Abstract
We had previously shown that endoplasmic reticulum (ER)-associated degradation (ERAD) of glycoproteins in mammalian cells involves trimming of three to four mannose residues from the N-linked oligosaccharide Man9GlcNAc2. A possible candidate for this activity, ER mannosidase I (ERManI), accelerates the degradation of ERAD substrates when overexpressed. Although in vitro, at low concentrations, ERManI removes only one specific mannose residue, at very high concentrations it can excise up to four α1,2-linked mannose residues. Using small interfering RNA knockdown of ERManI, we show that this enzyme is required for trimming to Man5–6GlcNAc2 and for ERAD in cells in vivo, leading to the accumulation of Man9GlcNAc2 and Glc1Man9GlcNAc2 on a model substrate. Thus, trimming by ERManI to the smaller oligosaccharides would remove the glycoprotein from reglucosylation and calnexin binding cycles. ERManI is strikingly concentrated together with the ERAD substrate in the pericentriolar ER-derived quality control compartment (ERQC) that we had described previously. ERManI knockdown prevents substrate accumulation in the ERQC. We suggest that the ERQC provides a high local concentration of ERManI, and passage through this compartment would allow timing of ERAD, possibly through a cycling mechanism. When newly made glycoproteins cannot fold properly, transport through the ERQC leads to trimming of a critical number of mannose residues, triggering a signal for degradation.
INTRODUCTION
The N-glycosylated glycoproteins comprise a majority of the proteins traversing the secretory pathway in eukaryotic cells. An elaborate quality control mechanism ensures that only correctly folded and processed glycoproteins exit the endoplasmic reticulum (ER) (Trombetta and Parodi, 2003; Sayeed and Ng, 2005). If a defect is found, ER chaperones assist in repair of the glycoproteins and if they fail to correct the defect within a certain time interval, the glycoproteins are targeted for degradation. In recent years, it has become clear that the timing of this interval is linked to trimming of mannose residues from the precursor Glc3Man9GlcNAc2, after the glucose residues have been excised (Hebert et al., 2005; Lederkremer and Glickman, 2005; Moremen and Molinari, 2006). Although generation of the ERAD targeting signal was initially thought to involve the trimming of only one specific mannose residue (mannose b in Figure 1D) to Man8GlcNAc2 isomer b (M8b), recent evidence in mammalian cells implicates further trimming of three to four α1,2-linked mannose residues, to form Man6GlcNAc2 (M6) or Man5GlcNAc2 (M5) (Ermonval et al., 2001; Frenkel et al., 2003; Hosokawa et al., 2003; Kitzmuller et al., 2003; Lederkremer and Glickman, 2005). In contrast to M8b, the M6 and M5 species lack mannose residue a (Figure 1D); therefore, they cannot be reglucosylated by the folding-sensor enzyme UDP-Glc:glycoprotein glucosyltransferase (UGGT), a step that promotes binding of incompletely folded glycoproteins to the chaperones calnexin (CNX) and calreticulin (Parodi, 2000; Moremen and Molinari, 2006). Interestingly, it was recently shown that several lower eukaryotes transfer oligosaccharides with a reduced number of mannose residues, but containing mannose-a, which is also subject to trimming in these organisms (Banerjee et al., 2007). The additional mannose trimming in mammalian cells would remove the glycoproteins from rescue attempts by the ER folding machinery and deliver them to ER-associated degradation (ERAD). ERManI was implicated in this process, because its overexpression leads to increased mannose trimming and accelerated ERAD (Hosokawa et al., 2003; Wu et al., 2003). Nevertheless, it has not been established whether these effects are due to overexpression of ERManI, whether ERManI is indeed required, and whether its endogenous activity accounts for trimming to M5 and M6 in vivo. Trimming to the smaller oligosaccharides might also be mediated by other enzymes. Possible candidates could be Golgi mannosidase IA (Golgi Man IA, also called Man9-mannosidase) that was reported to be either in the Golgi or in the ER, depending on the cell type (Velasco et al., 1993; Bieberich et al., 1997; Igdoura et al., 1999). This enzyme and two other Golgi α1,2 mannosidases were recently shown to accelerate the degradation and mannose trimming of an ERAD substrate, NHK α1-antitrypsin, when overexpressed, probably through recycling of this protein through the Golgi complex (Hosokawa et al., 2007). Other candidates could be ER-degradation enhancing α-mannosidase-like (EDEM) proteins. Recently, both EDEM1 and EDEM3 were reported to accelerate ERAD and to increase mannose trimming to M7-6 when overexpressed in vivo (Hirao et al., 2006; Olivari et al., 2006), even though none of the EDEMs have been shown to have mannosidase activity in vitro (Hosokawa et al., 2001; Mast et al., 2005; Olivari et al., 2005). So far, it has not been shown whether endogenous levels of any of these class I α-mannosidases cause extensive mannose trimming.
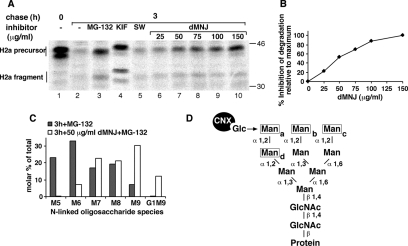
Figure 1.
Involvement of a class I α-mannosidase in ERAD of ASGPR H2a. Partial inhibition of mannosidase activity blocks ERAD while still allowing trimming to M8. (A) NIH 3T3 cells stably expressing H2a (2-18 cell line) were pulse labeled with [35S]Cys for 20 min followed by a 3-h chase in complete medium with or without addition of 20 μM MG-132 or of saturating amounts of the mannosidase inhibitors KIF (100 μM) and SW (4 μg/ml) or of the indicated concentrations of dMNJ. (Cells were preincubated for 30 min with the mannosidase inhibitors, which were also present during the labeling period.) Cells were lysed, H2a was immunoprecipitated, and the immunoprecipitates were separated in 10% SDS-PAGE followed by fluorography. Bands corresponding to the H2a precursor and the naturally occurring cleaved fragment are indicated on the left. Molecular mass markers (kilodaltons) are indicated on the right. The bands below the upper H2a precursor and fragment species are underglycosylated molecules (lacking one of the three sugar chains). (B) Percentage of inhibition of degradation of H2a relative to maximum inhibition obtained with 150 μg/ml dMNJ (=100) was calculated from a phosphorimager quantitation of the gel in A. Note that although reaching a plateau, dMNJ only inhibits degradation of H2a by 50% compared with 80% with KIF. (C) The same cells as in A were labeled with 2-[3H]Man for 1 h in glucose-free medium and chased for 3 h in complete medium in the presence of 20 μM MG-132 with or without 50 μg/ml dMNJ. Cells were lysed, H2a was immunoprecipitated, and the immunoprecipitates were treated with endo H. The N-linked oligosaccharides were separated by HPLC, and fractions were counted in a beta counter. Relative molar amounts of each oligosaccharide species were calculated based on mannose content. The percentage of each species relative to the sum of the molar amounts of all species present was then plotted. (D) Structure of M9, highlighting the α1,2-linked mannose (Man) residues (a–d) (boxed) that undergo trimming in the ER and the position (arrow) of the glucose (Glc) residue on the original precursor (after its two outer glucoses were excised) or after its readdition by UGGT. Note that trimming of mannose-c and/or mannose-b does not prevent reglucosylation but loss of mannose-a does.
