Abstract
The sorting and assembly machinery (SAM) complex functions in the assembly of β-barrel proteins into the mitochondrial outer membrane. It is related to the Omp85/YaeT machinery in bacterial outer membranes, but the eukaryotic SAM complex is distinguished by two peripheral subunits, Sam37 and Sam35, that sit on the cytosolic face of the complex. The function of these subunits in β-barrel protein assembly is currently unclear. By screening a library of sam35 mutants, we show that 13 distinct alleles were each specifically suppressed by overexpression of SAM37. Two of these mutants, sam35-409 and sam35-424, show distinct phenotypes that enable us to distinguish the function of Sam35 from that of Sam37. Sam35 is required for the SAM complex to bind outer membrane substrate proteins: destabilization of Sam35 inhibits substrate binding by Sam50. Sam37 acts later than Sam35, apparently to assist release of substrates from the SAM complex. Very different environments surround bacteria and mitochondria, and we discuss the role of Sam35 and Sam37 in terms of the problems peculiar to mitochondrial protein substrates.
INTRODUCTION
The mitochondrial outer membrane defines the physical barrier between mitochondria and the cytoplasm of a eukaryotic cell. It provides a selective entry gate for mitochondrial-targeted proteins into the double membrane-bound organelle, because almost all proteins that function inside mitochondria are encoded by nuclear genes. An important group of mitochondrial membrane proteins known as β-barrel proteins, are uniquely found in the outer membrane, and they function in the biogenesis of mitochondria. Structures determined for many β-barrel proteins from bacterial outer membranes have shown these proteins adopt barrel structures formed by anti-parallel β-strands embedded in the lipid bilayer, and they can form functional complexes with other outer membrane proteins (Buchanan, 1999; Gabriel et al., 2001; Schleiff and Soll, 2005).
The assembly of β-barrel proteins into the mitochondrial outer membrane requires the sorting and assembly machinery (SAM) complex (Pfanner et al., 2004; Paschen et al., 2005). Like other mitochondrial-targeted proteins, β-barrel proteins are first translocated across the outer membrane via the translocase of the outer membrane (TOM) complex, and, in a manner dependent on chaperones in the intermembrane space, they are then passed on to the SAM complex for their final assembly into the outer membrane (Pfanner et al., 2004; Paschen et al., 2005). Studies in yeast have identified key components of the SAM complex: Sam50 (also called Tob55; Kozjak et al., 2003; Paschen et al., 2003; Gentle et al., 2004), Sam35 (also called Tom38 and Tob38; Ishikawa et al., 2004; Milenkovic et al., 2004; Waizenegger et al., 2004), and Sam37 (also called Mas37; Gratzer et al., 1995; Wiedemann et al., 2003). Sam50, the membrane-embedded subunit of the SAM complex, is an essential protein predicted to have a β-barrel topology, and it is related to the Omp85 family of proteins that mediate protein assembly into bacterial outer membranes (Dolezal et al., 2006; Gentle et al., 2004, 2005). The other two subunits, Sam35 and Sam37, are peripheral membrane proteins that are assumed to associate with the outer membrane via direct contact with Sam50 (Kozjak et al., 2003; Milenkovic et al., 2004). The complex formed between Sam50, Sam35, and Sam37 is responsible for the assembly of all β-barrel proteins in yeast, and it is referred to as the SAMcore complex. Mdm10, another β-barrel protein involved in maintaining mitochondrial morphology and distribution (Sogo and Yaffe, 1994; Meisinger et al., 2004), has been shown to interact with the SAMcore complex and to have a specific role in assembling Tom40 into the TOM complex (Meisinger et al., 2004). Mdm10, and perhaps other proteins, form modules that give rise to a SAM supercomplex.
Recent studies show that although the SAMcore complex assists assembly of all β-barrel proteins, additional factors are required to mediate Tom40 assembly into a TOM complex (Ishikawa et al., 2004; Waizenegger et al., 2005). The identification of the SAM complex together with these new components mediating more select aspects of membrane protein assembly, has set the basic framework for a detailed characterization of the mechanisms driving the pathway of β-barrel protein assembly. Recently, it has been shown that the N-terminal domain of Sam50 exposed to the intermembrane space has receptor-like function for β-barrel proteins and may assist translocation of substrates from the trans side of the TOM complex to the SAM complex (Habib et al., 2007). Tom7, a conserved subunit of the TOM complex, mediates the segregation of Mdm10 from its interaction with the SAMcore complex (Meisinger et al., 2006). Mdm10 was very recently shown to associate with the Mdm12/Mmm1 complex, and both Mdm12 and Mmm1 are important for β-barrel protein assembly (Meisinger et al., 2007).
Despite the recent advances, little yet is known about how the two peripheral components of the SAMcore complex, Sam35 and Sam37, function. Their general involvement in constituting the SAMcore complex makes them important in β-barrel protein assembly, but their individual contributions to the function of the SAMcore complex remains unclear. We find a codependent relationship between Sam37 and Sam35. Mutations in Sam35 leads to decreased levels of Sam37, and deletion of Sam37 causes decrease in Sam35 levels. By maintaining the levels of Sam35 in Δsam37 mitochondria, we show that the Sam35-Sam50 is fully capable of assembling β-barrel precursors into their functional complexes. Two yeast mutants, sam35-409 and sam35-424, show distinct phenotypes that enable us to distinguish the function of Sam35 from that of Sam37. Sam35 is required in order for the Sam50 subunit to bind outer membrane substrate proteins: destabilization of Sam35 inhibits substrate binding by the SAM complex. Sam37 acts later than Sam35, apparently to assist release of substrates from the SAM complex.
MATERIALS AND METHODS
Yeast Strains and Growth
Saccharomyces cerevisiae cells are grown in rich medium with (1% yeast extract, 2% peptone, and 2% glucose [YPD] or 2% glycerol [YPG]) or synthetic complete medium lacking appropriate amino acid(s) for plasmids selection with glucose [0.67% yeast nitrogen base, 2% glucose) or lactate (0.3% yeast nitrogen base, 0.05% glucose, 0.05% CaCl2, 0.06% MgCl2, 0.1% KH2PO4, 0.1% NH4Cl, and 2.2% (vol/vol) lactic acid; adjust pH to 5.5 with NaOH]. NCY0601 (his3-11/his3-11 leu2-3/leu2-3 ura3-1/ura3-1 ade2-1/ade2-1 trp1-1/trp1-1 can1-100/can1-100 SAM35/Δsam35::His3-MX6) was generated by direct gene replacement with one copy of SAM35 open reading frame (ORF) by using methods described previously (Longtine et al., 1998). NCY0603 (MATα his3-11 leu2-3 ura3-1 ade2-1 trp1-1 can1-100 Δsam35::His3-MX6 URA3::YEpSam35) was generated by sporulation and dissection of NCY0601 transformed with YEpSam35. Δsam37 strain (MATα his3-11 leu2-3 ura3-1 ade2-1 trp1-1 can1-100 Δsam37::His3-MX6) was from Ian Gentle (Department of Biochemistry and Molecular Biology, University of Melbourne) and was generated as described previously (Longtine et al., 1998). All yeast strains used in this study were derived from W303 background.
