Abstract
Saccharomyces cerevisiae mating pheromones trigger dissociation of a heterotrimeric G protein (Gαβγ) into Gα-guanosine triphosphate (GTP) and Gβγ. The Gβγ dimer regulates both mitogen-activated protein (MAP) kinase cascade signaling and cell polarization. Here, by independently activating the MAP kinase pathway, we studied the polarity role of Gβγ in isolation from its signaling role. MAP kinase signaling alone could induce cell asymmetry but not directional growth. Surprisingly, active Gβγ, either alone or with Gα-GTP, could not organize a persistent polarization axis. Instead, following pheromone gradients (chemotropism) or directional growth without pheromone gradients (de novo polarization) required an intact receptor–Gαβγ module and GTP hydrolysis by Gα. Our results indicate that chemoattractant-induced cell polarization requires continuous receptor–Gαβγ communication but not modulation of MAP kinase signaling. To explore regulation of Gβγ by Gα, we mutated Gβ residues in two structurally distinct Gα–Gβ binding interfaces. Polarity control was disrupted only by mutations in the N-terminal interface, and not the Switch interface. Incorporation of these mutations into a Gβ–Gα fusion protein, which enforces subunit proximity, revealed that Switch interface dissociation regulates signaling, whereas the N-terminal interface may govern receptor–Gαβγ coupling. These findings raise the possibility that the Gαβγ heterotrimer can function in a partially dissociated state, tethered by the N-terminal interface.
INTRODUCTION
Cell polarization involves the asymmetric distribution of intracellular materials in a way that is usually directed toward localized instructional cues, which can be internal or external to the cell. External cues govern chemotaxis in cells such as Dictyostelium discoideum and mammalian neutrophils, such that a gradient of chemoattractant from a localized source stimulates cell polarization and directed cell movement along the gradient (Iijima et al., 2002; Bagorda et al., 2006; Franca-Koh et al., 2006). The ability to coordinate the direction of cell movement with the location of the external signal implies that the initial sensing of chemoattractant and at least some of the subsequent intracellular signal transduction events do not occur uniformly within the cell but instead occur in an asymmetric, spatially restricted manner.
The mating reaction of the budding yeast Saccharomyces cerevisiae provides a model system to study how cells generate an asymmetric response to polarization stimuli. Here, the two haploid cell types (a or α) secrete cell type-specific mating pheromones (a factor or α factor, respectively), which serve as chemoattractants that stimulate mating responses in cells of the opposite type (Dohlman and Thorner, 2001). Each pheromone binds a specific G protein-coupled receptor (GPCR) (Ste2 or Ste3, which recognize α factor and a factor, respectively) that couples to a heterotrimeric G protein (Gαβγ, composed of Gpa1 [Gα], Ste4 [Gβ], and Ste18 [Gγ]), which is not cell type specific. Binding of pheromone to the receptor catalyzes exchange of guanosine triphosphate (GTP) for guanosine diphosphate (GDP) on the Gα subunit, causing dissociation of Gα-GTP from the Gβγ dimer. Once released from the inhibitory Gα subunit, Gβγ can trigger signaling through a mitogen-activated protein (MAP) kinase cascade; this involves membrane recruitment of the scaffold protein Ste5, which leads to activation of the Ste5-associated kinases, Ste11 (MAP kinase kinase kinase), Ste7 (MAP kinase kinase), and Fus3 (MAP kinase [MAPK]) (Pryciak and Huntress, 1998; Mahanty et al., 1999; van Drogen et al., 2000, 2001; Winters et al., 2005). These signaling events induce cell cycle arrest and the transcription of mating specific genes (Dohlman and Thorner, 2001).
Mating pheromones also regulate cell morphology. The actin cytoskeleton and membrane trafficking systems are rearranged to form a polarized mating projection, which then grows toward the partner in a process termed chemotropism (Segall, 1993; Arkowitz, 1999; Pruyne and Bretscher, 2000). These polarization events highlight another role for Gβγ. Current evidence suggests that Gβγ recruits the polarity proteins Far1 and Cdc24 (Valtz et al., 1995; Butty et al., 1998; Nern and Arkowitz, 1998, 1999) to the region of plasma membrane receiving the highest pheromone dose. Cdc24 is the guanine-nucleotide exchange factor (GEF) for the Rho-family GTPase Cdc42, which controls actin organization and cell polarity (Chant, 1999; Etienne-Manneville, 2004). Far1 is an adaptor protein that binds both Gβγ and Cdc24 (Butty et al., 1998; Nern and Arkowitz, 1999). Hence, communication between Gβγ and Far1/Cdc24 is thought to direct Cdc42 activity to the proper site to allow polarized growth along the gradient of pheromone. This process may be facilitated by additional interactions between Gα and Fus3 (Metodiev et al., 2002) or between Gβγ and Rho1 (Bar et al., 2003).
For chemotactic cells (Dictyostelium and neutrophils) and chemotropic cells (yeast), a gradient of chemoattractant normally serves as a spatial cue for the direction of polarization. However, these cells will still polarize when exposed to a uniform concentration of chemoattractant, implying the existence of “symmetry breaking” mechanisms that can generate asymmetric responses to symmetric signals (Sohrmann and Peter, 2003; Wedlich-Soldner and Li, 2003). In yeast, prior work reveals two ways in which a cell can polarize when pheromone is provided uniformly rather than as a gradient, which we will refer to as “default” and “de novo” polarization. Default polarization uses preexisting polarity information provided by the bud site selection proteins as a spatial cue, resulting in the formation of a mating projection at the presumptive bud site (Madden and Snyder, 1992; Dorer et al., 1995; Nern and Arkowitz, 1999). This default polarization is independent of Far1–Cdc24 communication, but it depends on bud site selection proteins such as Rsr1/Bud1 (Nern and Arkowitz, 1999). When these default sites are absent, cells can still polarize in a uniform field of pheromone, but now the axis of polarization forms de novo at random positions that bear no relationship to previous polarization sites. This de novo polarization is independent of bud site selection proteins, but it requires Far1–Cdc24 binding; when this binding is disrupted, cells initiate polarization but fail to organize a stable axis of polarized growth (Nern and Arkowitz, 1999, 2000). Thus, interactions that are required for asymmetric response to a gradient of pheromone are also required when asymmetric growth is initiated de novo.
To better understand how external signals couple with intracellular factors to generate an asymmetric response, we investigated the regulation of yeast Gβγ activity in response to a gradient or uniform field of pheromone. We show that in addition to Gβγ, the receptor and Gα are required not only to sense pheromone gradients during chemotropism but also for directionally persistent growth during de novo polarization. In addition, by using a series of mutations in Gβ (Ste4) to disrupt regulation of Gβγ by Gα, we observed qualitatively different polarity phenotypes depending on which Gα-Gβ interaction interface is disrupted. Our results suggest that one Gα–Gβ interface controls signaling, whereas the other facilitates directional responses by allowing receptor–Gαβγ coupling.
MATERIALS AND METHODS
Yeast Strains
Yeast strains are listed in Table 1. PGAL1-STE5ΔN-CTM was integrated at the HIS3 locus by transformation with NheI-digested pPP1268 to create strains PPY1303, PPY1304, PPY1306, and PPY1307. PGAL1-STE11ΔN-STE7 was integrated at the HIS3 locus by transformation with NheI-digested pPP1270 to create strains PPY1309, PPY1310, PPY1311, PPY1312, PPY1313, PPY1314, PPY1951, and PPY1952.
Table 1.
Yeast strains used in this study
| Strain backgrounda | Strain name | Genotype | Source |
|---|---|---|---|
| (a) | PPY498 | MATa FUS1::FUS1-lacZ::LYS2 fus3::LEU2 kss1::ura3FOA | This study |
| (a) | PPY577 | MATa FUS1::FUS1-lacZ::LYS2 ADE3 | This study |
| (a) | PPY662 | MATa FUS1::FUS1-lacZ::LYS2 ste4::ura3FOA | This study |
| (a) | PPY663 | MATa FUS1::FUS1-lacZ::LYS2 | Strickfaden et al. (2007) |
| (a) | PPY817 | MATaFUS1::FUS1-lacZ::LYS2 far1::ADE2 | Strickfaden et al. (2007) |
| (a) | PPY820 | MATaFUS1::FUS1-lacZ::LYS2 fus3::LEU2 kss1::ura3FOA far1::ADE2 | This study |
| (a) | PPY824 | MATaFUS1::FUS1-lacZ::LYS2 fus3::LEU2 | This study |
| (a) | PPY827 | MATaFUS1::FUS1-lacZ::LYS2 fus3::LEU2 far1::ADE2 | This study |
| (a) | PPY836 | MATaHIS4 far1::ADE2 | This study |
| (b) | PPY398 | MATa | Harris et al. (2001) |
| (b) | PPY794 | MATa ste4::ura3FOA | Pryciak and Huntress (1998) |
| (b) | PPY842 | MATaste4::ura3FOA ste5::ADE2 ste20-1::TRP1 | Winters et al. (2005) |
| (b) | PPY856 | MATa FUS1::FUS1-lacZ::LEU2 ste4::ura3FOA | Winters et al. (2005) |
| (b) | PPY858 | MATaFUS1::FUS1-lacZ::LEU2 ste5::ADE2 | Pryciak and Huntress (1998) |
| (b) | PPY863 | MATaste5::ADE2 ste20-1::TRP1 | This study |
| (b) | PPY867 | MATaste4::ura3FOA ste5::ADE2 | This study |
| (b) | PPY886 | MATa FUS1::FUS1-lacZ::LEU2 ste5::ADE2 ste4::ura3FOA | Pryciak and Huntress (1998) |
| (b) | PPY978 | MATaFUS1::FUS1-lacZ::LEU2 ste5::ADE2 gpa1::URA3 | This study |
| (b) | PPY979 | MATaFUS1::FUS1-lacZ::LEU2 ste5::ADE2 ste2::URA3 | This study |
| (b) | PPY989 | MATaFUS1::FUS1-lacZ::LEU2 ste5::ADE2 ste18::URA3 | This study |
| (b) | PPY1228 | MATa ste4::ura3FOA gpa1::ura3FOA | This study |
| (b) | PPY1230 | MATaste4::ura3FOA gpa1::ura3FOA ste5::ADE2 | This study |
| (b) | PPY1248 | MATaste4::ura3FOA rsr1::ura3::HIS3 | This study |
| (b) | PPY1259 | MATarsr1::ura3::HIS3 | This study |
| (b) | PPY1303 | MATaHIS3::PGAL1-STE5ΔN-CTM | This study |
| (b) | PPY1304 | MATaHIS3::PGAL1-STE5ΔN-CTM ste5::ADE2 | This study |
| (b) | PPY1306 | MATaHIS3::PGAL1-STE5ΔN-CTM rsr1::URA3 | This study |
| (b) | PPY1307 | MATaHIS3::PGAL1-STE5ΔN-CTM rsr1::URA3 ste5::ADE2 | This study |
| (b) | PPY1309 | MATaHIS3::PGAL1-STE11ΔN-STE7 | This study |
| (b) | PPY1310 | MATaHIS3::PGAL1-STE11ΔN-STE7 ste5::ADE2 | This study |
| (b) | PPY1311 | MATaHIS3::PGAL1-STE11ΔN-STE7 ste5::ADE2 ste4::ura3FOA | This study |
| (b) | PPY1312 | MATaHIS3::PGAL1-STE11ΔN-STE7 rsr1::URA3 | This study |
| (b) | PPY1313 | MATaHIS3::PGAL1-STE11ΔN-STE7 rsr1::URA3 ste5::ADE2 | This study |
| (b) | PPY1314 | MATa HIS3::PGAL1-STE11ΔN-STE7 rsr1::URA3 ste5::ADE2 ste4::ura3FOA | This study |
| (b) | PPY1380 | MATa gpa1::ura3FOA ste4::ura3FOA rsr1::ura3FOA | This study |
| (b) | PPY1662 | MATaFUS1::FUS1-lacZ::LEU2 ste4::ura3FOA ste7::ADE2 | This study |
| (b) | PPY1663 | MATa FUS1::FUS1-lacZ::LEU2 ste4::ura3FOA gpa1::ura3FOA | This study |
| (b) | PPY1937 | MATa fus3::LEU2 ste4::ura3FOA | This study |
| (b) | PPY1942 | MATaste4::ura3FOA gpa1::ura3FOA sst2::LEU2 | This study |
| (b) | PPY1951 | MATa HIS3::PGAL1-STE11ΔN-STE7 ste5::ADE2 gpa1::ura3FOA | This study |
| (b) | PPY1952 | MATaHIS3::PGAL1-STE11ΔN-STE7 ste5::ADE2 gpa1::ura3FOA rsr1::URA3 | This study |
| (c) | AMR70 | MATα URA3::(lexAop)8-lacZ lys2 met2 | Hollenberg et al. (1995) |
| (c) | L40 | MATaLYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ | Hollenberg et al. (1995) |
| (c) | PPY762 | MATaLYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ ste11::ADE2 | Butty et al. (1998) |
| (c) | PPY1158 | MATaLYS2::(lexAop)4-HIS3 ura3FOA::(lexAop)8-lacZ ste11::ADE2 gpa1::URA3 | This study |
| (d) | PT2α | MATα hom3 ilv1 can1 | Pryciak and Huntress (1998) |
a Strain background: (a) 381G (cry1 ade2-1oc ade3 his4-580am leu2-3,112 lys2oc trp1am ura3-52 SUP4-3ts).(b) W303 (ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1). (c) S288c (ade2 his3-Δ200 leu2-3,112 trp1-901). (d) Other.
