Abstract
The 70-kDa heat-shock cognate protein (Hsc70) chaperone is an ATP-dependent “disassembly enzyme” for many subcellular structures, including clathrin-coated vesicles where it functions as an uncoating ATPase. Hsc70, and its cochaperone auxilin together catalyze coat disassembly. Like other members of the Hsp70 chaperone family, it is thought that ATP-bound Hsc70 recognizes the clathrin triskelion through an unfolded exposed hydrophobic segment. The best candidate is the unstructured C terminus (residues 1631–1675) of the heavy chain at the foot of the tripod below the hub, containing the sequence motif QLMLT, closely related to the sequence bound preferentially by the substrate groove of Hsc70 (Fotin et al., 2004b). To test this hypothesis, we generated in insect cells recombinant mammalian triskelions that in vitro form clathrin cages and clathrin/AP-2 coats exactly like those assembled from native clathrin. We show that coats assembled from recombinant clathrin are good substrates for ATP- and auxilin-dependent, Hsc70-catalyzed uncoating. Finally, we show that this uncoating reaction proceeds normally when the coats contain recombinant heavy chains truncated C-terminal to the QLMLT motif, but very inefficiently when the motif is absent. Thus, the QLMLT motif is required for Hsc-70–facilitated uncoating, consistent with the proposal that this sequence is a specific target of the chaperone.
INTRODUCTION
Clathrin-coated vesicles, the best known carrier of intracellular membrane traffic, transport proteins and lipids from the plasma membrane to endosomes and between endosomes and the trans-Golgi network. Polymerization of the principal coat protein clathrin into a lattice-like assembly by sequential addition of individual, cytosolic clathrin trimers to a growing shell shapes the budding coated pit (Ehrlich et al., 2004; for review, see Kirchhausen, 2000). Intermediary proteins, known as adaptors, form the interface between the outer clathrin coat and the incorporated membrane bilayer. These adaptors selectively recruit membrane-anchored proteins (“cargo”). The most prominent adaptors are the heterotetrameric AP-2 and its cousins AP-1, AP-3, and AP-4, but there are other, presumably more specialized, adaptors, such as β-arrestins, epsin, GGAs, and dishevelled (Yu et al., 2007; for review, see Owen et al., 2004; Robinson, 2004).
Coats can assemble in vitro without incorporated membrane, either from clathrin alone (“cages”, stable only at reduced pH) or from clathrin plus heteroteterameric APs (“coats”, stable at neutral pH) (Keen et al., 1979; Kirchhausen and Harrison, 1981; Vigers et al., 1986a,b; Kirchhausen, 2000; Fotin et al., 2004b). Coat assembly in vitro can proceed to completion without intervention of other factors. In vivo, coat assembly occurs only on membrane surfaces, and fission (pinching off) of the incorporating bilayer requires the large GTPase, dynamin (Sever, 2002; Praefcke and McMahon, 2004; Kruchten and McNiven, 2006; Macia et al., 2006). Removal of the coat (uncoating), essential for subsequent vesicle fusion and cargo delivery, is the last step in the clathrin cycle (Ehrlich et al., 2004; Merrifield et al., 2005; Lee et al., 2006; Massol et al., 2006). This step proceeds rapidly in vivo (∼5 s) immediately after membrane fission.
The cytosolic, heat-shock cognate 70-kDa protein (Hsc70), as shown many years ago using in vitro biochemical assays, has clathrin uncoating activity (Schmid et al., 1985; Schmid and Rothman, 1985; Ungewickell, 1985; Chappell et al., 1986; Greene and Eisenberg, 1990). Convincing demonstration of its in vivo function followed identification of the protein auxilin as its specific cochaperone (Ungewickell et al., 1995). Inactivation of yeast auxilin (Swa2) leads to anomalous accumulation of coated vesicles (Gall et al., 2000; Pishvaee et al., 2000), and RNA interference with auxilin in nematodes causes similar defects (Greener et al., 2001). Mammalian cells express a brain-specific auxilin1 and a ubiquitous auxilin2 (also called cyclin G-associated kinase; GAK) (Greener et al., 2000; Umeda et al., 2000). Auxilins have a region similar to the phosphatase and C2 domains of phosphatase and tensin homolog deleted on chromosome ten (PTEN), a central segment with binding sites for dynamin, AP-2, and clathrin, and a C-terminal J-domain that recruits Hsc70 (Haynie and Ponting, 1996; Holstein et al., 1996; Ungewickell et al., 1997; Scheele et al., 2001; Sever et al., 2005). In addition, GAK has an N-terminal Ser/Thr kinase domain of unknown in vivo function. Recruitment of auxilin to assembling coats at the plasma membrane requires the PTEN-like domain and occurs only after pinching off of the vesicle (Massol et al., 2006). This timing ensures that uncoating will follow vesicle release.