Generation of sam35 Random Mutant Library
Conditional alleles of sam35 were generated by low-fidelity polymerase chain reaction (PCR) to mutate a fragment of DNA containing SAM35, followed by recombination of mutant alleles with linearized pRS314 (CEN6, TRP1; Sikorski and Hieter, 1989) in vivo upon transformation into NCY0603 by using similar procedures as described previously (Sikorski and Hieter, 1989; Gabriel et al., 2003). Ura+ Trp+ transformants were selected at 25°C. Plasmids encoding the wild-type copy of SAM35 (YEpSam35) was ejected by selection of transformants on minimal glucose media containing 5-fluroorotic acid and appropriate supplements at 25°C. Approximately 600 transformants were collected and screened for growth defects at 25, 30, and 37°C on rich media containing glucose (YPAD) or glycerol (YPG). Plasmids were isolated from the yeast mutants that showed conditional phenotypes and mutations in the SAM35 ORF confirmed by DNA sequencing. Specificity of the mutant sam35 induced phenotypes were confirmed by isolating plasmids from the conditional mutants, followed by retransforming into NCY0603, plasmid shuffling as described above to eject pRS-SAM35, and growth phenotype was tested at 25, 30, and 37°C on rich media containing glucose (YPAD) or glycerol (YPG).
Plasmid Construction
All plasmid manipulations were carried out using protocols described previously (Sambrook and Russell, 2001). Details of each construct and sequences of all oligonucleotides used are available upon request. In brief, constructs used in the multicopy suppression experiments were generated by PCR amplification of genomic fragments encompassing the gene of interest from yeast genomic DNA and cloned into YEplac181 (LEU2, 2μ). To construct YEpSam35, the genomic fragment containing the SAM35 ORF plus 530-bp upstream and 466-bp downstream sequences was amplified from yeast genomic DNA and cloned into the BamHI, HindIII sites of the multicopy yeast expression vector YEplac195 (URA3, 2 μ). pRS-SAM35 (wild-type control for sam35 mutant alleles) was generated by subcloning the BamHI, HindIII fragment from YEpSam35 into pRS314 (Sikorski and Hieter, 1989).
Growth Assays
Cells were grown in rich media (YPD) or synthetic complete media (with glucose) lacking appropriate amino acids for plasmid selection to mid-logarithmic phase. Cells were diluted to OD600 of 0.04 followed by a series of fivefold dilutions and spotted onto the indicated plates and incubated at the indicated temperatures for 2–5 d.
Isolation of Mitochondria and In Vitro Protein Import
Mutant yeast strains and their corresponding wild-type control strains were grown in parallel in lactate medium at 25°C. Mitochondria were isolated by differential centrifugation as described previously (Daum et al., 1982). For in vitro transcription, pSP65 or pGEM vectors carrying ORFs of mitochondrial precursor proteins were linearized by restriction digest at a unique site downstream of the ORFs and used for in vitro transcription by using SP6 polymerase (Promega, Madison, WI) according to the manufacturer's instructions. Radiolabeled precursor proteins were in vitro translated in rabbit reticulocyte lysates (Promega) in the presence of [35S]methionine/cysteine (MP Biomedicals, Irvine, CA) at 30°C for 30 min before import experiment. For in vitro import, isolated mitochondria (25 μg/time point for SDS-polyacrylamide gel electrophoresis [PAGE]; 50 μg/time point for blue native [BN]-PAGE) were incubated in import buffer (0.6 M sorbitol, 50 mM HEPES, pH 7.4, 2 mM KPi, pH 7.4, 25 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, and 1 mM dithiothreitol) supplemented with 5 mM NADH and 1–4 mM ATP. We added 5% (vol/vol) 35S-labeled precursors to the mitochondria and incubated them at 25°C for the indicated time. Import reactions were stopped by dilution of import reaction in ice-cold import buffer with 100 μM carbonyl cyanide m-chlorophenyl-hydrazone (CCCP) and incubation on ice for matrix and inner membrane-targeted precursors. Proteins not imported into mitochondria were removed by treatment with 50 μg/ml proteinase K for 10 min. Proteinase K digestion was terminated by addition of 1 mM phenylmethylsulfonyl fluoride (PMSF). Mitochondria are isolated, boiled in sample loading buffer, and analyzed by SDS-PAGE. Phosphorimage analysis was carried out using a Typhoon TRIO variable mode imager (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), and quantification of radioactive signal was accomplished using ImageQuant software (GE Healthcare). For assembly assays analyzed by BN-PAGE, import reactions are terminated by incubation on ice without addition of CCCP or proteinase K. Mitochondria are isolated by centrifugation and solubilized as described below.
Blue Native Polyacrylamide Gel Electrophoresis
Mitochondria (50–100 μg of proteins) were solubilized by resuspension in 50 μl of ice-cold digitonin-containing buffer (0.8–1% digitonin, 20 mM Tris-Cl, 0.1 mM EDTA, 50 mM NaCl, 10% glycerol, and 1 mM PMSF, pH 7.4) and incubated on ice for 15 min with intermittent vortexing. Insoluble materials were pelleted by centrifugation at 10,000 rpm for 10 min, and the supernatant containing the protein complexes was transferred to a new tube. Loading buffer (13 μl of 5% Coomassie Blue G and 500 mM amino caproic acid in 100 mM bis-Tris, pH 7.0) was added to the supernatant, and the protein complexes were separated by BN-PAGE on a 6–16.5% polyacrylamide gel (Nijtmans et al., 2002; Wittig et al., 2006). For immunoblotting, the protein complexes were transferred onto polyvinylidene difluoride (PVDF) membrane and detected by antibodies using enhanced chemiluminescence methods. For import experiments, the BN gel was dried, and radiosignals were detected by phosphorimage analysis (GE Healthcare).
Confocal Microscopy
Wild-type and mutant strains were grown in synthetic complete media at 30°C with lactate as a carbon source to OD600 ∼0.8. Cells were stained with MitoTracker Red (Invitrogen, Carlsbad, CA), and mitochondria were visualized using a TCS SP2 imaging system (Leica, Wetzlar, Germany). In each experiment, 100 cells were randomly selected, and their mitochondrial morphology was assessed individually.
RESULTS
Sam37 Suppresses Lethality of All sam35 Alleles at Restrictive Temperature
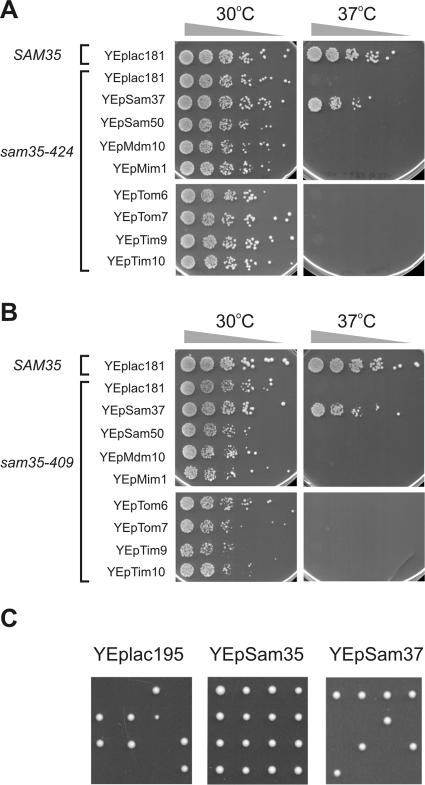
We used error-prone PCR to generate a random mutant library of sam35 mutants. From the initial six hundred mutants generated, 13 temperature-sensitive alleles were isolated by selecting cells that are inhibited in growth at increased temperature. We screened all 13 mutants for multicopy suppression by using plasmids encoding various proteins implicated in protein import and outer membrane protein assembly. The results for the sam35-424 (Figure 1A) and sam35-409 (Figure 1B) mutants are shown. Overexpression of the SAM37 gene suppressed the lethality of the sam35-424 and sam35-409 mutants at 37°C. Indeed, in each of the 13 sam35 mutants, overexpression of SAM37 restored growth at 37°C, and in no case did we find other components of the outer membrane that could do so.
