Abstract
Mutations in the UNI2 locus in Chlamydomonas reinhardtii result in a “uniflagellar” phenotype in which flagellar assembly occurs preferentially from the older basal body and ultrastructural defects reside in the transition zones. The UNI2 gene encodes a protein of 134 kDa that shares 20.5% homology with a human protein. Immunofluorescence microscopy localized the protein on both basal bodies and probasal bodies. The protein is present as at least two molecular-weight variants that can be converted to a single form with phosphatase treatment. Synthesis of Uni2 protein is induced during cell division cycles; accumulation of the phosphorylated form coincides with assembly of transition zones and flagella at the end of the division cycle. Using the Uni2 protein as a cell cycle marker of basal bodies, we observed migration of basal bodies before flagellar resorption in some cells, indicating that flagellar resorption is not required for mitotic progression. We observed the sequential assembly of new probasal bodies beginning at prophase. The uni2 mutants may be defective in the pathways leading to flagellar assembly and to basal body maturation.
INTRODUCTION
Centrioles are evolutionarily ancient organelles, possibly extant in a common ancestor of all eukaryotic cells (Cavalier-Smith, 2002). They are required for the organization of interphase and mitotic microtubule arrays and as basal bodies they serve as templates for the growth of nine doublet microtubules in axonemes of cilia and flagella (for review see Marshall, 2007). Although flowering plants and more derived fungi have reduced or eliminated centrioles and cilia, metazoans have diversified their functions in numerous sensory and developmental processes (for reviews see Singla and Reiter, 2006; Dawe et al., 2007). Several mammalian genetic disorders have been linked to defects in centrioles or cilia (for review see Afzelius, 2004; Pan et al., 2005). For example, the proteins associated with Bardet-Biedl syndrome, a debilitating pleiotropic human disorder characterized by obesity, renal abnormalities, polydactyly, retinal degeneration, and genitourinary tract malformations have been shown to localize to or around centrioles and cilia (for review see Beales, 2005; Blacque and Leroux, 2006).
Centrioles in actively dividing cells show conservative replication and semiconservative segregation during division (for review see Dawe et al., 2007). As a result, each pair of centrioles consists of an older and a younger centriole that differ in age by at least one cell generation. The “mother” centriole differs both in structure and function from the “daughter” centriole. For example, in the single pair of centrioles found in most types of mammalian cells, the mother centriole nucleates the growth of the primary cilium and contains proteins not present in the daughter centriole. The daughter centriole becomes competent for ciliary assembly only in the ensuing cell cycle (for review see Bornens, 2002). Extension of the pathway for sequential maturation of basal bodies has been revealed in studies of unicellular algae possessing two basal body pairs that nucleate assembly of two flagella that can differ in form and function (anisokont). A basal body that assembles one type of flagellum in one cell cycle will transform to assemble a different flagellum in the following cell cycle, thus prolonging its maturation pathway (for review see Beech et al., 1991). A “mature” basal body was defined by Beech et al. (1991) as one “that will not, in subsequent generations, change its interphase position in the cell… or produce a flagellum and/or roots that differ [from] those already present on that basal body.”
The unicellular green alga Chlamydomonas reinhardtii serves as a valuable genetic model system to better understand the molecular structure and regulation of centrioles (for review see Dutcher, 2003). In interphase cells, centrioles serve as basal bodies to nucleate the assembly of two flagella; in dividing cells, the centrioles are positioned near the poles of the mitotic spindle (Coss, 1974). Despite these important roles, centrioles are nonessential organelles in Chlamydomonas. For example, bld10 mutants lack most of the centriolar structure but the bld10 cells are viable (Matsura et al., 2004). Other mutations identify genes encoding proteins involved in assembly or positioning of basal bodies. The bld2 mutation in the gene encoding ε-tubulin (Dutcher et al., 2002) and the uni3 mutation in the gene encoding δ-tubulin (Dutcher and Trabuco, 1998) reveal the roles of these tubulins in assembly of B tubules and C tubules in triplet microtubules of the basal body wall. Mutations in the VFL1 gene encoding a basal body protein (Silflow et al., 2001) and in the VFL2 gene encoding centrin (Taillon et al., 1992) show the roles of these gene products in basal body positioning and segregation. Genomic and proteomic approaches have provided information on the protein composition of the flagella and basal bodies (Li et al., 2004; Keller et al., 2005; Pazour et al., 2005).
Genetic studies of basal body function in Chlamydomonas are supported by detailed ultrastructural characterization of the basal bodies and associated fibers (Ringo, 1967; Cavalier-Smith, 1974; O'Toole et al., 2003; Geimer and Melkonian, 2004). In interphase Chlamydomonas cells, the basal body apparatus contains two basal bodies that nucleate flagellar growth and two probasal bodies without flagella (Gould, 1975). Through a semiconservative segregation during mitosis, each daughter cell receives one basal body and its daughter basal body, both of which will assemble a flagellum. A marker for identifying the age of the basal body is provided by the eyespot, which has been shown by Holmes and Dutcher (1989) to reside on the same side of the cell as the younger basal body. Although the two flagella in Chlamydomonas appear morphologically identical (isokont), they differ in motility as shown by differential sensitivity to Ca2+ levels (Kamiya and Witman, 1984).
Previous studies in Chlamydomonas have identified mutations that result in a high number of uniflagellate cells (uni1, uni2, and uni3 mutations) in clonal populations. Flagellar assembly in each of these strains occurs preferentially from the older basal body, positioned trans to the eyespot (Huang et al., 1982; Dutcher and Trabuco, 1998). Therefore, uniflagellar mutants in Chlamydomonas provide a genetic means of studying pathways to flagellar assembly and basal body maturation.
In this study we characterize the protein encoded by the Chlamydomonas UNI2 gene. Two mutant alleles result in ultrastructural defects in the transition zone (TZ), a structure positioned just distal to the region where the triplet microtubules of the basal body convert into doublet microtubules of the axoneme microtubules. Using a strain rescued with a gene construct encoding an hemagglutinin (HA)-epitope tagged protein, we determined the Uni2 protein is posttranslationally modified by phosphorylation and is up-regulated in dividing cells. Because of its association with basal bodies and probasal bodies, the Uni2 protein provides a marker for examining the migration of basal bodies and the development of probasal bodies during the cell cycle.
MATERIALS AND METHODS
Strains and Culture Conditions
Mutants 12A10 (uni2-2) and 31E3 (uni2-3) were obtained from a visual screen for motility mutants among transformed cells expressing the NIT1 selectable marker gene (Tam and Lefebvre, 1993). The mutants were back-crossed to wild-type (WT) strains L5 and L8 (Tam and Lefebvre, 1993) and to WT strains CC-124 and CC-125 from the Chlamydomonas Resource Center, University of Minnesota. Strains with the uni2-2 and uni2-3 mutations were deposited to the Center as numbers CC4161 and CC4162, respectively. Phenotypically rescued strain UNI2-3-HA (CC4163) expresses a Uni2 protein tagged with the HA-epitope (see below). Cultures were grown axenically in minimal medium I (Sager and Granick, 1953) or Tris-acetate-phosphate (TAP) medium (Gorman and Levine, 1965), each supplemented with 0.005% arginine as required. Cultures were maintained at 24°C by bubbling continuously with filtered air and were illuminated by fluorescent white light at ∼60 μmol photons/m2/s1. For experiments with cells in G1 phase, cultures were grown on a 14:10-h light:dark cycle. For experiments with dividing cells, cultures were synchronized on a 12:12-h light:dark cycle in TAP medium at 24°C for at least 5 d, maintaining the cell density between 105 and 106 cells/ml by serial dilution as required. For long-term culture storage and mating, cultures were maintained on solid agar medium. Tetrad analysis was performed at 24°C using standard techniques (Levine and Ebersold, 1960). Diploid strains were constructed using the arg2 and arg7 mutations as described by James et al. (1988).
Quantitation of Flagellar Number, Percent Division, and cis-trans Eyespot Analysis
For conducting flagellar counts or scoring the percent of cells in division, cells were fixed in culture medium containing 2% glutaraldehyde. A compound microscope (Leica Microsystems Inc., Bannockburn, IL) equipped with differential interference contrast (DIC) optics was used to count the number of flagella per cell or to score for cell division. Cells were prepared for cis-trans analysis of the eyespot using a rapid fixation method as described in Mitchell (2003). For visualizing the eyespot, microscope settings were altered such that the DIC analyzer was used with the brightfield condenser. This combination enhanced the combined view of flagella and eyespot. Deflagellation was induced using pH shock as described by Lefebvre et al. (1978).