In vitro, ERManI cleaves only mannose residue b at low concentrations producing M8 (Tremblay and Herscovics, 1999; Herscovics, 2001). Only very small amounts of M7 are produced after extended, 24-h incubations. However, at very high concentrations it can cleave additional α1,2-linked mannose residues (Herscovics et al., 2002). In the cell, this would require a high local concentration of ERManI. Here, we show by knockdown experiments that ERManI is indeed involved in mannose trimming to M6-5 leading to ERAD. The enzyme is concentrated in a pericentriolar compartment that we had termed the ER-derived quality control compartment (ERQC), which would provide a high local concentration. We had shown that upon proteasomal inhibition this microtubule-dependent membrane-enclosed compartment accumulates ERAD substrates and the chaperones CNX and calreticulin but not BiP, protein disulfide isomerase, UGGT, or ERp57 (Kamhi-Nesher et al., 2001; Frenkel et al., 2004). We proposed that the ERQC is a staging ground for substrate delivery to the ubiquitin–proteasome machinery (Lederkremer and Glickman, 2005). We find that ERManI activity is needed for an ERAD substrate to be released from reglucosylation cycles and to accumulate in the ERQC on its way to ERAD.
MATERIALS AND METHODS
Materials
Rainbow 14C-labeled methylated protein standards were obtained from GE Healthcare (Little Chalfont, Buckinghamshire, United Kingdom). Promix cell labeling mixtures ([35S]Met and Cys, 1000 Ci/mmol) and Man (d-[2-3H(N)], 21 Ci/mmol) were from PerkinElmer Life and Analytical Sciences (Boston, MA). Protein A-Sepharose was from Repligen (Needham, MA). Lactacystin (Lac), N-carbobenzoxyl-leucinyl-leucinyl-leucinal (MG-132), swainsonine (SW), kifunensine (KIF), and deoxymannojirimycin (dMNJ) were from Calbiochem (La Jolla, CA). Leupeptin and other common reagents were from Sigma-Aldrich Corp. (St. Louis, MO). Endo-β-N-acetylglucosaminidase H (endo H) and ProtoScript First Strand cDNA Synthesis kit were obtained from New England Biolabs (Ipswich, MA).
Primers and Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total cell RNA was extracted with EZ-RNA kit (Biological Industries, Beit Haemek, Israel). ReddyMix (ABgene, Epsom, United Kingdom) was used for PCR. Reverse transcription was performed with a ProtoScript First Strand cDNA Synthesis kit using random primers. An aliquot (5%) of the RT product was used for PCR with the following primers: CCTTCAGTGAGTGGTTTGG and GTGGTCCATCTTGGCACTG for ERManI and ATAGTAGATGCCCTGGATAC and CAGATAGTTAGGATAAAGGC for Golgi ManIA.
Plasmids
H2a subcloned in pCDNA1 (Kamhi-Nesher et al., 2001; Invitrogen, Carlsbad, CA). Inserts for pSUPER and pSUPER-retro-green fluorescent protein (GFP) short hairpin RNA (shRNA) vectors were constructed as described in Brummelkamp et al. (2002) by using target sequences as follows: ACCAGCAAATCCACCCGTC for human ERManI, GAATTTAAGCAAGCCAGGA for mouse ERManI, and AGGAGGCCATTCAAGCAGT for human Golgi ManIA. pSUPER encoding anti-lacZ shRNA was a kind gift of S. Lavi (Tel Aviv University). GalT-YFP (β1,3-galactosyltransferase linked to yellow fluorescent protein [YFP]) in peYFP was a kind gift from K. Hirschberg (Tel Aviv University) Human ERManI-hemagglutinin (HA) (Hosokawa et al., 2003) and Golgi ManIC-HA (Tremblay and Herscovics, 2000) were cloned into pMH (Roche Diagnostics, Basel, Switzerland).
Antibodies
Rabbit polyclonal anti-human ERManI was described in Hosokawa et al. (2003). Rabbit polyclonal anti-H2 carboxy-terminal was used as in previous studies (Tolchinsky et al., 1996) as well as rabbit anti-ERp57 (Frenkel et al., 2004). Mouse anti-HA-tag was from Cell Signaling Technology (Danvers, MA); Mouse anti-γ-tubulin, rabbit anti-ERGIC53, and anti-lamp1 were from Sigma-Aldrich Corp. Rabbit anti-CNX C terminus was from Assay Designs (Ann Arbor, MI). Secondary goat anti-mouse immunoglobulin (Ig)G-cyanine (Cy)5 or fluorescein isothiocyanate (FITC), and goat anti-rabbit IgG-Cy5 or -Cy3 antibodies were from The Jackson Laboratory (Bar Harbor, ME).
Cell Culture and Transfection
NIH 3T3 cells and a stable transfectant expressing H2a (Tolchinsky et al., 1996) were grown in DMEM supplemented with 10% newborn calf serum. Transfections of NIH 3T3 cells were performed using the X-tremeGene small interfering RNA (siRNA) transfection reagent (Roche Diagnostics), with 1 μg of DNA per well in 24-well plates, according to the manufacturer's manual. Transfections of human embryonic kidney (HEK) 293 cells (grown in DMEM plus 10% fetal calf serum) were performed using a calcium phosphate procedure. Samples were processed 48-h posttransfection.
[35S]Cys Metabolic Labeling and Immunoprecipitation
Subconfluent (90%) cell monolayers in 60-mm dishes were labeled with [35S]Cys, lysed, and immunoprecipitated with anti-H2 antibodies, as described previously (Tolchinsky et al., 1996; Shenkman et al., 1997).
Gel Electrophoresis, Fluorography, and Quantitation
Reducing SDS-polyacrylamide gel electrophoresis (PAGE) was performed in 10% Laemmli gels. The gels were subjected to fluorography by using 20% 2,5-diphenyloxazole in acetic acid, and then they were exposed to Kodak BioMax MR film (Vancouver, BC, Canada). Quantitation was performed in a Fuji BAS 2000 phosphorimager (Fuji, Tokyo, Japan), by using Tina 2.1 software.
[2-3H]Man Labeling and Analysis of N-linked Oligosaccharides
Subconfluent (90%) monolayers of cells in 100-mm tissue culture dishes were metabolically labeled for 45–60 min with 350 μCi/ml [2-3H]Man, as described previously (Frenkel et al., 2003). Cells were rinsed and chased with normal DMEM plus 10% fetal calf serum. Cell lysis, immunoprecipitation, and endo H treatment were performed as for the 35S-labeled samples. High mannose N-linked oligosaccharide isolation and separation by high-performance liquid chromatography (HPLC) was as described previously (Frenkel et al., 2003) or using an NH2P-50E column from Shodex (Kawasaki, Japan) at a flow rate of 1 ml/min in acetonitrile:water (60:40, vol/vol ratio); fractions were monitored using a scintillation counter (Beckman Coulter, Fullerton, CA).
