural architecture of human tissue, which contains multiple cell types. We have shown that cervical tissue is permissive for infection by cell-free and cell-associated HCMV strains and co-infection by an HIV-1 macrophage-tropic (R5) strain. The results of our quantitative co-infection studies, although not yet definitive, suggest that HIV-1 may enhance HCMV infection. To gain a broader understanding of HCMV and HIV-1 interaction in the female genital compartment, co-infection with other R5 strains as well as CXCR4 and dual-tropic strains needs to be examined. Using this model, it will be possible to: 1) identify the specific types of infected cells; 2) analyze virus genotype in relationship to cell tropism; 3) determine the tissue and virus response to infection; and 4) evaluate the effects of antiviral agents and microbicides. Thus, these explants provide a relevant “in vivo-like” model to study infection and interaction of HCMV and HIV-1 in the female genital tract.
Materials and Methods
Virus Strains
A recombinant extracellular HCMV strain was constructed from a low-passage HCMV clinical isolate PT30 (Lurain et al., 1999) and the green fluorescent protein (GFP) expression plasmid MP234 derived from the vector pIRES-hrGFP1a (Stratagene, La Jolla, CA) (a gift from Mark Peeples, Ohio State University, Columbus, OH). The recombinant strain CMVPT30-gfp expresses the humanized Renilla GFP under the control of the CMV major immediate early promoter and produces extracellular virus. CMVPT30-gfp replicates to similar titers (approximately 1 × 106) and with the same growth kinetics as the parental strain PT30 (Lurain et al., 1999). There was no deletion of any of the open reading frames UL133 through UL148, which are conserved in all clinical isolates but are deleted in some higher passage laboratory strains (Cha et al., 1996).
The HCMV clinical strains NW-23-2, CH-14, and CH-22 (Lurain et al., 2006) were obtained from patient isolates at Loyola University Medical Center, Maywood, IL through an Institutional Review Board (IRB)-approved protocol. All HCMV strains were propagated in human foreskin fibroblasts (HFF) (Viromed, Minneapolis, MN) in Eagle’s minimum essential medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 20 mM HEPES buffer, 2 mM L-glutamine, amphotericin B 2.5 μg/ml, and gentamicin 50 μg/ml. The clinical strains were maintained as cell-associated virus and were passaged three to five times in HFF. Infected HFF to be used as a source of virus for infection of tissue were harvested at approximately 80%CPE, frozen at −80 C, and transferred to liquid nitrogen for storage. The HIV-1Ba-L strain (R5) was obtained through the NIH Division of AIDS Viral Quality Assurance Laboratory at Rush University Medical Center, Chicago, IL.
Tissue samples
Ectocervical tissue samples from premenopausal women undergoing hysterectomies for benign disease were obtained with patient consent at Northwestern University Prentice Women’s Hospital, Chicago, IL. The tissues were placed immediately in medium containing 2.5 μg/ml amphotericin B (Invitrogen), and transferred without identifiers within 4 h after surgery for processing in the laboratory at Rush University Medical Center. The experimental protocol was approved by the IRB at both institutions.
The tissues were washed in an antibiotic solution containing Dulbecco’s phosphate buffered saline (Invitrogen), supplemented with penicillin-streptomycin 200 units-200 μg/ml, nystatin 500 units/ml, amphotericin B 5 μg/ml, and gentamicin 100 μg/ml. The tissue was subsequently washed three times in RAFT medium: Dulbecco’s minimal essential medium (Invitrogen) containing 24% Ham’s F12 nutrient mixture, 10% fetal bovine serum, 20 mM HEPES buffer, penicillin-streptomycin 100 units-100 μg/ml, insulin 5 μg/ml, gentamicin 50μg/ml, and 1 mM sodium pyruvate. The tissues were cut into approximately 3 mm3 (10–60 mg) blocks, which retained the epithelial surface. Three explants were placed into individual wells of a 48-well tissue culture plate that contained 500 μl RAFT medium. Explants for HCMV infection were inoculated immediately with either a cell-associated clinical HCMV strain (approximately 5 × 103 infected cells per well) or the cell-free CMVPT30-gfp strain (1 × 105 pfu/well) and maintained completely submerged in the inoculum-containing medium. The HCMV inoculum was removed after 72 h. Tissues were washed twice in RPMI (Invitrogen) without FBS. At this time all explants for HIV-1 infection were inoculated with HIV-1 Ba-L (3.0 × 104 TCID50 per well). The HIV-1 inoculum was removed after an additional 72 h. The explants were washed vigorously two times in RPMI medium, transferred to fresh RAFT medium in new 48-well plates, and cultured up to 28 days post infection (p.i.) with complete changes of medium every 2 to 3 days.