An auxilin fragment comprising residues 547-910, which includes the clathrin-binding region and the J-domain, is sufficient for binding to clathrin coats in vitro, recruiting Hsc70 and for facilitating the uncoating reaction (Holstein et al., 1996; Umeda et al., 2000). The clathrin-binding region seems to be largely unstructured on the isolated fragment in solution, but ∼25 residues at its C-terminal end contribute two additional helices to the usual three of the globular J-domain (Ungewickell et al., 1997; Fotin et al., 2004a; Gruschus et al., 2004).
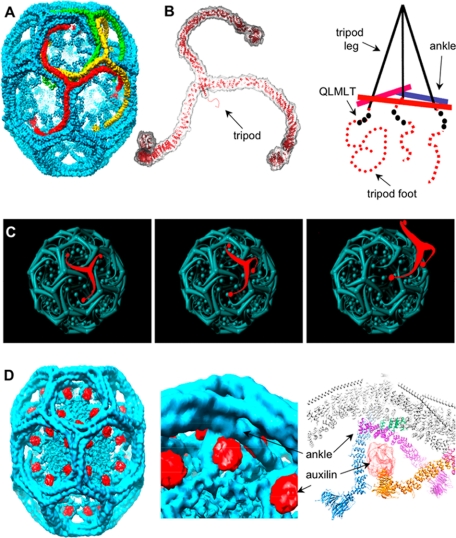
The clathrin trimer is a spider-like “triskelion,” which assembles by forming an elaborately interdigitated network (Figure 1A) (Kirchhausen and Harrison, 1981; Ungewickell and Branton, 1981; Smith et al., 1998; Musacchio et al., 1999; Fotin et al., 2004b). A triskelion hub lies at each vertex of the lattice. The proximal segments of the legs radiate toward the neighboring vertices and slope gently inward. The legs bend smoothly at the knees, and the distal segments extend toward the next vertex, where the ankles of three converging chains cross each other ∼75Å beneath the apex of the triskelion that is centered there. A tripod-like structure, formed by the C-terminal elements of the three converging clathrin heavy chains (HCs), projects inward from the apex and contacts the crossed ankles (Figure 1B) (Fotin et al., 2004b, 2006). The legs of the tripod are α-helices (clathrin residues 1590-1630), and its feet are less structured, proline-rich segments (residues 1631-1675). Thus, the C-terminal regions of one trimer project inward to interact with the ankle regions of trimers centered two vertices away.
Figure 1.
The clathrin triskelion and spatial relationships within a coat. (A) Three-dimensional image reconstruction of a clathrin coat at 8-Å resolution (Fotin et al., 2004b). The colored triskelions show three symmetry-independent molecules. (B) Backbone model of a triskelion for residues 1-1630, highlighting the location of its tripod underneath the hub, and schematic representation of the tripod as it relates to the ankles of legs from adjacent triskelions. (C) Schematic representation of how a relatively rigid rotation of a triskelion about its axis could “unlock” it from the coat, allowing withdrawal from the lattice without colliding with the remaining elements. (D) Three-dimensional image reconstruction of a clathrin coat with bound auxilin (547-910) at 12-Å resolution (Fotin et al., 2004a).