Cloning and Tagging the UNI2 Gene
The 12A10 and 31E3 strains were obtained from an insertional mutagenesis screen using plasmid pMN24 containing the NIT1 gene as a selectable marker (Tam and Lefebvre, 1993). Genomic DNA blot analysis with the 12A10 strain showed that one complete and one partial copy of the plasmid integrated in tandem into the genome, causing at least a 10-kb deletion at one of the integration junctions. A size-specific partial genomic library was constructed and the genomic fragment adjacent to one integration junction was cloned. When used as a hybridization probe on a blot of DNA from WT and mutant strains, the junction fragment identified restriction fragment polymorphisms in DNA from strains 12A10 and 31E3. The junction fragment was used to screen a lambda phage library of WT Chlamydomonas DNA to obtain nine overlapping clones covering a region of 29 kb. A 9.7-kb SalI fragment of genomic DNA containing the UNI2 gene was cloned into the plasmid vector pBlueScript II KS− (Stratagene, La Jolla, CA) to obtain the plasmid pML9.7. The DNA was sequenced on both strands and the DNA sequences were assembled using Genetics Computer Group software (Devereux et al., 1984). Because no expressed sequence tag (EST) support for the gene was available, cDNA sequences were identified by using reverse transcriptase PCR amplification of UNI2 RNA as described by Silflow et al. (2001). The 5′ end of the RNA transcript extends at least 272 base pairs upstream of the putative start codon. A putative polyadenylation signal (TGTAA) for Chlamydomonas (Silflow et al., 1985) is located 207 base pairs downstream of the TAG stop codon. The UNI2 gene sequence is available at GenBank under the accession number EF675992. The pML9.7 plasmid was modified by inserting a sequence encoding three copies of the HA epitope (Silflow et al., 2001) into a BamHI site located 13 codons upstream of the stop codon to create the pML9.7–3xHA plasmid. Isolation of nucleic acids from Chlamydomonas and RNA blotting were carried out as described by Silflow et al. (2001). Phenotypic rescue of the mutant strains was accomplished using the glass bead transformation with the pARG7.8 plasmid (Debuchy et al., 1989) together with lambda phage or linearized plasmid DNA as described by Silflow et al. (2001). Both pML9.7 and pML9.7–3xHA plasmids were linearized with SalI.
Immunoblotting and Densitometry
Immunoblotting was performed as described by Silflow et al. (2001). Protein extracts from ∼3 × 106 cells were loaded per lane on SDS-PAGE minigels. The HA-tagged protein was detected with a rat anti-HA (3F10) high-affinity antibody (Roche Molecular Biochemical, Indianapolis, IN) at a 1:1200 dilution. The primary antibody was detected using a secondary goat anti-rat IgG-peroxidase (Sigma Aldrich, St. Louis, MO) at a 1:10,000 dilution. For loading control a mouse anti-β-tubulin (2–10-B6) mAb (a gift from Dr. G. Piperno, Mount Sinai School of Medicine, New York, NY) was used at a 1:300 dilution. To detect the primary antibody, a goat anti-mouse IgG-peroxidase (Sigma Aldrich) was used at 1:25,000 dilution. Densitometry of the scanned HA-tagged protein blot was performed using Image J (U.S. National Institutes of Health, Bethesda, MD) by measuring the integrated density of a whole gel band and subtracting out the background blot intensity.
Protein Dephosphorylation
A pellet of 5 × 107 cells from a strain expressing the HA-tagged Uni2 protein was suspended in 100 μl of a solution containing 1× calf alkaline intestinal phosphatase (CIP) buffer (Roche Molecular Biochemical, Indianapolis, IN; no. 1 097 075), 3× protease inhibitors (Sigma Aldrich; P8340) and 10% SDS (Sigma Aldrich). The lysate was incubated at 37°C for 5 min and diluted by the addition of 300 μl of 1× CIP buffer and 1× protease inhibitors. The lysate was aliquoted into four tubes. The first tube received 15 μl 1× CIP buffer. The second tube received 10 μl CIP inhibitors (50 mM NaF, 25 mM β-glycerol phosphate and 100 μM sodium orthovanadate) and 5 μl of 1× CIP buffer. The third tube received 10 μl CIP inhibitors, 4 μl (80 U) CIP, and 1 μl of 1× CIP buffer. The fourth tube received 11 μl CIP buffer and 4 μl (80 U) CIP. All tubes were incubated at 37°C for 30 min. An equal volume of SDS sample buffer was added and the samples were boiled.
Indirect Immunofluorescence Labeling
Indirect immunofluorescence in whole cells and nucleoflagellar apparatuses (NFAps) was conducted as described by Silflow et al. (2001) based on methods described in Sanders and Salisbury (1995) and Wright et al. (1985) with the following modifications. The rat anti-HA (3F10) high-affinity antibody (Roche Molecular Biochemical) was used at a 1:200 dilution and detected using a 488-conjugated goat anti-rat secondary antibody (Sigma Aldrich) at a 1:400 dilution. Tubulin was detected using a Chlamydomonas-specific rabbit anti-α-tubulin antibody (Silflow and Rosenbaum, 1981) at a 1:1000 dilution and using a Texas red (TR)-conjugated goat anti-rabbit secondary antibody at a 1:400 dilution (ICN Pharmaceuticals, Aurora, OH). The rabbit anti-centrin (MC1) antibody (a gift from Dr. J. L. Salisbury, Mayo Clinic, Rochester, MN) was used at a 1:200 dilution and detected using a TR-conjugated rat anti-rabbit secondary antibody at a 1:400 dilution (ICN Pharmaceuticals). DNA was stained with DAPI at 0.05 mg/ml during incubation with secondary antibodies.
Preparations were viewed using a Nikon Eclipse E800 photomicroscope (Melville, NY) equipped with DIC and fluorescence optics including a 100 W mercury lamp epi-fluorescence illumination with UV (excitation filter 330–380 nm, barrier 420 nm), blue (excitation filter 470–490 nm, barrier 520–580 nm) and green (excitation filter 510–560 nm, barrier 570–620 nm) filter sets. The samples were viewed using a 60×, 1.40 NA plan apo lens with a 1.5× optivar. Digital images, 16-bit, were collected using a Roper CoolSnap HQ monochrome camera (Tucson, AZ) and captured to a Pentium IV 2.6-GHz personal computer using Image ProPlus AMS version 5.1 software (Media Cybernetics, Silver Springs, MD). Z-motor movements, filters, and shutters were managed using a Ludl MAC 3000 controller (Ludl Electronic Products, Hawthorne, NY) interfaced to the ImagePro software.
Digital images were obtained and image stacks collected from each individual illumination wavelength were deconvoluted using a Huygens Maximum Likelihood estimation algorithm in batch on computers at the Minnesota Supercomputing Institute (University of Minnesota). Using Image J, the brightness and contrast from the image stacks at each individual wavelength was adjusted and color channels were assigned as the images were combined. The focal planes containing the desired cellular features were projected using maximum intensity in the Z-dimension. The projected images from each wavelength and the combined images were then imported into Adobe Photoshop (Adobe Systems, San Jose, CA) and combined into a single panel. Individual RGB channel levels were adjusted and scale bars were added. Basal body regions were enlarged for cutout panels by enlarging the selected region four times using bicubic smoother. Images were moved into Adobe Illustrator (Adobe Systems) for labeling and sizing adjustments.
Ultrastructural Analysis
Cells in G1 phase of the cell cycle were prepared for electron microscopy (EM) according to Porter et al. (1999). Ultrathin sections were cut and stained with 1% uranyl acetate in 50% methanol for 20 min at room temperature, followed by staining with lead citrate for 5 min. Sections were examined using a transmission electron microscope (CM-12; Philips, Mahwah, NJ) at 60 kV.
Pre-embedding immunogold EM of NFAps was performed as described by Silflow et al. (2001). Ultrathin sections were imaged at (60,000×) using a transmission electron microscope (TEM; CM-12; Philips) at 60 kV. The gold particles were quantified on the entire image of 21 negatives, each containing a sectioned NFAp.
