Abstract
RTT107 (ESC4, YHR154W) encodes a BRCA1 C-terminal domain protein that is important for recovery from DNA damage during S phase. Rtt107 is a substrate of the checkpoint kinase Mec1, and it forms complexes with DNA repair enzymes, including the nuclease subunit Slx4, but the role of Rtt107 in the DNA damage response remains unclear. We find that Rtt107 interacts with chromatin when cells are treated with compounds that cause replication forks to arrest. This damage-dependent chromatin binding requires the acetyltransferase Rtt109, but it does not require acetylation of the known Rtt109 target, histone H3-K56. Chromatin binding of Rtt107 also requires the cullin Rtt101, which seems to play a direct role in Rtt107 recruitment, because the two proteins are found in complex with each other. Finally, we provide evidence that Rtt107 is bound at or near stalled replication forks in vivo. Together, these results indicate that Rtt109, Rtt101, and Rtt107, which genetic evidence suggests are functionally related, form a DNA damage response pathway that recruits Rtt107 complexes to damaged or stalled replication forks.
INTRODUCTION
Evolutionarily conserved DNA repair and checkpoint processes ensure accurate DNA replication in eukaryotic cells. The DNA damage checkpoint pathway is a signal transduction pathway that responds to DNA damage to facilitate cell cycle delay, promote DNA repair, and induce transcription when DNA lesions are present (Melo and Toczyski, 2002; Carr, 2002; McGowan and Russell, 2004). Mutation of DNA damage response pathways causes a failure to accurately transmit the genome from one generation to the next, resulting in genomic instability, which is a hallmark of cancerous cells (Kastan and Bartek, 2004).
In Saccharomyces cerevisiae, the DNA damage checkpoint is made up of a signaling cascade that includes the essential protein kinase Mec1 (Weinert et al., 1994; Sanchez et al., 1996). When DNA damage is encountered in S phase, stalling, uncoupling, and collapse of replication forks can result (Tercero and Diffley, 2001; Katou et al., 2003; Lopes et al., 2006). Mec1 together with its binding partner Ddc2 is thought to localize to stalled forks by binding to single-stranded (ss) DNA coated with the ssDNA-binding protein replication protein A (Zou and Elledge, 2003). Recruitment of Mec1 and Ddc2 to stalled replication forks leads to activation of the S phase checkpoint, and it is thought to delay the onset of mitosis until replication is complete (Osborn et al., 2002). Several other DNA replication and repair proteins are also thought to act at stalled replication forks to prevent fork collapse, remove or bypass DNA lesions, and ultimately restart DNA replication (for review, see Tourriere and Pasero, 2007).
Rtt107, a target of Mec1 phosphorylation, is one protein that has been proposed to promote the restart of DNA replication after DNA damage in S phase (Rouse, 2004; Chin et al., 2006; Roberts et al., 2006). RTT107 (ESC4; YHR154W) was originally identified in a genetic screen for increased Ty transposon mobility and contains several BRCA1 C-terminal (BRCT) homology domains (Scholes et al., 2001). The BRCT domain was first noted in the protein encoded by the human breast cancer susceptibility gene BRCA1 (Koonin et al., 1996; Bork et al., 1997), and it is a protein–protein interaction domain frequently found in proteins that have roles in the response to DNA damage, in cell-cycle control, and in checkpoint-mediated DNA repair (Callebaut and Mornon, 1997; Bork et al., 1997). BRCT domains are thought to facilitate assembly of signaling complexes in response to DNA damage, and they may direct repair proteins to the site of damage, resulting in removal of DNA lesions (El-Khamisy et al., 2003). Mutations in BRCT proteins frequently confer sensitivity to a variety of DNA-damaging agents (Callebaut and Mornon, 1997).
Deletion of rtt107 confers sensitivity to the DNA alkylating agent methyl methane sulfonate (MMS), the replication inhibitor hydroxyurea (HU), and camptothecin (CPT), an agent that induces replication dependent double-strand breaks (Chang et al., 2002; Hanway et al., 2002; Rouse, 2004). Rtt107 is required for normal DNA synthesis in the presence of DNA damage and for the resumption of DNA replication during recovery from DNA damage (Chang et al., 2002; Rouse, 2004; Roberts et al., 2006). Rtt107 binds the Slx4/Slx1 nuclease complex through the N-terminal, BRCT-containing half of Rtt107, and Slx4 is required for the Mec1 phosphorylation of Rtt107 (Roberts et al., 2006; Zappulla et al., 2006). Slx4, like Rtt107, is important for the resumption of DNA synthesis after DNA damage (Rouse, 2004; Roberts et al., 2006). Two-hybrid and high-throughput studies have found Rtt107 in complex with a number of DNA repair proteins, including Mms22, the DNA helicase Sgs1, and the homologous recombination proteins Rad55 and Rad57 (Ho et al., 2002; Chin et al., 2006; Zappulla et al., 2006). The fission yeast homologue of Rtt107, Brc1, is thought to act in the postreplication repair pathway through the Slx1 and Mus81 nucleases (Sheedy et al., 2005) and in a pathway with Mms22 to manage DNA damage in S phase (Dovey and Russell, 2007). These data indicate that Rtt107 functions with distinct DNA repair proteins to tolerate DNA damage that stalls replication forks. One possibility is that Rtt107 acts as a scaffold for the assembly of DNA damage response and repair proteins at stalled DNA replication forks, thereby promoting replication fork restart.
Here, we report that Rtt107 is recruited to chromatin in the presence of agents that cause replication fork stalling. The binding of Rtt107 to chromatin after DNA damage is regulated by a pathway comprising the acetyltransferase Rtt109 and the cullin Rtt101. Results from chromatin immunoprecipitation on tiling microarray analysis were consistent with Rtt107 functioning at stalled replication forks in vivo. We propose that Rtt109 acetylation of an unidentified target results in the Rtt101-dependent recruitment of Rtt107 to stalled replication forks. The interaction between Rtt107 and Slx4, and perhaps other DNA repair proteins, promotes their assembly at sites of replication fork stalling, leading to resumption of DNA synthesis.
MATERIALS AND METHODS
Yeast Strains and Media
Yeast strains used in this study were derivatives of W303 or BY4741 (Thomas and Rothstein, 1989; Brachmann et al., 1998), and they are listed in Supplemental Table 1. Nonessential haploid deletion strains marked with the kanamycin (G418) resistance gene were made by the Saccharomyces Gene Deletion Project (Winzeler et al., 1999). Deletion strains marked with nourseothricin (nat) resistance gene were constructed by switching the kanamycin resistance gene with the nat resistance gene as described previously (Tong et al., 2001). Standard yeast media and growth conditions were used (Sherman, 1991).