Immunofluorescence Microscopy
Cells were grown on glass coverslips overnight before transfection; 48 h after transfection the cells were fixed with methanol and methanol:acetone for 5 min at −20°C and processed as in [Kamhi-Nesher et al., 2001]. The samples were analyzed using a Zeiss laser scanning confocal microscope (LSM 510; Carl Zeiss, Jena, Germany). The thickness of the optical slices was 0.6–0.9 μm.
For deconvolution pictures were taken with a Zeiss Axiovert 200M inverted microscope, coupled to a CoolSnap HQ2 camera with a 63×, 1.4 numerical aperture lens, with 2 × 2 binning. The acquisition and analysis were carried out with Slidebook 4.1 software (Intelligent Imaging Innovations, Denver, CO). Twenty to 35 optical slices were taken of each cell (depending on cell shape). Images were deconvolved with the Constrained Iterative algorithm making use of measured point spread functions. Identification of the different intracellular compartments was carried out by intensity-based thresholding. For quantitation, the background signal (calculated from untransfected cells) was subtracted.
RESULTS
A Class I α-Mannosidase and Mannose Trimming to M6, M5 Are Required for ERAD of Asialoglycoprotein Receptor (ASGPR) H2a
Our previous studies, using the uncleaved precursor of ASGPR H2a and unassembled CD3δ as model ERAD substrate glycoproteins, suggested that mannose trimming of the sugar chains to M6 and M5 is involved in their targeting to ERAD (Frenkel et al., 2003). To study which type of mannosidase is involved in this processing, we performed a pulse-chase analysis with [35S]Cys of H2a stably expressed in NIH 3T3 cells, in the absence or presence of inhibitors. In hepatocytes, the membrane-bound precursor of H2a is cleaved (by signal peptidase; Yuk and Lodish, 1993) to a 35-kDa carboxy-terminal fragment, corresponding to its ectodomain, which matures to a soluble secreted form of the receptor, but it is inefficiently cleaved and mostly degraded in other cell types (Tolchinsky et al., 1996). In 3T3 cells H2a was extensively degraded after 3-h chase (Figure 1A). This degradation was inhibited by the proteasomal inhibitor MG-132 and by the mannosidase inhibitors KIF and dMNJ (Figure 1A). In contrast to the proteasomal inhibitor, the mannosidase inhibitors abrogated the shift in migration of the bands. We had shown that the shift in migration is due to mannose trimming (Frenkel et al., 2003). These effects were not observed with the class II mannosidase inhibitor SW (Figure 1A). The results indicate that this mannose trimming is mediated by a class I α-mannosidase.
Given that we had observed a relationship between the degree of mannose trimming to M6 and M5 and the stability of the glycoprotein (Frenkel et al., 2003), it was of interest to test whether inhibition of trimming to M6 and M5 (without affecting trimming to M8) would decrease degradation. For this purpose, we incubated cells expressing H2a with increasing concentrations of dMNJ, which showed a progressive inhibition of the trimming that correlated with a progressive inhibition of degradation (Figure 1, A and B). We analyzed the N-linked sugar chains of H2a, released by endo H after pulse labeling with [2-3H]Man, including MG-132 during the chase to prevent H2a degradation. We studied which species accumulate upon incubation of cells with a low nonsaturating concentration of dMNJ that would have little effect on trimming to M8. Incubation with 50 μg/ml dMNJ completely blocked oligosaccharide trimming to M5, greatly reduced trimming to M6 by more than fourfold but had relatively little inhibitory effect on the formation of M7, and it did not affect the formation of M8 (Figure 1C). Nevertheless, this concentration of dMNJ inhibited the degradation of H2a to half the extent obtained with a saturating amount of the inhibitor (150 μg/ml) after 3-h chase (Figure 1, A and B). Together, these results suggest that trimming of α1,2-mannose residues (Figure 1D) by a class I α1,2-mannosidase to M6 and M5 is a prerequisite for maximal degradation and that formation of M8 cannot be the only determinant leading to ERAD, as has been suggested in previous studies (Cabral et al., 2001).
ERManI Is Required for ERAD of Glycoprotein Substrates and for Mannose Trimming of Their Sugar Chains to M6 and M5
Several class I α1,2-mannosidases could be candidates for the mannose trimming activity leading to ERAD, all inhibited by dMNJ and KIF and insensitive to swainsonine (Herscovics, 2001; Mast and Moremen, 2006) as in the experiment of Figure 1A. In vitro ERManI efficiently converts M9 to M8 by removing residue b (Figure 1D), but it can remove additional α1,2-linked mannose residues at high concentrations (Herscovics et al., 2002). Other candidates, as explained above, could be Golgi α-mannosidases, e.g., Man IA and/or EDEM 1-3.
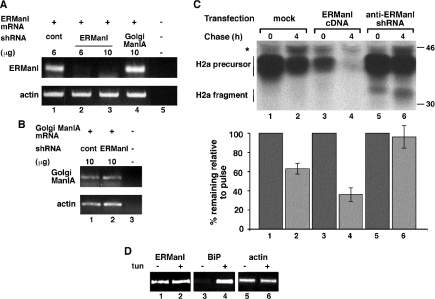
Although overexpression of ERManI leads to accelerated substrate degradation (Hosokawa et al., 2003; Wu et al., 2003), it was important to determine whether endogenous levels of ERManI are sufficient to promote ERAD. We therefore knocked down ERManI expression using an shRNA in a pSUPER vector. This plasmid was cotransfected with a vector carrying H2a cDNA into HEK 293 cells, which were used to achieve much more efficient transfection than in NIH 3T3 cells. We chose a region of the ERManI sequence with little or no homology to other class I mannosidases, obtaining very efficient knockdown of ERManI as analyzed by RT-PCR. Transfection with 10 μg of pSUPER anti-ERManI decreased the ERManI mRNA to undetectable levels, whereas an shRNA directed against Golgi Man IA had no effect (Figure 2A). As another control we determined that anti-ERManI shRNA had no effect on the level of Golgi Man IA mRNA (Figure 2B). The effect of anti-ERManI shRNA on H2a degradation was tested in a pulse-chase experiment. Degradation of H2a is slower in HEK 293 cells than in NIH 3T3 cells, but anti-ERManI shRNA blocked it completely (Figure 2C, lanes 5 and 6). In contrast, overexpression of ERManI accelerated the degradation (Figure 2C, compare lanes 1 and 2 with 3 and 4). ERManI overexpression also caused 40% reduction in the level of the pulse-labeled H2a (Figure 2C, compare lane 1 with lane 3), probably by reducing the initial lag in degradation (Tolchinsky et al., 1996).
Figure 2.