RESULTS
New Uni-flagellar (uni) Mutant Strains Identify a Novel Gene
Two independent mutant strains with a uniflagellar phenotype were obtained in an insertional mutagenesis screen (Tam and Lefebvre, 1993). Characterization of strains 12A10 and 31E3 showed that most of the cells in a population are aflagellate (>50%), a high percentage of cells assemble a single flagellum (>35%), and only a few cells assemble two (6–7%) or more (<1%) flagella (Table 1). The motility phenotypes of the mutant strains cosegregated with the selectable marker gene on the plasmid used for insertional mutagenesis, suggesting that the defects were caused by plasmid DNA insertion (Tam and Lefebvre, 1993; unpublished data).
Table 1.
Distribution of flagella in uniflagellar strains
| Straina | Flagellar number |
|||
|---|---|---|---|---|
| 0 | 1 | 2 | >2 | |
| % | % | % | % | |
| WT, 21gr | 2 | 2 | 96 | 0 |
| 12A10, uni2-2 | 52 | 40 | 7 | 1 |
| 31E3, uni2-3 | 55 | 38 | 6 | 1 |
| uni2-3 R −HA | 8 | 4 | 89 | 0 |
| uni2-3 R +HA a | 8 | 3 | 89 | 0 |
| uni2-3 R +HA b | 3 | 3 | 95 | 0 |
| uni2-3 R +HA c | 10 | 4 | 86 | 0 |
a n > 400 cells.
Mutant strains 12A10 and 31E3 had disruptions in the same region of the genome. Blotting experiments using a probe to a genomic DNA fragment flanking the plasmid insertion in strain 12A10 showed that the DNA from both strains had restriction fragment polymorphisms when compared with WT DNA. The probe was used to screen a lambda phage library to obtain WT DNA clones from the genomic region (Figure 1A). To identify the smallest genomic region required for rescue of the mutant phenotype, cloned WT DNA fragments were transformed into 31E3 mutant cells. The mutation in this strain was shown to be recessive to WT in heterozygous diploid strains (unpublished data). Some DNA fragments were able to rescue the uniflagellar phenotype to produce WT swimming cells (Figure 1 and Table 1). Interestingly, the same clones conferred only a partial rescue of the phenotype of strain 12A10, with 80% of transformed cells becoming biflagellate. The smallest genomic DNA fragment able to rescue the phenotype of 31E3 cells was a 9.7-kb subclone. It was mapped to Linkage group IX within 3 centiMorgans (cM) of the PF16 locus (Kathir et al., 2003). This map location and the mutant phenotype were in common with the uni2-1 mutation described by Dutcher and Trabuco (1998). We designated the mutant alleles in the 12A10 and 31E3 strains as uni2-2 and uni2-3, respectively.
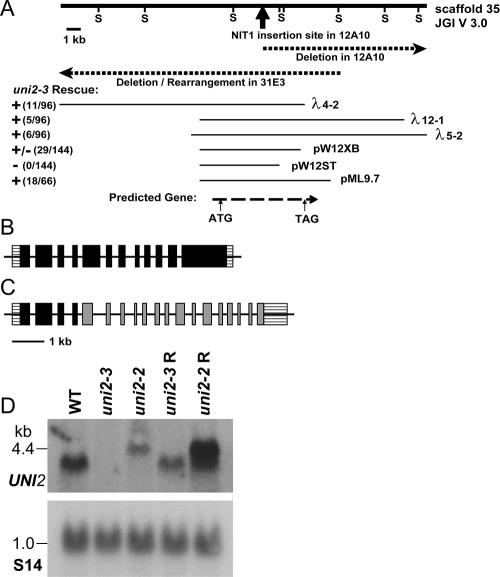
Figure 1.
UNI2 genomic region and transcripts. (A) The heavy solid line depicts a 25-kb genomic segment. Sac1 sites (S) were used in the mapping of genomic DNA from the 12A10 and 31E3 strains. The dark arrow indicates the site of insertion of the pMN24 plasmid DNA in the 12A10 strain. Dashed lines show the genomic region disrupted in the 12A10 and 31E3 strains. Thin lines below show cloned genomic fragments capable (+) and incapable (−) of rescuing the phenotype of the 31E3 strain. The bottom dashed line indicates the location of the coding region of the UNI2 gene as identified by RT-PCR experiments. (B) Schematic showing the structure of the WT UNI2 gene with 12 exons. (C) Schematic showing uni2-2 lesion in which the fourth intron of the UNI2 gene (black) is fused to the second intron of the NIT1 gene (gray). (D) Autoradiographs of RNA blots. Polyadenylated RNA (10 μg/lane) from the indicated cells was electrophoresed on a denaturing gel and transferred to a membrane. (top) The RNA was hybridized with a labeled probe consisting of the second exon of the UNI2 gene. (bottom) A labeled fragment of the RPS14 gene encoding the ribosomal protein S14 was used as a hybridization control for equal loading (Nelson et al., 1994).
Using reverse transcriptase PCR, we determined that the UNI2 gene consists of 12 exons (Figure 1B) that when spliced together result in one large open reading frame of 1331 codons. Lesions in the two uni2 alleles were examined using DNA blotting and sequencing. The uni2-2 allele contains one complete and one partial copy of the NIT1 selectable marker gene and at least a 10-kb deletion at one of the integration junctions. DNA sequencing of the cloned junction fragment revealed that the uni2-2 mutation results in a fusion gene containing the first four exons of the UNI2 gene fused to the NIT1 gene lacking its first exon. The putative fusion protein might account for the inability to obtain full rescue of the phenotype for the uni2-2 strain. DNA blot experiments showed that the uni2-3 allele consists of a deletion overlapping with the uni2-2 deletion in a region of ∼6 kb, but extending in the opposite direction (Figure 1A).
Expression of the UNI2 transcript was examined in RNA isolated from WT, mutant, and phenotypically rescued strains (Figure 1D). A probe containing DNA from exon 2 of the UNI2 gene hybridized with a 4-kb transcript in RNA from WT cells. This transcript was missing in RNA from uni2-3 cells, suggesting this is a null allele, but was present in cells rescued with the 9.7-kb genomic fragment. In RNA from uni2-2 cells, the probe hybridized to a larger transcript of ∼5 kb, presumably representing the transcript of the fusion gene. In rescued uni2-2 cells, the probe hybridized to a transcript of WT length in addition to the larger transcript.
The Uni2 Protein Has Potential Homologues in Vertebrate Genomes
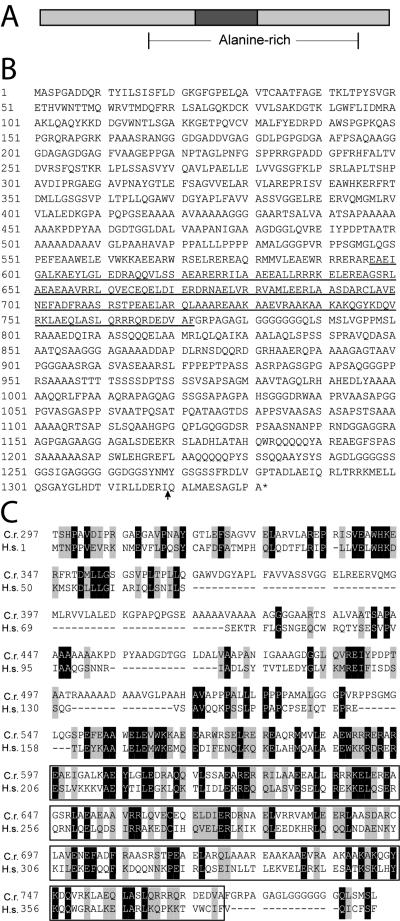
The UNI2 gene encodes a novel alanine-rich (22% overall) protein that has a predicted molecular mass of 134.3 kDa with an isoelectric point of 6.19 (Figure 2, A and B). It is predicted to contain a large central coiled-coil domain. A human protein predicted from genome sequence (FLJ36090) has 20.5% homology and 38.2% similarity with the Uni2 protein over a 497-amino acid region (Figure 2C). The human protein is the smallest of three predicted splicing variants, each of which contains this region of homology. Support for this splice variant is found in an EST from testis (Ota et al., 2004). The human gene is located at 5q23.2 (Venter et al., 2001). The region of homology includes sequence upstream of and within the coiled-coil domain and is conserved in other invertebrate and vertebrate organisms including purple sea urchin, rat, and chicken.
Figure 2.
(A) Schematic of the predicted Uni2 protein containing a central coiled-coil domain (dark box) and alanine-rich region (28% between brackets). (B) Deduced amino acid sequence of the Uni2 protein. The predicted coiled-coil domain is underlined; site of the inserted triple HA-epitope is indicated by an arrow. (C) Alignment of part of the Uni2 amino acid sequence (top) with the full-length predicted Homo sapiens sequence FLJ36090 (bottom) that shows 20.5% identity and 38.2% similarity within this region of Uni2. A box marks the predicted coiled-coil region.