Chromosome Spreads and Microscopy
Cells were grown in YPD at 30°C to early-log phase, and they were treated with no drug, 0.03% MMS (Sigma-Aldrich, St. Louis, MO), 150 mM HU (Bioshop, Toronto, ON, Canada), or 15 μg/ml CPT (Sigma-Aldrich) for 1 h. For α factor-treated cells, cultures were treated with 8 μg/ml α factor for 2 h, and then 0.03% MMS and 8 μg/ml α factor were added for an additional hour. Cells were harvested and prepared as described previously (Michaelis et al., 1997). To detect proteins immunofluorescently, microscope slides were treated with mouse anti-vesicular stomatitis virus (VSV) antibodies (Roche Diagnostics, Indianapolis, IN) and goat anti-mouse Alexa Fluor 546 (Invitrogen, Carlsbad, CA). Before examination, VECTASHIELD mounting media with 4,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA) was added. Fluorescence intensity was measured relative to background intensity for at least 100 nucleoids in two independent experiments by using Openlab 5.0.2 (Improvision, Coventry, United Kingdom).
Immunoblotting and Immunoprecipitation
Cultures were grown in the absence or presence of 100 mM HU or 0.03% MMS. Cells (1 × 108) were collected and treated with 10% trichloroacetic acid, and extracts were prepared essentially as described previously (Pellicioli et al., 1999). Proteins were resolved on 7.5% or 15% SDS-polyacrylamide gels, and they were subjected to immunoblot analysis with rabbit anti-VSV-G (Bethyl Laboratories, Montgomery, TX), anti-H3-K56-Ac (a kind gift from Alain Verreault), or rabbit anti-histone H3 antibody 1791 (Abcam, Cambridge, MA). Immunoblots were developed using SuperSignal ECL (Pierce Chemical, Rockford, IL). Immunoprecipitation was performed essentially as described previously (Roberts et al., 2006). Anti-hemagglutinin (HA) antibody (Covance Research Products, Princeton, NJ) and protein G-agarose (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) were used to immunoprecipitate HA-tagged proteins. After extensive washing, proteins were subjected to immunoblot analysis with anti-HA or anti-myc (9E10; Santa Cruz Biotechnology, Santa Cruz, CA).
In Situ Kinase Assay and Ddc2-Yellow Fluorescent Protein (YFP) Foci
Rad53 in situ kinase assays were carried out as described previously (Pellicioli et al., 1999), by using extracts from cells growing in mid-logarithmic phase in the absence of DNA-damaging agents. Imaging of Ddc2-YFP foci was carried out as described previously (Roberts et al., 2006). At least 200 cells were counted in two independent experiments.
Growth Measurements
For temperature sensitivity assays, cells were grown in YPD or SD-leucine, diluted serially, spotted onto YPD or SD−leucine plates, and incubated at 30°C or 37°C for 2–3 d. For DNA damage sensitivity assays, plates contained 0.03% (vol/vol) MMS, 150 mM HU, or 40 μM CPT in YPD, and they were used within 24 h of preparation.
Chromatin Immunoprecipitation (ChIP)-Chip
Chromatin immunoprecipitation on microarray (ChIP-chip) experiments were performed essentially as described previously (Katou et al., 2003; Katou et al., 2006). Cells were arrested for 2 h in 200 mM HU before cross-linking. After labeling, bound and unbound DNA was hybridized to whole-genome tiling arrays (P/N 520055; Affymetrix, Santa Clara, CA), and arrays were washed and scanned as described previously (Lee et al., 2007). For the analysis in Figure 7A, raw data from the Affymetrix GCOS software (.CEL format) was analyzed to judge the statistical significance of regions enriched in the bound fraction based on the continuity of significant change of -fold change value (Katou et al., 2006; Bermejo et al., 2007). For the analysis in Figure 7, B and C, raw data from Affymetrix GCOS software (.CEL format) were analyzed with Affymetrix Tiling Analysis software, version 1.1, and they were visualized with Affymetrix Integrated Genome Browser. The data were normalized with the built-in quantile normalization and probe-level analysis with perfect match probes, and they were run with a bandwidth of 60. Each probe was given a value calculated from the average of all probe intensities within the specified window (defined by the bandwidth parameter). Then, a ratio was calculated for the two-sample analysis. The resulting log2 ratio was then output for each probe position, where the probe position is defined as the center (13th) base coordinate for each 25-mer probe.
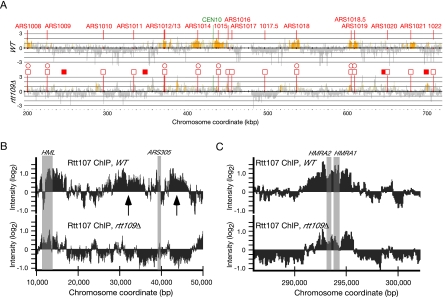
Figure 7.
Rtt107 localizes to origin-proximal regions after replication fork arrest. (A) ChIP-on-chip analysis was performed following synchronous arrest of wild-type (top) or rtt109Δ (bottom panel) cells in early S phase by releasing α factor–arrested cells into 200 mM HU for 2 h at 23°C. After cross-linking and DNA fragmentation, Rtt107 was precipitated, and enrichment of DNA fragments in the Rtt107-bound fraction relative to the unbound fraction is shown along chromosome X. The signal intensity ratio on a log2 scale is shown on the y-axis and the position along the chromosome is shown on the x-axis. Positive signal represents occupancy by Rtt107, and regions where the positive signal is statistically significant (Katou et al., 2006) are shown in yellow. Annotated, confirmed replication origins (Nieduszynski et al., 2007) are indicated by open squares. Potential origins marked by MCM protein binding (Wyrick et al., 2001; Xu et al., 2006) are indicated by closed squares. Origins that are active in HU as measured by accumulation of ssDNA (Feng et al., 2006) are indicated by open circles. (B) Distribution of Rtt107 near HML and ARS305 in wild type and rtt109Δ. Data from the ChIP-chip experiment in A was analyzed across the indicated chromosome coordinates of chromosome III. Arrows indicate two regions of Rtt107 binding flanking ARS305. The positions of HML and ARS305 are indicated by gray bars. (C) Distribution of Rtt107 near HMR in wild type and rtt109Δ. Data from the ChIP-chip experiment in A was analyzed across the indicated chromosome coordinates of Chromosome III. The position of HMR is indicated by gray bars.