Plasmids
Plasmids used in this study are listed in Table 2. Ste4 mutants defective at binding Gpa1 (see next section) were originally isolated as variants of the two-hybrid construct pPP268 (identical to pGAD-STE4 except for an extra SphI site following STE4) and then were transferred as EcoRI–BamHI fragments to a two-hybrid construct with a stronger promoter, pGADXP-STE4. All mutations were also transferred into pPP2968 (CEN URA3 STE4) as PstI–XhoI or BglII–BsaI (for W411R) fragments; pPP2968 contains STE4 on a 2.4-kb AflII–BspHI fragment cloned into blunted XbaI and XhoI sites of a derivative of pRS316 in which the URA3 gene lacks a PstI site. Mutations causing the most severe disruption of Gpa1–Ste4 binding (i.e., K126E, L117R, W136R L138F, and L154R N156K) were also moved to other contexts for further study. First, they were transferred as MscI–XhoI fragments from the pPP268 derivatives into the PGAL1-STE4 construct pGT-STE4 (Klein et al., 2000). Then, the mutants were placed under control of the native STE4 promoter by transferring MscI–BspEI fragments from these PGAL1-STE4 constructs into pPP1340. In addition, the mutations were transferred as MscI–XhoI fragments into a PGAL1-STE4-GPA1 fusion construct pSTE4-GPA1-b (Klein et al., 2000). These STE4–GPA1 fusions were then placed under control of the native STE4 promoter by transferring MscI–BspEI fragments from the PGAL1-STE4–GPA1 fusion constructs into pPP226, which contains STE4 on a 4-kb EcoRI fragment, from −2045 to +2003, in the EcoRI site of pRS316.
Table 2.
Plasmids used in this study
| Name | Alias | Description | Source |
|---|---|---|---|
| pPP120 | pRD-STE11-H3 | CEN URA3 PGAL1-GST-STE11ΔN | Neiman and Herskowitz (1994) |
| pPP126 | YCpGPA1 | CEN LEU2 GPA1 | Stone and Reed (1990) |
| pPP134 | pNC252 | 2 μm URA3 PGAL1-STE12 | Pryciak and Hartwell (1996) |
| pPP226a | p316-ST4-a | CEN URA3 PSTE4-STE4 (-2045 to +2003) | This study |
| pPP244 | pGAD424 | 2 μm LEU2 GAL4-AD vector | Bartel and Fields (1995) |
| pPP247 | pBTM-GPA1 | 2 μm TRP1 lexA-DBD-GPA1 | Pryciak and Hartwell (1996) |
| pPP249 | pGAD-STE4 | 2 μm LEU2 GAL4-AD-STE4 | Pryciak and Hartwell (1996) |
| pPP255 | pBTM-STE18 | 2 μm TRP1 lexA-DBD-STE18 | Pryciak and Hartwell (1996) |
| pPP268a | pGAD-YS4 | 2 μm LEU2 GAL4-AD-STE4 (w/extra SphI site) | This study |
| pPP271 | pGS12-T | 2 μm TRP1 PGAL1-STE12 | This study |
| pPP305 | pM276p16 | 2 μm TRP1 lexA-DBD-STE5(1-214) | Whiteway et al. (1995) |
| pPP446 | pRD53-2 μm | 2 μm URA3 PGAL1 vector | This study |
| pPP452 | pGS5 | CEN TRP1 PGAL1-STE5 | Pryciak and Huntress (1998) |
| pPP473 | pGS5ΔN-CTM | CEN TRP1 PGAL1-ste5ΔN-CTM (= Snc2 TM domain) | Pryciak and Huntress (1998) |
| pPP479 | pH-GS5ΔN-CTM | CEN HIS3 PGAL1-ste5ΔN-CTM (= Snc2 TM domain) | Pryciak and Huntress (1998) |
| pPP513 | pGFP-GS5ΔN-CTM | CEN TRP1 PGAL1-GFP-ste5ΔN-CTM (= Snc2 TM domain) | Pryciak and Huntress (1998) |
| pPP524 | pGFP-GS5ΔN-SEC22 | CEN TRP1 PGAL1-GFP-ste5ΔN-Sec22 TM domain | Pryciak and Huntress (1998) |
| pPP575 | pGS11ΔN-T | CEN TRP1 PGAL1-GST-STE11ΔN | Moskow et al. (2000) |
| pPP636 | pGADXP | 2 μm LEU2 strong PADH1-GAL4-AD vector | Butty et al. (1998) |
| pPP643a | pGADXP-STE4 | 2 μm LEU2 strong PADH1-GAL4-AD-STE4 | Butty et al. (1998) |
| pPP679 | pRS314 | CEN TRP1 vector | Sikorski and Hieter (1989) |
| pPP681 | pRS316 | CEN URA3 vector | Sikorski and Hieter (1989) |
| pPP741 | pNC252-HIS3 | 2 μm HIS3 PGAL1-STE12 | This study |
| pPP743 | pBTM-FAR1 174-285 | 2 μm TRP1 lexA-DBD-FAR1(174-285) | This study |
| pPP854 | p3058 | CEN LEU2 PFUS1-lacZ reporter | Roberts et al. (2000) |
| pPP1150a | pSTE4-GPA1-b | CEN URA3 PGAL1-STE4-GPA1 fusion (1 a.a. linker) | Klein et al. (2000) |
| pPP1151a | pGT-STE4 | CEN URA3 PGAL1-STE4 | Klein et al. (2000) |
| pPP1175 | pH-GFP-GS5ΔN-SEC22 | CEN HIS3 PGAL1-GFP-ste5ΔN-Sec22 TM domain | This study |
| pPP1268 | pIH-GS5ΔN-CTM | integrating HIS3 PGAL1-ste5ΔN-CTM (= Snc2 TM domain) | This study |
| pPP1270 | pIH-G11ΔN.S7 | integrating HIS3 PGAL1-STE11ΔN-STE7 fusion | This study |
| pPP1340a | pS4GA-WT | CEN URA3 PSTE4-STE4-GPA1 fusion | This study |
| pPP1501 | YCpGPA1-E28K | CEN LEU2 GPA1(E28K) | This study |
| pPP1502 | pBTM-GPA1-E28K | 2 μm TRP1 lexA-DBD-GPA1(E28K) | This study |
| pPP1503 | YCpGPA1-E28A | CEN LEU2 GPA1(E28A) | This study |
| pPP1505 | pBTM-GPA1-E28A | 2 μm TRP1 lexA-DBD-GPA1(E28A) | This study |
| pPP1621 | YCplac22-GPA1-WT | CEN TRP1 GPA1 | Stratton et al. (1996) |
| pPP2711 | p314-GPA1-WT | CEN TRP1 GPA1 | This study |
| pPP2743 | p314-GPA1-K21ER22E | CEN TRP1 GPA1(K21E R22E) | This study |
| pPP2773 | pRS314-G7 | CEN TRP1 PGAL1-STE7 | This study |
| pPP2775 | p314-GPA1-HA-M | CEN TRP1 HA3-GPA1 | This study |
| pPP2802 | p314-GPA1-Q323L | CEN TRP1 GPA1(Q323L) | This study |
| pPP2838a | p316P*-mycS4 | CEN URA3 PSTE4-myc13-STE4 | This study |
| pPP2839a | p316P*-mycS4GA-WT | CEN URA3 PSTE4-myc13-STE4-GPA1 fusion | This study |
| pPP2968a | p316P*-ST4AB | CEN URA3 PSTE4-STE4 (-780 to +1616) | This study |
a.a., amino acids; TM, transmembrane.
a These plasmids, containing wild-type STE4, serve as the parental constructs for multiple mutant derivatives used in this study encompassing 71 additional plasmids, which consist of 15 mutant derivatives each of pPP268 and pPP643, 13 derivatives of pPP2968, six derivatives each of pPP1150 and pPP1340, and four derivatives each of pPP226, pPP1151, pPP2838, and pPP2839.
To construct STE4–GPA1 fusions harboring the GPA1-Q323L mutation, the Q323L mutation was first transferred on a SphI–SphI fragment from pRS316-GPA1-Q323L (Apanovitch et al., 1998) into the STE4–GPA1 fusion construct pPP1340, creating pPP1859. Then, the BglII–BspEI fragment from pPP1859 was transferred into pPP1343 to create pPP2801. To place these mutant STE4–GPA1 fusions under control of the GAL1 promoter, the MscI–BspEI fragments from pPP1859 and pPP2801 were transferred into pSTE4-GPA1-b (Klein et al., 2000), creating pPP2806 and pPP2807.
Plasmid pPP2711 (CEN TRP1 GPA1) was created by polymerase chain reaction (PCR) amplification of GPA1 (−201 to +1707) and ligation as a SacI–KpnI fragment into pRS314 (Sikorski and Hieter, 1989). The GPA1 mutations Q323L and K21E R22E were transferred into pPP2711 as BsrGI–BstBI fragments from pRS316-GPA1-Q323L (Apanovitch et al., 1998) and YCplac22-GPA1-K21ER22E (Metodiev et al., 2002), to create pPP2802 and pPP2743, respectively. The GPA1 mutations E28K and E28A were generated by site-directed mutagenesis of YCpGPA1 (Stone and Reed, 1990) and pPP247 (Pryciak and Hartwell, 1996), creating plasmids pPP1501, pPP1502, pPP1503, and pPP1505. The PGAL1-STE7 construct pPP2773 was made by transferring STE7 as a BamHI–PstI fragment from pG7 (Harris et al., 2001) into the CEN TRP1 PGAL1 vector pPP449 (Pryciak and Huntress, 1998).
To create epitope-tagged forms of Ste4 and Gpa1, we first introduced unique NotI sites into their coding sequences. For STE4, the NotI site was added between a repeat of the first two codons (ATG GCA agc ggc cgc ATG GCA), and then a myc13 NotI–NotI cassette was inserted to create pPP2838 (myc13-STE4) and pPP2839 (myc13-STE4-GPA1 fusion). Although these derivatives were functional in vivo, the myc13 epitope caused reduced signaling in FUS1-lacZ and halo assays, and thus they were used only to test effects of Ste4 mutations on protein levels and Gpa1 coimmunoprecipitation. For GPA1, intact N and C termini of the protein are functionally important. Hence, the NotI site was added within a yeast-specific insert region (Lambright et al., 1996), overlapping codons 161–163 (AgC GGc CGc) and introducing a conservative residue change (T161S). An HA3 NotI–NotI cassette was then inserted, creating pPP2775. These derivatives were fully functional in vivo.
Isolation of Ste4 Mutants
A genetic screen was conducted using a two-hybrid assay to isolate Ste4 mutants with specific defects in binding Gpa1. Libraries of ste4 mutants (made by error-prone PCR) were created in a two-hybrid activation-domain vector (pGAD424) as described previously (Pryciak and Hartwell, 1996), and they were transformed into a MATa two-hybrid reporter strain (L40). Transformants were mated by replica plating to a MATα strain (AMR70) harboring DNA-binding domain (DBD) fusions to either Gpa1 (pBTM-GPA1) or Ste18 (pBTM-STE18). Clones were identified that showed reduced interaction with Gpa1 but not with Ste18. Secondary screens tested interaction with Ste5 (pM276p16) and Far1 (pPP743). All sequenced isolates harbored mutations in residues that contact Gα in crystal structures (Lambright et al., 1996; Sondek et al., 1996). These isolates (and their mutations) were as follows: Gm3-E41 (L154S), Gm3-C7 (K126E), Gm5-B47 (K126E D370G), Gm5-E73 (L117R), Gm4-H32 (D224V), Gm4-J38 (D224E D402E), and Gm4-I22 (S164W K213R D272A V375A). The D272A mutation in Gm4-I22 was separated from the other mutations by swapping restriction fragments; although largely responsible for the Gpa1-binding defect, the single D272A mutant was not as severe as the original quadruple mutant, and therefore all assays reported here for D272A used the stronger quadruple mutant. The isolate Gm3-E41 (L154S) was briefly reported previously (Pryciak and Hartwell, 1996). Because this L154S mutant did not show the strong chemotropism defects seen with N-terminal interface mutants (see Results), and because both Leu154 and the neighboring Asn156 residue are predicted to contact Gpa1 (Lambright et al., 1996; Sondek et al., 1996), we made an additional mutant (L154R N156K) in an attempt to cause a more severe disruption. In fact, this mutant and the original L154S mutant were effectively indistinguishable, and so only the mutant with the more-drastic substitution (L154R N156K) is analyzed here. In addition, for comparison to the phenotypically strong mutants L117R and K126E, we made less drastic substitutions at the same positions (L117A, L117G, K126A, K126N).
Another screen was designed to isolate Ste4 mutants with reduced mating efficiency but normal pathway signaling. A ste4 mutant library was created in a PGAL1 vector (pYES-R) as described previously (Pryciak and Hartwell, 1996), and it was transformed into a MATa ste4Δ FUS1-lacZ strain. Transformants were mated (on galactose medium) to a mixed lawn of pheromone-producing and pheromoneless MATα cells, and screened for clones showing reduced mating only with the pheromone-producing MATα partner. The same clones were also screened for intact FUS1-lacZ induction (on galactose plates), by using a filter-based β-galactosidase assay (Bartel and Fields, 1995). Five isolates with the strongest mating defects were sequenced. One had multiple mutations and was not pursued further. Each of the others harbored mutations at Lys126 (three were K126E, one was K126N). One isolate, called m3-D10 (T76S K126E) was described previously (Pryciak and Hartwell, 1996). Because their mutations and phenotypes were similar to other K126E mutants (see 1), further analysis of these isolates is not presented.