Despite the extensive interdigitation, assembly and disassembly are rapid (Ehrlich et al., 2004). The pattern of contacts is such that rotation of a triskelion about its axis could “unlock” it from the coat, and its shape would then permit it to withdraw from the lattice without colliding with the remaining elements (Figure 1C). If this description is correct, how might Hsc70 accelerate such a process? The structure of a clathrin coat complexed stoichiometrically with auxilin (547-910) provides an important clue (Fotin et al., 2004a). The auxilin fragment associates with the lattice at an inner radius, such that its J-domain faces toward a vertex (Figure 1D). Moreover, within the C-terminal segment at the foot of the tripod is the sequence motif QLMLT, closely related to the sequence bound preferentially by the substrate groove of Hsc70 (Takenaka et al., 1995). Thus, we have proposed that the auxilin J-domain recruits Hsc70 in an orientation that allows it to grab this segment and thereby to destabilize the coat (Fotin et al., 2004a).
To test this hypothesis, we have worked out how to coexpress clathrin heavy and light chains (LCs) in insect cells. We show that the recombinant triskelions assemble into lattices, forming clathrin cages and clathrin/AP-2 coats exactly like those assembled from native clathrin purified from bovine brain-coated vesicles. We then show that coats assembled from recombinant clathrin are, like those from brain-derived clathrin, good substrates for ATP- and auxilin-dependent, Hsc70 catalyzed uncoating. Finally, we show that this uncoating reaction proceeds normally when the coats contain recombinant heavy chains truncated C-terminal to the QLMLT motif, but very inefficiently when the truncation is N-terminal to it, when the motif is removed by internal deletion or when the motif is mutated. Thus, the QLMLT motif is required for Hsc-70 facilitated uncoating, consistent with the proposal that this sequence is indeed a specific target of the chaperone.
MATERIALS AND METHODS
Production of Recombinant Clathrin
A cDNA encoding rat clathrin heavy chain (Kirchhausen et al., 1987a) was used as a template to generate full-length (1675 HC), nested C-terminal truncations (1661 HC, 1643 HC, 1637 HC, 1630 HC, and 1596 HC), internal deletions (1675ΔPIVYGQ HC, 1643ΔPIVYGQ HC, and 1675ΔQLMLTA HC), and mutations (1643LML-AAA HC) of the heavy chain; each was then subcloned into the insect cell expression vector pFastBac1 (Invitrogen, Carlsbad, CA). A cDNA encoding rat liver clathrin light chain LCa (Kirchhausen et al., 1987b) was used as the template to subclone the region encoding the full light chain (residues 1-256) into the insect cell expression vector pFastBacHTb. The final construct (rLCa1i) contains at its N terminus a 6x-His-tag followed by a linker of 20 residues. Baculoviruses suitable for infection and expression were generated with the Bac-to-Bac system (BD Biosciences, San Jose, CA). Virus stocks were obtained after four rounds of amplification, and they were kept in the dark at 4°C. The open reading frame of rat brain clathrin light chain LCa1 was also used as a template to subclone it into the bacterial expression vector pET28b (Novagen, Madison, WI) between the NcoI and EcoRI restriction sites so as to generate a native, nontagged light chain. All constructs were verified by DNA sequencing.
Clathrin heavy chains together with light chain were expressed in Hi5 insect cells (1L, ∼1–1.5 × 106 cells/ml) grown for 2–3 d in spinner flasks at 27°C in Excell 420 medium after coinfection with the appropriate viruses. Alternatively, clathrin heavy chain only was expressed in a similar way. The cells were centrifuged at 1000 rpm for 10 min at room temperature by using an H6000A rotor (Sorvall, Newton, CT), and the pellets were resuspended in 20 ml lysis buffer (50 mM Tris, pH 8.0, 300 mM NaCl, 1 mM EDTA, 3 mM β-mercaptoethanol, and half of a tablet of Complete Protease Inhibitor Cocktail [Roche Applied Science, Indianapolis, IN]). The resuspended pellets were sonicated for 1 min on ice (Flat tip at 20% power, Ultrasonic processor XL; Heat Systems, Farmingdale, NY), cell debris was removed by centrifugation at 90,000 rpm for 20 min at 4°C by using a TLA 100.4 rotor (Beckman Coulter, Fullerton, CA), and the supernatant (∼20 ml) was dialyzed at 4°C for 12 h against 2 × 2 l of cage buffer (20 mM [2-(N-morpholino)ethanesulfonic acid] MES, pH 6.2, 2 mM CaCl2, 0.02% NaN3, and 0.5 mM dithiothreitol [DTT]). The sample was then centrifuged at 4°C, first at low speed (1000 rpm for 10 min) to remove large aggregates and then at high speed (54,000 rpm for 1 h) by using a Ti rotor (Beckman Coulter). The pellet, primarily containing clathrin (presumably assembled as cages) was resuspended in 6 ml of 100 mM MES, pH 6.5, 3 mM EDTA, 0.5 mM MgCl2, 0.02% NaN3, 0.5 mM DTT, and 0.5 mM phenylmethylsulfonyl fluoride) followed by addition of 3 ml of 2.4 M Tris, pH 7.4, 1 mM DTT, and incubation for 20 min at room temperature, a condition used to dissociate native clathrin assemblies. The sample was centrifuged at 90,000 rpm for 20 min at 4°C by using a TLA 100.4 rotor, and most of the clathrin was recovered in the supernatant.