RESULTS
Rtt107 Is Recruited to Chromatin in Response to Replication Fork Stalling
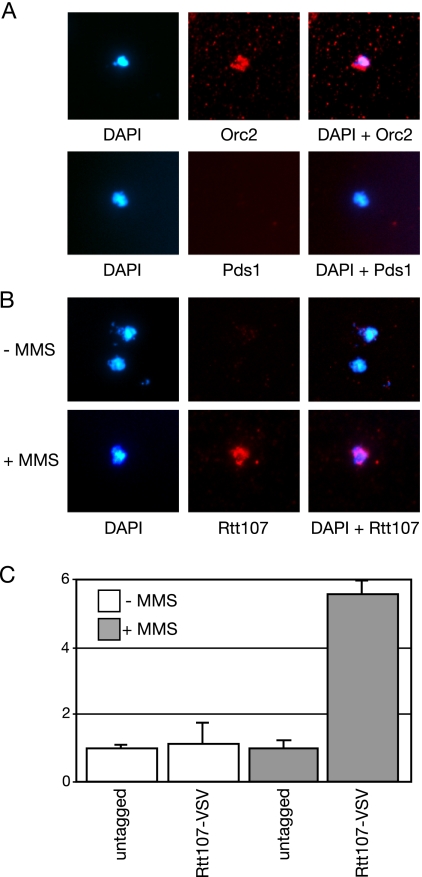
The interaction of Rtt107 with a number of DNA repair proteins (Ho et al., 2002; Chin et al., 2006; Roberts et al., 2006; Zappulla et al., 2006), and the role of Rtt107 in recovery from DNA damage (Rouse, 2004; Roberts et al., 2006), suggested that Rtt107 might function as a scaffold to assemble repair proteins onto sites of DNA damage. We tested this hypothesis by asking whether Rtt107 is recruited to chromatin after DNA damage, by using chromatin spreads (Figure 1). Spreads were prepared by spheroplasting the cells, cross-linking them with formaldehyde, and lysing the spheroplasts on glass slides. Unbound proteins were washed away, and the resulting nucleoids were visualized by staining the DNA with DAPI. The association of Pds1, Orc2, and Rtt107 with the spread chromatin nucleoids was detected by indirect immunofluorescence, by using antibodies directed against the epitope tag fused to each protein (Figure 1, A and B). Pds1 is a soluble nuclear protein, and it was included as a negative control. Orc2 binds to replication origins throughout the cell cycle (Diffley et al., 1994; Aparicio et al., 1997), and it was included as a positive control. As expected, Pds1 was not detected on the spread chromatin, whereas Orc2 localization to the nucleoids was clear (Figure 1A). When spread chromatin was probed for bound Rtt107, no significant localization of Rtt107 to spread chromatin was evident in the absence of DNA damage (Figure 1B, −MMS). By contrast, when cells were treated with MMS, which stalls DNA replication forks, localization of Rtt107 to chromatin was evident (Figure 1B, +MMS). We quantified the intensity of the Rtt107-VSV signal on 100 nucleoids for untagged and Rtt107-VSV strains, in the presence and absence of MMS (Figure 1C). The qualitative result in Figure 1B was borne out by this quantification, with MMS inducing an almost sixfold increase in Rtt107 localization to spread chromatin relative to the untagged control. Rtt107 localization in the absence of MMS was not significantly different from the background seen in the untagged control. Thus, DNA damage causes recruitment of Rtt107 to chromatin.
Figure 1.
Rtt107 is recruited to chromatin in response to MMS. (A and B) Logarithmically growing cultures expressing the indicated epitope-tagged proteins were treated with 0% (−) or 0.03% (+) MMS for 1 h, before preparation of chromosome spreads. DNA was stained with DAPI (left), and the indicated epitope-tagged proteins were detected with α-VSV (middle). The merged image is shown in the right panel. (C) VSV signal intensity minus background was quantified and expressed relative to the untagged control. The signal intensity was measured for at least 100 nucleoids in each experiment, and the average of two independent experiments is plotted, with error bars spanning 1 SD.
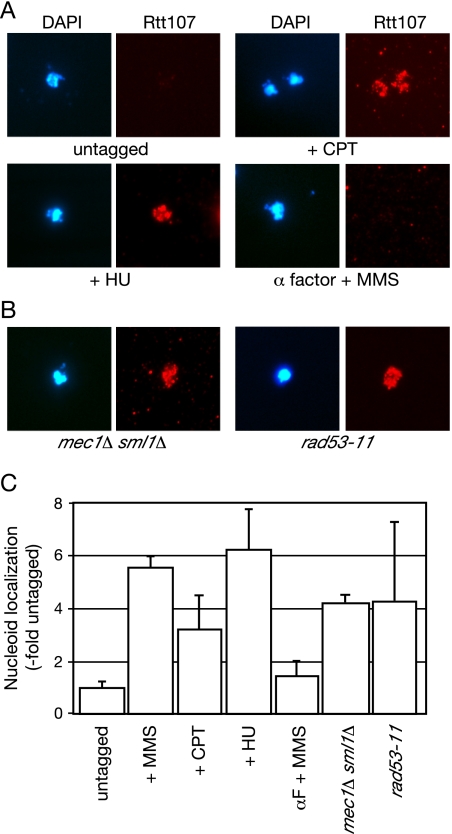
We next asked whether Rtt107 recruitment was specific for alkylation damage. We found that both HU and CPT caused a similar recruitment of Rtt107 to spread chromatin (Figure 2, A and B). Like MMS, HU and CPT cause replication forks to stall, although they do so by different mechanisms. This suggested that stalled replication forks, rather than DNA damage per se, caused recruitment of Rtt107 to chromatin. In agreement with this, Rtt107 was no longer associated with the nucleoids when cells arrested in G1 phase were treated with MMS (Figure 2, A and C, α factor + MMS). These findings are consistent with a proposed role for Rtt107 at the site of stalled replication forks (Chin et al., 2006; Roberts et al., 2006). Although experimental evidence indicates that Rtt107 also functions during an unperturbed S phase (Rouse, 2004; Roberts et al., 2006), we were unable to detect binding of Rtt107 to nucleoids in the absence of treatment with DNA-damaging agents. Perhaps the level of spontaneous replication fork stalling in the absence of DNA damage does not lead to sufficient Rtt107 recruitment to allow detection in our assay.
Figure 2.
Rtt107 is recruited to chromatin in response to replication fork stalling. (A) Logarithmically growing cultures were treated with 15 μg/ml camptothecin (+CPT) or 150 mM hydroxyurea (+HU) for 1 h, or they were treated with 8.3 μg/ml α factor for 2 h before addition of 8.3 μg/ml α factor and 0.03% MMS for 1 h (α factor + MMS), before preparation of chromosome spreads. DNA was stained with DAPI (left) and VSV-tagged Rtt107 was detected with α-VSV (right). (B) The role of checkpoint pathways in the localization of Rtt107 was analyzed by treating rad53-11 or mec1Δ sml1Δ cells with 0.03% MMS for 1 h, before preparation of chromosome spreads. DNA was stained with DAPI (left) and VSV-tagged Rtt107 was detected with α-VSV (right). (C) VSV signal intensity minus background was quantified and expressed relative to the untagged control. The signal intensity was measured for at least 100 nucleoids in each experiment, and the average of two independent experiments is plotted, with error bars spanning 1 SD. The analysis of MMS-treated cells from Figure 1C is included for comparison.
Rtt107 is a target of the checkpoint kinase Mec1 (Rouse, 2004). We examined the effect of the DNA damage checkpoint on the localization of Rtt107. Rtt107 recruitment to chromatin in the presence of MMS was not dependent on the checkpoint kinases Rad53 or Mec1 (Figure 2, B and C), indicating that although Rtt107 is a target of Mec1 phosphorylation the localization of Rtt107 to chromatin in response to replication fork stalling was independent of checkpoint activation.