A Ste4 mutant with Gpa1-binding defects, W136R L138F, which was isolated in a previous screen (Whiteway et al., 1994), was also studied here. This mutation was transferred by PCR from its original context (CEN HIS3 PGAL1-STE4Hpl 21–1) into pPP268, and then it was transferred to other contexts in parallel with the new mutations.
Site-directed mutagenesis was used to target Ste4 residues that were predicted to contact Gpa1 but that were not uncovered in unbiased screens. Structural data predict that Gα is contacted by 16 Gβ residues that lie in nine regions (Lambright et al., 1996; Sondek et al., 1996). The mutations identified from genetic screens (see 1–3) affected six of these nine regions. Therefore, the other three regions (composed of 6 residues) were mutated, using drastic residue changes to disrupt Gpa1 binding as severely as possible. These included the triple mutation N92G K94E S96A, the double mutation T181R Y183N, and the single mutation W411R. The T181R Y183N mutant was defective in all assays, and it was defective at binding all partners tested (including Ste18), suggesting it may be misfolded, and so it was not pursued further. The other mutations had only weak effects on Gpa1 binding (see Results), consistent with their absence from genetic screens.
Additional mutants used as controls were isolated in screens for Ste4 mutants with Ste18-binding defects (e.g., Gm3-F30 [D97G K380E]) (Pryciak and Hartwell, 1996) or with defects in binding Ste5 and Far1 (e.g., Gm3-J70 [L49P]). The D97G K380E mutation disrupts binding to the Gγ subunit Ste18 and hence to Gβγ-binding partners; each mutation lies close to a predicted Gγ-contact residue (W100 and L379) (Sondek et al., 1996), although it is not known which mutation disrupts Ste18 binding (or whether both contribute). The L49P mutant was isolated in a screen to be described elsewhere, but it harbors a mutation identical to a mutation found previously (Dowell et al., 1998).
Microscopy
For de novo and default polarization assays, transformants were grown in selective raffinose media, and expression of PGAL1-driven constructs was induced by addition of galactose to 2%; where indicated, 10 μM α factor was also added simultaneously. After 2- to 8-h induction, cells were visualized without fixation. Differential labeling of old and new cell surface growth with fluorophore-conjugated concanavalin A (ConA) used methods described previously (Nern and Arkowitz, 2000; Matheos et al., 2004). Cells were grown first in synthetic raffinose medium, labeled with 0.1 mg/ml fluorescein isothiocyanate (FITC)-ConA, induced with 2% galactose ± 10 μM α factor for 4 h, and then labeled with 0.05 mg/ml tetramethylrhodamine B isothiocyanate (TRITC)-ConA.
Mating Assays
For patch mating assays, a cells were patched directly onto a lawn of partner PT2α cells, mated overnight at 30°C, and then diploids were selected by replication to minimal media. After an immediate (1°) replica was made, more dilute replicas were made by repeating the replication of the master mating plate twice more, using a fresh velvet each time, to create 2° and 3° replicas (Harris et al., 2001). Replicas were incubated for 2 d at 30°C. For pheromone confusion assays, α factor was spread on one plate to give a final concentration of 30 μM (unless indicated otherwise) and allowed to dry before the PT2α lawn was spread.
Quantitative mating assays followed previous methods (Pryciak and Huntress, 1998; Lamson et al., 2002). In brief, plasmid-transformed cells were grown overnight in selective raffinose medium, and then 0.5–5 × 106 a cells were mixed with an equal number of wild-type α cells (strain PT2α) and collected onto filters. The filters were placed on SC/raffinose/galactose plates and incubated for 6–7 h (unless indicated otherwise) at 30°C. Afterward, filters were suspended in phosphate-buffered saline (PBS), and serial dilutions were plated on minimal media to select for diploids. Mating efficiency was calculated as the percentage of input haploid cells (before mating) that formed diploid colonies (after mating). Where indicated, α factor was spread on the mating plate (to give a final concentration of 30 μM) before filters were added.
To analyze zygote formation, mated cells were suspended in PBS, sonicated, and fixed in 5% formaldehyde. Cells were imaged by differential interference contrast microscopy (DIC) and green fluorescent protein (GFP) microscopy.
Pheromone Response, β-Galactosidase, and Two-Hybrid Assays
Halo assays of growth arrest were performed by plating cells on −Trp-Ura plates and overlaying with filters containing 20 μl of 1 mM α factor (Lamson et al., 2002). Plates were incubated for 2 d at 30°C.
FUS1-lacZ transcriptional induction assays were performed as described previously (Pryciak and Huntress, 1998; Lamson et al., 2002). Induction by pathway-activating constructs under control of the GAL1 promoter was measured 4 h after addition of 2% galactose ± 10 μM α factor to cultures grown in raffinose medium. For FUS1-lacZ assays comparing mutant derivatives of PGAL1-STE4 in ste4Δ cells, or STE4 derivatives in ste4Δ ste7Δ + PGAL1-STE7 cells, cultures in raffinose medium were pretreated with 2% galactose for 1 h and then treated for 2 h with 2% galactose ± 10 μM α factor.
β-Galactosidase and two-hybrid assays were performed as described previously (Pryciak and Hartwell, 1996; Lamson et al., 2002).
Cell Lysates and Protein Analysis
To test coimmunoprecipitation of myc13-Ste4 with HA3-Gpa1, clarified cell lysates were prepared by glass bead lysis, incubated with mouse anti-myc (9E10; Santa Cruz Biotechnology, Santa Cruz, CA) or mouse anti-hemagglutinin (HA) (HA.11; Covance Research Products, Princeton, NJ) antibodies and precipitated with protein G-agarose beads by using methods described previously (Lamson et al., 2002). After separation by SDS-polyacrylamide gel electrophoresis (PAGE) and transfer to polyvinylidene difluoride membranes, blots were probed with rabbit anti-myc (A-14; Santa Cruz Biotechnology) or rabbit anti-HA (HA.11; Covance Research Products). To analyze steady-state levels of Ste4 and Ste4-Gpa1 fusion proteins, crude whole-cell extracts were prepared by pelleting 1.5 ml of culture (OD660 = 0.5), resuspension in 45 μl of SDS-PAGE loading buffer supplemented with protease inhibitors (Lamson et al., 2002), and boiling for 10 min. After pelleting cell debris for 2 min, 10 μl was analyzed by SDS-PAGE, blotted, and probed with rabbit anti-myc (A-14; Santa Cruz Biotechnology).
RESULTS
Separating the Polarity Role of Gβγ from Its Signaling Role
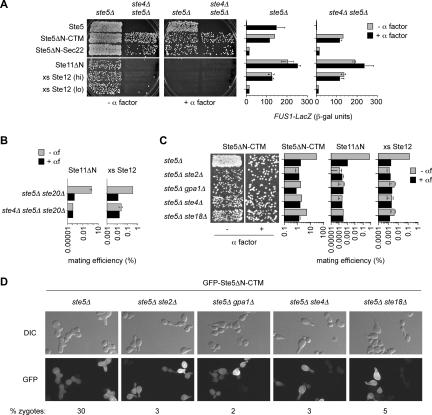
The Gβγ dimer normally performs two roles in the mating pathway: it activates MAP kinase cascade signaling and it regulates proteins that control cell polarity. Our goal in this work was to study the polarity role of the receptor–Gαβγ module in isolation from its requirement in activating the MAP kinase cascade. Therefore, we used a variety of methods to activate signaling independent of pheromone and Gβγ, and then studied how perturbing Gαβγ function affects chemotropism and cell polarization. This strategy is an extension of one used previously in which overexpression of the transcription factor Ste12 was used to study the mating role of various MAP kinase pathway components while bypassing their role in transcriptional induction (Schrick et al., 1997). Here, we used several newer reagents such as membrane-targeted versions of Ste5 (Ste5ΔN-CTM and Ste5ΔN-Sec22), which can promote robust MAP kinase cascade signaling, wild-type levels of mating, and normal polarized morphogenesis (Pryciak and Huntress, 1998; Harris et al., 2001). To ensure that communication between Gβγ and Ste5 was severed, these reagents used a truncated form of Ste5 that lacks the Gβγ-binding site (Ste5ΔN), and all assays were performed in strains where the genomic STE5 locus was deleted (ste5Δ). For comparison, we also used a constitutively active form of Ste11 (Ste11ΔN) (Cairns et al., 1992; Neiman and Herskowitz, 1994) and overexpressed Ste12 (Dolan and Fields, 1990; Schrick et al., 1997). When signaling output was measured by induction of a transcriptional reporter construct (FUS1-lacZ), we found that some bypass methods caused stronger activation than others, but all were independent of Ste4 (Gβ) and pheromone (Figure 1A, right).
Figure 1.
Chemotropism role of Gαβγ and receptor is separate from signaling. (A) Chemotropism requires Ste4 even when its signaling role is bypassed. Strains PPY858 (ste5Δ) and PPY886 (ste5Δ ste4Δ) harbored galactose-inducible forms of Ste5 (pPP452), Ste5ΔN-CTM (pPP513), Ste5ΔN-Sec22 (pPP524), Ste11ΔN (pPP575), or Ste12 (pPP741 or pPP271). Chemotropic proficiency (left) was assessed by patch matings performed in the absence (−) or presence (+) of exogenous α factor. The mating results using Ste5 derivatives show the more-dilute 2° replica, whereas the others show the 1° replica (see Materials and Methods). Results were similar in both a and α cells, and in both W303 and 381G strain backgrounds (unpublished observations). Transcriptional activation of FUS1-lacZ (right) is shown for the same strains and plasmids after galactose induction ± α factor. Bars, mean ± SD (n = 3). To emphasize that transcription levels were not the primary determinant of mating efficiency, results are shown using Ste12 overexpression constructs that yield high (hi) or low (lo) levels of transcriptional induction (due to different vector contexts). (B) Ste4 can perform its chemotropism role without Ste5 and Ste20. Quantitative matings were performed ± exogenous α factor. Strains PPY863 and PPY842 harbored galactose-inducible Ste11ΔN (pRD-STE11-H3) or Ste12 (pNC252). Bars, mean ± range (n = 2). (C) An intact receptor–Gαβγ module is required for chemotropism. Signaling was activated by galactose-inducible constructs (pPP513, pPP575, and pPP741) in the indicated strains (PPY858, PPY979, PPY978, PPY886, and PPY989). Chemotropism was monitored by patch or quantitative mating assays in the absence (−) or presence (+) of α factor. Bars, mean ± SD (for Ste5ΔN-CTM; n = 4) or mean ± range (n = 2). Transcriptional activation of FUS1-lacZ by the same constructs after galactose induction ± α factor was similar in all strains (data not shown). (D) Zygote formation. Strains from C, harboring galactose-inducible GFP-Ste5ΔN-CTM (pPP513), were mated with PT2α partner cells for 5.5 h. Representative fields of DIC and fluorescence (GFP) images are shown. We counted 500 cells for each mixture, and the percentage that were zygotes is shown.
We then used these signaling bypass methods to address the role of Gβγ in chemotropic mating. Chemotropism was monitored using a “pheromone confusion” assay (Dorer et al., 1995; Nern and Arkowitz, 1998), in which mating success is compared in the absence versus presence of excess exogenously added pheromone (α factor), which obscures natural pheromone gradients emanating from partner cells. Chemotropically proficient cells can use pheromone gradients to locate mating partners, and thus they mate with higher efficiency when pheromone gradients are left intact (−α factor) than when gradients are obscured (+α factor); cells defective at chemotropism cannot follow pheromone gradients, and thus they mate at the lower efficiency under either condition. Thus, this assay does not directly monitor directional growth, but rather it infers the ability of cells to detect and use pheromone gradients. Unlike earlier measures of “mating partner discrimination” (Jackson and Hartwell, 1990; Jackson et al., 1991), chemotropic proficiency in the pheromone confusion assay requires both Far1 and Far1–Cdc24 binding (Dorer et al., 1995; Valtz et al., 1995; Nern and Arkowitz, 1998), and it accurately reflects the ability of cells to establish a new polarization axis along pheromone gradients (Valtz et al., 1995; Nern and Arkowitz, 1998). Using this assay, we found that cells expressing Ste4 were proficient at chemotropism, whereas those lacking Ste4 were defective (Figure 1A, left). Among the different bypass methods, the membrane-targeted Ste5 reagents promoted the most efficient mating, but a consistent behavior emerged regardless of the bypass method or the absolute signaling level; namely, removal of Ste4 or addition of excess pheromone disrupted chemotropic mating without altering MAP kinase pathway signaling.
Gβγ activates MAP kinase cascade signaling via interactions with Ste5 and Ste20 (Whiteway et al., 1995; Inouye et al., 1997; Feng et al., 1998; Leeuw et al., 1998; Pryciak and Huntress, 1998). To unequivocally determine whether these interactions are dispensable for the chemotropic role of Gβγ, we performed quantitative assays of chemotropic mating in ste5Δ ste20Δ cells, by using activated Ste11 (Ste11ΔN) or excess Ste12 to induce signaling or transcription. Indeed, these cells were proficient at chemotropism, whereas strains that also lacked Ste4 (ste4Δ ste5Δ ste20Δ) were not (Figure 1B). Thus, the chemotropism role of Gβγ does not require it to interact with Ste5 or Ste20. These findings provided a framework from which to further probe the chemotropism and polarity functions of the receptor–Gαβγ module without concern for their effects on MAP kinase pathway signaling.