Knockdown of ERManI stabilizes H2a, whereas its overexpression enhances H2a degradation. (A) HEK 293 cells were transfected with the indicated amount of pSUPER plasmid encoding anti-lacZ shRNA (lane 1) or the indicated amounts of the same plasmid encoding anti-ERManI (lanes 2 and 3) or anti-Golgi Man IA shRNA (lane 4). RNA was extracted 48 h posttransfection and used for RT-PCR with primers for ERManI mRNA (25 cycles, top) compared with actin (20 cycles, bottom). No template was present in the control PCR reaction in lane 5. (B) Similar to A but with primers for Golgi Man IA. (C) In parallel with A, HEK 293 cells cotransfected with an H2a cDNA encoding vector together with either empty pMH (mock contransfection, lanes 1 and 2) or with pMH plasmid containing ERManI-HA cDNA (lanes 3 and 4) or with pSUPER encoding anti-ERManI shRNA (lanes 4 and 5) were pulse labeled for 20 min with [35S]cysteine and chased for 0 or 4 h in complete medium. The cells were then lysed, H2a was immunoprecipitated, and immunoprecipitates were separated in 10% SDS-PAGE, which was then subjected to fluorography. The bar graph shows percentage of H2a remaining after chase relative to the pulse, calculated from phosphorimager quantitations of the gels from the average of three independent experiments. (D) RT-PCR as in A but including cell pretreatment for 3 h with and without 10 μg/ml tunicamycin (tun). RT-PCR was also performed with primers for BiP mRNA (25 cycles; middle).
We hypothesized that the strong dependence of ERAD of H2a on the expression levels of ERManI might reflect a physiological situation where ERManI expression is up-regulated during the unfolded protein response. However, this was not the case. There was no change in the mRNA level of ERManI, as measured by RT-PCR, upon cell treatment with tunicamycin (an inducer of the unfolded protein response), compared with a strong up-regulation in the level of BiP mRNA (Figure 2D).
To study the effect of ERManI knockdown on the trimming of the sugar chains, we compared the N-linked glycans of H2a in cells transfected with pSUPER anti-ERManI with those in cells transfected with pSUPER carrying the irrelevant anti-lacZ shRNA. The cells were pulse-labeled with [2-3H]Man and chased for 4 h. ERManI knockdown nearly completely blocked trimming of the sugar chains to M5, and partially to M6 and M8, with the level of M7 remaining almost unchanged after 4 h of chase (Figure 3, A–E). Concomitantly there was a large relative increase of M9 and of the glucosylated species G1M9. Glycoprotein molecules that were spared from degradation by ERManI knockdown accumulated with these larger sugar chains. When comparing the relative molar amounts of the smaller species (M5 + M6) with the sum of the larger species (M8 + M9 + G1M9), it is clear that ERManI knockdown blocked the trimming to M5 and M6 during the chase (Figure 3F). This is even more evident when comparing cells treated with a proteasomal inhibitor, MG-132, which led to a lesser relative amount of larger species and accumulation of the smaller species, but not in the presence of ERManI shRNA (Figure 3F). Although there was a very efficient knockdown of ERManI (Figure 2A), residual activity might still account for partial trimming of up to 3 mannose residues (Figure 3, D and E). Alternatively, other α1,2-mannosidases may be responsible for this partial trimming. However, the partial trimming to M6-M8 in the presence of ERManI shRNA is not sufficient to target the glycoprotein to ERAD (Figure 2C), perhaps pointing to the requirement for further processing to M5. Another possibility is that all three sugar chains of each molecule of the substrate H2a must be trimmed to M5-M6 for ERAD to take place, and the presence of one untrimmed chain might be sufficient for CNX association and prevention of degradation.
Figure 3.
Knockdown of ERManI blocks mannose trimming to M6 and M5 and causes accumulation of M9 and G1M9. (A–D) HEK 293 cells cotransfected with an H2a-encoding vector together with pSUPER encoding anti-lacZ shRNA (A and B) or anti-ERManI shRNA (C and D) were pulse-labeled for 45 min with 2-[3H]Man in glucose-free medium and chased for 0 or 4 h in complete medium. Cells were lysed, H2a was immunoprecipitated and treated with endo-H. The N-linked oligosaccharides were separated by HPLC, and fractions were counted in a beta counter. (E) Relative molar amounts of each oligosaccharide species were calculated and plotted as in Figure 1C. (F) Plot of the molar percentages of M5 plus M6 from each panel in A–D compared with the sums of those of M8, M9, and G1M9. In addition, a similar experiment was done with addition of 20 μM MG-132 during the chase period, and quantified in the same way.
The accumulation of G1M9 upon knockdown of ERManI suggests reglucosylation of M9, because there is much more G1M9 present after chase than after the pulse (compare Figure 3, C and D). Also, in cells treated with dMNJ or KIF there was a large increase in the amount of the glucosylated species (data not shown). This suggests that normally ERManI removes H2a from cycles of deglucosylation by glucosidase II and readdition by UGGT of the glucose residue that binds to CNX (Figure 1D).
ERManI Is Concentrated in the ERQC Compartment
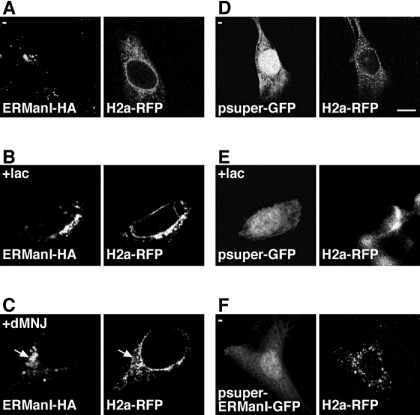
ERManI can cleave several mannose residues from the M9 precursor only at very high concentrations, as shown in vitro (Herscovics et al., 2002). We investigated the subcellular localization of the enzyme to see whether it could point to relatively high local concentrations. We could not detect the enzyme at its endogenous level with the antibody in our possession, although it recognized specifically exogenously expressed ERManI (Supplemental Figure 1). Therefore, we expressed an HA-tagged version of ERManI and compared its location with a red fluorescent protein (RFP) fusion protein of the ERAD substrate H2a (H2a-RFP). H2a-RFP showed up in an ER pattern in untreated cells and concentrated in the ERQC compartment upon proteasomal inhibition (Figure 4, A and B, middle). As mentioned, we previously demonstrated the existence of this compartment in the centrosomal region of the cell (Kamhi-Nesher et al., 2001; Frenkel et al., 2004). Surprisingly, ERManI seemed concentrated in the centrosomal region with or without treatment with the proteasomal inhibitor lactacystin, with relatively little visible in the peripheral regions (Figure 4, A–C). In the presence of lactacystin H2a-RFP colocalized with ERManI in the ERQC (Figure 4B). We had seen previously that H2a and H2a-RFP accumulate in this pericentriolar compartment, together with calnexin and calreticulin upon proteasomal inhibition, but that the chaperones and the ERAD substrate are found throughout the ER in untreated cells (Kamhi-Nesher et al., 2001; Frenkel et al., 2004). Indeed, ERManI colocalized with accumulated CNX only after cell treatment with lactacystin (Figure 4, D and E). To rule out that ERManI localizes in untreated cells to the ERGIC or Golgi (also in the centrosomal region), we performed double labeling of cells with markers of these compartments and found basically no colocalization (Figure 4, F and G). There was no colocalization either with the ER-resident oxidoreductase ERp57 (Figure 4G), which we had shown to be distributed throughout the ER and not to concentrate in the ERQC (Frenkel et al., 2004). The pattern of ERManI is most likely not the result of overexpression, because under the same conditions of expression HA-tagged Golgi mannosidase I C (homologous to ERManI) was located entirely in the Golgi, colocalizing with the Golgi marker β1,3-galactosyltransferase linked to YFP (GalT-YFP) (Figure 4H). ERManI showed no overlap with GalT-YFP (Figure 4G). Furthermore, ERManI occurred in the juxtanuclear pattern under a wide range of expression levels (Supplemental Figure 2).