Flagellar Assembly in uni2 Mutants
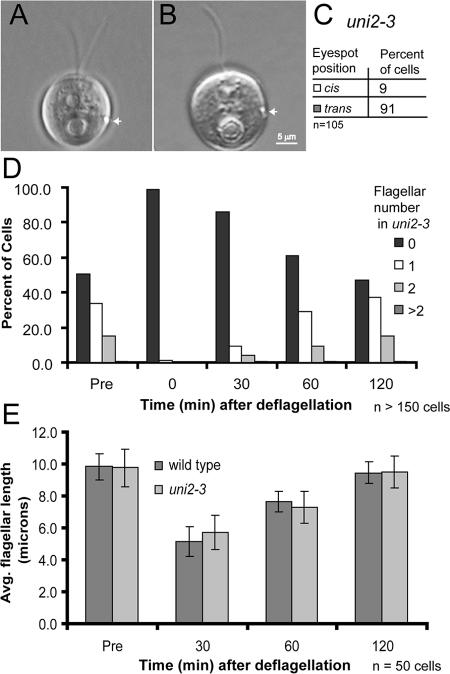
In uni mutants previously described, flagellar assembly occurs preferentially from the older of the two basal bodies, located trans to the eyespot (Huang et al., 1982; Dutcher and Trabuco 1998). We performed a cis-trans analysis of the flagellum relative to the eyespot in uniflagellate cells of the uni2-3 mutant. The majority (91%) of the flagella were positioned trans to the eyespot, indicating the chronologically older basal body is used preferentially to nucleate flagellar growth (Figure 3, A–C).
Figure 3.
Flagellar assembly in uni2-3 cells. (A and B) DIC images of the eyespot position (white arrow) in a biflagellate cell and a uniflagellate cell with the flagellum trans to the eyespot. (C) Number of uniflagellate cells in a population of uni2-3 cells that contain a flagellum cis or trans to the eyespot. (D and E) Flagellar number and length before deflagellation and during flagellar regeneration of uni2-3 cells. For comparison, the flagellar length in WT 21gr cells was determined. Both cultures were deflagellated by pH shock, and cells were allowed to regenerate flagella. Cells were fixed at 0, 30, 60, and 120 min after deflagellation. (D) The percent of uni2-3 cells in different flagellar number classes was determined at each time point. (E) The average flagellar length was determined in flagellated cells of both WT and mutant populations.
To determine whether the uni2 mutation affects the kinetics of flagellar assembly, we compared the flagella of uni2 cells before and after deflagellation. The distribution of flagellar numbers in a population of predeflagellated cells persists after flagellar excision and regeneration (Figure 3D). The rate of flagellar regeneration in mutant cells is indistinguishable from that of WT cells (Figure 3E). We conclude that regeneration probably occurs from the same basal body that was previously flagellated and that the uni2 mutation has no noticeable affect on the rate of flagellar assembly.
Structure of the TZ Is Abnormal in uni2 Mutants
We examined the basal bodies and associated structures of uni2 mutant cells using thin-section TEM. Previous analyses of uni1 cells showed defects in the TZ (Huang et al., 1982), whereas uni3 cells showed defects in the formation of triplet microtubules in basal bodies (Dutcher and Trabuco, 1998). Cross-sections of basal bodies and probasal bodies in uni2-2 and uni2-3 cells revealed normal triplet microtubules (unpublished data). The great majority of basal bodies appeared to be properly “docked,” with the plasma membrane at the surface of the cell and not misplaced within the cytoplasm. Rare longitudinal sections including both basal bodies as seen for a WT cell (Figure 4A) and for a uni2-2 mutant cell (Figure 4B) showed that fibrous components of the basal body apparatus such as the distal striated fiber linking the two basal bodies are present in mutant cells.
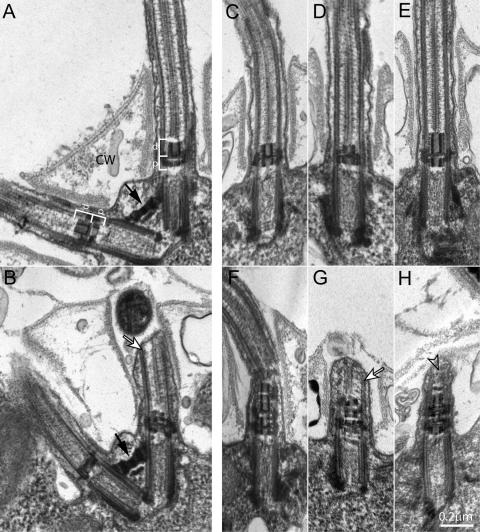
Figure 4.
Ultrastructural analysis of WT and mutant cells reveals TZ defects in uni2 mutants. Thin sections of WT or mutant strains were viewed using TEM. (A and C) WT cells have uniform TZs composed of proximal (p) and distal (d) electron dense cylinders forming an H-shaped region in longitudinal section (white brackets). The flagella project through openings in the cell wall (CW). (B) Mutant uni2-2 cells have the normal arrangement of two basal bodies connected by a distal striated fiber (black arrow) but most basal bodies have a (D) truncated and/or (B) disrupted distal TZ cylinder. (E–H) Mutant uni2-3 cells have (E) elongated distal TZ cylinders or (F–H) extra stacked TZ material. Basal bodies unable to assemble a complete flagellum sometimes show a flagellar stump with some axonemal microtubules (white arrows, B and G). In some cases, the tip of the stump is filled with amorphous material (open arrow, H).
The most consistent defect we observed in uni2 mutant cells was in the TZ, a region just distal to the basal body proper that contains two prototypical electron-dense cylinders. The distal cylinder observed in a WT TZ is slightly longer than the proximal cylinder (Figure 4, A and C). In TZs from uni2-2 mutant cells the distal cylinders are reduced in length and/or distorted (Figure 4, B and D). The basal body on the left in Figure 4B appears to assemble a flagellum, whereas the basal body on the right nucleates a flagellar stump less than 1 μm in length, which does not project through the opening of the “collar” in the cell wall. Sections from uni2-2 mutant cells showed that 94% (n = 37) of TZs were abnormal, with defects ranging from a shortened distal cylinder (Figure 4D) to disrupted TZ material (Figure 4B). A different defect in the TZ was observed consistently in the uni2-3 cells, where 78% (n = 56) of TZs were abnormal, with defects ranging from an elongated distal cylinder (Figure 4E) to disrupted and stacked TZ material (Figure 4, F–H). In favorable sections, it was possible to correlate the morphology of the TZ with the presence of a flagellum. The abnormal TZs did not prevent flagellar assembly (Figure 4, D–F). In other cases, a defective TZ was associated with a flagellar stub containing short microtubules (Figure 4, B and G) or amorphous material (Figure 4H). In uni2-3 mutants, 21% (n = 33) of defective TZs assembled a flagellum, whereas 64% (n = 11) of apparently normal TZs assembled a flagellum. Axonemal cross-sections from uni2-2 and uni2-3 mutants revealed a normal arrangement of outer doublet and central pair microtubules (data not shown).
The Uni2 Protein Is Modified by Phosphorylation
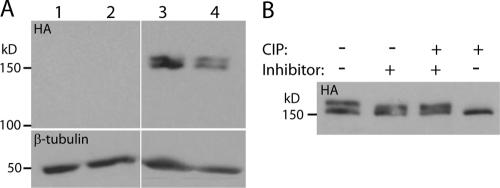
To determine the localization of the Uni2 protein in cells, an epitope-tagged protein was expressed in cells. The WT UNI2 gene was modified by the addition of a sequence encoding three copies of the HA epitope (Silflow et al., 2001). Transformation with the tagged gene construct rescued the flagellar number phenotype of the uni2-3 mutant (Table 1). Protein extracts from WT and mutant cells rescued with the tagged or untagged gene construct were analyzed by SDS-PAGE and immunoblotting. Antibodies against the HA epitope identified at least two distinct molecular-weight variants in multiple independent transformants (Figure 5A; top, lanes 3 and 4). The Uni2 protein runs at ∼155 kDa, which is slightly larger than the 139-kDa expected size of the epitope-tagged protein. Protein extracts from cells that did not contain the epitope-tagged gene produced no signal on the immunoblot (Figure 5A; top, lanes 1 and 2).