Figure 1.
Sam37 is a multicopy suppressor for sam35 alleles. (A) Δsam35 cells expressing wild-type SAM35 or sam35-424 from a plasmid and transformed with the indicated multicopy plasmids, were grown to mid-logarithmic phase. Five-fold serial dilutions were spotted onto synthetic complete medium (with glucose as a carbon source) lacking tryptophan and leucine, and plates were incubated at the indicated temperatures for 3 d. (B) Overexpression of Sam37 suppresses sam35-409 phenotype at restrictive temperature. Same as in A, except Δsam35 cells were transformed with plasmids encoding sam35-409 instead. (C) Sam37 and Sam35 are not functionally redundant. NCY0601 (heterozygous diploid SAM35/Δsam35) cells were transformed with a multicopy vector alone (YEplac195) or with the same vector containing SAM35 (YEpSam35) or SAM37 (YEpSam37). Transformed cells were sporulated and tetrads dissected on rich medium containing glucose. Results for four dissected asci are shown.
A trivial, although unexpected, explanation for this result would be that Sam37 and Sam35 are functionally redundant and that overexpression of Sam37 can compensate for a loss of Sam35 function. To test this possibility, heterozygous (Δsam35/SAM35) diploid yeast lacking one copy of the SAM35 gene were transformed with a control (multicopy) plasmid YEplac195, or the same plasmid containing the SAM35 gene (YEpSam35), or the same plasmid but containing instead the SAM37 gene (YEpSam37). After sporulation, the progeny of meiosis were dissected onto rich medium containing glucose as a carbon source: overexpression of the SAM37 gene cannot compensate for the absence of SAM35 (Figure 1C).
Characterization of sam35-424
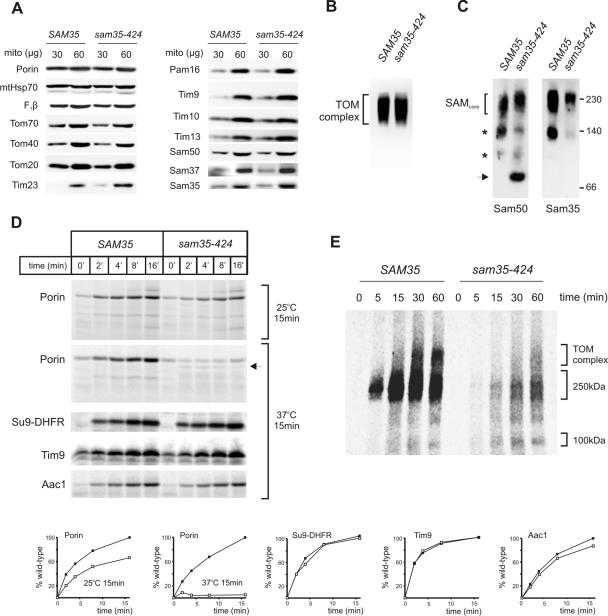
To investigate the defects in sam35-424, mitochondria were isolated from Δsam35 cells transformed with either a plasmid carrying the wild-type SAM35 gene (“SAM35”), or the mutant sam35-424 gene, after growth of the transformed cells at 25°C in lactate medium. Steady-state levels of various mitochondrial proteins as analyzed by immunoblotting showed no difference in any other mitochondrial proteins, including the level of the mutant sam35 protein observed in the sam35-424 strain (Figure 2A). The steady-state level of assembled TOM complex in sam35-424 is similar to wild type (Figure 2B), but immunoblotting from the same BN-PAGE gels with anti-Sam50 antibodies revealed defects in the assembly state of the SAM complex (Figure 2C). An additional complex of ∼80 kDa, containing Sam50, is absent in the wild-type control, but it is observed in sam35-424, suggesting that the SAM complex in sam35-424 is less stable and more sensitive to detergent treatment. This 80-kDa complex does not contain Sam35, as judged from immunoblotting by using anti-Sam35 antibodies (Figure 2C).
Figure 2.
Characterization of sam35-424. (A) Cells expressing wild-type SAM35 or sam35-424 were grown in lactate medium at 25°C, and mitochondria were isolated. Mitochondrial proteins were separated by SDS-PAGE, transferred onto nitrocellulose membrane, and immunodecorated with antisera against the indicated proteins. (B) Mitochondria (100 μg) from the indicated strains were solubilized in 1% digitonin, and protein complexes were separated by BN-PAGE and blotted onto PVDF membranes. Anti-Tom40 antisera were used to immunoblot for the TOM complex. (C) Mitochondria (100 μg) from the indicated strains were solubilized in 1% digitonin, and protein complexes were separated by BN-PAGE and blotted onto PVDF membranes. Antisera against Sam50 or Sam35 were used to immunoblot for the SAM complex. Asterisks, unidentified subcomplexes of the SAM complex obtained during solubilization; arrow, Sam50-containing complex specifically enriched in sam35-424. (D) Mitochondria from cells expressing wild-type SAM35 or sam35-424 were grown in lactate medium at 25°C, mitochondria were isolated and incubated at 25 or 37°C for 15 min in import buffer, followed by an equilibration at 25°C for 5 min. 35S-labeled porin was added to the mitochondria and incubated at 25°C for the indicated time, treated with 50 μg/ml proteinase K to remove unimported precursors, and analyzed by SDS-PAGE and digital autoradiography. Arrow, altered proteinase K cleavage profile of porin upon import into sam35-424 mitochondria preincubated at 37°C for 15 min. Su9-DHFR (matrix), Aac1 (inner membrane) and Tim9 (intermembrane space) precursors were imported into mitochondria from wild-type or sam35-424 cells, with 15 min preincubation at 37°C. Quantification of the results is shown below. Filled boxes, SAM35; open boxes, sam35-424. (E) 35S-labeled Tom40 was incubated with mitochondria for the indicated time. Mitochondria were reisolated and solubilized in 1% digitonin, and protein complexes were analyzed by BN-PAGE and digital autoradiography.
The temperature-sensitive defect in the sam35-424 is evident in vitro, because incubation of mitochondria at 37°C for 15 min before the import experiment was sufficient to dramatically decrease the ability to import porin (Figure 2D). A relatively small defect in porin import is seen if the mitochondria from sam35-424 cells are maintained at 25°C, but loss of Sam35 function is almost complete with a 15-min preincubation at 37°C. The altered proteolytic cleavage profile of porin when imported into heat-treated mutant mitochondria also suggests that the incorrect assembly of porin into the lipid bilayer of the outer membrane can be induced (Figure 2D). The heat treatment does not affect the rate of import into the matrix, the intermembrane space, and the inner membrane, as shown by import of Su9-DHFR, Tim9, and Aac1, respectively (Figure 2D).
To assess the assembly kinetics of Tom40 into the TOM complex, 35S-labeled Tom40 was imported and analyzed by BN-PAGE (Figure 2E). A major defect is seen in the total amount of Tom40 taken up by mutant mitochondria, with some accumulation of Tom40 at the 100-kDa assembly intermediate II, where Tom40 is in contact with Tom5 and/or Tom6 (Model et al., 2001; Wiedemann et al., 2003; Figure 2E). Whether the 100-kDa intermediate is found free in the outer membrane or represents a species that is more readily solubilized with digitonin during the assembly of Tom40 into the TOM complex by the SAM complex remains to be determined (see Discussion).