Figure 4.
ERManI is localized in the ERQC. (A and B) NIH 3T3 cells transiently expressing H2a-RFP and ERManI-HA were incubated for 3 h with or without 10 μM lactacystin (lac), fixed, and subjected to immunofluorescent staining with rabbit anti-ERManI antibodies and Cy5-conjugated goat anti-rabbit IgG. For better visualization, confocal images were pseudocolored red (H2a-RFP) and green (ERManI) in the merged images; colocalization is yellow. (C) ERManI was stained as in A, and centrosomes were stained with mouse anti-γ-tubulin and FITC-conjugated goat anti-mouse IgG (pseudocolored red; arrows). (D and E) Cells transiently expressing ERManI-HA were treated as described in A and B, and they were stained with mouse anti-HA tag and FITC-conjugated goat anti-mouse IgG, along with rabbit anti-CNX followed by Cy3-conjugated goat anti-rabbit IgG. (F) Cells transiently expressing ERManI-HA were stained as described in D and E but using rabbit anti-ERGIC53 instead of rabbit anti-CNX antibody. (G and H) Cells were transiently cotransfected with plasmids encoding the Golgi marker GalT-YFP (green in the merged image) and either ERManI-HA (G) or Golgi Man IC-HA (H). Both mannosidases were stained with mouse anti-HA tag antibody and Cy5-conjugated goat anti-mouse IgG (pseudocolored red). ER-localized ERp57 was visualized with rabbit anti-ERp57 and Cy3-conjugated goat anti-rabbit IgG (pseudocolored blue). (I and J) Cells transiently expressing ERManI-HA were stained with mouse anti-HA tag and FITC-conjugated goat anti-mouse IgG, together with rabbit anti-lamp1 (for visualization of lysosomes), followed by Cy3-conjugated goat anti-rabbit IgG. Cells in J were incubated for 3 h with leupeptin (20 μg/ml) before fixation. (K–M) Three-dimensional rendering of a cell transiently expressing GalT-YFP and ERManI-HA visualized with rabbit anti-ERManI primary and Cy3-conjugated goat anti-rabbit IgG secondary antibodies. Nuclear staining was performed with 4,6-diamidino-2-phenylindole. In this experiment, 35 optical slices of each cell were pictured at intervals of 0.3 μm, the stacks were then subjected to constrained iterative deconvolution using Slidebook 4.1 software. Two different angles are shown (K and M), and in L an overexposure of the image in K see provided to better visualize the faint ERManI staining outside of the ERQC. See also Supplemental Video.
Because it was recently reported that ERManI undergoes degradation in lysosomes (Wu et al., 2007), we investigated whether there is any colocalization of the enzyme with a lysosomal marker. No colocalization of ERManI with the lysosomal protein lysosome-associated membrane protein 1 (lamp1) was found in untreated cells (Figure 4I). However, incubation of cells with leupeptin, to inhibit lysosomal degradation, led to partial accumulation of ERManI in lamp1-positive lysosomes, consistent with the degradation of the enzyme in this organelle (Figure 4J).
We studied the three-dimensional distribution of ERManI compared with GalT-YFP by deconvolution analysis. ERManI showed a juxtanuclear concentration, surrounding the Golgi and with some continuity with the nuclear envelope (Figure 4, K–M, and Supplemental Video). This imaging allowed a quantitative analysis of the localization of ERManI. We calculated that the volume occupied by ERManI in the ERQC is on average ∼6% of the total volume that it occupies in the cell, namely, the ERQC plus the peripheral ER. Nevertheless, this relatively small compartment concentrates almost half of the total amount of ERManI in the cell. Therefore, we estimate that the concentration in the ERQC in NIH 3T3 cells is ∼10-fold higher than that in the peripheral ER (Table 1). The peripheral ERManI is distributed in a large volume, spreading that makes it hardly visible outside of the ERQC. The volume occupied by concentrated ERManI (and consequently by the ERQC) gave an average of 29 μm3 (Table 2). This was compared with the volume of the region occupied by GalT (medial- and trans-Golgi), which gave an average of 13 μm3, the same order of magnitude as that determined by stereological measurement from ultrathin sections by electron microscopy (Mironov and Mironov, 1998). As expected, there was variation in the volume of the ERQC and of the Golgi between cells, but the ratio of the volumes of these compartments in any given cell was surprisingly constant, ERManI occupying a little over double the volume occupied by GalT (2.26 ± 0.14-fold; Table 2). The confinement of ERManI with ERAD substrates in the ERQC could provide the required relatively high concentration for the removal of all α1,2-linked mannose residues from the precursor oligosaccharide.
Table 1.
Relative concentration of ERManI in the ERQC
| Volume occupied by ERManI in the ERQCa (voxels) | Volume occupied by ERManI not in the ERQCb (voxels) | Volume occupied by ERManI in the ERQC relative to that in the rest of the cellc (%) | Relative ERManI amount in the ERQCd (a.u.) | Relative ERManI amount not in the ERQCd (a.u.) | ERManI amount in the ERQC relative to that not in the ERQCe (%) | ERManI conc. in the ERQC relative to that in the rest of the cellf (-fold) | |
|---|---|---|---|---|---|---|---|
| Value | 5716.50 | 78,281.00 | 7.26 | 2,599,862.32 | 3,347,995.87 | 72.03 | 10.63 |
| SD | 1824.29 | 42,639.93 | 2.46 | 1,567,921.28 | 1,591,841.24 | 20.47 | |
| SEM | 644.98 | 15,075.49 | 0.93 | 554,343.88 | 562,800.86 | 7.74 |
a Average number of voxels occupied by ERManI accumulated in the ERQC measured from images similar to those in Figure 4K.
b Average number of voxels occupied by ERManI that is not accumulated in the ERQC (a population that is mostly in the peripheral ER).
c Volume occupied by ERManI in the ERQC relative to the volume that it occupies in other regions (column 2/column 3 × 100).
d Intensity (arbitrary units) of ERManI signal per voxel multiplied by total number of voxels occupied.
e Intensity of ERManI signal in the ERQC relative to the intensity of ERManI in other regions (column 5/column 6 × 100).
f Concentration of ERManI in the ERQC relative to that in other regions. Concentration in each region is defined as intensity of ERManI signal per volume it occupies in that region. (column 5/column 2)/(column 6/column 3).
Table 2.
Volume of the ERQC compared with the Golgi
| ERQC volumea (μm3) | Golgi volumeb (μm3) | Volume ratioc (-fold) | |
|---|---|---|---|
| Value | 29.14 | 12.97 | 2.26 |
| SD | 12.04 | 5.04 | 0.41 |
| SEM | 4.01 | 1.68 | 0.14 |
a Average volume occupied by ERManI accumulated in the ERQC measured from images similar to those in Figure 4I.
b Average volume occupied by GalT-YFP in the Golgi measured from images similar to those in Figure 4I.
c Volume occupied by ERManI in the ERQC divided by that occupied by GalT in the Golgi.