MTT assay
MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays were performed to monitor viability (Fletcher et al., 2006). Single explants were added to individual wells of a 48-well plate containing 500 μg/ml of MTT in Dulbecco’s minimal essential medium (Invitrogen). The explants were incubated at 37 °C for 3 h and then transferred to new 48-well plates containing 1 ml methanol per well. The plates were incubated and rotated at room temperature overnight in the dark. For analysis, explants were removed to preweighed tubes to record the weight. The absorbance of 3 replicates of the MTT supernatant fluid was determined at 570 nm on a CERES 900 UV HDi plate reader (Bio-Tek Instruments, Winooski, VT).
DNA Extraction
HCMV DNA from infected explants was extracted with the QIAamp DNA Mini Kit (Qiagen, Valencia, CA) using the tissue protocol. Before extraction, individual tissues were weighed, cut into smaller pieces, and lysed using the buffer provided. Volumes after lysis were approximately 200 μl. The QIAamp DNA Mini Kit was also used to extract HCMV DNA from 200 μl of tissue culture supernatants using the Blood and Body Fluid Spin Protocol. All samples were eluted in a final volume of 100 μl.
Replication of CMVPT30-gfp and HCMV clinical strains
Infected explants were monitored for GFP-expressing cells by fluorescence microscopy daily until termination of the experiment. Values of 1+ through 5+ were used to score the number of fluorescent cells (1+, 1–20 cells; 2+, 21–50 cells; 3+, 51–100 cells; 4+, 101–200 cells; 5+, >200 cells).
HCMV replication was measured quantitatively by real-time PCR using an in-house assay and the Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA). The primer pair: forward (5′-CTCGTGCGTG TGCTACGAGA-3′) and reverse (5′-GCCGATCGTRAAGAGATGAAGAC-3′) was used to amplify a 132 kb product from the conserved N-terminus of the DNA polymerase gene, which was detected with the probe 5′-6FAM-AGTGCAGCCCCGRCCATCGTTC-TAMRA-3′. The reaction was performed in a total volume of 25 μL, using Taqman 2X Universal master mix (Applied Biosystems). Amplification consisted of one cycle of 50° C for 2 min, and 95° C for 10 min; and fifty cycles of 95° C for 15 sec, and 60° C for 1 min. Dilutions of HCMV DNA of known copy number (Advanced Biotechnologies Inc., Columbia, MD) were used to generate a standard curve. Analysis was performed using the Applied Biosystems 7500 System Sequence Detection Software version 1.3. HCMV DNA copy number was normalized to the tissue weight recorded before the DNA extraction.
HIV-1Ba-L detection and quantitation
For HIV-1Ba-L quantitation studies the inoculum was removed from the explants at day 3 p.i. The explants were washed 3 times with serum-free RPMI, and transferred to new plates. Samples of supernatant fluid were taken at this time point for p24 antigen quantitation to determine the residual inoculum background. Additional samples were quantitated at 7, 14 and 21 days p.i.. The Alliance HIV-1 p24 Antigen ELISA kit (PerkinElmer, Wellesley, MA) was used for quantitation. All supernatants were diluted 1:8 to obtain readings within the linear range of the assay. Samples with values above this range were further diluted and re-tested. The p24 antigen data were analyzed using the Laboratory Data Management System (LDMS) (Frontier Science and Technology Research Foundation, Inc., Amherst, NY). HIV-1 DNA was extracted from the tissues using the QIAamp DNA Mini Kit (QIAGEN), and detected using the AMPLICOR HIV-1 DNA test, version 1.5 (Roche Molecular Systems, Inc., Branchburg, NJ), which gives only qualitative results.