Recruitment of Rtt107 to Chromatin Requires the Cullin Rtt101 and the Acetyltransferase Rtt109
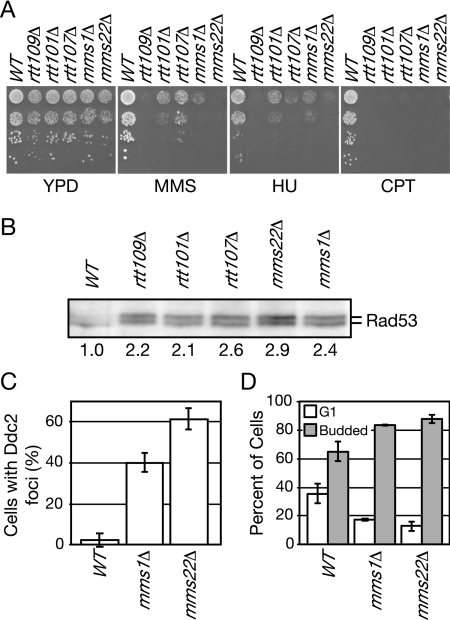
To identify genes that act in the same pathway as RTT107 and that might regulate the recruitment of Rtt107 to chromatin, we chose candidates based on two-dimensional hierarchical clustering of genetic interaction data in which RTT107 clustered closely with RTT109, RTT101, MMS22, and MMS1 (Collins et al., 2007). These genes displayed the most similar genetic interaction profiles among a set of ∼400 genes with roles in DNA replication, DNA repair, and other aspects of chromosome dynamics, suggesting that the products of these genes may form a functional pathway or complex (Collins et al., 2007). Additional data link these genes, including the role of Rtt101 in promoting replication fork progression through DNA lesions (Luke et al., 2006), which is similar to that of Rtt107, and protein–protein interactions between Mms22 and Rtt101 and Mms22 and Rtt107 in high-throughput analysis (Ho et al., 2002). SLX4 was also chosen as a candidate, because Rtt107 and Slx4 form a complex (Roberts et al., 2006; Zappulla et al., 2006). We tested whether additional phenotypes were shared by strains carrying deletions of these genes (Figure 3). All were sensitive to agents that stall DNA replication forks, MMS, HU, and CPT (Figure 3A). We also found that the checkpoint kinase Rad53 was hyperactive in rtt109Δ, rtt107Δ, rtt101Δ, mms1Δ, and mms22Δ during normal cell cycle progression, displaying levels of autophosphorylation 2–3 times that observed in wild-type cells (Figure 3B), suggesting the presence of elevated levels of spontaneous DNA damage. Consistent with this, DNA damage response foci formed by the checkpoint protein Ddc2 were elevated in mms1Δ and mms22Δ, by 8- and 12-fold relative to the wild type (Figure 3C), and mms1Δ and mms22Δ displayed a cell cycle delay in G2 phase (Figure 3D). Elevated levels of spontaneous DNA damage were noted previously for rtt107Δ (Roberts et al., 2006), rtt101Δ (Luke et al., 2006), and rtt109Δ (Han et al., 2007a). Thus, in addition to having highly similar genetic interaction profiles (Collins et al., 2007), these genes share a number of phenotypes relevant to maintenance of genome integrity, suggesting that they function in the same pathway.
Figure 3.
Deletion of RTT107, RTT109, RTT101, MMS1, or MMS22 confers similar phenotypes. (A) Ten-fold serial dilutions of the indicated strains were spotted on media containing 0.03% MMS, 150 mM HU, 40 μM CPT, or no drug (YPD), and strains were grown for 2–3 d at 30°C. (B) Spontaneous Rad53 activation was measured in logarithmically growing cells of the indicated strains by using an in situ kinase assay. An autoradiograph of Rad53 is shown, and the amount of autophosphorylation relative to the wild-type control is indicated. (C) The percentage of cells with spontaneous Ddc2 foci was measured for the indicated strains. At least 200 cells were counted in each of two experiments, and the average of the two experiments is plotted with error bars spanning 1 SD. (D) The percent of unbudded and budded cells was measured for the indicated strains. At least 200 cells were counted in each of two experiments, and the average of the two experiments is plotted with error bars spanning 1 SD.
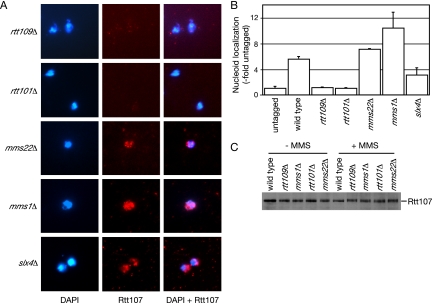
Recruitment of Rtt107 to chromatin was examined in the absence of each of these genes (Figure 4, A and B). Localization of Rtt107 required the acetyltransferase Rtt109 and the cullin Rtt101, but it was not dependent on Mms22, Mms1, or Slx4. Although localization of Rtt107 to chromatin in slx4Δ was clearly evident (Figure 4A), the fluorescence intensity was less than that observed in wild type (Figure 4B). Conversely, the signal intensity was higher in mms1Δ and mms22Δ (Figure 4B). The significance of this, if any, remains unclear. Rtt107 protein levels were not affected by deletion of any of these genes, indicating that the observed effects were not due to depletion of Rtt107 (Figure 4C). Thus, these data indicate that Rtt107 functions downstream of Rtt109 and Rtt101.
Figure 4.
Rtt107 recruitment to chromatin requires RTT109 and RTT101, but not MMS1, MMS22, or SLX4. (A) Logarithmically growing cultures of the indicated strains were treated with 0.03% MMS for 1 h, before preparation of chromosome spreads. DNA was stained with DAPI (left) and VSV-tagged Rtt107 was detected with α-VSV (middle). The merged image is shown in the right panel. (B) VSV signal intensity minus background was quantified and expressed relative to the untagged control. The signal intensity was measured for at least 100 nucleoids in each experiment, and the average of two independent experiments is plotted, with error bars spanning 1 SD. (C) Deletion of RTT109 or RTT101 does not affect Rtt107 expression. Extracts of logarithmically growing cultures of the indicated yeast strains expressing Rtt107-VSV, grown in the presence or absence of MMS, were fractionated by SDS-PAGE, and immunoblots were probed with α-VSV antibodies.
Rtt101 Recruitment to Chromatin in the Presence of MMS
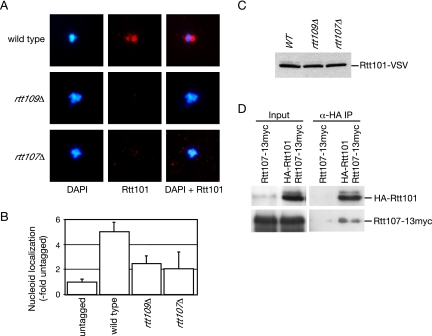
To order the function of the components of this putative pathway, we analyzed the association of Rtt101 with chromatin in spreads (Figure 5). Like Rtt107, Rtt101 was recruited to chromatin in the presence (Figure 5A), but not the absence, of MMS (data not shown). Deletion of RTT109 or RTT107 abrogated the chromatin binding of Rtt101 (Figure 5, A and B). The levels of Rtt101 were not affected in the absence of RTT109 or RTT107, indicating that the observed effect was not due to depletion of Rtt101 (Figure 5C). The simplest interpretation of these data is that Rtt109, Rtt101, and Rtt107 form a pathway that responds to stalled replication forks, with Rtt109 upstream of Rtt101 and Rtt107. Mms1, Mms22, and Slx4 were not detected on chromatin spreads (data not shown); so, their positions within the putative pathway could not be determined.