ERManI Activity Is Required for the Accumulation of the ERAD Substrate in the ERQC
We had seen that mannosidase inhibition with dMNJ prevents the accumulation of the ERAD substrate in the ERQC compartment (Frenkel et al., 2003). Incubation of cells transfected with both ERManI and H2a-RFP with dMNJ had only a minor effect on the distribution of ERManI, most remaining concentrated in the ERQC, whereas the ERAD substrate was depleted from this region (Figure 5C, arrows). This is in contrast to the colocalization of the proteins in the ERQC upon treatment with lactacystin (Figure 5B). The pattern of H2a-RFP after treatment with dMNJ is punctate, different from that in untreated cells (compare Figure 5, A and C). To observe cells undergoing ERManI knockdown, we transfected an anti-ERManI shRNA encoding plasmid that also encodes for GFP. Knockdown of ERManI had an effect similar to dMNJ. In the presence of anti-ERManI shRNA (GFP-expressing cells), H2a-RFP did not concentrate in the ERQC, but it was distributed in a punctate pattern (Figure 5F), more discrete than that seen upon treatment with dMNJ (Figure 5C). A control using a similar vector, expressing GFP but not encoding for ERManI shRNA had no deleterious effect on H2a-RFP localization in untreated or in lactacystin-treated cells (Figure 5, D and E). Because knockdown of ERManI prevents degradation of the ERAD substrate, an effect similar to that of the proteasome inhibitor would have been expected, that is concentration of H2a-RFP in the ERQC. Instead, the lack of concentration of H2a-RFP in the centrosomal region suggests that ERManI activity is needed for the localization of the ERAD substrate glycoprotein in the ERQC.
Figure 5.
ERManI activity is required for ERAD substrate concentration in the ERQC. (A–C) NIH 3T3 cells transiently expressing H2a-RFP and ERManI-HA were pretreated without (A), with 10 μM lactacystin (B), or with 100 μM dMNJ (C) for 3 h before fixation and subjected to immunofluorescent staining of ERManI-HA with mouse anti HA-tag antibody and FITC-conjugated goat anti-mouse IgG. (D–F) H2aRFP was transiently coexpressed in NIH 3T3 cells with pSUPER-retro-GFP (expressing GFP from a PGK promoter, D and E) or the same vector encoding anti-ERManI shRNA in addition to GFP (F). Bar, 10 μm.
DISCUSSION
Mounting recent evidence suggests that, in mammalian cells, progressive mannose trimming of all or most α1,2-linked mannose residues from N-linked oligosaccharide precursors targets glycoproteins that are misfolded or otherwise misprocessed to ERAD (Frenkel et al., 2003; Hosokawa et al., 2003; Kitzmuller et al., 2003; Lederkremer and Glickman, 2005; Moremen and Molinari, 2006; Ruddock and Molinari, 2006). An indication that ERManI is involved in this process was obtained because its overexpression leads to concomitant accelerated degradation of ERAD substrates and increased trimming of mannose residues (Hosokawa et al., 2003; Wu et al., 2003). Here, we present additional evidence that trimming to M6 and M5 promotes ERAD and that endogenous levels of ERManI are required to generate the signal for ERAD targeting. Although we cannot rule out that other class I α-mannosidases might also be involved, evidence for their function at endogenous levels is still lacking. The ERManI knockdown experiments clearly indicate that endogenous ERManI can trim the oligosaccharides to species smaller than M8 in vivo, removing more than one and possibly all four α1,2-linked mannose residues (Figure 3). Its action is therefore not restricted to removing only the middle branch terminal mannose-b (see scheme in Figure 1D), as previously thought (Vallee et al., 2000; Cabral et al., 2001; Helenius and Aebi, 2004). Most likely, the additional trimming is accomplished by the high concentration of ERManI in the ERQC (Figure 4 and Table 1). We had previously shown that CNX and calreticulin traffic from the ER to this compartment with the ERAD substrate (Kamhi-Nesher et al., 2001; Frenkel et al., 2004). High concentrations of ERManI are not required for initial trimming to M8 at early stages, before glycoprotein delivery to the ERQC, because there was trimming to M8 on pulse-labeled H2a, produced by residual ERManI activity or by other mannosidases, even after ERManI knockdown (Figure 3C).
Figure 6 shows a working model of the early trafficking and sugar chain processing events for an N-glycosylated protein and that result either in its successful folding and exit to the Golgi or in its targeting to ERAD. These events start in the ER with binding of the newly synthesized glycoprotein to CNX or calreticulin, after removal of two glucose residues by glucosidases I and II. Some mannose residues may be removed in the ER by the relatively low concentration of ERManI. The incompletely folded (or misfolded) glycoprotein is then targeted to the pericentriolar ERQC. Removal of the last glucose residue by glucosidase II may take place just before or upon entry into the ERQC, where the relatively high concentration of ERManI can trim additional mannose residues. The glycoprotein might recycle to the peripheral ER, where it is reglucosylated by UGGT, an enzyme that is not located in the ERQC (Kamhi-Nesher et al., 2001). It can then reassociate with CNX and return to the ERQC for another round of mannose trimming by ERManI. Proper folding releases the glycoprotein from this cycle, because it is no longer a substrate for UGGT and the glycoprotein can then be delivered to the Golgi. However, for a misfolded glycoprotein, mannose-a (Figure 1D) is ultimately trimmed to form M6 and M5, which prevents it from being reglucosylated and from reassociating with CNX; thus, it is marked for degradation. This process would be determined not simply by time of ER residence because when we had analyzed a more stable ERAD substrate (H1i5), it was trimmed more slowly to M6 and M5 (Frenkel et al., 2003) and showed prolonged association with CNX (Shenkman et al., 1997), compared with H2a (Frenkel et al., 2003, 2004). Therefore, the rate of mannose trimming and release from the CNX cycle is somehow linked to the stability of the glycoprotein. While remaining in the CNX cycle, the glycoprotein is spared from ERAD.
Figure 6.
Model illustrating the relation between the trimming of N-linked sugar chains by ERManI, early glycoprotein trafficking events and ERAD in mammalian cells. After cleavage from the precursor oligosaccharide of two glucose residues by glucosidases I and II (GlcI-II), the newly synthesized glycoprotein binds to CNX or calreticulin (CRT). Trimming of mannose residues could occur in the ER lumen but inefficiently, given the low relative concentration of ERManI. The glycoprotein then moves to the ERQC where before or upon entry it is deglucosylated and CNX is released. It is then trimmed by the relatively high concentration of ERManI in the ERQC and recycles back to the peripheral ER where it is reglucosylated by the folding sensor UGGT to reassociate with CNX. These cycles are repeated until proper folding is achieved, and the glycoprotein is no longer a substrate of UGGT and exits to the Golgi or until a critical number of three mannose residues are excised (after which the glycoprotein cannot be reglucosylated), and the glycoprotein is delivered to ERAD, with the participation, through an undetermined mechanism, of EDEM1-3 and possibly of a putative mammalian orthologue of yeast YOS9.