Foscarnet inhibition assay
In a 48-well plate four replicate wells each containing three tissue blocks, were pre-treated with foscarnet concentrations ranging from 50μM to 800μM for 72 h. After the pre-incubation period, the drug-containing medium was changed and the tissues were inoculated with CMVPT30-gfp as previously described. Thereafter, medium with drug was completely replaced twice per week. HCMV DNA was quantitated by real-time PCR at day 14 p.i.
Cytokine analysis
Supernatants were collected and assessed for various cytokines at 7, 14, and 21 days p.i. Human IL-6, IL-8, IL-1β, IFN-α, IFN-β and IFN-γ were assayed using ELISA kits (Invitrogen), and human GRO-α was determined by the Quantikine Immunoassay for Human CXCL1/GRO-alpha (R & D Systems, Minneapolis, MN). Standard curves were constructed based on the optical density of the standards provided with each kit. Samples producing signals greater than the highest standard were further diluted using the standard diluent buffer provided and were reanalyzed. The ELISA plates were read at 450 nm on a DuPont Kinetic Microplate Reader (Molecular Devices, Sunnyvale, CA).
Immunofluorescence
Tissue specimens for immunofluorescence staining were placed in OCT medium (Ted Pella, Redding, CA) or Neg50 (Richard Allan Scientific, Kalamazoo, MI), immediately frozen, and stored at −80° C. Ten micron sections were made using a Microm HM550 cryostat (Richard Allan Scientific). Sections were fixed in PIPES buffer solution containing 3.7% formaldehyde, 2 mM MgCl2, 1 mM EGTA, and 100 mM PIPES. Slides were washed between incubation steps four times in Dulbecco’s phosphate-buffered saline at 4° C (Invitrogen). Sections were blocked using 20% (v/v) rabbit serum (Sigma-Aldrich, St. Louis, MO) containing 0.1% Triton X-100 and 0.01 % Sodium Azide (Sigma-Aldrich). Unconjugated mouse anti-human CD68 or HLA-DR (Invitrogen) monoclonal antibodies (mAb) were added to tissue sections from cervical tissue that had been infected with either CMVPT30-gfp or HCMV cell-associated clinical strains. A mouse anti-human CD1a hybridoma supernatant (OKT6), was also used as a primary antibody. Alexa-fluor 594 F(ab′)2 rabbit anti-mouse IgG (Invitrogen) and Cy5-conjugated donkey anti-mouse whole IgG (Jackson ImmunoResearch, West Grove, PA) were used as secondary antibodies. Nuclei were visualized using Slow-Fade Gold Antifade reagent with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen). Tissues that had been infected with the cell-associated clinical strains were directly stained for the delayed early DNA binding protein, p52, using a mouse FITC-conjugated anti-cytomegalovirus mAb, Clone CCH2 (Dako, Glostrup, Denmark). HIV-1 infection in tissues was detected using mAb p17 (Capricorn, Portland, ME) and p24 (AG 3.0, NIH AIDS Research and Reference Reagent Program). Slides were imaged with a DeltaVision RT epifluorescent microscope (Applied Precision Inc., Issaquah, WA). Images were captured as z-series on a CCD digital camera. Out of focus light was digitally removed using the Softworks deconvolution software (Applied Precision Inc.). Alternatively slides were imaged with a Nikon eclipse TE 2000-S inverted fluorescence microscope equipped with a SPOT RT KE 7.3 Three Shot Color camera (Diagnostic Instruments, Inc, Sterling Heights, MI) using MetaMorph version 6.0 imaging software (Molecular Devices Corp.)
Flow cytometry
Cells were dissociated from intact tissues for flow cytometric analysis (Baig et al., 2002). Approximately twelve explant tissues were collected in 10 ml complete RPMI with 5% FBS (RPMI-5). The tissues were pelleted at 400 × g, resuspended in 30 ml 1X Hanks Balanced Salt Solution (Invitrogen) with 5% FBS and 0.5 mM EDTA and rotated at 300 RPM at 37° C in a Model G25 Controlled Environmental Incubator Shaker (New Brunswick Scientific Company, Inc., Edison, NJ) at 37° C to loosen the epithelium. The tissues were pelleted again and resuspended in RPMI-5 containing 60 units/ml Type II Collagenase (Sigma-Aldrich) and rotated for 1 h at 37° C. The tissues were pelleted, resuspended in 10 ml RPMI-5, and passed approximately ten times through a 20 cc syringe with a 16 gauge feeding needle to dissociate the cells. The cell suspension was passed through a Falcon 70 μm cell strainer (BD Biosciences, San Jose, CA). The filtered suspension was pelleted and resuspended in 10 ml RPMI-5. The suspension was layered over a 35%-65% discontinuous Percoll (Sigma-Aldrich) gradient and centrifuged for 20 min at 1000 × g. Cells were collected from above the 35% layer as well as from the interface between the 35% and 65% layers, washed and resuspended in RPMI-5. Approximately 1×106 dissociated cells were obtained from up to 12 explants from the same specimen.