Multicopy Suppression of sam35-424 Cells Suggests Sam37 Acts Downstream of Sam35
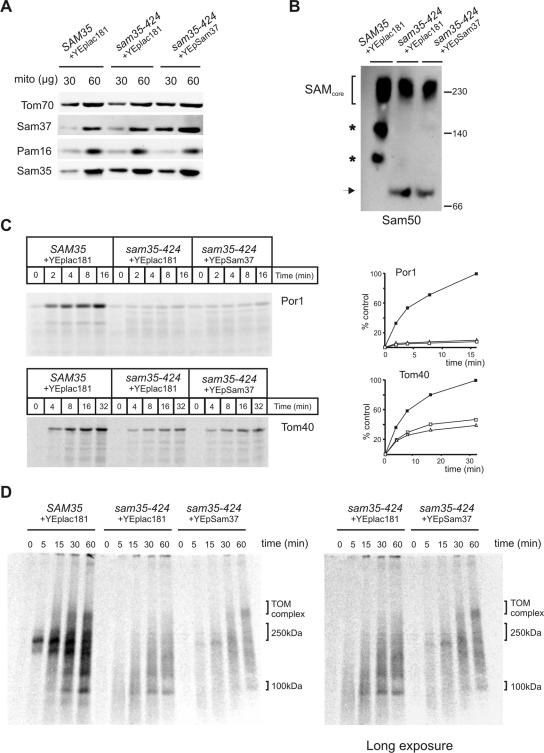
Mitochondria were isolated from Δsam35 cells transformed with plasmids either carrying the wild-type SAM35, or sam35-424, or sam35-424 and overexpressing SAM37, and immunoblots confirmed overexpression of Sam37 (Figure 3A). Because the SAM complex of sam35-424 is destabilized (Figure 2C), we investigated whether overexpression of Sam37 in sam35-424 resulted in stabilization of the SAM complex, which could in turn enable more efficient assembly of Tom40 into the TOM complex and the assembly of other β-barrel proteins, explaining the suppressed growth phenotype of sam35-424. To test this, mitochondria from sam35-424 and sam35-424 overexpressing Sam37 were solubilized in digitonin-containing buffer, and protein complexes were analyzed by BN-PAGE. Anti-Sam50 was used to immunoblot for the SAM complexes (Figure 3B). Overexpression of Sam37 does not stabilize the SAM complex in sam35-424, because the SAM complex in sam35-424 overexpressing Sam37 is similar to sam35-424 mitochondria alone.
Figure 3.
Overexpression of Sam37 suppresses the phenotypes of sam35-424 by functioning as an assembly factor downstream of Sam35. (A) Mitochondria from Δsam35 cells transformed with plasmid containing wild-type SAM35 and vector YEplac181, sam35-424 transformed with the vector YEplac181, and sam35-424 expressing multiple copies of SAM37 from YEplac181 were analyzed by SDS-PAGE, and immunoblot analysis was completed using antisera against the indicated proteins. (B) Mitochondria (100 μg) from wild-type SAM35 or sam35-424 cells, transformed with the indicated plasmids, were solubilized in 1% digitonin buffer. Protein complexes analyzed by BN-PAGE, followed by blotting onto PVDF membrane and immunodecoration by using anti-Sam50 antisera. Asterisks, unidentified subcomplexes of the SAM complex obtained during solubilization; arrow, Sam50-containing complex specifically enriched in sam35-424. (C) Mitochondria from the indicated strains were preincubated at 37°C for 15 min, followed by equilibration at 25°C for 5 min before incubation with 35S-labeled porin or Tom40 at 25°C for the indicated time. Unimported precursors were removed by treatment with 50 μg/ml proteinase K, and imported proteins were analyzed by SDS-PAGE and digital autoradiography. Quantification of the results shown on the right. Filled boxes, SAM35; open boxes, sam35-424; open triangles, sam35-424 overexpressing SAM37. (D) 35S-labeled Tom40 was imported. into mitochondria from the indicated strains. Shown on the right is a longer exposure of a portion from the same gel.
Protein import was monitored into mitochondria of Δsam35 cells transformed with plasmids either carrying the wild-type SAM35, or sam35-424, or sam35-424 and overexpressing SAM37, but no improvement of the amount of porin or Tom40 imported into sam35-424 when Sam37 was overexpressed (Figure 3C). Because the imported Tom40 shown in Figure 3C represents Tom40 molecules that are in a protease inaccessible and membrane-protected environment, it does not discriminate between forms of Tom40 in assembly intermediates from those that are properly assembled into the TOM complex, as both are protease insensitive (Model et al., 2001). To address this, we analyzed the import of Tom40 by BN-PAGE (Figure 3D). Consistent with the results in Figure 2E, where the overall signal from the radiolabeled Tom40 is highly reduced in both sam35-424 and when it is overexpressing Sam37, a clear difference can be seen in the distribution of Tom40 on BN-PAGE. Overexpression of Sam37 promotes the assembly of Tom40 molecules into the TOM complex, without improving the capacity of sam35-424 mitochondria to import more Tom40.
Overexpression of Sam37 Stabilizes the SAM Complex in sam35-409, and Steady-State Levels of Sam35 and Sam37 Are Codependent
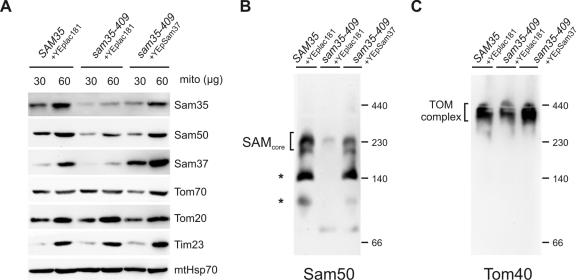
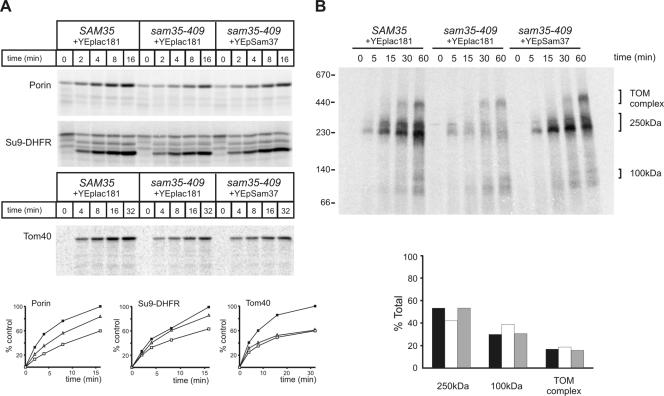
The sam35-409 mutant strain is also temperature sensitive, and, like sam35-424, its phenotype is suppressed by overexpression of SAM37. However, the mechanism of suppression of the sam35-409 phenotype by Sam37 is distinct from that seen in sam35-424. Mitochondria were isolated from Δsam35 cells transformed with plasmids either carrying the wild-type SAM35, or sam35-409, or sam35-409 and overexpressing SAM37. Immunoblotting for Sam35 by using two independent polyclonal sera raised against Sam35 shows that the steady-state level of the mutant protein in sam35-409 is significantly reduced, leading to a moderately reduced level of Sam50 and a highly reduced level of Sam37 (Figure 4A). The levels of outer membrane protein Tom70 and Tom20, inner membrane protein Tim23, and matrix protein mtHsp70 remain similar to wild type. By overexpressing Sam37 in sam35-409, the levels of the mutant sam35 protein and Sam50 are largely restored (Figure 4A). The steady-state level of the SAM complex, too, is restored as judged by immunoblots from BN-PAGE (Figure 4B). Concomitantly, the steady-state level of assembled TOM complex is like wild type in mitochondria from sam35-409 cells, provided SAM37 is overexpressed (Figure 4C).