Figure 5.
Recruitment of Rtt101 to chromatin requires RTT109 and RTT107. (A) Logarithmically growing cultures were treated with 0.03% MMS for 1 h, before preparation of chromosome spreads. DNA was stained with DAPI (left), and VSV-tagged Rtt101 was detected with α-VSV (middle). The merged image is shown in the right panel. (B) VSV signal intensity minus background was quantified and expressed relative to the untagged control. The signal intensity was measured for at least 100 nucleoids in each experiment, and the average of two independent experiments is plotted, with error bars spanning 1 SD. (C) Deletion of RTT109 or RTT107 does not affect Rtt101 expression. Extracts of logarithmically growing cultures of the indicated yeast strains expressing Rtt101-VSV, grown in the presence or absence of MMS, were fractionated by SDS-PAGE, and immunoblots were probed with α-VSV antibodies. (D) Rtt101 forms a complex with Rtt107. Extracts from cells expressing the indicated tagged proteins were immunoprecipitated with anti-HA antibody. The input and immunoprecipitate fractions were analyzed on immunoblots with anti-HA antibodies to detect HA-Rtt101 and with anti-myc antibodies to detect Rtt107-13myc.
Because Rtt107 was required for the chromatin binding of Rtt101 and vice versa, Rtt101 and Rtt107 might be recruited as a complex in response to replication fork stalling. To examine this possibility, epitope-tagged Rtt101 was immunoprecipitated, and the bound fraction was assayed for the presence of Rtt107 by immunoblotting. We found that Rtt101 specifically coimmunoprecipitated Rtt107 (Figure 5D). The interaction between Rtt101 and Rtt107 was independent of DNA damage (data not shown). These data are consistent with Rtt101 and Rtt107 being recruited to chromatin as a complex.
Rtt107 Recruitment to Chromatin Does Not Require Histone H3 Lysine 56 Acetylation
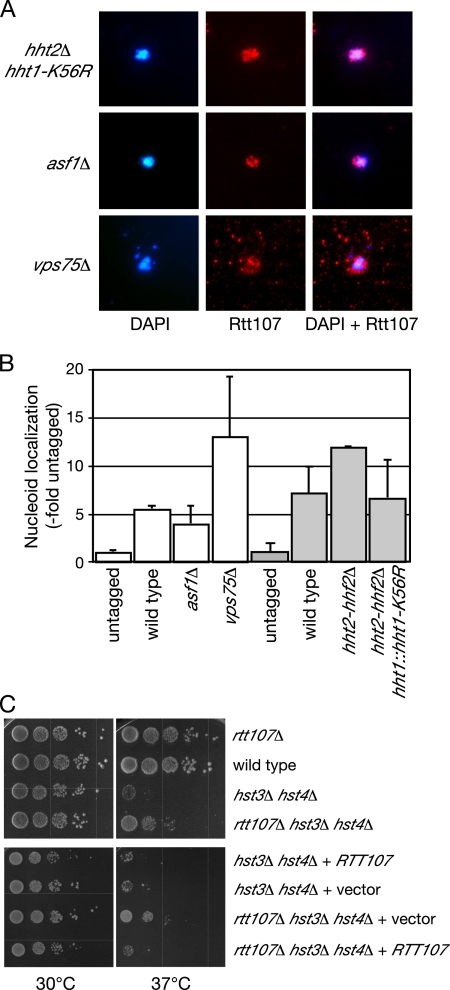
Because the histone acetyltransferase Rtt109 was required for recruitment of Rtt107 to chromatin in the presence of replication fork stalling, we tested whether acetylation of known Rtt109 target histone H3 lysine 56 (Collins et al., 2007; Han et al., 2007a; Driscoll et al., 2007) is important for the association of Rtt107 with DNA. Recruitment of Rtt107 to chromatin in the absence of H3-K56 acetylation was monitored by performing chromosome spread analysis of Rtt107-VSV in the absence of Asf1, a histone chaperone required for K56 acetylation (Recht et al., 2006), and in a strain expressing only mutant histone H3 that cannot be acetylated on K56 (H3-K56R) (Figure 6, A and B). Rtt107 localization to chromatin was not affected by the absence of H3-K56 acetylation, indicating that this modification was not required for the recruitment of Rtt107 to chromatin. Rtt109 copurifies with a histone chaperone, Vps75, which is not required for acetylation of H3-K56 (Collins et al., 2007; Han et al., 2007c; Selth and Svejstrup, 2007; Tsubota et al., 2007), suggesting that Rtt109 might have additional acetylation targets. Rtt107 was still localized to chromatin in vps75Δ cells (Figure 6, A and B), indicating that Vps75 was not required for the recruitment of Rtt107 to chromatin. Thus, although the recruitment of Rtt107 to chromatin in response to stalled replication forks required the acetyltransferase Rtt109, it did not require the acetylation of the known Rtt109 target histone H3-K56.
Figure 6.
Rtt107 recruitment to chromatin does not require acetylation of H3-K56. (A) Logarithmically growing cultures of the indicated strains were treated with 0.03% MMS for 1 h, before preparation of chromosome spreads. DNA was stained with DAPI (left) and VSV-tagged Rtt107 was detected with α-VSV (middle). The merged image is shown in the right panel. (B) VSV signal intensity minus background was quantified and expressed relative to the untagged control. The signal intensity was measured for at least 100 nucleoids in each experiment, and the average of two independent experiments is plotted, with error bars spanning 1 SD. (C) Deletion of RTT107 suppresses the temperature sensitivity of hst3Δ hst4Δ. Ten-fold serial dilutions of cultures of the indicated strains were spotted onto YPD (top) or SD-Leu (bottom) and incubated at 30°C or 37°C for 3 d.
rtt107Δ Suppresses the Temperature Sensitivity of hst3Δ hst4Δ
Hst3 and Hst4 are the deacetylases that act in opposition to Rtt109 by deacetylating histone H3-K56 (Celic et al., 2006; Maas et al., 2006; Miller et al., 2006). hst3Δ hst4Δ mutant cells are temperature sensitive at 37°C, and they are sensitive to DNA damage (Celic et al., 2006; Maas et al., 2006). Several lines of evidence suggest that these phenotypes are largely due to hyperacetylation of H3-K56 (Celic et al., 2006; Maas et al., 2006). It remains formally possible, however, that hyperacetylation of additional targets contributes to these phenotypes, so we tested whether deletion of RTT107 suppresses the temperature sensitivity of hst3Δ hst4Δ cells. As shown in Figure 6C, rtt107Δ partially suppressed the temperature sensitivity of hst3Δ hst4Δ, whereas rtt107Δ had no effect on cell growth in a wild-type background. The temperature sensitivity of rtt107Δ hst3Δ hst4Δ was restored when Rtt107 was expressed from a plasmid, indicating that the partial suppression of the temperature sensitivity of hst3Δ hst4Δ cells was due to deletion of RTT107. These data are consistent with hyperacetylation of some target in the absence of Hst3 and Hst4 activating a pathway that contains Rtt107, resulting in temperature and DNA damage sensitivity. Because we also found that Rtt107 recruitment is independent of H3-K56 acetylation, it is likely that Rtt109 acetylation of a distinct target is required for Rtt107 recruitment, although it remains formally possible that the role of Rtt109 in recruiting Rtt107 is independent of acetylation.