Figure 5.
Immunoblot analysis of HA-tagged Uni2 protein and phosphatase-treated Uni2 protein. Proteins were separated by SDS-PAGE and transferred to a PVDF membrane. A high-affinity anti-HA antibody was used to identify the HA-tagged Uni2 protein. (A) Protein extracts were lane 1, WT; 2, uni2-3; and 3–4, two independent uni2-3 strains phenotypically rescued with the HA-tagged UNI2 gene. Top, a high-affinity anti-HA antibody was used to identify the HA-tagged Uni2 protein; bottom, an antibody against β-tubulin was used as a loading control. (B) Protein from an HA-tagged transformant after a 30-min treatment at 37°C with CIP buffer and the indicated reagents. The blot was probed with the anti-HA antibody.
Uni2 protein is present in at least two forms in whole cell protein extracts, suggesting that the protein might undergo posttranslational modification. Analysis of Uni2 protein with NetPhos 2.0 (Blom et al., 1999) predicted numerous sites with greater than 95% chance of being phosphorylated including 21 serine, one threonine, and one tyrosine residues. To determine whether the higher molecular-weight band of the Uni2 protein is due to phosphorylation, protein extracts from whole cells were treated with CIP (Figure 5B). The results show that Uni2 protein in untreated extract, in mock-treated extract with phosphatase inhibitors, or in mock-treated extract with phosphatase inhibitors and CIP migrates as a doublet. In extracts incubated with active CIP the HA-tagged protein migrates as a single isoform, suggesting that the higher molecular-weight isoform is converted to the lower molecular-weight isoform by CIP.
To determine whether the two Uni2 protein isoforms are located in cell bodies or in flagella, we analyzed the Uni2 protein from nondeflagellated cells and from cells <5 min after deflagellation. No noticeable changes in the quantity or ratio of the two major Uni2 protein isoforms were observed (unpublished data), suggesting that the majority of the Uni2 protein resides proximal to the flagellar deflagellation point, which is at the distal end of the TZ (Sanders and Salisbury, 1989).
The Uni2 Protein Localizes to Basal Bodies and Probasal Bodies
The cellular localization of the Uni2 protein was examined using immunofluorescence microscopy. Cells expressing the HA-tagged protein were doubly stained for Uni2 protein and for α-tubulin, which provides a useful whole cell marker. A Z-series of images from fixed and labeled whole cells was collected, deconvoluted, and projected to show a single image. In a cluster of C. reinhardtii cells all viewed from above the flagella, α-tubulin antibodies stain the flagella, basal bodies, and cytoplasmic microtubules (Figure 6, A and C). Localization of the Uni2 protein was observed consistently at four distinct spots (green) at the base of the flagella (red; Figure 6, A and B). Nuclear DNA and chloroplastic DNA stained with DAPI (blue) were localized within the cell body (Figure 6, A and D). The localization of Uni2 protein coincides with the expected location of basal bodies and probasal bodies. Side views of similarly stained cells sometimes showed three or four spots, depending on the angle of observation (see Figure 8, A and C). A control using cells labeled in the absence of the primary antibodies showed no significant staining (unpublished data).
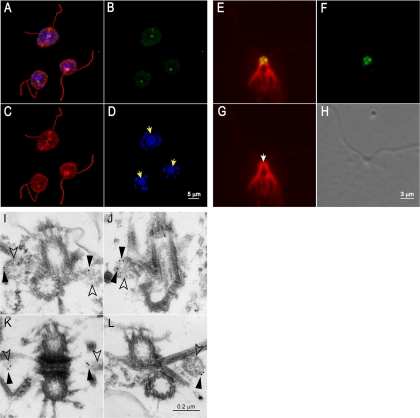
Figure 6.
Indirect immunofluorescence and immunoelectron microscopy of the Uni2 protein. (A–D) Immunofluorescence of fixed whole cells stained with rat anti-HA and rabbit anti-α-tubulin antibodies. Secondary anti-rat 488 (green) and anti-rabbit TR (red) antibodies were used to detect primary antibodies. DNA was localized with DAPI (blue). (A) Staining of Uni2 protein, α-tubulin and DAPI; (B) Uni2 protein; (C) α-tubulin; (D) DNA in nuclei and chloroplasts with yellow arrows marking the nuclei. (E–G) Fixed NFAps were stained with rat anti-HA and rabbit anti-centrin antibodies, followed by secondary anti-rat 488 (green) and anti-rabbit TR (red) antibodies. A deconvoluted Z-series of images are shown. (E) Staining of Uni2 protein and centrin; (F) Uni2 protein; (G) centrin with the white arrow marking the distal striated fiber; and (H) DIC image of the NFAp. (I–L) Immunogold labeling of NFAps stained with rat anti-HA antibodies, followed by secondary goat anti-rat antibody conjugated with 12-nm gold particles. The two basal bodies are shown at the top and bottom of each image; the two probasal bodies (open arrowheads) lie to the left and right of the basal bodies. Gold particles are indicated by filled arrowheads.
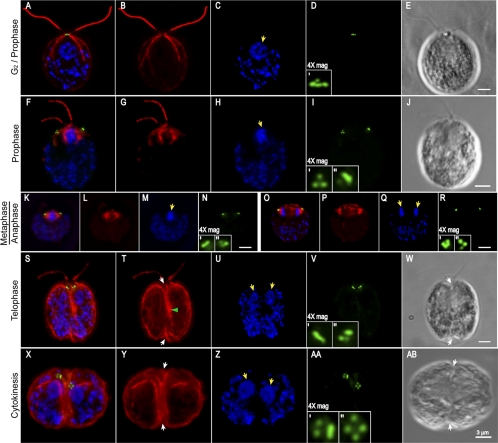
Figure 8.
Indirect immunofluorescence microscopy of the Uni2 protein in dividing cells. Fixed whole cells were stained with rat anti-HA and rabbit anti-α-tubulin antibodies. Secondary anti-rat 488 (green) and anti-rabbit TR (red) antibodies were used to detect primary antibodies. DNA was localized with DAPI (blue). Images from specific cell cycle stages including (A–E) G2/prophase, (F–J) late prophase, (K–N) metaphase, (O–R) anaphase, (S–W) telophase, and (X–AB) cytokinesis are depicted. (A, F, K, O, S, and X) Triple staining of α-tubulin, DAPI, and Uni2 protein. (B, G, L, P, T, and Y) Staining of α-tubulin, (C, H, M, Q, U, and Z) staining of DAPI, and (D, I, N, R, V, and AA) staining of the Uni2 protein are shown independently for all stages, with the left and right basal body regions magnified. (E, J, W, and AB) DIC images of the stained cells. Yellow arrows mark nuclei/chromosomes, green arrows mark the cleavage furrow, and white arrows mark the phycoplast. Scale bar in each series, 3 μm.
Because costaining with tubulin suggested that the Uni2 protein is associated with basal bodies and probasal bodies, we compared the position of Uni2 protein with centrin. In Chlamydomonas, centrin is a major component of the distal striated fiber, which connects the two flagellated basal bodies in interphase cells (Salisbury et al., 1988). Additional centrin-containing fibers descend from the basal bodies toward the nucleus ending in fimbrae that drape around the nucleus. NFAps were obtained by lysis of whole cells with nonionic detergent (Wright et al., 1985). A DIC image of the NFAp depicted in Figure 6H shows a successful extraction of cytoplasmic components. The Uni2 protein remained associated with the NFAps during purification. Staining of the Uni2 protein (green) with centrin (red) revealed two spots residing at the ends of the distal striated fiber in the expected location of basal bodies; two additional spots reside on either side of distal striated fiber in the expected location of probasal bodies (Figure 6E). The localization pattern of the Uni2 protein with centrin is the same as that reported previously for the basal body–specific protein Vfl1 (Silflow et al., 2001).
Using pre-embedding immunogold EM in NFAps, we determined the Uni2 protein localizes to probasal bodies (Figure 6, I–L). We labeled NFAps from the rescued strain or a WT strain as a control using the anti-HA antibody followed by a secondary antibody conjugated with 12-nm gold particles. On thin sections from the tagged strain we identified 21 images, each containing a single NFAp representing ∼0.34 μm2 (18%) of a total area of 1.89 μm2. On these images we found 40 gold particles, of which 95% localized on probasal body structures (Figure 6, I, J, and L) or in the expected location of probasal bodies (Figure 6K). Staining of similar material from a non-HA-tagged strain showed no probasal body staining (unpublished data). Although immunofluorescence microscopy indicated that basal bodies also are stained with the anti-HA antibody, we did not detect staining with the gold-conjugated antibody, perhaps because it did not have access to the epitope in basal bodies.