The resulting sample was subjected to gel filtration chromatography (90 cm × Ø = 3 cm column containing Sephacryl-S 500 [GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom] in 0.5 M Tris, pH 7.4, 0.04% NaN3, and 0.5 mM DTT) at room temperature and with a flow of 2 ml/min. Fractions of 5.5 ml containing the clathrin peak (∼100 ml) were pooled and then subjected to adsorption chromatography (5 ml, hydroxyapatite, Econo-Pac CHT-II; Bio-Rad, Hercules, CA); the column was pre-equilibrated with low phosphate buffer (10 mM NaH2PO4, pH 7.1, 100 mM NaCl, 0.02% NaN3, and 0.5 mM DTT) and eluted with a linear gradient from low to high phosphate concentration (500 mM NaH2PO4, pH 7.1, 100 mM NaCl, 0.02% NaN3, and 0.5 mM DTT) at room temperature with a flow of 1 ml/min. Fractions (1 ml) were collected into microcentrifuge tubes containing 2 μl of 0.5 M EDTA. Typical clathrin yields were in the range of 3–40 mg per 1 l of cell culture. Western blot analysis was used to confirm the expression of clathrin heavy and light chains.
The rat clathrin light chain rLCa1b was expressed in Escherichia coli strain BL21(DE3). The bacteria were grown in Luria-Bertani (LB) medium containing 30 mg/l kanamycin at 37°C with shaking (250 rpm) to an optical density of 0.5. Expression was induced by addition of isopropyl β-d-thiogalactoside (IPTG) (final concentration, 0.6 mM). After 3 h, the cell were harvested by centrifugation at 5000 rpm for 10 min at 4°C by using an H6000A rotor (Sorvall) and resuspended in ice-cold lysis buffer (20 mM Bis-Tris adjusted to pH 6.0 at room temperature, 0.5 mM dithiothreitol, 1 mM EDTA, and Complete Protease Inhibitor Cocktail) by using 20 ml of lysis buffer per 3.5 g of wet cell weight. The suspension was placed into a glass vessel, and the vessel was immersed in boiling water for 4 min and then chilled on ice. The boiled suspension was centrifuged at 54,000 rpm for 30 min at 4°C by using a 60Ti rotor (Beckman Coulter) to remove the precipitated material. rLCa1b was purified from the filtered supernatant (0.2-μm syringe filter) by anion exchange chromatography at 4°C on a HiTrap MonoQ column equilibrated with buffer A (20 mM Bis-Tris, adjusted to pH 6.0 at room temperature, and 0.5 mM dithiothreitol) and eluted using a linear gradient from 0 to 32% buffer B (20 mM Bis-Tris, adjusted to pH 6.0 at room temperature, 0.5 mM dithiothreitol, and 1 M NaCl).
For the in vitro reconstitution of clathrin, recombinant heavy chain (expressed in insect cells without light chain) was mixed with excess rLCa1b (expressed in bacteria) by using a weight ratio of 3:1 (equivalent to a molar ratio HC:LC of 1:2.4) just before cage or coat assembly for 40 min at room temperature.