Figure 4.
Overexpression of Sam37 suppresses the phenotype of sam35-409 by maintaining the level of sam35 in the SAM complex. (A) Mitochondria from Δsam35 cells transformed with plasmid encoding wild-type SAM35 and vector YEplac181, sam35-409 transformed with the vector YEplac181, and sam35-409 expressing multiple copies of SAM37 from YEplac181 were isolated from lactate medium at 25°C. The indicated amount of total protein (micrograms) was analyzed by SDS-PAGE, blotted onto nitrocellulose membrane, and immunodecorated using antisera against the indicated proteins. (B) Mitochondria (100 μg) were solubilized in 1% digitonin buffer, and protein complexes were analyzed by BN-PAGE, followed by blotting onto PVDF membrane and immunodecoration using anti-Sam50 antisera. Asterisks, unidentified subcomplexes of the SAM complex obtained during solubilization. (C) Mitochondria (100 μg) were subject to BN-PAGE and immunoblot analysis using anti-Tom40 antisera.
The sam35-409 mutant shows a defect in the import of porin and the matrix-targeted Su9-DHFR (Figure 5A). This is consistent with a small decrease in the steady-state level of the TOM complex (Figure 4C). Overexpression of SAM37 restores the level of the TOM complex (Figure 4C), restores import of Su9-DHFR into the matrix, and partially restores the import of porin (Figure 5A). However, the sam53-409 mutant also shows a defect in the level of 35S-labeled Tom40 accumulated within the outer membrane and/or intermembrane space that is not restored by overexpression of SAM37 (Figure 5A). We interpret this to reflect that only some of the imported 35S-labeled Tom40 is productively bound to the SAM complex and that, although SAM37 can suppress the defect in productive binding to the SAM complex (Figure 5B), it does not restore proteinase K protection to 35S-labeled Tom40 not bound to the SAM complex.
Figure 5.
Restoration of the TOM and SAM complexes in sam35-409 by overexpression of Sam37 cure import and assembly defects into various mitochondrial subcompartments. (A) 35S-labeled porin, Su9-DHFR, and Tom40 were incubated with mitochondria isolated from Δsam35 cells transformed with plasmid encoding wild-type SAM35 and vector YEplac181, sam35-409 transformed with the vector YEplac181, and sam35-409 expressing multiple copies of SAM37 from YEplac181 for the indicated time at 25°C. Precursors not imported were removed by treatment with 50 μg/ml proteinase K and analyzed by SDS-PAGE, followed by digital autoradiography. Quantification of the results is shown below. Filled boxes, SAM35; open boxes, sam35-409; open triangles, sam35-409 overexpressing Sam37. (B) 35S-labeled Tom40 precursor was incubated with the mitochondria from the indicated strains for the indicated time at 25°C. Mitochondria were reisolated and solubilized in 0.8% digitonin buffer. Insoluble materials were pelleted, and protein complexes were analyzed by BN-PAGE and digital autoradiography. Quantification of the signals from each assembly intermediate as a percentage of the total signal from 250-kDa, 100-kDa intermediates, and the TOM complex is shown below. Black bars, SAM35; white bars, sam35-409; gray bars, sam35-409–overexpressing Sam37.
There is a defect in the amount of 35S-labeled Tom40 productively incorporated into a 250-kDa assembly intermediate (consisting of nascent Tom40 associated with the SAMcore complex) in the sam35-409 mutant (Figure 5B). However, the kinetics of Tom40 assembly seems undiminished: of the 35S-labeled Tom40 that gets into this first intermediate, equivalent proportions occur subsequently in the assembly intermediate and mature TOM complex (Figure 5B). Overexpression of SAM37 fully restores the amount of 35S-labeled Tom40 that gets into the 250-kDa assembly intermediate, without changing the kinetics of the assembly reaction. Both the capacity to assemble TOM complexes (Figure 5B) and the concomitant import of precursors such as Su9-DHFR through the TOM complex (Figure 5A) are fully restored by overexpression of SAM37, which stabilizes the SAM complex in sam35-409. The sam35-409 mutant reflects a case in which increased copy number of a partner protein (Sam37) can compensate for a destabilizing mutation such that levels and functionality of the SAM complex are maintained.
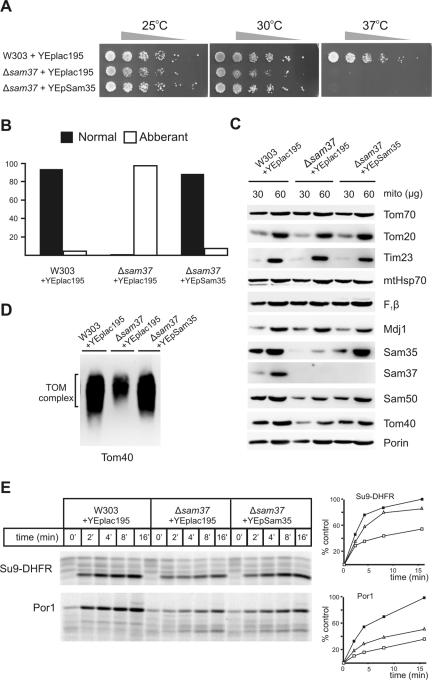
Decreased Levels of Sam35 Contribute to Phenotypes of Δsam37 Cells
Given the apparent codependence on Sam37 for the function of Sam35, we asked whether overexpression of Sam35 in Δsam37 cells suppresses any aspects of the phenotype of Δsam37 cells. The growth of wild-type (W303), Δsam37, and Δsam37 cells overexpressing Sam35 was compared. The Δsam37 mutants are temperature sensitive (Figure 6A; Gratzer et al., 1995; Meisinger et al., 2007). Overexpression of Sam35 in Δsam37 cells can suppress the growth defects at 25 and 30°C, but not the lethality at 37°C (Figure 6A). Because Δsam37 cells also display a mitochondrial morphology defect (Meisinger et al., 2004), we asked whether overexpression of Sam35 can suppress this defect as well. Confocal microscopy was used to compare mitochondrial morphology from 100 cells randomly selected from each of wild-type and mutant strains grown in lactate medium at 30°C. In wild-type cells, almost all cells contain the normal reticulated network of mitochondria (Figure 6B), whereas almost all Δsam37 cells contained mitochondria with aberrant, aggregated mitochondria. Overexpression of Sam35 in Δsam37 cells restored ∼90% of the cells to wild-type mitochondrial morphology (Figure 6B).
Figure 6.