Rtt107 Binds Regions Flanking Early Replication Origins When Replication Forks Stall
Although Rtt107 is hypothesized to act at stalled replication forks, and we have found that Rtt107 binds to chromatin when replication forks stall, the chromosomal sites of Rtt107 action have not been identified. We used a chromatin immunoprecipitation strategy, with tiling microarrays, to determine whether Rtt107 is bound to specific regions of chromosomes when replication forks are stalled by nucleotide depletion. Cells were arrested in G1 phase and then released synchronously into S phase in the presence of HU to stall replication forks close to early-firing replication origins. After cross-linking with formaldehyde, Rtt107–DNA complexes were isolated and the bound DNA was hybridized to a whole-genome tiling microarray. When compared with whole genomic DNA from the unbound fraction a distinctive pattern of enrichment at particular genome sites was observed. Figure 7A shows peaks of enrichment along the length of chromosome X, with significant enrichment indicated by yellow bars. Peaks of enrichment coincided with the positions of replication origins known to “fire” in HU (Feng et al., 2006), indicated by open circles. Enrichment of sequences flanking late firing origins (marked by open squares only), which are dormant in HU, was not observed. This pattern of enrichment of sites adjacent to early-firing replication origins was evident on all chromosomes, and it was reminiscent of the pattern displayed by replication fork proteins such as Cdc45 (Katou et al., 2003) and Ctf4 (Lengronne et al., 2006). Enrichment of origin proximal regions was not observed when Rtt107 was immunoprecipitated from cells that had not been treated with HU (data not shown). The replication origin proximal pattern of Rtt107 binding, and the similarity to the pattern of the known replication fork components support the conclusion that Rtt107 is bound at stalled replication forks after treatment of cells with HU.
Because we had observed that the recruitment of Rtt107 to chromatin required RTT109, we tested whether the binding of Rtt107 to origin proximal regions upon fork stalling had a similar requirement. When a chromatin immunoprecipitation of Rtt107 was performed using rtt109Δ cells, the enrichment at replication origin proximal regions was absent, as shown for chromosome X (Figure 7A). Interestingly, we also observed binding of Rtt107 to the silent mating loci HML and HMR on chromosome III (Figure 7, B and C), consistent with a proposed role for Rtt107 in transcriptional silencing (Zappulla et al., 2006). In this analysis, the enrichment of sequences flanking ARS305 was evident in the Rtt107-bound fraction (Figure 7B, top, arrows). This enrichment was entirely absent when Rtt107 was immunoprecipitated from rtt109Δ cells (Figure 7B, bottom). In contrast to the binding of Rtt107 to ARS305 flanking regions, the binding of Rtt107 to the silent mating loci remained, albeit at lower levels, when rtt109 was deleted (Figure 7, B and C, bottom). Binding to the silent loci, then, serves as a positive control for Rtt107 activity in the rtt109Δ experiment. Thus, the binding of Rtt107 to stalled replication forks required Rtt109, as did recruitment of Rtt107 to chromatin.
DISCUSSION
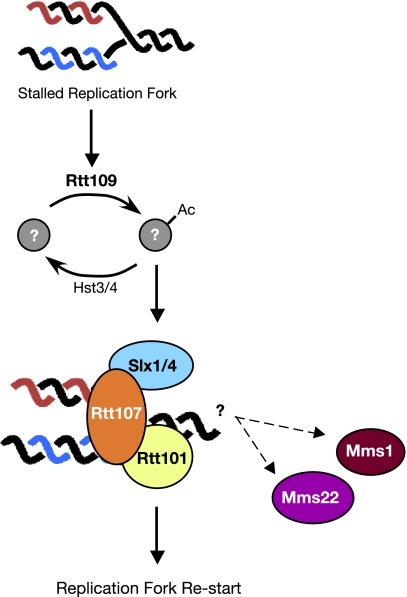
We find that Rtt107 functions as part of a DNA damage response pathway, composed of Rtt109, Rtt101, and Rtt107, which preserves genome integrity when replication forks stall. Rtt107 is recruited to chromatin when cells are treated with DNA-damaging agents that cause replication forks to arrest. This damage-dependent chromatin binding does not require checkpoint activation or complex formation between Rtt107 and the DNA repair protein Slx4. Rtt107 recruitment depends on the acetyltransferase Rtt109 and the cullin Rtt101, which, like Rtt107, is recruited to chromatin in a DNA damage- and Rtt109-dependent manner. Finally, we find that Rtt107 is bound at or near stalled replication forks in vivo. Together, these results indicate that Rtt109, Rtt101, and Rtt107 form a pathway that recruits Rtt107 complexes to damaged or stalled replication forks (Figure 8). Furthermore, our data indicate that Rtt109 sits at the top of the pathway, likely recruiting an Rtt101/Rtt107 complex to stalled replication forks.
Figure 8.
Model of the Rtt107 replication fork restart pathway. See text for details.
One striking feature of our study is that it indicates that Rtt109 function is not exclusively dependent on acetylation of histone H3-K56. Several lines of evidence support this assertion. First, RTT109 but not ASF1, VPS75, or K56 was required for chromatin binding of Rtt107. Second, the temperature sensitivity of hst3Δ hst4Δ was partially suppressed by deletion of RTT107, indicating that Rtt107 functions downstream of an acetylation event. Third, the rtt109Δ mutant was more damage sensitive than rtt107Δ, consistent with Rtt109 having a damage resistance role not shared by Rtt107, presumably histone H3-K56 acetylation. Finally, although K56 acetylation was not required for Rtt107 recruitment, Rtt109 was. Because our data indicate that Rtt107 acts downstream of an acetylation event, we propose that the role of Rtt109 in recruiting Rtt107 to stalled replication forks involves the acetylation of an unidentified target. However, until the active site residues of Rtt109 are identified with certainty, it remains possible that the role of Rtt109 in recruiting Rtt107 does not involve acetylation.
We envisage two possible models for Rtt109 function in regulating the recruitment of Rtt107 to stalled replication forks. In the first model, Rtt109 acetylation of a protein target provides a binding site for an Rtt101–Rtt107 complex at stalled replication forks. Candidates for this putative target include replication fork proteins and Rtt101 and Rtt107 themselves. In the second model, Rtt109 is required to maintain the integrity of stalled replication forks rather than directly regulating recruitment of Rtt101 and Rtt107. In the absence of Rtt109, replication proteins that normally provide a binding site for Rtt101-Rtt107 dissociate from the stalled replication fork, thereby abrogating Rtt107 recruitment. Although evidence is emerging that loss of Rtt109 affects replisome stability (Han et al., 2007b), loss of Asf1 has similar effects on replisome stability (Franco et al., 2005), yet it does not eliminate Rtt107 recruitment. Irrespective of the mechanism of Rtt109 action, our data indicate that it is required, directly or indirectly, for recruitment of Rtt101 and Rtt107 to chromatin when replication forks stall, and for the binding of Rtt107 to stalled replication forks. The role of Rtt101 in the recruitment of Rtt107 to stalled replication forks seems to be direct, because Rtt101 and Rtt107 were found in complex together and chromatin binding by Rtt101 and Rtt107 was mutually dependent.