During or after mannose trimming, the glycoprotein ERAD substrate might be recognized by any of the recently identified putative lectins: EDEM 1-3, and a possible orthologue (OS9) of yeast YOS9 (Moremen and Molinari, 2006; Ruddock and Molinari, 2006). Yeast YOS9 has a mannose 6-phosphate homology domain that would bind to precursor N-linked chains (Bhamidipati et al., 2005; Kim et al., 2005; Szathmary et al., 2005), although it is still unknown whether it has a functional mammalian orthologue. The EDEM proteins and YOS9 promote ERAD substrate glycoprotein degradation, but it is still not understood how they function in the process. In mammalian cells, EDEM1 and EDEM3 were shown to have some α1,2-mannosidase activity in vivo (Hirao et al., 2006; Olivari et al., 2006; Olivari and Molinari, 2007), and EDEM1 was also found to act as a chaperone (Hosokawa et al., 2006). The localization of EDEM1 was recently reported to be in vesicular structures, and it is unknown whether these are associated with the ERQC or another ER subcompartment (Zuber et al., 2007). In yeast, YOS9 was found in complex with the E3 ligase HRD1, which would link it to the ubiquitination machinery (Carvalho et al., 2006; Denic et al., 2006; Gauss et al., 2006).
Despite the appeal of the mannose trimming model, one cannot rule out that ERManI might also have another role in routing the glycoprotein to ERAD, possibly as a lectin, as was proposed in a study in Schizosaccharomyces pombe (Movsichoff et al., 2005). This would be consistent with the location of ERManI in the ERQC, where we have found several components of the ERAD machinery, such as Derlin-1, HRD1, and p97/VCP, on the cytosolic side (Kondratyev et al., 2007).
Contrary to what was proposed in a previous model (that mannose trimming leads to persistence of the monoglucosylated oligosaccharides due to decreased glucosidase activity (Cabral et al., 2001), at least in our experimental system ERManI activity decreases reglucosylation (Figure 3), which would promote ERAD by removing the glycoprotein from the CNX cycle.
Knockdown of ERManI dramatically stabilizes the substrate (Figure 2) and prevents its accumulation in the ERQC (Figure 5). This would suggest that ERManI activity is necessary for retention of the substrate in the ERQC and its delivery to the final stages of ERAD. We can speculate that in the absence of ERManI the substrate would rapidly recycle back to the peripheral ER and remain trapped in the CNX cycle, thus avoiding degradation.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Stuart Kornfeld for help and advice in the initial stages of this work. Work was supported by grants from the U.S.-Israel Binational Science Foundation and from the Israel Cancer Association to G.Z.L. and by a grant from the Canadian Institutes of Health Research to A.H.
Abbreviations used:
- dMNJ
1-deoxymannojirimycin
- endo H
endo-β-N-acetylglucosaminidase H
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated degradation
- ERManI
ER mannosidase I
- ERQC
endoplasmic reticulum-derived quality control compartment
- KIF
kifunensine
- lac
lactacystin
- MG-132
N-carbobenzoxyl-leucinyl-leucinyl-leucinal
- UGGT
UDP-Glc:glycoprotein glucosyltransferase.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-05-0505) on November 14, 2007.
REFERENCES
- Banerjee S., Vishwanath P., Cui J., Kelleher D. J., Gilmore R., Robbins P. W., Samuelson J. The evolution of N-glycan-dependent endoplasmic reticulum quality control factors for glycoprotein folding and degradation. Proc. Natl. Acad. Sci. USA. 2007;104:11676–11681. doi: 10.1073/pnas.0704862104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhamidipati A., Denic V., Quan E. M., Weissman J. S. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol. Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Bieberich E., Treml K., Volker C., Rolfs A., Kalz-Fuller B., Bause E. Man9-mannosidase from pig liver is a type-II membrane protein that resides in the endoplasmic reticulum. cDNA cloning and expression of the enzyme in COS 1 cells. Eur. J. Biochem. 1997;246:681–689. doi: 10.1111/j.1432-1033.1997.t01-1-00681.x. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T. R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Cabral C. M., Liu Y., Sifers R. N. Dissecting glycoprotein quality control in the secretory pathway. Trends Biochem. Sci. 2001;26:619–624. doi: 10.1016/s0968-0004(01)01942-9. [DOI] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Denic V., Quan E. M., Weissman J. S. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Ermonval M., Kitzmuller C., Mir A. M., Cacan R., Ivessa N. E. N-glycan structure of a short-lived variant of ribophorin I expressed in the MadIA214 glycosylation-defective cell line reveals the role of a mannosidase that is not ER mannosidase I in the process of glycoprotein degradation. Glycobiology. 2001;11:565–576. doi: 10.1093/glycob/11.7.565. [DOI] [PubMed] [Google Scholar]
- Frenkel Z., Gregory W., Kornfeld S., Lederkremer G. Z. Endoplasmic reticulum-associated degradation of mammalian glycoproteins involves sugar chain trimming to Man6–5GlcNAc2. J. Biol. Chem. 2003;278:34119–34124. doi: 10.1074/jbc.M305929200. [DOI] [PubMed] [Google Scholar]
- Frenkel Z., Shenkman M., Kondratyev M., Lederkremer G. Z. Separate roles and different routing of calnexin and ERp57 in endoplasmic reticulum quality control revealed by interactions with asialoglycoprotein receptor chains. Mol. Biol. Cell. 2004;15:2133–2142. doi: 10.1091/mbc.E03-12-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R., Jarosch E., Sommer T., Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat. Cell Biol. 2006;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- Hebert D. N., Garman S. C., Molinari M. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 2005;15:364–370. doi: 10.1016/j.tcb.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Helenius A., Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Herscovics A. Structure and function of Class I alpha 1,2-mannosidases involved in glycoprotein synthesis and endoplasmic reticulum quality control. Biochimie. 2001;83:757–762. doi: 10.1016/s0300-9084(01)01319-0. [DOI] [PubMed] [Google Scholar]
- Herscovics A., Romero P. A., Tremblay L. O. The specificity of the yeast and human class I ER alpha 1,2-mannosidases involved in ER quality control is not as strict as previously reported. Glycobiology. 2002;12:14G–15G. [PubMed] [Google Scholar]
- Hirao K., et al. EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J. Biol. Chem. 2006;281:9650–9658. doi: 10.1074/jbc.M512191200. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Tremblay L. O., You Z., Herscovics A., Wada I., Nagata K. Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J. Biol. Chem. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., Wada I., Hasegawa K., Yorihuzi T., Tremblay L. O., Herscovics A., Nagata K. A novel ER {alpha}-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2001;2:415–422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Wada I., Natsuka Y., Nagata K. EDEM accelerates ERAD by preventing aberrant dimer formation of misfolded alpha1-antitrypsin. Genes Cells. 2006;11:465–476. doi: 10.1111/j.1365-2443.2006.00957.x. [DOI] [PubMed] [Google Scholar]