The Pheromone Receptor and All Three Gαβγ Subunits Are Required for Chemotropism
The ability of Gβγ to mediate chemotropism without regulating MAP kinase cascade signaling is consistent with the fact that Gβγ interacts directly with polarity proteins via Far1 (Butty et al., 1998; Nern and Arkowitz, 1998, 1999). However, because chemotropism is a directional phenomenon, it would be logical that Gβγ–Far1 binding could help guide polarization in the proper direction only if Gβγ was activated in a spatially asymmetric manner, congruent with the pheromone gradient. Because Gβγ activation is regulated by the receptor and Gα subunit, we directly compared the requirement for the receptor and all three G protein subunits in chemotropic mating assays. As mentioned above, MAP kinase signaling, transcription, or both were activated independent of Gβγ, so that genetic perturbation of the receptor–Gαβγ module would affect only chemotropism. We found that only the cells with an intact receptor–Gαβγ module could use pheromone gradients to increase their mating success, whereas cells lacking the receptor (ste5Δ ste2Δ) or any one of the G protein subunits (ste5Δ gpa1Δ, ste5Δ ste4Δ, or ste5Δ ste18Δ) could not (Figure 1C). It is important to note that under these signaling bypass conditions the receptor–Gαβγ module can only contribute to mating success when pheromone gradients are intact, whereas it performs no detectable role when gradients are absent (i.e., the wild-type and null alleles of each component are indistinguishable).
Microscopic analysis (Figure 1D) confirmed that the intact receptor–Gαβγ module allowed cells to locate and fuse with mating partners, as judged by the formation of dumbbell-shaped zygotes, although the mating-defective cells could still form polarized mating projections. Therefore, under these conditions the receptor-Gαβγ module is not required for polarization per se, but for properly guiding cell polarization toward a mating partner. Note that these findings are consistent with the expectation that polarization in the “correct” direction (i.e., toward the source of pheromone) should require spatial regulation of Gβγ activity, and so they do not necessarily imply that the receptor and/or Gα perform separate polarization functions.
Free Gβγ Is Insufficient for De Novo Polarization
To study the polarity function of the receptor–Gαβγ module in a setting where cells do not have to detect the direction of a localized stimulus, we assayed de novo polarization in response to a uniform field of pheromone. Polarization was restricted to the de novo pathway by using rsr1Δ mutant strains, in which the default pathway is inactivated (Nern and Arkowitz, 1999, 2000). First, we tested whether pheromone had a role in de novo polarization beyond activating the MAP kinase cascade. Pathway signaling was activated by expressing either Ste5ΔN-CTM (PGAL1-STE5ΔN-CTM) or an activated form of Ste11 fused to Ste7 (PGAL1-STE11ΔN-STE7), which permits normal mating morphology by reducing cross-activation of other signaling pathways (Harris et al., 2001). Despite being able to trigger default polarization (i.e., in RSR1 cells), pathway activation by Ste5ΔN-CTM or Ste11ΔN-Ste7 could not trigger de novo polarization (i.e., in rsr1Δ cells) (Figure 2A). This inability was not due to interference from excess MAP kinase pathway signaling, as cells harboring these activators (ste5Δ + PGAL1-STE5ΔN-CTM or PGAL1-STE11ΔN-STE7) could undergo de novo polarization when pheromone was added (Figure 2B). Importantly, we found that de novo polarization requires Gβγ activity, as cells lacking the Gβ subunit (ste4Δ ste5Δ + PGAL1-STE11ΔN-STE7) did not polarize even when pheromone was added (Figure 2B). Notably, however, communication between Gβγ and Ste5 was not required, because pheromone could stimulate polarization in cells lacking Ste5 (ste5Δ + PGAL1-STE11ΔN-STE7) (Figure 2B). As expected, transcriptional induction levels and kinetics in these strains were uninfluenced by the factors that governed polarization, such as RSR1, STE4, or pheromone (Figure 2, A and D). Therefore, as with chemotropism, the ability of pheromone and Gβγ to regulate de novo polarization is separable from any regulatory effects on the MAP kinase cascade.
Figure 2.
De novo polarization requires pheromone and Gα, in addition to Gβγ. (A) Polarization was examined in RSR1 or rsr1Δ cells, after signaling was activated by α factor (strains PPY398 and PPY1259) or by galactose induction of PGAL1-STE5ΔN-CTM (strains PPY1303 and PPY1306) or PGAL1-STE11ΔN-STE7 (strains PPY1309 and PPY1312). Right, the same strains were transformed with a FUS1-lacZ reporter (p3058), and transcriptional induction (mean ± SD; n = 4) was monitored 0, 2, and 4 h after addition of galactose with (i and ii) or without (iii–vi) α factor. (B) Pheromone, Ste4, and Gpa1 are required for de novo polarization. The indicated rsr1Δ strains harboring PGAL1-STE5ΔN-CTM or PGAL1-STE11ΔN-STE7 (PPY1307, PPY1313, PPY1314, and PPY1952) were treated with galactose ± α factor. Congenic RSR1 strains (PPY1304, PPY1310, PPY1311, and PPY1951) show that the galactose-inducible constructs can activate polarization by the default pathway. Right, old cell wall was labeled with FITC-ConA (green), and new cell wall formed during the period of mating pathway activation was labeled with TRITC-ConA (red). (C) Free Gβγ is not sufficient for de novo polarization. Strains PPY794, PPY1228, PPY1248, and PPY1380 harboring PGAL1-STE4 (pGT-STE4) were induced with galactose ± α factor. At right, old and new cell wall was labeled as in B. (D) Conditions affecting de novo polarization do not affect transcription. The eight strains in B were transformed with reporter plasmid p3058, and FUS1-lacZ induction (mean ± SD; n = 4) was monitored 2–4 h after addition of galactose ± α factor.
Because de novo polarization does not require cells to sense the direction from which pheromone emanates, and because Gβγ interacts directly with Far1 and Cdc24, (Butty et al., 1998; Nern and Arkowitz, 1998, 1999), it seemed possible that Gβγ alone would be sufficient to promote de novo polarization, with pheromone serving only to generate free Gβγ by dissociating the Gαβγ heterotrimer. To test this view, Gβγ was activated without using pheromone, by deletion of GPA1 or by overexpression of STE4 (PGAL1-STE4). To avoid persistent growth arrest due to constitutive MAP kinase pathway signaling, GPA1 was deleted in a ste5Δ strain harboring PGAL1-STE11ΔN-STE7 (Figure 2B) or in a ste4Δ strain harboring PGAL1-STE4 (Figure 2C). Remarkably, although each method of Gβγ activation (i.e., gpa1Δ or PGAL1-STE4) could induce cell cycle arrest and cell polarization by the default pathway (i.e., in RSR1 cells), neither method could induce de novo polarization (i.e., in rsr1Δ cells) (Figure 2, B and C). Furthermore, the ability of pheromone to trigger de novo polarization was actually eliminated by the gpa1Δ mutation (Figure 2, B and C, right columns), and thus it requires Gα in addition to Gβγ. Therefore, although Gβγ can directly communicate with polarization proteins, free Gβγ is not sufficient for de novo polarization. This deficiency might reflect a separate role for ligand-bound receptors or GTP-loaded Gα, or it might indicate that receptors and Gα can promote an asymmetric distribution of Gβγ activity even when external pheromone is distributed uniformly.
To follow the pattern of cell surface growth, we used fluorophore-conjugated ConA to differentially label cell wall formed before versus during the period of mating pathway activation. It was previously shown that when de novo polarization failed due to disruption of Far1–Cdc24 interaction, cells could initiate polarized growth, but the polarization axis could not be maintained; consequently, it wandered about a broad region of the cell periphery (Nern and Arkowitz, 2000). Similarly, we observed that when cells failed de novo polarization due to absence of pheromone, Gpa1, or Ste4, they still showed asymmetric cell surface growth, but it was broadly distributed across one hemisphere (Figure 2, B and C, right). Thus, consistent with the requirement for Fus3 (Matheos et al., 2004), our results suggest that activation of the MAP kinase cascade is sufficient to initiate asymmetric growth, even in the absence of Gα or Gβγ. Nevertheless, organization of a well-focused and persistent polarization axis requires both Far1–Cdc24 interaction and an intact receptor–Gαβγ module.
Gβ Mutants Reveal Distinct Roles for Two Gα–Gβ Binding Interfaces
To further investigate the requirement for the intact receptor–Gαβγ module in chemotropism and de novo polarization, we used a series of Gβ mutants to disrupt regulation of Gβγ by the Gα subunit. Crystal structures of mammalian Gαβγ heterotrimers reveal two contact surfaces between Gα and Gβ, termed the “switch interface” and the “N-terminal interface” (Wall et al., 1995; Lambright et al., 1996). The switch (Sw) interface involves a region of Gα that undergoes a conformational switch upon GTP binding, whereas the N-terminal (Nt) interface involves an N-terminal helix of Gα that protrudes away from the globular GTPase domain (Figure 3A). The Gα and Gβ residues contacting one another in each interface are well conserved between mammalian and yeast counterparts (Lambright et al., 1996; Sondek et al., 1996); in Gβ these include 16 residues in nine regions. Here, we studied a series of Ste4 (Gβ) mutations that collectively affect all positions predicted to contact Gα. These mutants were derived from multiple sources; briefly, mutations in six of the nine Gα-contact regions were identified in three different genetic screens, and the remaining regions were changed by site-directed mutagenesis (see Materials and Methods).
Figure 3.
Ste4 mutations in Gα–Gβ binding interfaces. (A) Model for orientation of the G protein-coupled receptor rhodopsin, the heterotrimeric G protein transducin, and the membrane. Adapted from Hamm, 2001; copyright Proceedings of the National Academy of Sciences USA. (B) Effects of Ste4 mutations on Gpa1 binding. Mutated Ste4 residues are listed alongside their homologous residues in mammalian Gβ1. Activation domain fusions (AD-Ste4) were expressed in a gpa1Δ ste11Δ two-hybrid tester strain (PPY1158), with DNA-binding domain fusions to Gpa1 (pPP247) or to the Gβγ-binding regions of Ste5 (pPP305) or Far1 (pPP743). Binding was detected by growth on −His plates ± 3-aminotriazole (3-AT; an inhibitor of His3), or by quantitative β-galactosidase assay (bars, mean ± SD; n = 4). To reveal the full range of effects on Gpa1 binding, quantitative assays used AD–Ste4 fusions expressed from either a strong promoter (derivatives of pPP643) or a weak promoter (derivatives of pPP268); plate growth assays used pPP643 derivatives. As controls (at bottom), two additional Ste4 mutations outside the Gα–Gβ interfaces (see Materials and Methods) were analyzed in parallel; these disrupt binding to Ste5 and Far1, but not to Gpa1. (C) Ste4 mutations cause deregulated signaling. To avoid constitutive growth arrest, the Ste4 mutants were tested in ste4Δ ste7Δ cells (PPY1662) harboring PGAL1-STE7 (pPP2773). FUS1-lacZ induction (mean ± SD; n = 4) was measured 2 h after addition of galactose ± α factor. Ste4 was expressed from the native STE4 promoter (derivatives of pPP2968; vector = pRS316). (D) Ste4 mutations disrupt coimmunoprecipitation with Gpa1. HA-tagged Gpa1 (pPP2775) or vector (pRS314) was coexpressed with myc-tagged Ste4 (derivatives of pPP2838; vector = pRS316) in strain PPY1230 (gpa1Δ ste4Δ ste5Δ). Cell extracts were analyzed by immunoprecipitation (IP) and immunoblotting (blot).
By a two-hybrid assay (Figure 3B), the Ste4 mutations identified in genetic screens reduced Gpa1 binding to levels that were either undetectable (L117R, W136R L138F, and L154R N156K) or extremely faint (K126E, D224E, and D272A), whereas site-directed mutations at the remaining positions (N92G K94E S96A and W411R) had only mild effects (suggesting why they were not identified in screens). A similar rank order of defects was seen using a coprecipitation assay of these AD–Ste4 fusions with Gpa1-GST (unpublished observations). In comparison, these mutations had minimal effects on interaction with Ste5 and Far1 (Figure 3B); this is consistent with our findings (to be described in detail elsewhere) that binding to Ste5 and Far1 requires Ste4 (Gβ) and Ste18 (Gγ) residues in the coiled-coil region of the Gβγ dimer (Lamson and Pryciak, unpublished observations), which has been previously implicated in downstream signaling (Leberer et al., 1992; Grishin et al., 1994a,b; Leeuw et al., 1998; Winters et al., 2005). As expected for mutants released from repression by Gpa1, these Ste4 mutants caused constitutively active pathway signaling (Figure 3C), although some mutants that retained detectable Gpa1 binding also showed incomplete deregulation, because signaling could still be increased by α factor. Therefore, we focused most of our subsequent studies on four Ste4 mutants with the strongest Gpa1-binding defects and the most deregulated signaling, which include two in the Nt interface (L117R and K126E) and two in the Sw interface (W136R L138F and L154R N156K, hereafter termed WL/RF and LN/RK, respectively). These mutations did not affect Ste4 protein levels but they eliminated detectable coimmunoprecipitation with Gpa1 (Figure 3D).
Despite similar behavior in binding and signaling assays, mutations in the two Gα–Gβ interfaces had opposite effects on polarity control. Mutations in the Nt interface disrupted chemotropism, because the cells were largely insensitive to the presence or absence of pheromone gradients (Figure 4A). This phenotype is consistent with deregulated Gβγ activity. Surprisingly, however, the Sw interface mutants were chemotropism proficient (Figure 4A), even though by signaling and binding criteria they seemed to be as strongly dissociated from Gpa1 as the Nt interface mutants. Although it was possible that the individual Sw mutations have a less disruptive effect on Gpa1 interaction than the Nt mutations, two further observations suggested that the explanation was not this simple. First, chemotropism remained intact even when multiple Sw interface mutations were combined, such as W136R L138F L154R N156K (WL/RF + LN/RK; Figure 4A), as well as D224V D272A, L154S D272A, and W136R L138F D272A (unpublished observations). Second, by making less drastic substitutions at Nt interface residues (i.e., K126A, K126N, L117A, and L117G), we could detect residual Gpa1 binding for several of these new mutants (as well as the original K126E); yet, they all showed a stronger defect in chemotropic mating than the Sw mutants WL/RF and LN/RK (Figure 4B).