Uni2 Protein Expression during the Cell Cycle
In addition to two flagella-bearing basal bodies, interphase C. reinhardtii cells also have two disk-shaped probasal bodies. These structures lack TZs and do not nucleate flagellar growth. After their elongation during prophase, the probasal bodies are segregated along with mother and daughter basal bodies to daughter cells as the cells divide (Gaffal, 1988). The flagella and TZs are resorbed during the beginning stages of cell division (Cavalier-Smith, 1974). The cells then undergo one or more rounds of mitotic division coordinated with cycles of basal body duplication. When mitotic divisions are completed, the daughter cells assemble new TZs and flagella, enabling them to swim out of the mother cell wall.
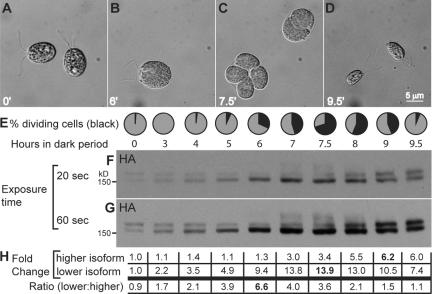
To explore the possibility that the phosphorylation state of Uni2 protein might be altered during the basal body cycle in dividing cells, we analyzed the abundance and posttranslational modification pattern of the Uni2 protein during the cell cycle. A strain expressing the HA-tagged Uni2 protein was synchronized on a 12:12-h light:dark cycle. Growth conditions included high light and carbon-rich growth medium. Under these conditions cells are stimulated to undergo multiple rounds of mitotic divisions and basal body replication cycles during each dark period. Cells were harvested at various time points after entry into the dark period. For each time point, cells were collected, fixed, and scored for division using DIC microscopy. Before division, cells were uniformly large and flagellated (Figure 7, A and B). Cells were considered to be in division if they had visible cleavage furrows or had completed one or more rounds of division but remained within their mother cell wall (Figure 7C). At the end of the dark period, after two or three rounds of cell division, four or eight small daughter cells formed flagella and were released from the mother cell wall (Figure 7D). The percentage of cells in division was determined at time points after entry into the dark period (Figure 7E). For each time point, protein extracts from an equal volume of culture were analyzed on an immunoblot using the anti-HA antibody. Both 20- and 60-s exposures of the same immunoblot are shown (Figure 7, F and G). The total pixel intensity of both isoforms from the shorter exposure time (Figure 7F) was quantified using densitometry (Figure 7H). The value at each time point was compared with the value at time t = 0, which was set at 1.0. The ratio of the lower molecular-weight isoform to the higher molecular-weight isoform was determined from the original densitometry data.
Figure 7.
Expression of the Uni2 protein during the cell cycle. Cells expressing the HA-tagged Uni2 protein were synchronized on a 12:12-h light:dark cycle. (A–D) During the dark period, cells were fixed for DIC microscopy; representative images are shown. (E) At the indicated time points, the percent of cells in division (black) was scored by observation of cleavage furrows or divided cells still within the parental cell wall. (F and G) Cells from equal volumes of culture were lysed in SDS sample buffer for immunoblot analysis using the anti-HA antibody. A 20- and 60-s exposure of an identical immunoblot are shown. (H) Densitometry was used to compare the levels of the two molecular-weight isoforms on the film exposed for 20 s. The fold change from t = 0 is shown for each time point. The ratio of the two isoforms was determined from the original densitometry data.
The Uni2 protein is up-regulated during the division cycle (Figure 7, E–G). The lower molecular-weight isoform increases in abundance more rapidly, reaching a ninefold increase during the first 6 h of the dark period. During this time when existing probasal bodies elongate and new probasal bodies assemble, little increase was detected in the higher molecular-weight isoform. In contrast, during the later stages of division, the abundance of the higher molecular-weight isoform increases to approximately sixfold that of the original, whereas the abundance of the lower molecular-weight isoform decreases, reaching a final level approximately sevenfold that of the zero time point. This increased abundance of the higher molecular-weight isoform coincides with the end of division as cells are assembling TZs and flagella. Even higher molecular-weight isoforms were detected in protein samples from time points most enriched for dividing cells (Figure 7G, hour 7–8), but were not detected in G1 cells (Figure 7G, hour 9.5). The transient larger isoforms might result from additional posttranslational modification.
The Uni2 Protein Provides a Marker for the Basal Body Cycle
Because expression of the Uni2 protein was shown to be up-regulated during mitosis and it appears to be a component of basal bodies, we analyzed the localization of the Uni2 protein during mitosis, which is coordinated with the basal body cycle (Gaffal, 1988). We compared the localization of Uni2 protein with that of tubulin as the dynamics of microtubules during the cell cycle were characterized in detail previously by Doonan and Grief (1987). Using synchronized cell cultures, we localized the HA-tagged Uni2 protein (green) with α-tubulin (red) and DAPI (blue) in whole cells and collected a Z-series of images (Figure 8). A cell in late interphase or early prophase (Figure 8, A–E) has brighter nuclear staining and subtly shorter flagella than most cells in G1 phase. It contains a single cluster of four HA spots, typical for a G1 cell. At late prophase, the nucleus is condensed and the flagella are shorter and of unequal length (Figure 8, F–J). Two clusters of HA spots have migrated away from the base of the flagella. Typically, each cluster contains more than the two spots of HA staining expected from distribution of the four interphase spots. For example in panels F and I, clusters of three or four spots were observed on both the right and left sides of the cell. In other prophase cells, the number of spots per cluster varied between two and four. By metaphase the condensed DNA and the mitotic spindle become visible, and the clusters of HA spots were observed on either side of the spindle in the expected location of basal bodies (Figure 8, K–N).
Metaphase cells contained two to four spots in each cluster (unpublished data). In a cell in anaphase, the chromosomes have separated, the spindle remains visible, and the three to four bright spots of HA staining remain associated with either end of the spindle (Figure 8, O–R). In a telophase cell, the DAPI staining in the two nuclear regions becomes more diffuse (Figure 8, S and U). The cytoplasmic microtubules become more visible and form the phycoplast along the cleavage furrow (Figure 8, S, T, and W). The cell still retains short unequal length flagella from the mother cell (Figure 8, S, T, and W). Distinct spots detected by the anti-HA antibody were observed in clusters of three to four (Figure 8, S and V). All other telophase cells observed showed clusters of four spots on each side of the cell when viewed from the appropriate angle (unpublished data).
In a cell in cytokinesis (Figure 8, X and AB) two distinct nuclei are apparent (Figure 8, X and Z). The cleavage furrow has formed completely, and the microtubule rootlets are more prominent. A cluster of four HA spots resides in each daughter cell, with each cluster arranged in the diamond-shaped pattern of spots observed in interphase cells (Figure 8, X and AA). Some dividing cells retained flagella throughout the first mitotic division (Figure 8, A–E, F–J, and S–W). We also observed cells in all stages of division that did not retain their flagella. Because we could not distinguish between cells that have naturally resorbed or excised flagella and cells that have lost their flagella as a result of the fixation and mounting process, we were not able to quantify the frequency of flagellated cells during division. However, our results raise the interesting possibility that flagella can be retained on dividing cells well after the time when basal bodies move away from the base of flagella.
DISCUSSION
The Uni2 protein provides a molecular marker that we have used to further elucidate the basal body cycle in Chlamydomonas. Key to its utility as a marker is the observation that the epitope-tagged Uni2 protein labels basal bodies and probasal bodies, which copurify with isolated NFAps. The accumulation of the Uni2 protein in distinct spots at both ends of the distal striated fiber and on both sides of the fiber is identical to the localization observed with the basal body protein Vfl1 (Silflow et al., 2001). Immunogold labeling showed that the Uni2 protein is associated with probasal bodies. It is not clear why we labeled only probasal bodies in these experiments. Perhaps the epitopes on this low-abundance protein are masked as the probasal bodies elongate to basal bodies. Further evidence that the Uni2 protein is a component of basal bodies is its localization at the poles of mitotic spindles (Coss, 1974; Matsuura et al., 2004). Despite the ultrastructural defect in TZs in uni2 mutant cells, the mitotic migration of the Uni2 protein to the spindle poles suggests it is not a TZ component. Previous ultrastructural studies have shown that TZs remain associated with remnants of flagella during mitosis (Johnson and Porter, 1968) and that the TZ is resorbed along with the flagellum during mitosis (Cavalier-Smith, 1974).