Cells were treated with FcR Blocking Reagent (Miltenyi Biotec, Auburn, CA) and stained with fluorochrome-conjugated mouse anti-human mAbs at room temperature for 20 min in the dark. The anti-CD14-PerCP (peridinin-chlorophyll-protein), anti-CD45-APC (allophycocyanin) and anti-HLA-DR-PE-Cy7 (phycoerythrin-cyanin7) mAb were obtained from BD Biosciences; and anti-CD1a-PE was obtained from Beckman Coulter (Miami, FL). Isotype-matched control antibodies were used as negative controls. Following incubation, the cells were washed twice with cold PBS containing BSA, fixed with 2% formaldehyde, and stored at 4° C until analysis. Samples were acquired and analyzed on a BD LSR II flow cytometer using FACSDiva™ v5.01 software (BD Biosciences). GFP was analyzed in the FL1 channel. The gate for GFP+ cells was determined by analysis of background fluorescence of uninfected control tissues and GFP fluorescence in CMVPT30-gfp-infected fibroblasts.
Sequencing HCMV open reading frames (ORF) UL128, UL130, and UL131
The genomic region containing HCMV ORFs UL128-UL131 was amplified from DNA extracts of virus stocks, supernatant fluid and infected tissue as a 2275 bp product using the following primers: 5′-CTCTATCGGCGATAAACACC-3′ (forward) and AACTGACCACCTCGGAAATG-3′ (reverse). The complete product was sequenced and sequences were compared to those of the HCMV strains Merlin (GenBank accession number AY446894) and AD169 (GenBank accession number NC001347) to identify mutations that affect predicted protein expression.
Statistical analysis
Real-time PCR quantitative results were compared using the Wilcoxan rank sum test.
Acknowledgments
This work was supported in part by HD40539 to A.L. and T.J.H., N01-A1-50044 to J.W.B., and AI48073 subcontract to N.S.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angel CE, George E, Brooks AE, Ostrovsky LL, Brown TL, Dunbar PR. Cutting edge: CD1a+ antigen-presenting cells in human dermis respond rapidly to CCR7 ligands. J Immunol. 2006;176(10):5730–4. doi: 10.4049/jimmunol.176.10.5730. [DOI] [PubMed] [Google Scholar]
- Arcasoy SM, Kotloff RM. Lung transplantation. N Engl J Med. 1999;340(14):1081–91. doi: 10.1056/NEJM199904083401406. [DOI] [PubMed] [Google Scholar]
- Baig J, Levy DB, McKay PF, Schmitz JE, Santra S, Subbramanian RA, Kuroda MJ, Lifton MA, Gorgone DA, Wyatt LS, Moss B, Huang Y, Chakrabarti BK, Xu L, Kong WP, Yang ZY, Mascola JR, Nabel GJ, Carville A, Lackner AA, Veazey RS, Letvin NL. Elicitation of simian immunodeficiency virus-specific cytotoxic T lymphocytes in mucosal compartments of rhesus monkeys by systemic vaccination. J Virol. 2002;76(22):11484–90. doi: 10.1128/JVI.76.22.11484-11490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bego MG, St Jeor S. Human cytomegalovirus infection of cells of hematopoietic origin: HCMV-induced immunosuppression, immune evasion, and latency. Exp Hematol. 2006;34(5):555–70. doi: 10.1016/j.exphem.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Boppana SB, Rivera LB, Fowler KB, Mach M, Britt WJ. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N Engl J Med. 2001;344(18):1366–71. doi: 10.1056/NEJM200105033441804. [DOI] [PubMed] [Google Scholar]
- Caux C, Dezutter-Dambuyant C, Schmitt D, Banchereau J. GM-CSF and TNF-alpha cooperate in the generation of dendritic Langerhans cells. Nature. 1992;360(6401):258–61. doi: 10.1038/360258a0. [DOI] [PubMed] [Google Scholar]
- Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70(1):78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke LM, Duerr A, Feldman J, Sierra MF, Daidone BJ, Landesman SH. Factors associated with cytomegalovirus infection among human immunodeficiency virus type 1-seronegative and -seropositive women from an urban minority community. J Infect Dis. 1996;173(1):77–82. doi: 10.1093/infdis/173.1.77. [DOI] [PubMed] [Google Scholar]
- Collins KB, Patterson BK, Naus GJ, Landers DV, Gupta P. Development of an in vitro organ culture model to study transmission of HIV-1 in the female genital tract. Nat Med. 2000;6(4):475–9. doi: 10.1038/74743. [DOI] [PubMed] [Google Scholar]
- Craigen JL, Yong KL, Jordan NJ, MacCormac LP, Westwick J, Akbar AN, Grundy JE. Human cytomegalovirus infection up-regulates interleukin-8 gene expression and stimulates neutrophil transendothelial migration. Immunology. 1997;92(1):138–45. doi: 10.1046/j.1365-2567.1997.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa CE, Annels NE, Faaij CM, Forsyth RG, Hogendoorn PC, Egeler RM. Presence of osteoclast-like multinucleated giant cells in the bone and nonostotic lesions of Langerhans cell histiocytosis. J Exp Med. 2005;201(5):687–93. doi: 10.1084/jem.20041785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng T, Ni A, Yang G, Galvin SR, Hoffman IF, Cohen MS. Distribution of the CCR5 gene 32-base pair deletion and CCR5 expression in Chinese minorities. J Acquir Immune Defic Syndr. 2003;32(2):131–4. doi: 10.1097/00126334-200302010-00003. [DOI] [PubMed] [Google Scholar]
- Fletcher P, Kiselyeva Y, Wallace G, Romano J, Griffin G, Margolis L, Shattock R. The nonnucleoside reverse transcriptase inhibitor UC-781 inhibits human immunodeficiency virus type 1 infection of human cervical tissue and dissemination by migratory cells. J Virol. 2005;79(17):11179–86. doi: 10.1128/JVI.79.17.11179-11186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher PS, Wallace GS, Mesquita PM, Shattock RJ. Candidate polyanion microbicides inhibit HIV-1 infection and dissemination pathways in human cervical explants. Retrovirology. 2006;3(1):46. doi: 10.1186/1742-4690-3-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F, Dieu-Nosjean MC, Dezutter C, Valladeau J, Kayal S, Leborgne M, Brousse N, Saeland S, Davoust J. Accumulation of immature Langerhans cells in human lymph nodes draining chronically inflamed skin. J Exp Med. 2002;196(4):417–30. doi: 10.1084/jem.20020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F, Lepelletier Y, Fraitag S, Valladeau J, Bodemer C, Debre M, Leborgne M, Saeland S, Brousse N. Differentiation of Langerhans cells in Langerhans cell histiocytosis. Blood. 2001;97(5):1241–8. doi: 10.1182/blood.v97.5.1241. [DOI] [PubMed] [Google Scholar]
- Gerna G, Percivalle E, Lilleri D, Lozza L, Fornara C, Hahn G, Baldanti F, Revello MG. Dendritic-cell infection by human cytomegalovirus is restricted to strains carrying functional UL131-128 genes and mediates efficient viral antigen presentation to CD8+ T cells. J Gen Virol. 2005;86(Pt 2):275–84. doi: 10.1099/vir.0.80474-0. [DOI] [PubMed] [Google Scholar]
- Greenhead P, Hayes P, Watts PS, Laing KG, Griffin GE, Shattock RJ. Parameters of human immunodeficiency virus infection of human cervical tissue and inhibition by vaginal virucides. J Virol. 2000;74(12):5577–86. doi: 10.1128/jvi.74.12.5577-5586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi P, Minoli L, Percivalle E, Irish W, Vigano M, Gerna G. Clinical and virological monitoring of human cytomegalovirus infection in 294 heart transplant recipients. Transplantation. 1995;59(6):847–51. [PubMed] [Google Scholar]