Figure 4.
Ste4 mutations in the Nt interface, but not the Sw interface, disrupt polarity control. (A) Chemotropism proficiency was assessed by patch mating assays performed in the absence (−) or presence (+) of exogenous α factor. PPY867 (ste4Δ ste5Δ) harbored PGAL1-STE5ΔN-SEC22 (pPP524) and either vector (pRS316) or Ste4 variants expressed from the native STE4 promoter (derivatives of pPP2968). The effects of the Ste4 mutations on Gpa1 binding are summarized for reference (see Figure 3B). (B) Analysis of additional Nt interface mutants. Left, two-hybrid binding assays (as in Figure 3B) using AD–Ste4 fusions (derivatives of pPP643; vector=pPP636) and DBD-Gpa1 (pPP247) in PPY1158. (Note: in quantitative assays using the lacZ reporter, the Gpa1 interaction signal of Ste4-K126A was measurably reduced to 27% of Ste4-WT.) Right, quantitative assays of Ste4 function during chemotropic mating. PPY867 (ste4Δ ste5Δ) harbored PGAL1-STE5ΔN-SEC22 (pPP524) and either vector (pRS316) or Ste4 variants (derivatives of pPP2968). As Ste4 affects mating under these conditions only when gradients are intact (see Figures 1, A–C, and 4A), these assays were conducted only in the absence of exogenous α factor. Results (mean ± SD; n = 4) were normalized to the average mating efficiency of wild-type Ste4. (C) Default and de novo polarization phenotypes. To avoid constitutive growth arrest, Ste4 mutants were expressed from the GAL1 promoter (derivatives of pPP1151) in PPY794 (ste4Δ RSR1) or PPY1248 (ste4Δ rsr1Δ), and cell morphology was examined after induction with galactose ± α factor.
The Ste4 mutants also segregated into two phenotypic classes in de novo polarization assays, in which the Sw interface mutants remained competent, whereas the Nt interface mutants were defective (Figure 4C, rsr1Δ). Notably, although the Sw interface mutants could mediate de novo polarization, they still required the addition of pheromone, as with wild-type Ste4. Thus, the Sw interface mutants remain capable of mediating a pheromone-dependent step, despite their strong dissociation from Gpa1 in binding and signaling assays. This raised the possibility that the Sw interface mutants can maintain a weak interaction between Gα and Gβ at the Nt interface (perhaps only when membrane-associated) and thus remain in regulatory communication with the pheromone receptor. This scenario, and others, was addressed by further experiments described below.
Suppression of an Nt Interface Mutant by a Compensatory Mutation in Gα
First, we wanted to determine whether the stronger phenotype of the Nt mutants was truly a consequence of disrupted interaction between Gβ and Gα, rather than between Gβ and some other protein involved in cell polarity. Although these Ste4 mutants could still bind Far1 (Figure 3B), in principle we could not rule out effects on binding to other, unknown partners. Therefore, we attempted to restore Gα–Gβ binding via a compensatory mutation in Gpa1. One of the Nt interface mutations (Ste4-K126E) involves a residue that, based on mammalian Gαβγ structures, is expected to form an ion pair between Lys126 in Ste4 and Glu28 in Gpa1 (Figure 5A). The Ste4 mutation changes Lys126 to Glu (K126E), thereby reversing the charge. To make a compensatory charge-reversal mutation in Gpa1, we changed Glu28 to Lys (E28K). This Gpa1–E28K mutation, but not a control mutation (Gpa1-E28A), restored measurable binding with Ste4-K126E (Figure 5B), although not to wild-type levels. Also, the Gpa1 E28K and E28A mutations each reduced binding to wild-type Ste4, although by a mild degree that was most noticeable when Ste4 was expressed at lower levels from a weak promoter (Figure 5B). It was not entirely surprising that Gpa1-E28K only partially restored binding to Ste4-K126E and that Ste4-K126E caused a stronger binding defect than Gpa1-E28K, because the mammalian Gβ residue homologous to Ste4 Lys126 contacts Gα not only through this ion pair but also through hydrogen bonding and van der Waals interactions (Lambright et al., 1996). Nevertheless, these binding effects were enough to confer informative phenotypes in mating assays.
Figure 5.
Allele-specific suppression of a Ste4 Nt interface mutant. (A) Red side chains highlight an ion pair predicted to form between Ste4(Gβ)-K126 and Gpa1(Gα)-E28 in the Nt interface, based on homologous residues (Gβ-K89 and Gα-E16) in transducin (Lambright et al., 1996). Structural rendering of Gαβγ used Protein Data Bank coordinate set #1GOT and Cn3D version 4.1 software (National Center for Biotechnology Information/National Library of Medicine). (B) Two-hybrid assay showing that the Gpa1-E28K mutation partially restores interaction with Ste4-K126E. DBD fusions to Gpa1 derivatives (pPP247, pPP1502, and pPP1505) were coexpressed in PPY762 with AD–Ste4 fusions under control of either a strong or weak promoter (pPP636, pPP643, pPP1121, or pPP249). Bars, mean ± SD (n = 3). (C) Gpa1-E28K partially suppresses the chemotropism defect of Ste4-K126E. Patch mating assays used ste4Δ gpa1Δ ste5Δ cells (PPY1230) harboring PGAL1-STE5ΔN-SEC22 (pPP524) and the indicated combinations of GPA1 (YCpGPA1, pPP1501, pPP1503), and STE4 (derivatives of pPP226). (D) Quantitative mating assay measuring suppression of Ste4-K126E by Gpa1-E28K. Cells were as in panel C, mated to partner strain PT2α in the absence of exogenous α factor. Mating frequencies from eight independent 7-h mating experiments were expressed relative to the average of the fully wild-type combination (Gpa1-WT + Ste4-WT). Bars, mean ± SEM (n = 8). For Ste4-K126E matings, the difference between Gpa1-WT and Gpa1-E28K (means, 4.1 vs. 18.7%) was ranked highly significant (p < 0.0005) by a two-tailed unpaired Student's t test.
Indeed, the chemotropism defect of the Ste4-K126E mutant was at least partially suppressed by the Gpa1-E28K mutant, because mating of cells harboring Ste4-K126E was more efficient when coexpressed with Gpa1-E28K than with Gpa1-wild type (WT) (Figure 5, C and D). Whereas the Gpa1–E28K mutation reduced mating when combined with Ste4-WT, it increased mating by an average of 4.6-fold when combined with Ste4-K126E (Figure 5D). Although mating was not restored to wild-type levels, it would be unreasonable to expect this given the incomplete restoration of Gpa1–Ste4 binding. Notably, the observed suppression was allele-specific, because Gpa1-E28A did not suppress Ste4-K126E, and neither Gpa1-E28K nor Gpa1-E28A could suppress the other Nt mutant, Ste4-L117R (Figure 5, C and D). Furthermore, although the Gpa1–E28K mutation improved mating by Ste4-K126E, it reduced mating by Ste4-WL/RF and Ste4-LN/RK, such that these Sw mutants were actually more defective than Ste4-K126E in cells expressing Gpa1-E28K (Figure 5C). This finding makes it highly unlikely that the phenotypic differences between Ste4 Nt and Sw mutations can be explained by their different impact on binding between Gβγ and an unknown factor. Instead, the pattern of allele-specific suppression and enhancement found with the Gpa1–E28K mutation supports a special role for Gα–Gβ binding via the Nt interface in chemotropism and cell polarization, and it suggests that this interface can remain functional when the Sw interface is dissociated by mutation. Because the Gpa1-E28K mutant is sensitized to disruption of the Sw interface, an intact Sw interface may help maintain Gα–Gβ association when the Nt interface is mildly disrupted.
Differences between Nt and Sw Interface Mutants Are Not Due to Altered Gpa1–Fus3 Interaction
Gpa1 can interact with the MAPK Fus3 via a docking motif in its N terminus, and mutations in this motif (Gpa1-K21E R22E, denoted Gpa1-EE) disrupt Fus3 binding and reduce mating (Metodiev et al., 2002). This raised the possibility that an intact Nt interface is required mainly to allow proper interaction between the Gpa1 N terminus and Fus3. To test this notion, we used the Gpa1-EE mutant to disrupt interaction between Gpa1 and Fus3, and we found that the Nt and Sw mutations in Ste4 still showed their characteristic phenotypic differences (Figure 6A). We also tested the Ste4 mutants in cells lacking Fus3 (ste4Δ fus3Δ). Here, mating by the Sw interface mutants was not as efficient as that of wild-type Ste4, but it was still more efficient than that of the Nt interface mutants (Figure 6A). Thus, both approaches suggest that the role of the Nt interface, and the different behavior of the Nt versus Sw mutants, cannot be explained by indirect effects on the Gpa1–Fus3 interaction.
Figure 6.
Qualitative differences between Ste4 Nt and Sw mutants are independent of Gpa1–Fus3 interaction. (A) Patch mating assays used the following strain and plasmid combinations. Left, strain PPY1228 (ste4Δ gpa1Δ) harbored the indicated Gpa1 derivatives (pPP2711 or pPP2743) and PGAL1-STE4 plasmids (derivatives of pPP1151). Center right, strain PPY1230 (ste4Δ gpa1Δ ste5Δ) harbored Gpa1 derivatives (pPP2711 or pPP2743) plus native STE4 plasmids (derivatives of pPP226) and either PGAL1-STE5ΔN-CTM (pPP479) or PGAL1-STE5ΔN-SEC22 (pPP1175). Far right, strain PPY1937 (fus3Δ ste4Δ) harbored the indicated PGAL1-STE4 plasmids. (B) Pheromone confusion assay showing that fus3Δ cells are chemotropically proficient. Strains, from top to bottom: PPY577, PPY824, PPY827, and PPY836. Similar results were also seen in S288C and W303 strain backgrounds (unpublished observations). (C) Quantitative mating assay of strains PPY663, PPY817, PPY498, and PPY820 expressing galactose-inducible Ste12 (pPP271). Cells were mated for 18 h, ± exogenous α factor. Bars, mean ± SD (n = 3).
It is notable that we did not detect a strong mating defect for the Gpa1-EE mutant alone, suggesting that the Gpa1–Fus3 interaction may not be required for chemotropism. To address this issue further, we compared the roles of Fus3 and Far1. Loss of either protein caused a mating defect, but a much greater defect occurred when both Fus3 and Far1 were absent (Figure 6B), indicating that although both proteins are required for maximum mating efficiency, each protein can perform its function without the other. Moreover, an important distinction between the roles of Fus3 and Far1 was apparent: the fus3Δ cells were proficient at chemotropism (i.e., sensitive to whether pheromone gradients were present or absent), whereas the far1Δ cells were defective (Figure 6B). To rule out the possibility that chemotropic proficiency in fus3Δ cells reflects redundancy between Fus3 and Kss1, we performed additional mating assays using fus3Δ kss1Δ cells (in which sterility was suppressed by PGAL1-STE12). Despite low overall mating efficiency, the fus3Δ kss1Δ cells could still use pheromone gradients, and this behavior required Far1 (Figure 6C). The simplest overall interpretation is that detecting gradients and using gradient information to locate mating partners does not require Fus3, whereas the role of Fus3 in polarized morphogenesis (Matheos et al., 2004) is distinct from gradient sensing per se. These results also imply that, unlike cell cycle arrest (Gartner et al., 1998), phosphorylation of Far1 by Fus3 is dispensable for chemotropism.
Fusion of Gβ to Gα Suggests a Role for the Nt Interface in Receptor Coupling
Finally, we addressed whether maintenance of the Nt interface was necessary for coupling of the heterotrimer (Gαβγ) to the receptor. Existing models for the coupling between GPCRs and heterotrimeric G proteins predict that the Nt interface lies tangential to the membrane (Hamm, 1998, 2001). In addition, the N terminus of Gα has been implicated in receptor recognition (Taylor et al., 1994; Itoh et al., 2001; Cabrera-Vera et al., 2003). These facts suggested that the Nt interface mutations may not only disrupt interaction between Gβ and Gα, but they may also disrupt the way Gαβγ interacts with the receptor. However, the ability to test this notion was hindered by the fact that the Ste4 mutations caused constitutive signaling, which obscured whether the receptor might still exert some regulatory control over the G protein. To circumvent this difficulty, we took advantage of a previously described Ste4–Gpa1 fusion protein (Klein et al., 2000), with the rationale that forced association to Gpa1 may inhibit constitutive signaling by the Ste4 mutants and thus allow us to assay receptor coupling.
Starting with the prior Ste4–Gpa1 fusion construct, we replaced the original GAL1 promoter with the native STE4 promoter, and then compared its function with wild-type (unfused) polypeptides. By multiple assays, we found this Gβ–Gα fusion (Ste4-Gpa1) to function in a manner that was virtually indistinguishable from separate Gβ and Gα polypeptides. This included growth arrest (Figure 7A, left), regulation by the RGS-family protein Sst2 (Figure 7A, right), and pheromone-induced transcription (Figure 7B). Furthermore, the Gβ–Gα fusion was able to mediate total mating levels and chemotropic mating behavior that was similar to the wild-type heterotrimer (Figure 7, C and D). Thus, fusion of Gβ to Gα does not interfere with Gαβγ function in either signaling or gradient detection.