The localization of Uni2 protein confirmed the timing for initiation of the basal body development cycle in Chlamydomonas. We found that the Uni2 protein accumulates in at least one new focus as early as prophase. Ultrastructural observations of serial sections through dividing cells showed new probasal body formation between late metaphase and early anaphase (Gaffal, 1988). Marshall et al. (2001) suggested that the initiation of basal body replication occurs during the S/G2 phase of the cell cycle. Our data show that the first appearance of Uni2 protein coincides with the assembly of probasal body structures during M phase, in agreement with Gaffal (1988).
Previous reports have suggested that basal bodies in C. reinhardtii dissociate from attached flagella at the onset of mitosis (Johnson and Porter, 1968; Gould, 1975). Subsequent reports, however, have suggested that flagellar resorption is completed before mitosis (Cavalier-Smith, 1974; Gaffal, 1988). We found that at least some cells are able to retain flagella throughout all mitotic stages of the first division, indicating that complete flagellar resorption is not a prerequisite for basal body migration. A similar observation was reported for the unicellular green alga Chlorogonium elongatum, which has been shown to retain fully functional flagella detached from basal bodies throughout mitosis (Hoops and Whitman, 1985).
The difference in chronological age between the two basal bodies may be reflected in the timing of replication. Gaffal (1988) suggested that basal body replication might be sequential between the two basal bodies because probasal body structures did not appear simultaneously along side the two basal bodies. Our immunolocalization results are consistent with this suggestion. In earlier stages of mitosis, typically only three Uni2 protein spots were observed on either side of the spindle, whereas in later stages, four spots were usually distinguishable. Furthermore, basal body replication may occur asymmetrically as we sometimes observed different numbers of Uni2 stained spots in the two basal body clusters in a dividing cell.
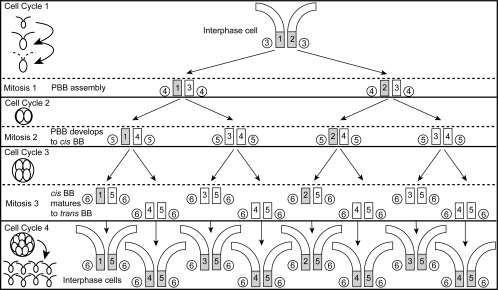
The maturation pathway for basal bodies in Chlamydomonas occurs over four cell cycles, including three mitotic events that may occur during a single cell-division period as depicted in Figure 9. Alternatively, after any of the mitotic cycles depicted in Figure 9, a cell could exit the cell division cycle and assemble flagella. In M phase of cell cycle one, the probasal bodies (3) present in the interphase cell elongate and are segregated with mother (1) and daughter (2) basal bodies to opposite poles of the spindle (Gaffal, 1988). New probasal bodies (4) are “born” during M phase and accumulate the Uni2 protein. After segregation to daughter cells, the probasal bodies (4) lie dormant until they elongate at prophase of cell cycle two and are segregated to daughter cells. In the third cell cycle, the basal bodies (4) are in the “cis” position with respect to the eyespot. During the third mitosis, the basal bodies (4) migrate to the final “trans” position. On assembling a flagellum in the ensuing cell cycle four, they are considered fully “mature.” Both cis (immature/daughter) and trans (mature/mother) basal bodies are capable of flagellar assembly in WT cells. The uni2 mutations affect flagellar assembly from both cis and trans basal bodies. However, the more pronounced defect in flagellar assembly seen for the cis basal body suggests that the UNI2 gene product also functions in the pathway for basal body maturation.
Figure 9.
Model of basal body developmental pathway and phosphorylation of the Uni2 protein over three successive mitotic events occurring in a single dark-phase period. Left, a schematic diagram of the cell bodies during multiple mitotic divisions. Dotted flagella indicate flagellar resorption at mitosis. Right, the basal bodies (BB, squares) and probasal bodies (PBB, circles) during these mitotic stages with their chronological age (1–6 oldest to youngest) are marked. The oldest basal body (trans to the eyespot) is always positioned on the left. Basal bodies shaded in gray contain phosphorylated Uni2 protein. Depending on growth conditions, cells may exit from the cell division cycle after going one, two, or three mitotic divisions. However, the maturation pathway from probasal body assembly to trans basal body requires four cell cycles.
Results from analysis of Uni2 protein expression in synchronized cultures suggest that the posttranslational modification of the Uni2 protein is correlated with cell cycle progression. A model to explain the results is that phosphorylation of the Uni2 protein coincides with formation of a TZ and assembly of a flagellum (Figure 9). This model predicts that Uni2 protein is first phosphorylated when a basal body assembles a TZ and flagellum and remains modified in all ensuing cell cycles. In interphase cells, approximately equal amounts of the two isoforms represent the basal body component (modified) and the probasal body component (unmodified). During early stages of the dark period, the lower molecular-weight isoform becomes more abundant as new basal body assembly is occurring in the absence of TZ assembly. Later in the dark period when cells are finishing division and beginning to assemble new TZs and flagella, an increase in the higher molecular-weight isoform is seen. When multiple cell cycles occur during one dark period (as seen in Figure 9), the phosphorylation occurs only at the completion of all division cycles when all the daughter cells assemble flagella (cell cycle 4). As cells complete the division period, equal amounts of modified and unmodified Uni2 protein are again observed. In this model, a basal body would first acquire the phosphorylated Uni2 isoform as a flagellated daughter basal body positioned cis to the eyespot.
This pathway may involve the gradual accumulation of phosphorylation during successive cell cycles because the phosphorylated Uni2 isoform never appears to decrease in the early stages of the division cycle. A threshold level of phosphorylation reached by the younger (cis) basal body in a WT cell at the end of the division cycle would permit flagellar assembly. In mutant cells, the absence of the Uni2 protein results in defects in flagellar assembly from both cis (daughter) and trans (mother) basal bodies. However, assembly from the mother basal body is less affected by the mutation, suggesting that additional components act in the maturation pathway.
Phenotypes of the uni2 mutants are strikingly similar to those reported for the uni1 mutants (Huang et al., 1982). Like uni2 mutants, cultures of uni1 mutants contain a large proportion of uniflagellate cells, in which flagellar assembly occurred from the older basal body. Ultrastructural studies of uni1 mutants revealed morphological defects in the TZ. These results suggest that the UNI1 gene, which has not been characterized, and the UNI2 gene both function in pathways leading to basal body maturation and the assembly of flagella.
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation Grant MCB-0344661 to C.D.S. Wei-Chien Wu conducted experiments to clone and characterize the UNI2 gene. We acknowledge the College of Biological Sciences' Imaging Center at The University of Minnesota, including assistance from Mark A. Sanders with fluorescence microscopy, Gilbert G. Ahlstrand with EM, and Tracy E. Anderson with image processing. Ravishankar Chityala at The Supercomputing Institute at The University of Minnesota provided assistance with batch deconvolution. Nedra F. Wilson provided assistance with protein dephosphorylation experiments.
Abbreviations used:
- CIP
calf alkaline intestinal phosphatase
- EST
expressed sequence tag
- NFAp
nucleoflagellar apparatus
- TAP
Tris-acetate-phosphate medium
- TEM
transmission electron microscopy
- TR
Texas red
- TZ
transition zone
- WT
wild type.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-08-0798) on October 17, 2007.