- Hosokawa N., You Z., Tremblay L. O., Nagata K., Herscovics A. Stimulation of ERAD of misfolded null Hong Kong alpha1-antitrypsin by Golgi alpha1,2-mannosidases. Biochem. Biophys. Res. Commun. 2007;362:626–632. doi: 10.1016/j.bbrc.2007.08.057. [DOI] [PubMed] [Google Scholar]
- Igdoura S. A., Herscovics A., Lal A., Moremen K. W., Morales C. R., Hermo L. Alpha-mannosidases involved in N-glycan processing show cell specificity and distinct subcompartmentalization within the Golgi apparatus of cells in the testis and epididymis. Eur. J. Cell Biol. 1999;78:441–452. doi: 10.1016/s0171-9335(99)80071-5. [DOI] [PubMed] [Google Scholar]
- Kamhi-Nesher S., Shenkman M., Tolchinsky S., Fromm S. V., Ehrlich R., Lederkremer G. Z. A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell. 2001;12:1711–1723. doi: 10.1091/mbc.12.6.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Spear E. D., Ng D. T. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol. Cell. 2005;19:753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Kitzmuller C., Caprini A., Moore S. E., Frenoy J. P., Schwaiger E., Kellermann O., Ivessa N. E., Ermonval M. Processing of N-linked glycans during endoplasmic-reticulum-associated degradation of a short-lived variant of ribophorin I. Biochem. J. 2003;376:687–696. doi: 10.1042/BJ20030887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratyev M., Avezov E., Shenkman M., Groisman B., Lederkremer G. Z. PERK-dependent compartmentalization of ERAD and unfolded protein response machineries during ER stress. Exp. Cell Res. 2007;313:3395–3407. doi: 10.1016/j.yexcr.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Lederkremer G. Z., Glickman M. H. A window of opportunity: timing protein degradation by trimming of sugars and ubiquitins. Trends Biochem. Sci. 2005;30:297–303. doi: 10.1016/j.tibs.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Mast S. W., Diekman K., Karaveg K., Davis A., Sifers R. N., Moremen K. W. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology. 2005;15:421–436. doi: 10.1093/glycob/cwi014. [DOI] [PubMed] [Google Scholar]
- Mast S. W., Moremen K. W. Family 47 alpha-mannosidases in N-glycan processing. Methods Enzymol. 2006;415:31–46. doi: 10.1016/S0076-6879(06)15003-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov A. A., Jr, Mironov A. A. Estimation of subcellular organelle volume from ultrathin sections through centrioles with a discretized version of the vertical rotator. J. Microsc. 1998;192:29–36. doi: 10.1046/j.1365-2818.1998.00392.x. [DOI] [PubMed] [Google Scholar]
- Moremen K. W., Molinari M. N-linked glycan recognition and processing: the molecular basis of endoplasmic reticulum quality control. Curr. Opin. Struct. Biol. 2006;16:592–599. doi: 10.1016/j.sbi.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movsichoff F., Castro O. A., Parodi A. J. Characterization of Schizosaccharomyces pombe ER alpha-mannosidase: a reevaluation of the role of the enzyme on ER-associated degradation. Mol. Biol. Cell. 2005;16:4714–4724. doi: 10.1091/mbc.E05-03-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivari S., Cali T., Salo K. E., Paganetti P., Ruddock L. W., Molinari M. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem. Biophys. Res. Commun. 2006;349:1278–1284. doi: 10.1016/j.bbrc.2006.08.186. [DOI] [PubMed] [Google Scholar]
- Olivari S., Galli C., Alanen H., Ruddock L., Molinari M. A novel stress-induced EDEM variant regulating endoplasmic reticulum-associated glycoprotein degradation. J. Biol. Chem. 2005;280:2424–2428. doi: 10.1074/jbc.C400534200. [DOI] [PubMed] [Google Scholar]
- Olivari S., Molinari M. Glycoprotein folding and the role of EDEM1, EDEM2 and EDEM3 in degradation of folding-defective glycoproteins. FEBS Lett. 2007;581:3658–3664. doi: 10.1016/j.febslet.2007.04.070. [DOI] [PubMed] [Google Scholar]
- Parodi A. J. Protein glucosylation and its role in protein folding. Annu. Rev. Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- Ruddock L. W., Molinari M. N-glycan processing in ER quality control. J. Cell Sci. 2006;119:4373–4380. doi: 10.1242/jcs.03225. [DOI] [PubMed] [Google Scholar]
- Sayeed A., Ng D. T. Search and destroy: ER quality control and ER-associated protein degradation. Crit. Rev. Biochem. Mol. Biol. 2005;40:75–91. doi: 10.1080/10409230590918685. [DOI] [PubMed] [Google Scholar]
- Shenkman M., Ayalon M., Lederkremer G. Z. Endoplasmic reticulum quality control of asialoglycoprotein receptor H2a involves a determinant for retention and not retrieval. Proc. Natl. Acad. Sci. USA. 1997;94:11363–11368. doi: 10.1073/pnas.94.21.11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmary R., Bielmann R., Nita-Lazar M., Burda P., Jakob C. A. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Tolchinsky S., Yuk M. H., Ayalon M., Lodish H. F., Lederkremer G. Z. Membrane-bound versus secreted forms of human asialoglycoprotein receptor subunits: role of a juxtamembrane pentapeptide. J. Biol. Chem. 1996;271:14496–14503. doi: 10.1074/jbc.271.24.14496. [DOI] [PubMed] [Google Scholar]
- Tremblay L. O., Herscovics A. Cloning and expression of a specific human alpha 1,2-mannosidase that trims Man9GlcNAc2 to Man8GlcNAc2 isomer B during N-glycan biosynthesis. Glycobiology. 1999;9:1073–1078. doi: 10.1093/glycob/9.10.1073. [DOI] [PubMed] [Google Scholar]
- Tremblay L. O., Herscovics A. Characterization of a cDNA encoding a novel human Golgi alpha 1, 2-mannosidase (IC) involved in N-glycan biosynthesis. J. Biol. Chem. 2000;275:31655–31660. doi: 10.1074/jbc.M004935200. [DOI] [PubMed] [Google Scholar]
- Trombetta E. S., Parodi A. J. Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 2003;19:649–676. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- Vallee F., Karaveg K., Herscovics A., Moremen K. W., Howell P. L. Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases. J. Biol. Chem. 2000;275:41287–41298. doi: 10.1074/jbc.M006927200. [DOI] [PubMed] [Google Scholar]
- Velasco A., Hendricks L., Moremen K. W., Tulsiani D. R., Touster O., Farquhar M. G. Cell type-dependent variations in the subcellular distribution of alpha-mannosidase I and II. J. Cell Biol. 1993;122:39–51. doi: 10.1083/jcb.122.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Swulius M. T., Moremen K. W., Sifers R. N. Elucidation of the molecular logic by which misfolded alpha 1-antitrypsin is preferentially selected for degradation. Proc. Natl. Acad. Sci. USA. 2003;100:8229–8234. doi: 10.1073/pnas.1430537100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Termine D. J., Swulius M. T., Moremen K. W., Sifers R. N. Human endoplasmic reticulum mannosidase I is subject to regulated proteolysis. J. Biol. Chem. 2007;282:4841–4849. doi: 10.1074/jbc.M607156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuk M. H., Lodish H. F. Two pathways for the degradation of the H2 Subunit of the asialoglycoprotein receptor in the endoplasmic reticulum. J. Cell Biol. 1993;123:1735–1749. doi: 10.1083/jcb.123.6.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber C., Cormier J. H., Guhl B., Santimaria R., Hebert D. N., Roth J. EDEM1 reveals a quality control vesicular transport pathway out of the endoplasmic reticulum not involving the COPII exit sites. Proc. Natl. Acad. Sci. USA. 2007;104:4407–4412. doi: 10.1073/pnas.0700154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.