Figure 7.
Phenotypes of Ste4–Gpa1 fusion proteins. (A) A Ste4–Gpa1 fusion expressed from the native STE4 promoter functions indistinguishably from separate Ste4 and Gpa1 polypeptides. Halo assays show growth arrest of ste4Δ gpa1Δ cells (PPY1228) or ste4Δ gpa1Δ sst2Δ cells (PPY1942) harboring Gpa1 and Ste4 as separate (YCplac22-GPA1-WT + pPP226) or fused (pPP1340 + pRS314 vector) polypeptides. (B) FUS1-lacZ assays (mean ± SD; n = 4) showing that the Ste4-Gpa1 fusion retains a normal dose response to pheromone. PPY1663 (ste4Δ gpa1Δ) harbored plasmids as in A. Cells were treated with the indicated concentration of α factor for 2 h. (C) Patch mating assay of strains PPY1230 (ste5Δ ste4Δ gpa1Δ) or PPY886 (ste5Δ ste4Δ GPA1) harboring PGAL1-STE5ΔN-CTM (pPP473) plus pRS316, pPP226, or pPP1340. (D) Patch mating assays showing that the Ste4-Gpa1 fusion protein remains proficient at chemotropism. Strain PPY1230 carried PGAL1-STE5ΔN-CTM (pPP479) and the following plasmid combinations: pPP2711 + pRS316, pRS314 + pPP226, pPP2711 + pPP226, or pRS314 + pPP1340. (E) The Ste4-Gpa1 fusion context reveals distinct signaling phenotypes for the Ste4 Nt and Sw mutants. Top, strain PPY1662 (ste4Δ ste7Δ) harbored a PGAL1-STE7 construct (pPP2773) plus either vector (pRS316) or the indicated STE4 or STE4-GPA1 fusion alleles, expressed from the native STE4 promoter. Bottom, strain PPY856 (ste4Δ) harbored either vector (pPP446) or the indicated PGAL1-STE4 or PGAL1-STE4-GPA1 fusion constructs. FUS1-lacZ activation was measured after induction with galactose ± α factor. Bars, mean ± SD (n = 4). (F) The Ste4 mutations do not affect protein levels. Lysates of cells harboring myc-tagged Ste4 (derivatives of pPP2838) or myc-tagged Ste4-Gpa1 (derivatives of pPP2839) were analyzed by anti-myc immunoblot. (Relevant portions of each blot are shown; as expected, myc-Ste4-Gpa1 runs considerably larger than myc-Ste4.)
Next, we incorporated the Nt and Sw interface mutations into the STE4 portion of the STE4-GPA1 fusion gene (in both the GAL1 promoter and native STE4 promoter contexts), and we used FUS1-lacZ assays to determine whether forced association with Gpa1 could suppress the constitutive signaling activity of the Ste4 mutants. Again, the Nt interface and Sw interface mutants showed distinct phenotypes, but here it was the Sw interface mutants that were more strongly deregulated; that is, fusion to Gpa1 could squelch the constitutive signaling of the Nt interface mutants but not that of the Sw interface mutants (Figure 7E, left and middle). This suggests that dissociation of the Sw interface is the primary regulator of downstream signaling. There was a slight difference between the two Sw interface mutants in the native promoter context, because fusion to Gpa1 partially reduced signaling by Ste4-LN/RK but not Ste4-WL/RF (Figure 7E, top). Protein levels were unaffected by these mutations (Figure 7F).
Further analysis of these mutant fusion proteins showed that although constitutive signaling by the Nt interface mutants was suppressed by fusion to Gpa1, signaling could not be efficiently reactivated by the addition of pheromone, in contrast to the fusion containing wild-type Ste4 (Figure 7E, right). The feeble pheromone response suggests that mutations in the Nt interface disrupt coupling between Gαβγ and the receptor, which may explain their defective behavior in both chemotropism and de novo polarization assays. Thus, although both Sw and Nt mutants show constitutive signaling when not fused to Gpa1, their different behaviors when fused to Gpa1 suggest the possibility that their different chemotropism/polarity phenotypes are a consequence of disrupted receptor–Gαβγ coupling in the Nt mutants, and by inference that this coupling can still occur in Sw mutants despite their constitutive signaling.
GTP Hydrolysis by Gα Is Required for De Novo Polarization and Chemotropism
The findings described above suggest that proper communication between Gαβγ and the receptor is necessary for directionally persistent polarization even when the pheromone stimulus is provided uniformly. In theory, this receptor-Gαβγ communication could be required solely to promote GTP–GDP nucleotide exchange on Gα, perhaps allowing GTP-bound Gα to perform a polarization role that acts synergistically with Gβγ. Alternatively, receptor–Gαβγ communication might be required to generate asymmetry in the distribution of Gβγ activity (and/or Gα-GTP), which otherwise would remain symmetric (e.g., in gpa1Δ cells or with constitutively active Ste4 mutants). To address these possibilities, we used an activated mutant form of the Gα subunit, Gpa1-Q323L (herein referred to as Gpa1-QL), which is defective at GTP hydrolysis (Dohlman et al., 1996; Apanovitch et al., 1998). We found that simultaneous activation of both Gα and Gβγ, by coexpressing Gpa1-QL with Ste4, was still not sufficient for de novo polarization in the absence of pheromone (Figure 8, A and B). This was true regardless of whether Gpa1-QL was expressed with Ste4-WT or with the constitutively active Sw interface mutant, Ste4-WL/RF. In fact, we found that trapping Gpa1 in the GTP-bound state was detrimental, because cells expressing Gpa1-QL could not polarize even after exposure to pheromone (Figure 8, A and B, + α factor). Therefore, polarization requires more than just the acquisition of both GTP-bound Gα and active Gβγ.
Figure 8.
Polarity control requires GTP hydrolysis by Gpa1. (A and B) De novo polarization was monitored using ste4Δ gpa1Δ rsr1Δ cells (PPY1380) expressing Ste4 and Gpa1 variants as either separate or fused polypeptides, after 4-h induction with galactose ± α factor. Separate Ste4 and Gpa1 subunits were expressed from pPP1151, pPP1228 (= WL/RF derivative of pPP1151), pPP2711, and pPP2802. Ste4–Gpa1 fusions were expressed from derivatives of pPP1150. The Gpa1-QL mutant is defective at GTP hydrolysis. (A) Representative images of the predominant morphologies. (B) Cell morphologies were quantified by blind counting of 200 cells per condition in three separate experiments. Cells were scored as “polarized” if they formed pear-shaped shmoos or elongated to a point at one end; cells were scored as “elongated” if one axis was clearly longer than the other but neither end was pointed. Bars, mean − SD (n = 3). (C) Transcriptional induction. PPY856 (ste4Δ) cells harboring the indicated PGAL1-STE4-GPA1 plasmids (derivatives of pPP1150) were induced with galactose ± α factor. Results for the WT–WT and WL/RF–WT fusion proteins are identical to those for the PGAL1-driven constructs in Figure 7E (bottom, middle and right), and they are shown here for comparison. Bars, mean ± SD (n = 4). (D) Fusion proteins harboring the Gpa1-QL mutation are defective at chemotropism. Mating was assayed ± 20 μM α factor by using strain PPY1230 (ste4Δ gpa1Δ ste5Δ) expressing PGAL1-STE5ΔN-CTM (pPP479) and Ste4-Gpa1 fusions (as in C). (E) Gpa1-QL eliminates the chemotropic advantage of Sw interface mutants over Nt interface mutants. Mating was assayed in strain PPY1230 (ste4Δ gpa1Δ ste5Δ) expressing PGAL1-STE5ΔN-SEC22 (pPP1175) and the indicated combination of Gpa1 (pPP2711, pPP2802) and Ste4 (derivatives of pPP226; vector = pRS316).
It seemed possible that activated Gα and Gβγ subunits might have to remain in close mutual proximity and that this might be accomplished during receptor-mediated activation but not by coexpression of mutationally activated subunits. Therefore, to force GTP-bound Gα to remain associated with Gβγ, we incorporated the Gpa1-QL mutation into the Ste4–Gpa1 fusion. In signaling assays, either the Ste4-WL/RF or Gpa1-QL mutations (or both) caused constitutive activity (Figure 8C); yet, none of these fusions could induce de novo polarization in the absence of pheromone (Figure 8, A and B, −α factor). Notably, the fusions containing the Gpa1-QL mutation did promote a detectable increase in elongation (and more so than when the same subunits were expressed as separate polypeptides), but these elongated cells did not form the pointed, pear-shaped shmoos seen during pheromone treatment. Thus, the cells were still missing some aspect of pheromone-induced polarization that can focus morphogenesis to a restricted portion of the cell perimeter.
Furthermore, the ability of pheromone to trigger de novo polarization remained intact when Ste4 was fused to Gpa1, but this was disrupted by the Gpa1-QL mutation (Figure 8, A and B, +α factor). Consistent with these findings, the fusions containing the Gpa1-QL mutation were also defective in chemotropic mating assays (Figure 8D), and the Gpa1-QL mutant (expressed as a separate polypeptide) eliminated the mating advantage of Sw interface mutants over Nt interface mutants (Figure 8E). Thus, interfering with GTP hydrolysis activates Gβγ signaling, but it disrupts cell polarization and chemotropism, regardless of whether Gα is fused to Gβ or kept separate. Finally, it should be noted that the chemotropism and polarity defects shown by the Gpa1-QL mutant are recessive to wild-type Gpa1 (unpublished observations). Together, these findings suggest that the ability of pheromone-bound receptor molecules to guide cell polarization, either along a de novo polarization axis or along pheromone gradients, requires normal coupling to the Gα GTP hydrolysis cycle.
DISCUSSION
This work examines how a GPCR and its coupled Gαβγ heterotrimer coordinate an asymmetric response to external stimuli. Our findings indicate that although yeast Gβγ is known to interact with downstream polarity factors (e.g., Far1, Cdc24), proper control of cell polarity in either the presence or absence of pheromone gradients requires an intact receptor-Gαβγ module and a normal GTP–GDP cycle on Gα. Furthermore, our results suggest distinct roles for the two Gα–Gβ interaction interfaces, whereby the Sw interface controls signaling and the Nt interface governs coupling to the receptor. As such, pheromone control of polarity requires maintenance of the Nt interface but not the Sw interface. Overall, these results suggest that continuous communication between the receptor and Gαβγ is important for chemotropism and polarized growth, perhaps to regulate Gαβγ in a spatially asymmetric manner.
Distinctions between Signaling and Polarity Roles of Gβγ
Bypassing the role of Gβγ in signaling revealed several interesting points regarding mating and cell polarization. First, the ability of pheromone and Gβγ to regulate chemotropism or de novo polarization does not require the ability to modulate MAP kinase cascade signaling or the presence of the scaffold protein Ste5 (see Figures 1 and 2). Second, the level of mating success is neither solely dictated by nor strongly dependent on transcription levels, because the two membrane-targeted Ste5 reagents activated transcription to either high (Ste5ΔN-CTM) or low (Ste5ΔN-Sec22) levels and yet yielded higher efficiency mating than the Ste11 and Ste12 reagents (Figure 1). Presumably, the Ste5 reagents promote the most efficient mating because they can activate the MAP kinase cascade (i.e., in contrast to Ste12 overexpression, which induces transcription only), but they do not cross-activate other pathways that inhibit mating (i.e., in contrast to Ste11ΔN, which also activates the HOG pathway) (Harris et al., 2001). Third, the ability of pheromone gradients to increase mating success is independent of the baseline level of mating, as this behavior was observed whether the baseline level of mating was relatively high (Ste5ΔN-CTM and Ste5ΔN-Sec22) or relatively low (Ste11ΔN and Ste12) (Figure 1).
Heterotrimeric G Protein Dissociation
Several of our findings are relevant to the mechanism of Gαβγ dissociation. Although it is generally accepted that nucleotide exchange causes Gα-GTP to dissociate from Gβγ (Gilman, 1987; Neer, 1995; Cabrera-Vera et al., 2003), some studies suggest that complete dissociation may not be necessary (Rebois et al., 1997; Klein et al., 2000; Levitzki and Klein, 2002; Bunemann et al., 2003; Gales et al., 2006). Indeed, prior work showed that the yeast heterotrimer remained functional when Gα and Gβ were expressed as a Gβ–Gα fusion protein (Klein et al., 2000), which should restrict dissociation. We extended these results by showing that this Gβ–Gα fusion protein performs both signaling and polarity functions indistinguishably from the wild-type (unfused) proteins, even when expressed at native levels. Moreover, the chemotropic and de novo polarization proficiency we observed with the Sw interface mutants indicates that despite being active for signaling, they can still mediate a response to the pheromone ligand whereas the Nt interface mutants cannot. This implies that the Sw mutants maintain a functional Nt interface and hence raises the possibility that the Gαβγ heterotrimer may remain tethered by the Nt interface, allowing continued communication with the receptor (Figure 9). By extension, it is conceivable that this “partial dissociation” state applies not only to the Sw interface mutants but also to receptor-activated Gαβγ. That is, GTP-induced conformational changes in the Switch region of Gα may trigger an initial state in which the Sw interface dissociates but the Nt interface does not.
Figure 9.