Production of Recombinant Auxilin
A protein chimera of glutathione transferase (GST) with bovine auxilin (spanning residues 547-910) was generated by fusion in the vector pGEX4T-1 and then used for expression in E. coli BL21 (Fotin et al., 2004a). The bacteria were grown in LB medium supplemented with ampicillin to an OD600 ∼ 0.5–0.6 at 37°C. Protein expression was induced by addition of 1 mM IPTG (final concentration) and the cells grown for another 4 h at 25°C. The cells (from 1 l of culture) were centrifuged at 5000 rpm for 15 min at 4°C, and the pellet was kept frozen overnight. The pellet was resuspended in 25 ml of pGEX lysis buffer (20 mM HEPES, pH 7.6, 100 mM KCl, 0.2 mM EDTA, 20% glycerol, 1 mM DTT, and half a tablet of Complete Protease Inhibitor Cocktail) and sonicated on ice using three consecutive sonication cycles of 60, 30, and 30 s (standard microtip, 20% power). The sample was centrifuged at 45,000 rpm for 1 h at 4°C by using a 60Ti rotor, and the supernatant mixed with 0.5 ml of a 50% (vol/vol) slurry of glutathione-Sepharose 4 beads (GE Healthcare). After 2 h of end-over-end rotation at 4°C, the beads were poured into a propylene Econo-Column (Bio-Rad), washed with 15 ml of pGEX lysis buffer, and then washed with 15 ml of 25 mM HEPES, pH 7.0, 100 mM NaCl, and 0.1 mM EGTA. Elution of GST-auxilin (in 2 ml) was achieved by supplementing the solution with 50 mM glutathione, adjusted to pH 8. These steps were carried out at 4°C. Release of the GST portion was achieved by incubation of 1 mg of GST-auxilin with 1 U of thrombin at room temperature for 6 h. Proteolysis was ended by addition of 1 mg of Pefabloc SC (Roche Applied Science). The ∼40-Da auxilin fragment was further purified using a Mono S column (Pharmacia, Peapack, NJ). The sample was first dialyzed overnight against MES buffer A (50 mM MES, pH 6.7, 1 mM EDTA, and 3 mM β-mercaptoethanol), and then it was loaded onto the column (pre-equilibrated with MES buffer A) and eluted with a linear gradient of buffer A and with MES buffer B (50 mM MES, pH 6.7, 500 mM NaCl, 1 mM EDTA, and 3 mM β-mercaptoethanol) at a flow of 1 ml/min. The auxilin sample was stored at −80°C with 20% glycerol (final concentration).
Production of Recombinant Hsc70
N-terminal 6x-His–tagged bovine Hsc70 (full length) cloned into the pET21a-vector was expressed in E. coli BL21. The bacteria were grown at 37°C in LB supplemented with 0.1 mg/ml ampicillin to an OD600 of ∼0.5, transferred to 28°C, and induced with 0.1 mM IPTG for 5 h. The cells were centrifuged at 5000 rpm for 15 min at 4°C, and the pellets from 1l culture resuspended in 25 ml 50 mM Tris, pH 8.0, 300 mM NaCl, 1 mM ATP, 2 mM MgCl2, 10 mM β-mercaptoethanol, and half a tablet of Complete Protease Inhibitor Cocktail without EDTA. The supernatant obtained after sonication and centrifugation (as with auxilin) was mixed with 1 ml of 50% (vol/vol) slurry of nickel-nitrilotriacetic acid-agarose beads (QIAGEN, Valencia, CA) for 4 h by end-over-end rotation at 4°C. The beads were placed into an Econo Pac column and then washed with 30 ml of 50 mM Tris, pH 8.0, 300 mM NaCl, 10 mM β-mercaptoethanol, 10 mM imidazole, 1 mM ATP, and 1 mM MgCl2). Hsc70 was then eluted at 4°C with 5–6 ml of the same solution supplemented with 200 mM imidazole. Fractions of 1 ml were collected into microcentrifuge tubes containing 40 μl of 0.1 M EGTA. The samples containing 20% glycerol (final concentration) were stored at −80°C.