The roles of Mms1 and Mms22 in this pathway remain mysterious. The mms1Δ and mms22Δ mutants share a number of phenotypes with rtt107Δ, including sensitivity to drugs that stall replication forks and elevated levels of spontaneous DNA damage, and they have similar genetic interaction profiles (Collins et al., 2007). However, neither MMS1 nor MMS22 were required for the recruitment of Rtt107 to stalled replication forks, suggesting that they may act downstream of Rtt107 or in a pathway parallel to Rtt107. Because high-throughput protein–protein interaction data and yeast two-hybrid analysis indicate that Mms22 interacts with both Rtt101 and Rtt107 (Ho et al., 2002; Zappulla et al., 2006), and DNA combing analysis indicates that mms1Δ and mms22Δ have defective replication fork recovery after stalling (Peter, unpublished data), we favor the possibility that Mms1 and Mms22 are also recruited to stalled replication forks, likely by an Rtt101–Rtt107 complex, where they carry out some as yet unidentified downstream function (Figure 8).
The most likely function of Rtt107 at stalled DNA replication forks is the assembly of DNA repair proteins to facilitate the efficient resumption of DNA synthesis. Rtt107 is found in complex with a number of DNA repair proteins, including Slx4, Rad55, and Rad57 (Chin et al., 2006; Roberts et al., 2006; Zappulla et al., 2006). The Rtt107–Slx4 complex is particularly relevant to the resumption of DNA synthesis after fork stalling, because mutants in RTT107 and SLX4 display replication defects after treatment with agents that stall DNA replication forks (Chang et al., 2002; Rouse, 2004; Chin et al., 2006; Roberts et al., 2006; Flott et al., 2007). DNA combing analysis has further indicated that unreplicated gaps remain after fork stalling in slx4Δ and rtt101Δ (Flott et al., 2007; Luke et al., 2006), and rtt107Δ displays an anaphase arrest morphology (Roberts et al., 2006) consistent with the presence of unreplicated regions (Torres-Rosell et al., 2007). We found that recruitment of Rtt107 to stalled replication forks did not require Slx4; yet, Rtt107 and Slx4 are found in complex (Roberts et al., 2006), suggesting that Rtt107 brings Slx4 to sites of replication fork stalling (Figure 8). Considerable evidence suggests that the Slx1–Slx4 nuclease is active on replication fork structures and that it could be a critical element in the restart of stalled forks (Kaliraman and Brill, 2002; Fricke and Brill, 2003; Coulon et al., 2004, 2006; Flott and Rouse, 2005; Roberts et al., 2006). In addition to Slx4, we suspect that Rtt107 assembles other repair proteins onto stalled replication forks, particularly because rtt107Δ mutants are considerably more sensitive to MMS and HU than slx4Δ mutants (Chang et al., 2002; Chin et al., 2006; Roberts et al., 2006; Zappulla et al., 2006), and rtt107Δ slx4Δ double mutants are slightly more MMS sensitive than single mutants (Roberts et al., 2006). Thus, we propose that the action of Rtt109 and Rtt101 leads to recruitment of Rtt107 to stalled replication forks where Rtt107 acts as a scaffold for the assembly of DNA repair enzymes such as Slx4 to facilitate the bypass or removal of the replication fork block, ultimately resulting in the restart of stalled replication forks (Figure 8).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jef Boeke (Johns Hopkins University), Jeffrey Fillingham (University of Toronto), Katsuhiko Shirahige (Tokyo Institute of Technology), Igor Stagljar (University of Toronto), and Alain Verrault (Université de Montréal) for strains and reagents. We also thank Brigitte Lavoie for advice on chromosome spreads, Katsuhiko Shirahige and Yuki Katou for teaching us the ChIP-chip procedure and for assistance with data analysis, Guri Giaever and Corey Nislow for tiling arrays, and Jeffrey Fillingham and Marta Davidson for helpful discussions. This work was supported by funds from the National Cancer Institute of Canada (to G.W.B.), the Swiss National Science Foundation, Oncosuisse, and ETH Zurich (to M.P.), and the Terry Fox Foundation (to T.M.R.). G.W.B. is a research scientist of the National Cancer Institute of Canada.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-09-0961) on October 31, 2007.
REFERENCES
- Aparicio O. M., Weinstein D. M., Bell S. P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Bermejo R., Doksani Y., Capra T., Katou Y. M., Tanaka H., Shirahige K., Foiani M. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007;21:1921–1936. doi: 10.1101/gad.432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Hofmann K., Bucher P., Neuwald A. F., Altschul S. F., Koonin E. V. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR–mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Callebaut I., Mornon J. P. From BRCA1 to RAP 1, a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- Carr A. M. DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair. 2002;1:983–994. doi: 10.1016/s1568-7864(02)00165-9. [DOI] [PubMed] [Google Scholar]
- Celic I., Masumoto H., Griffith W. P., Meluh P., Cotter R. J., Boeke J. D., Verreault A. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone H3 lysine 56 deacetylation. Curr. Biol. 2006;16:1280–1289. doi: 10.1016/j.cub.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Chang M., Bellaoui M., Boone C., Brown G. W. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl. Acad. Sci. USA. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J. K., Bashkirov V. I., Heyer W. D., Romesberg F. E. Esc4/Rtt107 and the control of recombination during replication. DNA Repair. 2006;5:618–628. doi: 10.1016/j.dnarep.2006.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. R., et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Coulon S., Gaillard P. H., Chahwan C., McDonald W. H., Yates J. R., 3rd, Russell P. Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol. Biol. Cell. 2004;15:71–80. doi: 10.1091/mbc.E03-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon S., Noguchi E., Noguchi C., Du L. L., Nakamura T. M., Russell P. Rad22Rad52-dependent repair of ribosomal DNA repeats cleaved by Slx1-Slx4 endonuclease. Mol. Biol. Cell. 2006;17:2081–2090. doi: 10.1091/mbc.E05-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H., Dowell S. J., Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Dovey C. L., Russell P. Mms22 preserves genomic integrity during DNA replication in Schizosaccharomyces pombe. Genetics. 2007;177:47–61. doi: 10.1534/genetics.107.077255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R., Hudson A., Jackson S. P. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science. 2007;315:649–652. doi: 10.1126/science.1135862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khamisy S. F., Masutani M., Suzuki H., Caldecott K. W. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Collingwood D., Boeck M. E., Fox L. A., Alvino G. M., Fangman W. L., Raghuraman M. K., Brewer B. J. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat. Cell Biol. 2006;8:148–155. doi: 10.1038/ncb1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S., Alabert C., Toh G. W., Toth R., Sugawara N., Campbell D. G., Haber J. E., Pasero P., Rouse J. Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Mol. Cell Biol. 2007;27:6433–6445. doi: 10.1128/MCB.00135-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flott S., Rouse J. Slx4 becomes phosphorylated after DNA damage in a Mec1/Tel1-dependent manner and is required for repair of DNA alkylation damage. Biochem. J. 2005;391:325–333. doi: 10.1042/BJ20050768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco A. A., Lam W. M., Burgers P. M., Kaufman P. D. Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev. 2005;19:1365–1375. doi: 10.1101/gad.1305005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke W. M., Brill S. J. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Zhou H., Horazdovsky B., Zhang K., Xu R. M., Zhang Z. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science. 2007a;315:653–655. doi: 10.1126/science.1133234. [DOI] [PubMed] [Google Scholar]