Model for how Sw and Nt mutants differentially affect dissociation of Gαβγ and receptor coupling. The schematic diagram of the receptor and heterotrimer is based on the molecular model shown in Figure 3A. Zig-zag lines signify membrane-anchoring groups attached to the N terminus of Gα and the C terminus of Gγ. Mutations in either the Sw or Nt interface cause constitutive activation of Gβγ, but the Sw interface mutants remain competent to mediate directional responses to chemoattractant (i.e., chemotropism and de novo polarization), whereas the Nt interface mutants are defective. Alone, contacts in the Nt interface may be insufficient to maintain Gα–Gβ association, but membrane insertion of the adjacent lipophilic groups may stabilize this weak interaction; this could explain why the Sw interface mutants show no Gα interaction in the absence of membranes (e.g., in coimmunoprecipitation and two-hybrid assays), and yet can still mediate responses to pheromone in vivo. Therefore, mutations in the Sw interface are proposed to result in a partially dissociated structure that remains tethered by the Nt interface, and hence remains in regulatory communication with the receptor. See text for further discussion.
Several structural considerations are pertinent to this partial dissociation model. First, the N-terminal helix of Gα is implicated in receptor recognition, and it is thought to lie tangential to the membrane (Taylor et al., 1994; Bourne, 1997; Hamm, 1998, 2001; Itoh et al., 2001; Cabrera-Vera et al., 2003). Second, Gαβγ is anchored to the membrane by addition of lipophilic groups to the Gα N terminus and the Gγ C terminus (Wedegaertner et al., 1995), which lie adjacent to each other at one end of the Nt interface (Wall et al., 1995; Lambright et al., 1996) (Figure 9). Membrane insertion of these groups stabilizes the intact heterotrimer (Bigay et al., 1994); thus, it may also stabilize the weak interaction at the Nt interface enough to keep the partially dissociated structure intact in vivo but not in detergent-solubilized cell extracts. Third, the behavior of the Gβ–Gα fusion protein is compatible with the partial dissociation model, because the link formed between the Gβ C terminus and the Gα N terminus would constrain separation of the Nt interface but not the Sw interface. Fourth, crystal structures of Gαβγ show that the N-terminal helix of Gα makes no tertiary contacts with the remainder of Gα (Wall et al., 1995; Lambright et al., 1996), and so the intervening linkage could freely rotate to allow the Gα and Gβ halves of the Sw interface to swivel apart. Cocrystal structures are also compatible with partial dissociation, because contacts between Gβγ and effectors involve residues in the Sw interface rather than the Nt interface (Gaudet et al., 1996; Lodowski et al., 2003).
Note that this partial dissociation model does not postulate that Gα and Gβγ are inseparable, but only that dissociation of the Sw interface does not automatically lead to dissociation of the Nt interface as a necessary or immediate consequence. Complete dissociation might follow if GTP hydrolysis is slow or when Gα-GTP and Gβγ bind their targets (Tesmer et al., 2005). Partial dissociation could be advantageous by allowing an additional level of heterotrimer regulation or by accelerating heterotrimer reformation after GTP hydrolysis by Gα. In our physiological context, it could facilitate chemotropism and de novo polarization by allowing activated Gβγ to remain in regulatory communication with the receptor.
Symmetry Breaking and De Novo Polarization
Symmetry breaking, in which cells or molecular modules self-polarize without a localized cue, is thought to involve the amplification of an initial asymmetry that occurs through stochastic variation (Kirschner et al., 2000; Wedlich-Soldner and Li, 2003). In yeast, symmetry breaking can produce polarized buds in the absence of directional cues (Chant and Herskowitz, 1991; Irazoqui et al., 2003). This has been proposed to result from stochastic variation in the local amount of activated (GTP-bound) Cdc42, which is then amplified by positive feedback loops via two possible pathways. One pathway involves self-assembly of the adaptor protein Bem1, which links Cdc24 and Cdc42 (Butty et al., 2002; Irazoqui et al., 2003; Wedlich-Soldner et al., 2004). The other pathway involves feedback between Cdc42-induced actin polymerization and polarized transport of secretory vesicles carrying Cdc42 as cargo (Gulli et al., 2000; Wedlich-Soldner et al., 2003; Wedlich-Soldner and Li, 2003).
Analogous mechanisms may apply during pheromone response to allow de novo polarization. In principle, breaking the symmetry inherent in a uniform field of pheromone could begin with small differences in receptor–Gαβγ activation. However, our results indicate that activation of the MAP kinase cascade is sufficient to trigger some asymmetry even in the absence of pheromone or G protein subunits. Furthermore, previous studies show that establishment and maintenance of the polarity axis are separable (Nern and Arkowitz, 2000) and that the MAP kinase Fus3 is required to establish asymmetry (Matheos et al., 2004). Hence, the most parsimonious model may be that the MAP kinase pathway imparts the polarity machinery with some inherent self-organizational ability, whereas the receptor–Gαβγ module controls orientation and directional persistence, either by influencing the initial directional choice or by providing continuous reinforcement of the initial asymmetry.
Our findings implicate a role for continuous communication between pheromone-bound receptors and Gαβγ, because activated Gβγ and Gα alone cannot promote directionally persistent polarization. Because pheromone receptors are rapidly internalized and redelivered by secretory transport along actin filaments (Jenness and Spatrick, 1986; Schandel and Jenness, 1994; Ayscough and Drubin, 1998), initial asymmetries might become amplified by polarized delivery of new pheromone receptors to the initial landmark, resulting in heightened pheromone response in that location which would reinforce the initial directional choice. The mutations that disrupt de novo polarization (i.e., gpa1-Q323L, or ste4 Nt interface mutations) may disable this positive reinforcement by disrupting the ability to coordinate receptor localization with the activity/localization of Gα, Gβγ, or both. In contrast, the behavior of the Sw interface mutants suggests that, despite constitutively active Gβγ signaling, these mutant heterotrimers remain in regulatory communication with the receptor. Although it remains unclear what this communication achieves in molecular terms, we envision three speculative scenarios: 1) interaction with asymmetrically localized receptors might influence localization of the heterotrimer, thereby causing asymmetrically localized Gβγ activity; 2) the Sw mutants might allow nucleotide exchange on the Gα subunit at sites of pheromone-bound receptors, potentially promoting localized Gα-GTP, which could control polarization in synergy with Gβγ; or 3) pheromone-bound receptors might interact directly with downstream polarity factors, but normal affinity of receptors for ligand might require coupling to a Gαβγ heterotrimer with an intact Nt interface and normal GDP-GTP exchange properties. Distinguishing among these scenarios will require further study.
GTP Hydrolysis and Localized Signaling
GTP hydrolysis is important for polarity control by Cdc42 in both yeast and mammalian cells (Stowers et al., 1995; Irazoqui et al., 2003). Similarly, we found that chemotropism and de novo polarization require GTP hydrolysis by the Gα subunit Gpa1. To provide a directional cue, GTP loading of Gα and the resultant activation of Gβγ may have to occur in a localized manner, which would be obscured if all Gα molecules were locked in the GTP-bound state and distributed uniformly. Thus, the spatial distribution of active signaling molecules is likely to be critical to their function in guiding cell polarity. To focus this spatial distribution, the activities of these proteins may also need to be rapidly extinguished by mechanisms such as by GTP hydrolysis, phosphorylation, or endocytosis. Indeed, chemotaxis is thought to depend on a combination of local excitation and global inhibition of polarization activities (Iijima et al., 2002). Endocytic clearing allows directionally transported membrane proteins to maintain a polarized distribution (Valdez-Taubas and Pelham, 2003; Marco et al., 2007), and pheromone-regulated polarization in yeast is facilitated by endocytosis signals in receptor C termini (Konopka et al., 1988; Vallier et al., 2002). Thus, localized signaling and delivery may generally work in concert with down-regulatory mechanisms to ensure proper polarity control.
CONCLUSIONS
The findings reported here reveal several unexpected lessons that may be applicable to other examples of cell polarity control, heterotrimeric G protein function, or both. First, the activated Gαβγ heterotrimer (and the resulting downstream signaling) is not inherently able to organize a directionally persistent polarization axis. Second, the requirements of the receptor–Gαβγ module are similar for gradient-controlled polarization and de novo polarization. Third, the two structurally distinct Gα–Gβ interfaces have functionally distinct roles. Fourth, the yeast heterotrimer may be able to function in a partially dissociated state that allows continued regulatory communication with the receptor. In future studies, it will be interesting to compare these behaviors shown by the yeast system to those of other systems.
ACKNOWLEDGMENTS
We thank Henrik Dohlman, Alexander Levitzki, David Stone, and Malcolm Whiteway for plasmids, as well as thank Danny Lew and Dan McCollum for comments on the manuscript. We are especially indebted to Rachel Lamson and Matt Winters for technical assistance during the completion and revision stages. This work was supported by a grant from the National Institutes of Health (GM57769) to P.M.P.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-04-0385) on October 31, 2007.
REFERENCES
- Apanovitch D. M., Iiri T., Karasawa T., Bourne H. R., Dohlman H. G. Second site suppressor mutations of a GTPase-deficient G-protein α-subunit. Selective inhibition of Gβγ-mediated signaling. J. Biol. Chem. 1998;273:28597–28602. doi: 10.1074/jbc.273.44.28597. [DOI] [PubMed] [Google Scholar]
- Arkowitz R. A. Responding to attraction: chemotaxis and chemotropism in Dictyostelium and yeast. Trends Cell Biol. 1999;9:20–27. doi: 10.1016/s0962-8924(98)01412-3. [DOI] [PubMed] [Google Scholar]
- Ayscough K. R., Drubin D. G. A role for the yeast actin cytoskeleton in pheromone receptor clustering and signalling. Curr. Biol. 1998;8:927–930. doi: 10.1016/s0960-9822(07)00374-0. [DOI] [PubMed] [Google Scholar]
- Bagorda A., Mihaylov V. A., Parent C. A. Chemotaxis: moving forward and holding on to the past. Thromb. Haemost. 2006;95:12–21. [PubMed] [Google Scholar]
- Bar E. E., Ellicott A. T., Stone D. E. Gbetagamma recruits Rho1 to the site of polarized growth during mating in budding yeast. J. Biol. Chem. 2003;278:21798–21804. doi: 10.1074/jbc.M212636200. [DOI] [PubMed] [Google Scholar]
- Bartel P. L., Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- Bigay J., Faurobert E., Franco M., Chabre M. Roles of lipid modifications of transducin subunits in their GDP-dependent association and membrane binding. Biochemistry. 1994;33:14081–14090. doi: 10.1021/bi00251a017. [DOI] [PubMed] [Google Scholar]
- Bourne H. R. How receptors talk to trimeric G proteins. Curr. Opin. Cell Biol. 1997;9:134–142. doi: 10.1016/s0955-0674(97)80054-3. [DOI] [PubMed] [Google Scholar]
- Bunemann M., Frank M., Lohse M. J. Gi protein activation in intact cells involves subunit rearrangement rather than dissociation. Proc. Natl. Acad. Sci. USA. 2003;100:16077–16082. doi: 10.1073/pnas.2536719100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butty A. C., Perrinjaquet N., Petit A., Jaquenoud M., Segall J. E., Hofmann K., Zwahlen C., Peter M. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. EMBO J. 2002;21:1565–1576. doi: 10.1093/emboj/21.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butty A. C., Pryciak P. M., Huang L. S., Herskowitz I., Peter M. The role of Far1p in linking the heterotrimeric G protein to polarity establishment proteins during yeast mating. Science. 1998;282:1511–1516. doi: 10.1126/science.282.5393.1511. [DOI] [PubMed] [Google Scholar]
- Cabrera-Vera T. M., Vanhauwe J., Thomas T. O., Medkova M., Preininger A., Mazzoni M. R., Hamm H. E. Insights into G protein structure, function, and regulation. Endocr. Rev. 2003;24:765–781. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- Cairns B. R., Ramer S. W., Kornberg R. D. Order of action of components in the yeast pheromone response pathway revealed with a dominant allele of the STE11 kinase and the multiple phosphorylation of the STE7 kinase. Genes Dev. 1992;6:1305–1318. doi: 10.1101/gad.6.7.1305. [DOI] [PubMed] [Google Scholar]
- Chant J. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 1999;15:365–391. doi: 10.1146/annurev.cellbio.15.1.365. [DOI] [PubMed] [Google Scholar]
- Chant J., Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;65:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- Dohlman H. G., Song J., Ma D., Courchesne W. E., Thorner J. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein α subunit) Mol. Cell Biol. 1996;16:5194–5209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman H. G., Thorner J. W. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- Dolan J. W., Fields S. Overproduction of the yeast STE12 protein leads to constitutive transcriptional induction. Genes Dev. 1990;4:492–502. doi: 10.1101/gad.4.4.492. [DOI] [PubMed] [Google Scholar]
- Dorer R., Pryciak P. M., Hartwell L. H. Saccharomyces cerevisiae cells execute a default pathway to select a mate in the absence of pheromone gradients. J. Cell Biol. 1995;131:845–861. doi: 10.1083/jcb.131.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell S. J., Bishop A. L., Dyos S. L., Brown A. J., Whiteway M. S. Mapping of a yeast G protein betagamma signaling interaction. Genetics. 1998;150:1407–1417. doi: 10.1093/genetics/150.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. Cdc42–the centre of polarity. J. Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- Feng Y., Song L. Y., Kincaid E., Mahanty S. K., Elion E. A. Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr. Biol. 1998;8:267–278. doi: 10.1016/s0960-9822(98)70108-3. [DOI] [PubMed] [Google Scholar]