Preparation of Native Clathrin and AP-2 Clathrin Adaptors
Clathrin and AP-2 were purified from coated vesicles isolated from calf brains as described previously (Gallusser and Kirchhausen, 1993; Boll et al., 1996). Contaminating auxilin was removed by using the same final hydroxylapatite chromatographic step used for the purification of recombinant clathrin. Absence of auxilin (detection limit of 0.0015 μg/μl) was established by Western blot analysis by using the monoclonal antibody 100/4 (gift from Dr. E. Ungewickell, Hanover Medical School, Germany) specific for auxilin. The samples containing 20% glycerol (final concentration) were stored at −80°C.
Assembly of Clathrin Cages
Clathrin cages were assembled at 4°C by overnight dialysis of clathrin triskelions (1 mg/ml) against cage formation buffer (Kirchhausen and Harrison, 1981). The sample was then centrifuged at low speed (12,000 rpm for 10 min at 4°C) to remove large aggregates, and the supernatant was centrifuged at high speed (65,000 rpm for 12 min at 4°C; TLA-100 rotor). The pellet (containing cages) was resuspended at room temperature to ∼1 mg/ml in cage formation buffer and then stored at 4°C.
Assembly of Clathrin Coats
Coats were assembled from a mixture of native or recombinant clathrin triskelions (∼1 mg/ml) and AP-2 adaptors (∼0.4 mg/ml) to a final 3:1 ratio by weight and dialyzed overnight at 4°C against coat buffer (80 mM MES, pH 6.5, 20 mM NaCl, 2 mM EDTA, and 0.4 mM DTT) followed by the low- and high speed centrifugation steps used during the generation of cages. The medium ionic strength is based on previous work (Fotin et al., 2004b), and it was chosen to prevent assembly of cages (lacking AP-2) but with a lower yield of coats (∼40–70% of total protein). The pellet (containing coats) was resuspended at room temperature to ∼1 mg/ml in uncoating buffer (20 mM imidazole, pH 6.8, 2 mM MgCl2, 100 mM KCl, and 1 mM DTT) to a concentration of ∼1 mg/ml and stored at 4°C for up to 5 days.
Uncoating Reaction
The uncoating reaction catalyzed by Hsc70, auxilin, and ATP was carried out with coat samples in a volume of 0.1 ml by using final concentrations of 0.5 mg/ml coats, 0.6 mg/ml Hsc70, 0.1 mg/ml auxilin (547-910), 30 U/ml creatine phosphate kinase (Sigma-Aldrich, St. Louis, MO), 15 mM creatine phosphate (Sigma-Aldrich), and 1 mM ATP-MgCl2 (Sigma-Aldrich). Similar results were obtained using the ammonium phosphate-potassium based buffer used earlier (Greene and Eisenberg, 1990). All the stock solutions were made in uncoating buffer. Hsc70 was preincubated for 8 min with ATP at 37°C and then kept on ice; the remaining components of the reaction were added including uncoating buffer as needed. The mixture was then transferred to 37°C (Eppendorf Thermomixer; 350 rpm) and incubated for 5 min. The uncoating reaction was ended by transfer of the mixture to ice; the sample was centrifuged at high speed (100,000 rpm for 10 min at 4°C; TLA-100 rotor). Then, 2 × 20 μl of supernatant was removed immediately after the end of centrifugation step, followed by removal of the remaining supernatant; the pellet was resupended with 100 μl of uncoating buffer. SDS-polyacrylamide gel electrophoresis (PAGE) and Coomassie Blue staining was used for analysis, with known amounts of native brain clathrin as an internal standard. The gels were imaged with a charge-coupled device camera, and the data analyzed using MacBas, version 2.1 (Fujifilm, Tokyo, Japan).
Electron Microscopy
Samples were examined by negative staining electron microscopy as described previously (Gallusser and Kirchhausen, 1993). Briefly, samples (∼0.05 mg/ml) were applied onto the surface of freshly glow discharged carbon-coated electron microscopy grids, and the adsorbed samples were then washed with several drops of freshly prepared of 1.2% (wt/vol) uranyl acetate, blotted, and air-dried. Images were acquired at 80 kV by using 1200EX-80kV (JEOL, Tokyo, Japan) or G2 Spirit Bio Twin (Fei, Hillsboro, OR) transmission electron microscopes.