- Grundy JE, Lawson KM, MacCormac LP, Fletcher JM, Yong KL. Cytomegalovirus-infected endothelial cells recruit neutrophils by the secretion of C-X-C chemokines and transmit virus by direct neutrophil- endothelial cell contact and during neutrophil transendothelial migration. J Infect Dis. 1998;177(6):1465–74. doi: 10.1086/515300. [DOI] [PubMed] [Google Scholar]
- Gupta P, Collins KB, Ratner D, Watkins S, Naus GJ, Landers DV, Patterson BK. Memory CD4(+) T cells are the earliest detectable human immunodeficiency virus type 1 (HIV-1)-infected cells in the female genital mucosal tissue during HIV-1 transmission in an organ culture system. J Virol. 2002;76(19):9868–76. doi: 10.1128/JVI.76.19.9868-9876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol. 2004;78(18):10023–33. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel L, Lacaille VG, Strobl H, Mellins ED, Mocarski ES. Susceptibility of immature and mature Langerhans cell-type dendritic cells to infection and immunomodulation by human cytomegalovirus. J Virol. 2003;77(13):7563–74. doi: 10.1128/JVI.77.13.7563-7574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell AL, Edkins RD, Rier SE, Yeaman GR, Stern JE, Fanger MW, Wira CR. Human immunodeficiency virus type 1 infection of cells and tissues from the upper and lower human female reproductive tract. J Virol. 1997;71(5):3498–506. doi: 10.1128/jvi.71.5.3498-3506.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Frank I, Williams V, Santos JJ, Watts P, Griffin GE, Moore JP, Pope M, Shattock RJ. Blockade of attachment and fusion receptors inhibits HIV-1 infection of human cervical tissue. J Exp Med. 2004;199(8):1065–75. doi: 10.1084/jem.20022212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humar A, St Louis P, Mazzulli T, McGeer A, Lipton J, Messner H, MacDonald KS. Elevated serum cytokines are associated with cytomegalovirus infection and disease in bone marrow transplant recipients. J Infect Dis. 1999;179(2):484–8. doi: 10.1086/314602. [DOI] [PubMed] [Google Scholar]
- Jacobson MA. Treatment of cytomegalovirus retinitis in patients with the acquired immunodeficiency syndrome. N Engl J Med. 1997;337(2):105–14. doi: 10.1056/NEJM199707103370207. [DOI] [PubMed] [Google Scholar]
- Jacobson MA. AIDS-related cytomegalovirus retinitis. Drugs Today (Barc) 1998;34(5):409–13. doi: 10.1358/dot.1998.34.5.485238. [DOI] [PubMed] [Google Scholar]
- Jarvis MA, Nelson JA. HCMV tropism for endothelial cells: Not all endothelial cells are created equal. J Virol. 2006 doi: 10.1128/JVI.01422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CA, Baillie J, Sinclair JH. Human cytomegalovirus modulation of CCR5 expression on myeloid cells affects susceptibility to human immunodeficiency virus type 1 infection. J Gen Virol. 2006;87(Pt 8):2171–80. doi: 10.1099/vir.0.81452-0. [DOI] [PubMed] [Google Scholar]
- Kovacs A, Wasserman SS, Burns D, Wright DJ, Cohn J, Landay A, Weber K, Cohen M, Levine A, Minkoff H, Miotti P, Palefsky J, Young M, Reichelderfer P. Determinants of HIV-1 shedding in the genital tract of women. Lancet. 2001;358(9293):1593–601. doi: 10.1016/S0140-6736(01)06653-3. [DOI] [PubMed] [Google Scholar]
- Lane BR, Lore K, Bock PJ, Andersson J, Coffey MJ, Strieter RM, Markovitz DM. Interleukin-8 stimulates human immunodeficiency virus type 1 replication and is a potential new target for antiretroviral therapy. J Virol. 2001a;75(17):8195–202. doi: 10.1128/JVI.75.17.8195-8202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane BR, Strieter RM, Coffey MJ, Markovitz DM. Human immunodeficiency virus type 1 (HIV-1)-induced GRO-alpha production stimulates HIV-1 replication in macrophages and T lymphocytes. J Virol. 2001b;75(13):5812–22. doi: 10.1128/JVI.75.13.5812-5822.