- Franca-Koh J., Kamimura Y., Devreotes P. Navigating signaling networks: chemotaxis in Dictyostelium discoideum. Curr. Opin. Genet Dev. 2006;16:333–338. doi: 10.1016/j.gde.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Gales C., Van Durm J. J., Schaak S., Pontier S., Percherancier Y., Audet M., Paris H., Bouvier M. Probing the activation-promoted structural rearrangements in preassembled receptor-G protein complexes. Nat. Struct. Mol. Biol. 2006;13:778–786. doi: 10.1038/nsmb1134. [DOI] [PubMed] [Google Scholar]
- Gartner A., Jovanovic A., Jeoung D. I., Bourlat S., Cross F. R., Ammerer G. Pheromone-dependent G1 cell cycle arrest requires Far1 phosphorylation, but may not involve inhibition of Cdc28-Cln2 kinase, in vivo. Mol. Cell Biol. 1998;18:3681–3691. doi: 10.1128/mcb.18.7.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet R., Bohm A., Sigler P. B. Crystal structure at 2.4 Å resolution of the complex of transducin βγ and its regulator, phosducin. Cell. 1996;87:577–588. doi: 10.1016/s0092-8674(00)81376-8. [DOI] [PubMed] [Google Scholar]
- Gilman A. G. G proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Grishin A. V., Weiner J. L., Blumer K. J. Biochemical and genetic analysis of dominant-negative mutations affecting a yeast G-protein γ subunit. Mol. Cell Biol. 1994a;14:4571–4578. doi: 10.1128/mcb.14.7.4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishin A. V., Weiner J. L., Blumer K. J. Control of adaptation to mating pheromone by G protein β subunits of Saccharomyces cerevisiae. Genetics. 1994b;138:1081–1092. doi: 10.1093/genetics/138.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulli M., Jaquenoud M., Shimada Y., Niederhauser G., Wiget P., Peter M. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell. 2000;6:1155–1167. doi: 10.1016/s1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- Hamm H. E. The many faces of G protein signaling. J. Biol. Chem. 1998;273:669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- Hamm H. E. How activated receptors couple to G proteins. Proc. Natl. Acad. Sci. USA. 2001;98:4819–4821. doi: 10.1073/pnas.011099798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K., Lamson R. E., Nelson B., Hughes T. R., Marton M. J., Roberts C. J., Boone C., Pryciak P. M. Role of scaffolds in MAP kinase pathway specificity revealed by custom design of pathway-dedicated signaling proteins. Curr. Biol. 2001;11:1815–1824. [PubMed] [Google Scholar]
- Hollenberg S. M., Sternglanz R., Cheng P. F., Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima M., Huang Y. E., Devreotes P. Temporal and spatial regulation of chemotaxis. Dev. Cell. 2002;3:469–478. doi: 10.1016/s1534-5807(02)00292-7. [DOI] [PubMed] [Google Scholar]
- Inouye C., Dhillon N., Thorner J. Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997;278:103–106. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- Irazoqui J. E., Gladfelter A. S., Lew D. J. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 2003;5:1062–1070. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Cai K., Khorana H. G. Mapping of contact sites in complex formation between light-activated rhodopsin and transducin by covalent crosslinking: use of a chemically preactivated reagent. Proc. Natl. Acad. Sci. USA. 2001;98:4883–4887. doi: 10.1073/pnas.051632998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C. L., Hartwell L. H. Courtship in S. cerevisiae: both cell types choose mating partners by responding to the strongest pheromone signal. Cell. 1990;63:1039–1051. doi: 10.1016/0092-8674(90)90507-b. [DOI] [PubMed] [Google Scholar]
- Jackson C. L., Konopka J. B., Hartwell L. H. S. cerevisiae α pheromone receptors activate a novel signal transduction pathway for mating partner discrimination. Cell. 1991;67:389–402. doi: 10.1016/0092-8674(91)90190-a. [DOI] [PubMed] [Google Scholar]
- Jenness D. D., Spatrick P. Down regulation of the α-factor pheromone receptor in S. cerevisiae. Cell. 1986;46:345–353. doi: 10.1016/0092-8674(86)90655-0. [DOI] [PubMed] [Google Scholar]
- Kirschner M., Gerhart J., Mitchison T. Molecular “vitalism.”. Cell. 2000;100:79–88. doi: 10.1016/s0092-8674(00)81685-2. [DOI] [PubMed] [Google Scholar]
- Klein S., Reuveni H., Levitzki A. Signal transduction by a nondissociable heterotrimeric yeast G protein. Proc. Natl. Acad. Sci. USA. 2000;97:3219–3223. doi: 10.1073/pnas.050015797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka J. B., Jenness D. D., Hartwell L. H. The C-terminus of the S. cerevisiae alpha-pheromone receptor mediates an adaptive response to pheromone. Cell. 1988;54:609–620. doi: 10.1016/s0092-8674(88)80005-9. [DOI] [PubMed] [Google Scholar]
- Lambright D. G., Sondek J., Bohm A., Skiba N. P., Hamm H. E., Sigler P. B. The 2.0 Å crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- Lamson R. E., Winters M. J., Pryciak P. M. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell Biol. 2002;22:2939–2951. doi: 10.1128/MCB.22.9.2939-2951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Dignard D., Harcus D., Thomas D. Y., Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuw T., Wu C., Schrag J. D., Whiteway M., Thomas D. Y., Leberer E. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- Levitzki A., Klein S. G-protein subunit dissociation is not an integral part of G-protein action. Chembiochemistry. 2002;3:815–818. doi: 10.1002/1439-7633(20020902)3:9<815::AID-CBIC815>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Lodowski D. T., Pitcher J. A., Capel W. D., Lefkowitz R. J., Tesmer J. J. Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gβγ. Science. 2003;300:1256–1262. doi: 10.1126/science.1082348. [DOI] [PubMed] [Google Scholar]
- Madden K., Snyder M. Specification of sites for polarized growth in Saccharomyces cerevisiae and the influence of external factors on site selection. Mol. Biol. Cell. 1992;3:1025–1035. doi: 10.1091/mbc.3.9.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanty S. K., Wang Y., Farley F. W., Elion E. A. Nuclear shuttling of yeast scaffold Ste5 is required for its recruitment to the plasma membrane and activation of the mating MAPK cascade. Cell. 1999;98:501–512. doi: 10.1016/s0092-8674(00)81978-9. [DOI] [PubMed] [Google Scholar]
- Marco E., Wedlich-Soldner R., Li R., Altschuler S. J., Wu L. F. Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity. Cell. 2007;129:411–422. doi: 10.1016/j.cell.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheos D., Metodiev M., Muller E., Stone D., Rose M. D. Pheromone-induced polarization is dependent on the Fus3p MAPK acting through the formin Bni1p. J. Cell Biol. 2004;165:99–109. doi: 10.1083/jcb.200309089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metodiev M. V., Matheos D., Rose M. D., Stone D. E. Regulation of MAPK function by direct interaction with the mating-specific Gα in yeast. Science. 2002;296:1483–1486. doi: 10.1126/science.1070540. [DOI] [PubMed] [Google Scholar]
- Moskow J. J., Gladfelter A. S., Lamson R. E., Pryciak P. M., Lew D. J. Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. Mol. Cell Biol. 2000;20:7559–7571. doi: 10.1128/mcb.20.20.7559-7571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer E. J. Heterotrimeric G proteins: organizers of transmembrane signals. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- Neiman A. M., Herskowitz I. Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homologue STE7 by STE11. Proc. Natl. Acad. Sci. USA. 1994;91:3398–3402. doi: 10.1073/pnas.91.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A., Arkowitz R. A. A GTP-exchange factor required for cell orientation. Nature. 1998;391:195–198. doi: 10.1038/34458. [DOI] [PubMed] [Google Scholar]
- Nern A., Arkowitz R. A. A Cdc24p-Far1p-Gβγ protein complex required for yeast orientation during mating. J. Cell Biol. 1999;144:1187–1202. doi: 10.1083/jcb.144.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A., Arkowitz R. A. G proteins mediate changes in cell shape by stabilizing the axis of polarity. Mol. Cell. 2000;5:853–864. doi: 10.1016/s1097-2765(00)80325-1. [DOI] [PubMed] [Google Scholar]
- Pruyne D., Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 2000;113:365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- Pryciak P. M., Hartwell L. H. AKR1 encodes a candidate effector of the Gβγ complex in the Saccharomyces cerevisiae pheromone response pathway and contributes to control of both cell shape and signal transduction. Mol. Cell Biol. 1996;16:2614–2626. doi: 10.1128/mcb.16.6.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryciak P. M., Huntress F. A. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebois R. V., Warner D. R., Basi N. S. Does subunit dissociation necessarily accompany the activation of all heterotrimeric G proteins? Cell Signal. 1997;9:141–151. doi: 10.1016/s0898-6568(96)00133-7. [DOI] [PubMed] [Google Scholar]
- Roberts C. J., et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- Schandel K. A., Jenness D. D. Direct evidence for ligand-induced internalization of the yeast α-factor pheromone receptor. Mol. Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrick K., Garvik B., Hartwell L. H. Mating in Saccharomyces cerevisiae: the role of the pheromone signal transduction pathway in the chemotropic response to pheromone. Genetics. 1997;147:19–32. doi: 10.1093/genetics/147.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall J. E. Polarization of yeast cells in spatial gradients of α mating factor. Proc. Natl. Acad. Sci. USA. 1993;90:8332–8336. doi: 10.1073/pnas.90.18.8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M., Peter M. Polarizing without a c(l)ue. Trends Cell Biol. 2003;13:526–533. doi: 10.1016/j.tcb.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Sondek J., Bohm A., Lambright D. G., Hamm H. E., Sigler P. B. Crystal structure of a GA protein βγ dimer at 2.1 Å resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- Stone D. E., Reed S. I. G protein mutations that alter the pheromone response in Saccharomyces cerevisiae. Mol. Cell Biol. 1990;10:4439–4446. doi: 10.1128/mcb.10.9.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers L., Yelon D., Berg L. J., Chant J. Regulation of the polarization of T cells toward antigen-presenting cells by Ras-related GTPase CDC42. Proc. Natl. Acad. Sci. USA. 1995;92:5027–5031. doi: 10.1073/pnas.92.11.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton H. F., Zhou J., Reed S. I., Stone D. E. The mating-specific Gα protein of Saccharomyces cerevisiae downregulates the mating signal by a mechanism that is dependent on pheromone and independent of Gβγ sequestration. Mol. Cell. Biol. 1996;16:6325–6337. doi: 10.1128/mcb.16.11.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden S. C., Winters M. J., Ben-Ari G., Lamson R. E., Tyers M., Pryciak P. M. A Mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M., Jacob-Mosier G. G., Lawton R. G., Remmers A. E., Neubig R. R. Binding of an α2 adrenergic receptor third intracellular loop peptide to Gβ and the amino terminus of Gα. J. Biol. Chem. 1994;269:27618–27624. [PubMed] [Google Scholar]
- Tesmer V. M., Kawano T., Shankaranarayanan A., Kozasa T., Tesmer J. J. Snapshot of activated G proteins at the membrane: the Gαq-GRK2-Gβγ complex. Science. 2005;310:1686–1690. doi: 10.1126/science.1118890. [DOI] [PubMed] [Google Scholar]
- Valdez-Taubas J., Pelham H. R. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 2003;13:1636–1640. doi: 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Vallier L. G., Segall J. E., Snyder M. The alpha-factor receptor C-terminus is important for mating projection formation and orientation in Saccharomyces cerevisiae. Cell Motil. Cytoskeleton. 2002;53:251–266. doi: 10.1002/cm.10073. [DOI] [PubMed] [Google Scholar]
- Valtz N., Peter M., Herskowitz I. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J. Cell Biol. 1995;131:863–873. doi: 10.1083/jcb.131.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen F., O'Rourke S. M., Stucke V. M., Jaquenoud M., Neiman A. M., Peter M. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 2000;10:630–639. doi: 10.1016/s0960-9822(00)00511-x. [DOI] [PubMed] [Google Scholar]
- van Drogen F., Stucke V. M., Jorritsma G., Peter M. MAP kinase dynamics in response to pheromones in budding yeast. Nat. Cell Biol. 2001;3:1051–1059. doi: 10.1038/ncb1201-1051. [DOI] [PubMed] [Google Scholar]
- Wall M. A., Coleman D. E., Lee E., Iñiguez-Lluhi J. A., Posner B. A., Gilman A. G., Sprang S. R. The structure of the G protein heterotrimer Giα1β1γ2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- Wedegaertner P. B., Wilson P. T., Bourne H. R. Lipid modifications of trimeric G proteins. J. Biol. Chem. 1995;270:503–506. doi: 10.1074/jbc.270.2.503. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Altschuler S., Wu L., Li R. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science. 2003;299:1231–1235. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Li R. Spontaneous cell polarization: undermining determinism. Nat. Cell Biol. 2003;5:267–270. doi: 10.1038/ncb0403-267. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Wai S. C., Schmidt T., Li R. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J. Cell Biol. 2004;166:889–900. doi: 10.1083/jcb.200405061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M., Clark K. L., Leberer E., Dignard D., Thomas D. Y. Genetic identification of residues involved in association of α and β G-protein subunits. Mol. Cell. Biol. 1994;14:3223–3229. doi: 10.1128/mcb.14.5.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M. S., Wu C., Leeuw T., Clark K., Fourest-Lieuvin A., Thomas D. Y., Leberer E. Association of the yeast pheromone response G protein βγ subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- Winters M. J., Lamson R. E., Nakanishi H., Neiman A. M., Pryciak P. M. A membrane binding domain in the Ste5 scaffold synergizes with Gβγ binding to control localization and signaling in pheromone response. Mol. Cell. 2005;20:21–32. doi: 10.1016/j.molcel.2005.08.020. [DOI] [PubMed] [Google Scholar]