REFERENCES
- Afzelius B. A. Cilia-related diseases. J. Pathol. 2004;4:470–477. doi: 10.1002/path.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beales P. L. Lifting the lid on Pandora's box: the Bardet-Biedl syndrome. Curr. Opin. Genet. Dev. 2005;3:315–323. doi: 10.1016/j.gde.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Beech P. L., Heimann K., Melkonian M. Development of the flagellar apparatus during the cell cycle in unicellular algae. Protoplasma. 1991;164:23–37. [Google Scholar]
- Blacque O. E., Leroux M. R. Bardet-Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell Mol. Life Sci. 2006;18:2145–2161. doi: 10.1007/s00018-006-6180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;5:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 2002;1:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J. Cell Sci. 1974;3:529–556. doi: 10.1242/jcs.16.3.529. [DOI] [PubMed] [Google Scholar]
- Coss R. A. Mitosis in Chlamydomonas reinhardtii basal bodies and the mitotic apparatus. J. Cell Biol. 1974;1:325–329. doi: 10.1083/jcb.63.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe H. R., Farr H., Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- Debuchy R., Purton S., Rochaix J. D. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 1989;10:2803–2809. doi: 10.1002/j.1460-2075.1989.tb08426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;1(Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan J. H., Grief C. Microtubule cycle in Chlamydomonas reinhardtii an immunofluorescence study. Cell Motil. Cytoskelet. 1987;4:381–392. [Google Scholar]
- Dutcher S. K. Elucidation of basal body and centriole functions in Chlamydomonas reinhardtii. Traffic. 2003;4:443–451. doi: 10.1034/j.1600-0854.2003.00104.x. [DOI] [PubMed] [Google Scholar]
- Dutcher S. K., Morrissette N. S., Preble A. M., Rackley C., Stanga J. ε-tubulin is an essential component of the centriole. Mol. Biol. Cell. 2002;11:3859–3869. doi: 10.1091/mbc.E02-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S. K., Trabuco E. C. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes δ-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell. 1998;6:1293–1308. doi: 10.1091/mbc.9.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffal K. P. The basal body-root complex of Chlamydomonas reinhardtii during mitosis. Protoplasma. 1988;143:139–148. [Google Scholar]
- Geimer S., Melkonian M. The ultrastructure of the Chlamydomonas reinhardtii basal apparatus: identification of an early marker of radial asymmetry inherent in the basal body. J. Cell Sci. 2004;117:2663–2674. doi: 10.1242/jcs.01120. [DOI] [PubMed] [Google Scholar]
- Gorman D. S., Levine R. P. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA. 1965;6:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould R. R. The basal bodies of Chlamydomonas reinhardtii. Formation from probasal bodies, isolation, and partial characterization. J. Cell Biol. 1975;1:65–74. doi: 10.1083/jcb.65.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes J. A., Dutcher S. K. Cellular asymmetry in Chlamydomonas reinhardtii. J. Cell Sci. 1989;94:273–285. doi: 10.1242/jcs.94.2.273. [DOI] [PubMed] [Google Scholar]
- Hoops H. J., Witman G. B. Basal bodies and associated structures are not required for normal flagellar motion or phototaxis in the green alga Chlorogonium elongatum. J. Cell Biol. 1985;1:297–309. doi: 10.1083/jcb.100.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Ramanis Z., Dutcher S. K., Luck D. J. Uniflagellar mutants of Chlamydomonas: evidence for the role of basal bodies in transmission of positional information. Cell. 1982;3:745–753. doi: 10.1016/0092-8674(82)90436-6. [DOI] [PubMed] [Google Scholar]
- James S. W., Ranum L. P., Silflow C. D., Lefebvre P. A. Mutants resistant to anti-microtubule herbicides map to a locus on the uni linkage group in Chlamydomonas reinhardtii. Genetics. 1988;1:141–147. doi: 10.1093/genetics/118.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson U. G., Porter K. R. Fine structure of cell division in Chlamydomonas reinhardi. Basal bodies and microtubules. J. Cell Biol. 1968;2:403–425. doi: 10.1083/jcb.38.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R., Witman G. B. Submicromolar levels of calcium control the balance of beating between the two flagella in demembranated models of Chlamydomonas. J. Cell Biol. 1984;1:97–107. doi: 10.1083/jcb.98.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathir P., LaVoie M., Brazelton W. J., Haas N. A., Lefebvre P. A., Silflow C. D. Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell. 2003;2:362–379. doi: 10.1128/EC.2.2.362-379.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L. C., Romijn E. P., Zamora I., Yates J. R., 3rd, Marshall W. F. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 2005;12:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Lefebvre P. A., Nordstrom S. A., Moulder J. E., Rosenbaum J. L. Flagellar elongation and shortening in Chlamydomonas. IV. Effects of flagellar detachment, regeneration, and resorption on the induction of flagellar protein synthesis. J. Cell Biol. 1978;1:8–27. doi: 10.1083/jcb.78.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine R. P., Ebersold W. T. The genetics and cytology of Chlamydomonas. Annu. Rev. Microbiol. 1960:197–216. doi: 10.1146/annurev.mi.14.100160.001213. [DOI] [PubMed] [Google Scholar]
- Li J. B., et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;4:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Marshall W. F. What is the function of centrioles? J. Cell Biochem. 2007;4:916–922. doi: 10.1002/jcb.21117. [DOI] [PubMed] [Google Scholar]
- Marshall W. F., Vucica Y., Rosenbaum J. L. Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr. Biol. 2001;5:308–317. doi: 10.1016/s0960-9822(01)00094-x. [DOI] [PubMed] [Google Scholar]
- Matsuura K., Lefebvre P. A., Kamiya R., Hirono M. Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J. Cell Biol. 2004;5:663–671. doi: 10.1083/jcb.200402022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D. R. Orientation of the central pair complex during flagellar bend formation in Chlamydomonas. Cell Motil. Cytoskelet. 2003;2:120–129. doi: 10.1002/cm.10142. [DOI] [PubMed] [Google Scholar]
- Nelson J. A., Savereide P. B., Lefebvre P. A. The CRY1 gene in Chlamydomonas reinhardtii: structure and use as a dominant selectable marker for nuclear transformation. Mol. Cell Biol. 1994;6:4011–4019. doi: 10.1128/mcb.14.6.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T., et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;1:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- O'Toole E. T., Giddings T. H., McIntosh J. R., Dutcher S. K. Three-dimensional organization of basal bodies from wild-type and δ-tubulin deletion strains of Chlamydomonas reinhardtii. Mol. Biol. Cell. 2003;7:2999–3012. doi: 10.1091/mbc.E02-11-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Wang Q., Snell W. J. Cilium-generated signaling and cilia-related disorders. Lab. Invest. 2005;85:452–463. doi: 10.1038/labinvest.3700253. [DOI] [PubMed] [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J., Witman G. B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005;1:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M. E., Bower R., Knott J. A., Byrd P., Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell. 1999;3:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringo D. L. Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J. Cell Biol. 1967;3:543–571. doi: 10.1083/jcb.33.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Granick S. Nutritional studies with Chlamydomonas reinhardi. Ann. NY Acad. Sci. 1953;5:831–838. doi: 10.1111/j.1749-6632.1953.tb30261.x. [DOI] [PubMed] [Google Scholar]
- Salisbury J. L., Baron A. T., Sanders M. A. The centrin-based cytoskeleton of Chlamydomonas reinhardtii: distribution in interphase and mitotic cells. J. Cell Biol. 1988;2:635–641. doi: 10.1083/jcb.107.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. A., Salisbury J. L. Immunofluorescence microscopy of cilia and flagella. Methods Cell. Biol. 1995;47:163–169. doi: 10.1016/s0091-679x(08)60805-5. [DOI] [PubMed] [Google Scholar]
- Sanders M. A., Salisbury J. L. Centrin-mediated microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 1989;5:1751–1760. doi: 10.1083/jcb.108.5.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow C. D., Chisholm R. L., Conner T. W., Ranum L. P. The two alpha-tubulin genes of Chlamydomonas reinhardi code for slightly different proteins. Mol. Cell Biol. 1985;9:2389–2398. doi: 10.1128/mcb.5.9.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow C. D., LaVoie M., Tam L. W., Tousey S., Sanders M., Wu W., Borodovsky M., Lefebvre P. A. The Vfl1 Protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J. Cell Biol. 2001;1:63–74. doi: 10.1083/jcb.153.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow C. D., Rosenbaum J. L. Multiple alpha- and beta-tubulin genes in Chlamydomonas and regulation of tubulin mRNA levels after deflagellation. Cell. 1981;1:81–88. doi: 10.1016/0092-8674(81)90503-1. [DOI] [PubMed] [Google Scholar]
- Singla V., Reiter J. F. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;5787:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- Taillon B. E., Adler S. A., Suhan J. P., Jarvik J. W. Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J. Cell Biol. 1992;6:1613–1624. doi: 10.1083/jcb.119.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam L. W., Lefebvre P. A. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics. 1993;2:375–384. doi: 10.1093/genetics/135.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J. C., et al. The sequence of the human genome. Science. 2001;5507:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wright R. L., Salisbury J., Jarvik J. W. A nucleus-basal body connector in Chlamydomonas reinhardtii that may function in basal body localization or segregation. J. Cell Biol. 1985;5(Pt 1):1903–1912. doi: 10.1083/jcb.101.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]