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CM, Drew L, Miner R, Jekic-McMullen D, Impraim C, Kao SY. Detection of cytomegalovirus in plasma and cerebrospinal fluid specimens from human immunodeficiency virus-infected patients by the AMPLICOR CMV test. J Clin Microbiol. 1998;36(9):2434–8. doi: 10.1128/jcm.36.9.2434-2438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurain NS, Fox AM, Lichy HM, Bhorade SM, Ware CF, Huang DD, Kwan SP, Garrity ER, Chou S. Analysis of the human cytomegalovirus genomic region from UL146 through UL147A reveals sequence hypervariability, genotypic stability, and overlapping transcripts. Virol J. 2006;3:4. doi: 10.1186/1743-422X-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurain NS, Kapell KS, Huang DD, Short JA, Paintsil J, Winkfield E, Benedict CA, Ware CF, Bremer JW. Human cytomegalovirus UL144 open reading frame: sequence hypervariability in low-passage clinical isolates. J Virol. 1999;73(12):10040–50. doi: 10.1128/jvi.73.12.10040-10050.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurain NS, Robert ES, Xu J, Camarca M, Landay A, Kovacs AA, Reichelderfer PS. HIV type 1 and cytomegalovirus coinfection in the female genital tract. J Infect Dis. 2004;190(3):619–23. doi: 10.1086/422533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher D, Wu X, Schacker T, Horbul J, Southern P. HIV binding, penetration, and primary infection in human cervicovaginal tissue. Proc Natl Acad Sci U S A. 2005;102(32):11504–9. doi: 10.1073/pnas.0500848102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari A, Smythies LE, Wu X, Novak L, Clements R, Eckhoff D, Lazenby AJ, Britt WJ, Smith PD. Cytomegalovirus blocks intestinal stroma-induced down-regulation of macrophage HIV-1 infection. J Leukoc Biol. 2006;80(5):1111–7. doi: 10.1189/jlb.0306230. [DOI] [PubMed] [Google Scholar]
- Mostad SB, Kreiss JK, Ryncarz AJ, Overbaugh J, Mandaliya K, Chohan B, Ndinya-Achola J, Bwayo JJ, Corey L. Cervical shedding of cytomegalovirus in human immunodeficiency virus type 1-infected women. J Med Virol. 1999;59(4):469–73. [PubMed] [Google Scholar]
- Murayama T, Kuno K, Jisaki F, Obuchi M, Sakamuro D, Furukawa T, Mukaida N, Matsushima K. Enhancement human cytomegalovirus replication in a human lung fibroblast cell line by interleukin-8. J Virol. 1994;68(11):7582–5. doi: 10.1128/jvi.68.11.7582-7585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowzari H, Jorgensen MG, Aswad S, Khan N, Osorio E, Safarian A, Shidban H, Munroe S. Human cytomegalovirus-associated periodontitis in renal transplant patients. Transplant Proc. 2003;35(8):2949–52. doi: 10.1016/j.transproceed.2003.10.026. [DOI] [PubMed] [Google Scholar]
- Oguariri RM, Brann TW, Imamichi T. Hydroxyurea and interleukin-6 synergistically reactivate HIV-1 replication in a latently infected promonocytic cell line via SP1/SP3 transcription factors. J Biol Chem. 2007;282(6):3594–604. doi: 10.1074/jbc.M608150200. [DOI] [PubMed] [Google Scholar]
- Paczesny S, Li YP, Li N, Latger-Canard V, Marchal L, Ou-Yang JP, Bordigoni P, Stoltz JF, Eljaafari A. Efficient generation of CD34+ progenitor-derived dendritic cells from G-CSF-mobilized peripheral mononuclear cells does not require hematopoietic stem cell enrichment. J Leukoc Biol. 2007 doi: 10.1189/jlb.0406296. [DOI] [PubMed] [Google Scholar]
- Palacio J, Souberbielle BE, Shattock RJ, Robinson G, Manyonda I, Griffin GE. In vitro HIV1 infection of human cervical tissue. Res Virol. 1994;145(3–4):155–61. doi: 10.1016/s0923-2516(07)80017-3. [DOI] [PubMed] [Google Scholar]