Highly reduced levels of Sam35 contribute to phenotypes of Δsam37 cells. (A) Serial fivefold dilutions of wild-type (W303) cells transformed with YEplac195, Δsam37 cells transformed with YEplac195, and Δsam37 cells transformed with YEpSam35 were spotted onto synthetic complete medium plates lacking uracil and incubated at the indicated temperatures for 2–4 d. (B) Cells from the three strains described in A were grown to mid-logarithmic phase in synthetic complete medium with lactate at 30°C and stained with Mitotracker Red; cells were analyzed by confocal microscopy. One hundred cells from each strain were randomly selected, and their mitochondrial morphology was recorded. “Normal” (filled) represents mitochondria that exist as reticulated network of tubules, “Abberant” (open) represents mitochondria that exist as condensed organelles, aggregated organelles, or both. (C) Mitochondria were isolated from the three strains described in A in lactate medium at 25°C and the indicated amount (micrograms of total protein) analyzed by SDS-PAGE, blotted on to nitrocellulose membranes, and immunodecorated using antisera against the indicated proteins. (D) Mitochondria (100 μg of protein) from the indicated strains were solubilized in 1% digitonin buffer. Protein complexes were analyzed by BN-PAGE and blotted onto PVDF membranes, followed by immunodecoration by using anti-Tom40 antisera. (E) 35S-labeled Su9-DHFR and porin precursors were incubated with the indicated strains (described in A) for the indicated time at 25°C. Precursors not imported were removed by treatment with 50 μg/ml proteinase K followed by analysis by SDS-PAGE and digital autoradiography. Quantification of the results is shown on the right. Filled boxes, W303; open boxes, Δsam37; and open triangles, Δsam37-overexpressing Sam35.
To see whether the steady-state level of Sam35 is affected in Δsam37 cells, mitochondria were isolated from wild-type, Δsam37, and Δsam37 cells overexpressing Sam35 from lactate medium at 25°C. Mitochondria were analyzed by SDS-PAGE and immunoblotted for SAM complex components and various mitochondrial proteins (Figure 6C). The levels of the TOM complex receptors (Tom70 and Tom20), the core subunit of the inner membrane TIM23 translocase (Tim23), and matrix proteins (mtHsp70, F1β, and Mdj1) are very similar between wild-type, Δsam37, and Δsam37 cells overexpressing Sam35. The level of Sam35, however, is dramatically reduced in Δsam37 mitochondria, and the level of Sam50 is moderately reduced. The level of Tom40 is also reduced in Δsam37 mitochondria, but we did not detect a reduction in the levels of porin. By overexpressing Sam35 in Δsam37 cells, the level of Sam35 restored, and it also largely restored the levels of Sam50 and Tom40 (Figure 6C).
Consistent with the decreased Tom40 levels in Δsam37 mitochondria, the TOM complex is also diminished (Figure 6D). By restoring the levels of Sam35 in Δsam37 mitochondria via overexpression of Sam35, the steady-state levels of the TOM complex is restored (Figure 6D). An improved import of Su9-DHFR and porin across the outer membrane when Sam35 is overexpressed in Δsam37 mitochondria (Figure 6E) is consistent with the observed increase in the steady-state level of TOM complex.
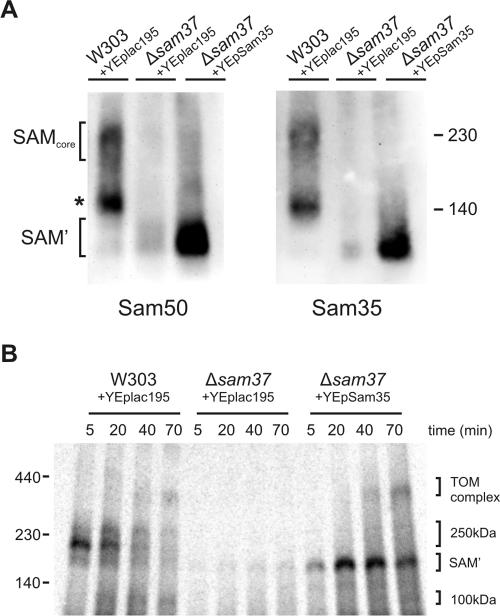
Sam35–Sam50 Complex Is the Functional Core Module of the SAM Complex
Because loss of Sam37 leads to reduced levels of Sam35, we used BN-PAGE to analyze this effect at the level of the SAM complex (Figure 7A). Immunoblotting for Sam50 and Sam35 shows that Δsam37 mitochondria cannot form the 230-kDa SAM complex but instead forms an ∼130-kDa complex consisting of Sam50 and Sam35 (SAM'; Figure 7A), as reported previously (Wiedemann et al., 2003; Waizenegger et al., 2004), but even this 130-kDa complex is present at reduced levels in Δsam37 mitochondria. Overexpression of Sam35 in Δsam37 mitochondria significantly stabilized the 130-kDa Sam35–Sam50 complex (Figure 7A).
Figure 7.
Sam35–Sam50 complex is sufficient for binding and assembly of β-barrel substrates. (A) Mitochondria (100 μg of total protein) isolated from wild-type (W303) cells transformed with YEplac195, Δsam37 cells transformed with YEplac195, and Δsam37 cells transformed with YEpSAM35 were solubilized in 1% digitonin buffer, and protein complexes were analyzed by BN-PAGE, blotted onto PVDF membranes, and immunodecorated with antisera against Sam50 and Sam35. (B) 35S-labeled Tom40 precursor was incubated with mitochondria from the indicated strains for the indicated time at 25°C, and import was stopped by incubation on ice. Mitochondria were reisolated and solubilized with 0.8% digitonin buffer. Insoluble materials were pelleted, and protein complexes analyzed by BN-PAGE and digital autoradiography.
We tested the ability of this SAM' complex to assemble Tom40 precursor into the TOM complex (Figure 7B). In contrast to Δsam37 mitochondria where very little 35S-labeled Tom40 precursor is bound, stabilization of the Sam35–Sam50 complex (via overexpression of Sam35) is sufficient to restore high levels of Tom40 precursors bound even in the absence of Sam37. Together, our data suggest that the Sam35–Sam50 complex is sufficient to assemble β-barrel proteins into the outer membrane. The assembly defect in Δsam37 mitochondria is partly due to the decreased levels of Sam35, and the function of Sam37 is to facilitate release of substrate β-barrels from the SAMcore complex.
DISCUSSION
Functions of Sam35 and Sam37
The two peripheral membrane proteins of the yeast SAM complex, Sam35 and Sam37, function codependently in the biogenesis of β-barrel proteins. Overexpression of Sam37, but not Sam50 or other proteins implicated in the β-barrel protein biogenesis pathway, was able to suppress the lethality of 13 independent temperature-sensitive sam35 alleles. We characterized two of these temperature-sensitive alleles, sam35-424 and sam35-409, and we showed that the mechanisms by which overexpression of Sam37 suppressed the mutant phenotypes are distinct. In sam35-424 mutants, the rate of assembly of Tom40 into the TOM complex was decreased as a result of the mutation in Sam35. This was reflected in a decreased amount of Tom40 in the assembly intermediate I (the 250-kDa complex of Tom40 substrate in contact with the SAMcore complex). Overexpression of SAM37 does not stabilize the SAMcore complex in sam35-424. The effect of increasing Sam37 levels is to accelerate the assembly of Tom40 into the TOM complex; but this occurs without an increase in the amount of β-barrel substrate bound by the SAMcore. This suggests the suppression of β-barrel protein assembly defects in this mutant is achieved via the specific function of Sam37 as an assembly factor, that acts downstream of Sam35, to mediate effective release of substrate β-barrels from the SAM complex.
In sam35-409 cells the steady-state levels of the mutant Sam35 protein is significantly decreased, leading to a strong decrease in the levels of the SAMcore complex as detected by BN-PAGE. Overexpression of SAM37 in the sam35-409 mutants suppressed the mutant phenotype by maintaining the levels of the mutant sam35-409 subunit in the SAMcore complex. Likewise, Δsam37 cells can be cured of protein import defects by overexpression of SAM35, which restores a stable SAM' complex (albeit without Sam37). The SAM' complex is fully competent for binding Tom40 substrate. Together, the data presented here suggest an important function of Sam37 is to maintain the stability of Sam35; but an additional, specific function is to affect the release of folded substrate proteins from the SAM complex.