- Han J., Zhou H., Li Z., Xu R. M., Zhang Z. Acetylation of lysine 56 of histone H3 catalyzed by Rtt109 and regulated by Asf1 is required for replisome integrity. J. Biol. Chem. 2007b;282:28587–28596. doi: 10.1074/jbc.M702496200. [DOI] [PubMed] [Google Scholar]
- Han J., Zhou H., Li Z., Xu R. M., Zhang Z. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J. Biol. Chem. 2007c;282:14158–14164. doi: 10.1074/jbc.M700611200. [DOI] [PubMed] [Google Scholar]
- Hanway D., Chin J. K., Xia G., Oshiro G., Winzeler E. A., Romesberg F. E. Previously uncharacterized genes in the UV- and MMS-induced DNA damage response in yeast. Proc. Natl. Acad. Sci. USA. 2002;99:10605–10610. doi: 10.1073/pnas.152264899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Kaliraman V., Brill S. J. Role of SGS1 and SLX4 in maintaining rDNA structure in Saccharomyces cerevisiae. Curr. Genet. 2002;41:389–400. doi: 10.1007/s00294-002-0319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M. B., Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Katou Y., Kaneshiro K., Aburatani H., Shirahige K. Genomic approach for the understanding of dynamic aspect of chromosome behavior. Methods Enzymol. 2006;409:389–410. doi: 10.1016/S0076-6879(05)09023-3. [DOI] [PubMed] [Google Scholar]
- Katou Y., Kanoh Y., Bando M., Noguchi H., Tanaka H., Ashikari T., Sugimoto K., Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Altschul S. F., Bork P. BRCA1 protein products. Functional motifs. Nat. Genet. 1996;13:266–268. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- Lee W., Tillo D., Bray N., Morse R. H., Davis R. W., Hughes T. R., Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Lengronne A., McIntyre J., Katou Y., Kanoh Y., Hopfner K. P., Shirahige K., Uhlmann F. Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol. Cell. 2006;23:787–799. doi: 10.1016/j.molcel.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Lopes M., Foiani M., Sogo J. M. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- Luke B., Versini G., Jaquenoud M., Zaidi I. W., Kurz T., Pintard L., Pasero P., Peter M. The cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr. Biol. 2006;16:786–792. doi: 10.1016/j.cub.2006.02.071. [DOI] [PubMed] [Google Scholar]
- Maas N. L., Miller K. M., DeFazio L. G., Toczyski D. P. Cell cycle and checkpoint regulation of histone H3 K56 acetylation by Hst3 and Hst4. Mol. Cell. 2006;23:109–119. doi: 10.1016/j.molcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- McGowan C. H., Russell P. The DNA damage response: sensing and signaling. Curr. Opin. Cell Biol. 2004;16:629–633. doi: 10.1016/j.ceb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Melo J., Toczyski T. A unified view of the DNA-damage checkpoint. Curr. Op. Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- Michaelis C., Ciosk R., Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Miller K. M., Maas N. L., Toczyski D. P. Taking it off: regulation of H3 K56 acetylation by Hst3 and Hst4. Cell Cycle. 2006;5:2561–2565. doi: 10.4161/cc.5.22.3501. [DOI] [PubMed] [Google Scholar]
- Nieduszynski C. A., Hiraga S., Ak P., Benham C. J., Donaldson A. D. OriDB: a DNA replication origin database. Nucleic Acids Res. 2007;35:D40–46. doi: 10.1093/nar/gkl758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn A. J., Elledge S. J., Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 2002;12:509–516. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- Pellicioli A., Lucca C., Liberi G., Marini F., Lopes M., Plevani P., Romano A., Di Fiore P. P., Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht J., et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. USA. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T. M., Kobor M. S., Bastin-Shanower S. A., Ii M., Horte S. A., Gin J. W., Emili A., Rine J., Brill S. J., Brown G. W. Slx4 regulates DNA damage checkpoint-dependent phosphorylation of the BRCT domain protein Rtt107/Esc4. Mol. Biol. Cell. 2006;17:539–548. doi: 10.1091/mbc.E05-08-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J. Esc4p, a new target of Mec1p (ATR), promotes resumption of DNA synthesis after DNA damage. EMBO J. 2004;23:1188–1197. doi: 10.1038/sj.emboj.7600129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y., Desany B. A., Jones W. J., Liu Q., Wang B., Elledge S. J. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- Scholes D. T., Banerjee M., Bowen B., Curcio M. J. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth L., Svejstrup J. Q. Vps75, a new yeast member of the NAP histone chaperone family. J. Biol. Chem. 2007;282:12358–12362. doi: 10.1074/jbc.C700012200. [DOI] [PubMed] [Google Scholar]
- Sheedy D. M., Dimitrova D., Rankin J. K., Bass K. L., Lee K. M., Tapia-Alveal C., Harvey S. H., Murray J. M., O'Connell M. J. Brc1-mediated DNA repair and damage tolerance. Genetics. 2005;171:457–468. doi: 10.1534/genetics.105.044966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Tercero J. A., Diffley J. F. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Tong A. H. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J., De Piccoli G., Cordon-Preciado V., Farmer S., Jarmuz A., Machin F., Pasero P., Lisby M., Haber J. E., Aragon L. Anaphase onset before complete DNA replication with intact checkpoint responses. Science. 2007;315:1411–1415. doi: 10.1126/science.1134025. [DOI] [PubMed] [Google Scholar]
- Tourriere H., Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair. 2007;6:900–913. doi: 10.1016/j.dnarep.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Tsubota T., Berndsen C. E., Erkmann J. A., Smith C. L., Yang L., Freitas M. A., Denu J. M., Kaufman P. D. Histone H3–K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. A., Kiser G. L., Hartwell L. H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wyrick J. J., Aparicio J. G., Chen T., Barnett J. D., Jennings E. G., Young R. A., Bell S. P., Aparicio O. M. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- Xu W., Aparicio J. G., Aparicio O. M., Tavare S. Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics. 2006;7:276. doi: 10.1186/1471-2164-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla D. C., Maharaj A. S., Connelly J. J., Jockusch R. A., Sternglanz R. Rtt107/Esc4 binds silent chromatin and DNA repair proteins using different BRCT motifs. BMC Mol. Biol. 2006;7:40. doi: 10.1186/1471-2199-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Elledge S. J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.