- Patrone M, Secchi M, Fiorina L, Ierardi M, Milanesi G, Gallina A. Human cytomegalovirus UL130 protein promotes endothelial cell infection through a producer cell modification of the virion. J Virol. 2005;79(13):8361–73. doi: 10.1128/JVI.79.13.8361-8373.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson BK, Landay A, Andersson J, Brown C, Behbahani H, Jiyamapa D, Burki Z, Stanislawski D, Czerniewski MA, Garcia P. Repertoire of chemokine receptor expression in the female genital tract: implications for human immunodeficiency virus transmission. Am J Pathol. 1998;153(2):481–90. doi: 10.1016/S0002-9440(10)65591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paya CV, Marin E, Keating M, Dickson R, Porayko M, Wiesner R. Solid organ transplantation: results and implications of acyclovir use in liver transplants. J Med Virol. 1993;(Suppl 1):123–7. doi: 10.1002/jmv.1890410524. [DOI] [PubMed] [Google Scholar]
- Poppe WA, Drijkoningen M, Ide PS, Lauweryns JM, Van Assche FA. Lymphocytes and dendritic cells in the normal uterine cervix. An immunohistochemical study. Eur J Obstet Gynecol Reprod Biol. 1998;81(2):277–82. doi: 10.1016/s0301-2115(98)00202-4. [DOI] [PubMed] [Google Scholar]
- Redman TK, Britt WJ, Wilcox CM, Graham MF, Smith PD. Human cytomegalovirus enhances chemokine production by lipopolysaccharide-stimulated lamina propria macrophages. J Infect Dis. 2002;185(5):584–90. doi: 10.1086/339007. [DOI] [PubMed] [Google Scholar]
- Ross SA, Fowler KB, Ashrith G, Stagno S, Britt WJ, Pass RF, Boppana SB. Hearing loss in children with congenital cytomegalovirus infection born to mothers with preexisting immunity. J Pediatr. 2006;148(3):332–6. doi: 10.1016/j.jpeds.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol. 2006;80(2):710–22. doi: 10.1128/JVI.80.2.710-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinzger C, Grefte A, Plachter B, Gouw AS, The TH, Jahn G. Fibroblasts, epithelial cells, endothelial cells and smooth muscle cells are major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J Gen Virol. 1995;76(Pt 4):741–50. doi: 10.1099/0022-1317-76-4-741. [DOI] [PubMed] [Google Scholar]
- Streblow DN, van Cleef KW, Kreklywich CN, Meyer C, Smith P, Defilippis V, Grey F, Fruh K, Searles R, Bruggeman C, Vink C, Nelson JA, Orloff SL. Rat Cytomegalovirus Gene Expression In Cardiac Allograft Recipients Is Tissue Specific And Does Not Parallel The Profiles Detected In Vitro. J Virol. 2007 doi: 10.1128/JVI.02425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm-Ramirez K, Gaye-Diallo A, Eisen G, Mboup S, Kanki PJ. High levels of tumor necrosis factor-alpha and interleukin-1beta in bacterial vaginosis may increase susceptibility to human immunodeficiency virus. J Infect Dis. 2000;182(2):467–73. doi: 10.1086/315713. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Yamada H, Minami M, Kataoka S, Numazaki K, Minakami H, Tsutsumi H. Screening for vaginal shedding of cytomegalovirus in healthy pregnant women using real-time PCR: correlation of CMV in the vagina and adverse outcome of pregnancy. J Med Virol. 2006;78(6):757–9. doi: 10.1002/jmv.20619. [DOI] [PubMed] [Google Scholar]
- Wang D, Shenk T. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J Virol. 2005;79(16):10330–8. doi: 10.1128/JVI.79.16.10330-10338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman GR, Asin S, Weldon S, Demian DJ, Collins JE, Gonzalez JL, Wira CR, Fanger MW, Howell AL. Chemokine receptor expression in the human ectocervix: implications for infection by the human immunodeficiency virus-type I. Immunology. 2004;113(4):524–33. doi: 10.1111/j.1365-2567.2004.01990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]