The phenotypes of the sam35 alleles and the mechanism and consequences for Sam37 multicopy suppression of the phenotypes of these alleles can be explained by two potential, related activities for Sam35. In the first, Sam35 is a receptor for Tom40 and other β-barrel proteins, being the first point of contact for substrates entering the SAM complex. However, because substrates enter the SAM complex from the intermembrane space (Model et al., 2001), and Sam35 is exposed on the mitochondrial surface (Ishikawa et al., 2004; Milenkovic et al., 2004; Waizenegger et al., 2004), any “receptor” domain of Sam35 would need to sit exposed to the intermembrane space. Mutations like that in the sam35-424 cells that decrease receptor activity, or a decrease in the level of Sam35 as seen in Δsam37 cells, would thereby inhibit import of Tom40.
Our data also suggest Sam35 directly assists Sam50 to form the 250-kDa assembly intermediate of Tom40 (or an equivalent substrate:SAM complex for other β-barrel substrates). If the precise function of Sam35 in this process is to assist binding of β-barrel substrates to Sam50, it would explain the subtle effects seen in sam35-424 cells overexpressing Sam37: mitochondria from these cells remain incompetent at binding high levels of Tom40 substrate because of the mutation in Sam35, but they exhibit an increased clearance of the bound Tom40 out of Sam50, provided enough Sam37 is present. We would argue that the sam35-424 mutant sits at a functional tipping point and that Sam37 can influence the structural stability of the mutant Sam35 protein to enhance the release of β-barrel substrates without a gross improvement of the binding capacity seen in the SAM complex.
Does this codependency hold for the metaxins, the putative human counterparts of Sam35 and Sam37? The SAM complex found in human cells is around 300 kDa (Humphries et al., 2005; Kozjak-Pavlovic et al., 2007), and it does not contain metaxins stably bound to it (Kozjak-Pavlovic et al., 2007). Although a proportion of Metaxin 1 and Metaxin 2 are found on the mitochondrial surface, the proteins have also been observed free in the cytosol (Armstrong et al., 1997). Metaxin 1 and Metaxin 2, which share sequence similarity to Sam37 and Sam35, respectively, are found together in a much larger complex of ∼600 kDa (Kozjak-Pavlovic et al., 2007). Consistent with the codependence of Sam35 and Sam37, RNAi knockdown of the expression levels of Metaxin 2 causes a concomitant decrease in Metaxin 1 levels. Mitochondria from these “metaxin-depleted” cells are defective in assembly of β-barrel proteins into their outer membrane (Kozjak-Pavlovic et al., 2007).
Has Assembly Intermediate II Already Left the SAM Complex?
After import, unfolded Tom40 precursors rapidly bind the SAMcore complex to form an assembly intermediate I that migrates on BN-PAGE at ∼250 kDa and contains Sam37, Sam35, and Sam50 as well as Tom40 substrate (Model et al., 2001; Paschen et al., 2003; Wiedemann et al., 2003; Ishikawa et al., 2004; Milenkovic et al., 2004; Waizenegger et al., 2004). Under the same solubilization conditions, Tom40 substrates can be seen to move subsequently into a form that contains Tom5 and perhaps other small Tom proteins (Model et al., 2001; Wiedemann et al., 2003), with this intermediate running at ∼100 kDa on BN-PAGE (Model et al., 2001). No SAM complex subunits comigrate with this solubilized complex. One possibility is that this assembly intermediate has been released from the SAM complex to exist independently in the outer membrane and that other Tom subunits (such as Tom7, Tom22, and Tom20) will be subsequently added, unassisted, to the assembly intermediate to eventually form a mature TOM complex.
However, an alternative possibility is that the 100-kDa assembly intermediate II is still bound to the SAM complex in outer membranes but that the nascent substrate complex is solubilized out of the SAM complex by the lysis conditions used for BN-PAGE. This distinction matters. It offers an explanation to our observation that Sam37 can function as an assembly factor for Tom40 downstream of the 250-kDa intermediate I, by facilitating the progression of Tom40 from the 100-kDa intermediate II to the mature TOM complex. It also helps rationalize observations made of Mdm10: that it mediates the late stage of TOM complex assembly (from 100-kDa intermediate II to mature TOM complex) and that it associates with the SAM complex. The functional interplay between Tom7 and Mdm10 identified recently by Meisinger et al. (2006) is also consistent with this alternative interpretation.
Why Do Mitochondria Need Sam35 and Sam37, When Bacteria Do Not?
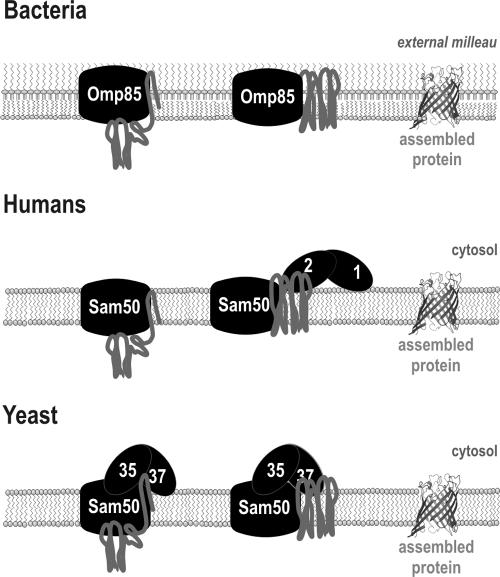
The mitochondrial SAM complex is derived from the bacterial Omp85 complex, but, to date, no detailed structural analysis of a mitochondrial β-barrel protein has been completed. Might differences in the mitochondrial β-barrel substrate proteins dictate a need for Sam35 and Sam37? One clear difference exists in the nature of the external environment of the mitochondrial and bacterial outer membranes. Bacterial outer membranes are built on an asymmetric bilayer, with phospholipids confined to the inner leaflet and glycolipids in the outer leaflet (Kamio and Nikaido, 1976). Bacterial β-barrel proteins have elongated interstrand loops that would likely sit within the glycolipid environment: mitochondrial β-barrels have interstrand loops that would be exposed to the cytosol and might be more difficult to fold, require protection from cytoplasmic proteases during the folding process, or both. Both the transient interaction of metaxins in human cells and the constant, sheltering presence of Sam35 and Sam37 in yeast would afford a protective environment for assembly of the extramembrane domains of mitochondrial β-barrel proteins (Figure 8). Certainly, the cytosol of a eukaryote presents a much more complex and protein-rich environment than the extracellular medium surrounding a bacterial cell, and factors that can assist substrate proteins into and out from the SAM complex would benefit the folding of β-barrels in the mitochondrial outer membrane.
Figure 8.
Schematic representation of β-barrel protein assembly in bacteria, humans, and yeast. In bacteria, Omp85, together with factors facing the periplasm, is necessary and sufficient for assisting correct assembly of β-barrel into the bacterial outer membrane (Voulhoux et al., 2003; Ruiz et al., 2005; Wu et al., 2005). In humans, Metaxin 1 and Metaxin 2 are associated with the mitochondrial outer membrane as part of an uncharacterized ∼600-kDa complex (Kozjak-Pavlovic et al., 2007) and assist Sam50 in assembling β-barrel proteins into the outer membrane. In yeast, Sam35 and Sam37 associate directly with Sam50 to from the SAMcore complex. Sam35 assists Sam50 to bind β-barrel precursors, and Sam37 is important for clearance of β-barrel precursors from the SAM complex, thereby assisting assembly. Sam35 and Sam37 are important for stabilizing each other at the SAM complex, most likely via direct interactions.
ACKNOWLEDGMENTS
We thank Chris Meisinger (Institute for Biochemistry and Molecular Biology, University of Freiburg, Germany) and Toshiya Endo (Department of Chemistry, Graduate School of Science, Nagoya University, Japan) for generously providing antibodies and Ian Gentle and Dejan Bursać for critical comments on the manuscript. This work was supported by grants from the Australian Research Council and National Health & Medical Research Council (to T.L.) and an Australian Postgraduate Award (to N.C.C.)
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-08-0796) on October 31, 2007.
REFERENCES
- Armstrong L. C., Komiya T., Bergman B. E., Mihara K., Bornstein P. Metaxin is a component of a preprotein import complex in the outer membrane of the mammalian mitochondrion. J. Biol. Chem. 1997;272:6510–6518. doi: 10.1074/jbc.272.10.6510. [DOI] [PubMed] [Google Scholar]
- Buchanan S. K. Beta-barrel proteins from bacterial outer membranes: structure, function and refolding. Curr. Opin. Struct. Biol. 1999;9:455–461. doi: 10.1016/S0959-440X(99)80064-5. [DOI] [PubMed] [Google Scholar]
- Daum G., Bohni P. C., Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Dolezal P., Likic V., Tachezy J., Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Gabriel K., Buchanan S. K., Lithgow T. The alpha and the beta: protein translocation across mitochondrial and plastid outer membranes. Trends Biochem. Sci. 2001;26:36–40. doi: 10.1016/s0968-0004(00)01684-4. [DOI] [PubMed] [Google Scholar]
- Gabriel K., Egan B., Lithgow T. Tom40, the import channel of the mitochondrial outer membrane, plays an active role in sorting imported proteins. EMBO J. 2003;22:2380–2386. doi: 10.1093/emboj/cdg229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle I., Gabriel K., Beech P., Waller R., Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle I. E., Burri L., Lithgow T. Molecular architecture and function of the Omp85 family of proteins. Mol. Microbiol. 2005;58:1216–1225. doi: 10.1111/j.1365-2958.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- Gratzer S., Lithgow T., Bauer R. E., Lamping E., Paltauf F., Kohlwein S. D., Haucke V., Junne T., Schatz G., Horst M. Mas37p, a novel receptor subunit for protein import into mitochondria. J. Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib S. J., Waizenegger T., Niewienda A., Paschen S. A., Neupert W., Rapaport D. The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial beta-barrel proteins. J. Cell Biol. 2007;176:77–88. doi: 10.1083/jcb.200602050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries A. D., Streimann I. C., Stojanovski D., Johnston A. J., Yano M., Hoogenraad N. J., Ryan M. T. Dissection of the mitochondrial import and assembly pathway for human Tom40. J. Biol. Chem. 2005;280:11535–11543. doi: 10.1074/jbc.M413816200. [DOI] [PubMed] [Google Scholar]
- Ishikawa D., Yamamoto H., Tamura Y., Moritoh K., Endo T. Two novel proteins in the mitochondrial outer membrane mediate beta-barrel protein assembly. J. Cell Biol. 2004;166:621–627. doi: 10.1083/jcb.200405138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamio Y., Nikaido H. Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- Kozjak V., Wiedemann N., Milenkovic D., Lohaus C., Meyer H. E., Guiard B., Meisinger C., Pfanner N. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem. 2003;278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- Kozjak-Pavlovic V., Ross K., Benlasfer N., Kimmig S., Karlas A., Rudel T. Conserved roles of Sam50 and metaxins in VDAC biogenesis. EMBO Rep. 2007;8:576–582. doi: 10.1038/sj.embor.7400982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Meisinger C., et al. The morphology proteins Mdm12/Mmm1 function in the major beta-barrel assembly pathway of mitochondria. EMBO J. 2007;26:2229–2239. doi: 10.1038/sj.emboj.7601673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C., et al. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell. 2004;7:61–71. doi: 10.1016/j.devcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Meisinger C., Wiedemann N., Rissler M., Strub A., Milenkovic D., Schonfisch B., Muller H., Kozjak V., Pfanner N. Mitochondrial protein sorting: differentiation of beta-barrel assembly by Tom7-mediated segregation of Mdm10. J. Biol. Chem. 2006;281:22819–22826. doi: 10.1074/jbc.M602679200. [DOI] [PubMed] [Google Scholar]
- Milenkovic D., Kozjak V., Wiedemann N., Lohaus C., Meyer H. E., Guiard B., Pfanner N., Meisinger C. Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability. J. Biol. Chem. 2004;279:22781–22785. doi: 10.1074/jbc.C400120200. [DOI] [PubMed] [Google Scholar]
- Model K., Meisinger C., Prinz T., Wiedemann N., Truscott K. N., Pfanner N., Ryan M. T. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat. Struct. Biol. 2001;8:361–370. doi: 10.1038/86253. [DOI] [PubMed] [Google Scholar]
- Nijtmans L. G., Henderson N. S., Holt I. J. Blue Native electrophoresis to study mitochondrial and other protein complexes. Methods. 2002;26:327–334. doi: 10.1016/S1046-2023(02)00038-5. [DOI] [PubMed] [Google Scholar]
- Paschen S. A., Neupert W., Rapaport D. Biogenesis of beta-barrel membrane proteins of mitochondria. Trends Biochem. Sci. 2005;30:575–582. doi: 10.1016/j.tibs.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Paschen S. A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., Neupert W. Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- Pfanner N., Wiedemann N., Meisinger C., Lithgow T. Assembling the mitochondrial outer membrane. Nat. Struct. Mol. Biol. 2004;11:1044–1048. doi: 10.1038/nsmb852. [DOI] [PubMed] [Google Scholar]
- Ruiz N., Falcone B., Kahne D., Silhavy T. J. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. Molecular Cloning: A Laboratory Manual. Cold Spring Habor, NY: Cold Spring Harbor Press; 2001. [Google Scholar]
- Schleiff E., Soll J. Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep. 2005;6:1023–1027. doi: 10.1038/sj.embor.7400563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo L. F., Yaffe M. P. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J. Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux R., Bos M. P., Geurtsen J., Mols M., Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- Waizenegger T., Habib S. J., Lech M., Mokranjac D., Paschen S. A., Hell K., Neupert W., Rapaport D. Tob38, a novel essential component in the biogenesis of beta-barrel proteins of mitochondria. EMBO Rep. 2004;5:704–709. doi: 10.1038/sj.embor.7400183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger T., Schmitt S., Zivkovic J., Neupert W., Rapaport D. Mim1, a protein required for the assembly of the TOM complex of mitochondria. EMBO Rep. 2005;6:57–62. doi: 10.1038/sj.embor.7400318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N., Kozjak V., Chacinska A., Schonfisch B., Rospert S., Ryan M. T., Pfanner N., Meisinger C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 2003;424:565–571. doi: 10.1038/nature01753. [DOI] [PubMed] [Google Scholar]
- Wittig I., Braun H. P., Schagger H. Blue native PAGE. Nat. Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Wu T., Malinverni J., Ruiz N., Kim S., Silhavy T. J., Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]