RESULTS AND DISCUSSION
Expression and Purification of Recombinant Clathrin
We developed two ways to generate recombinant clathrin triskelions, one way where both recombinant mammalian heavy and light chains are purified after coexpression in insect cells, and a second way where mammalian recombinant light chain expressed in bacteria is added to heavy chains purified after expression in insect cells.
Coexpression of Clathrin Heavy and Light Chain in Insect Cells
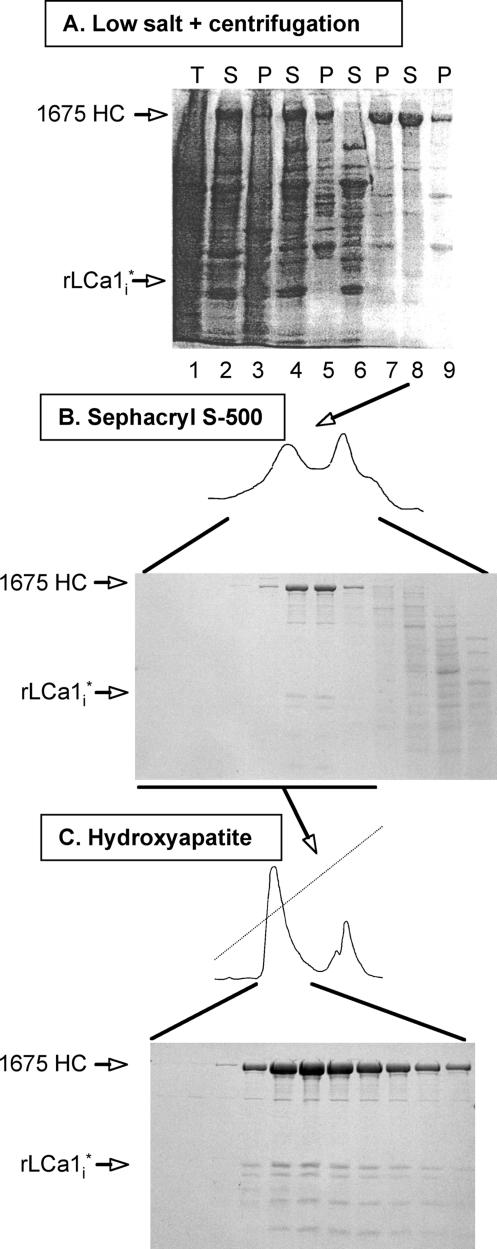
Rat clathrin heavy and light chains were expressed for 2–3 d in insect cells coinfected with baculoviruses encoding heavy and light chain. Most of the recombinant clathrin was recovered in the supernatant fraction after high-speed centrifugation of the cell lysate (Figure 2A, lanes 1–3). For the next purification step, we assumed that the recombinant clathrin would form triskelions (three heavy chains each bound to a light chain), which in turn would spontaneously assemble into lattices if transferred into low-salt conditions. Indeed, after overnight dialysis against low ionic strength conditions, >90% of the recombinant protein was recovered in the pellet fraction of a high-speed centrifugation step (Figure 2A, lane 7). Transfer of the pellet fraction to 0.8 M Tris-HCl pH 7.4 (conditions under which clathrin cages are known to disassemble; Keen et al., 1979) followed by high-speed centrifugation released clathrin into the supernatant fraction (Figure 2A, lane 8), again in good accord with the properties of authentic clathrin cages (Kirchhausen and Harrison, 1981). Sizing chromatography of this material shows that it elutes as a major species (Figure 2B) with a retention volume identical to that of native triskelions. The fractions enriched in clathrin were pooled and eluted from a hydroxyapatite column with a linear gradient of phosphate-containing buffer (Figure 2, panel C); typical yields were ∼3–40 mg clathrin/l of culture.
Figure 2.
Purification of recombinant clathrin. Analysis by SDS-PAGE and Coomassie Blue staining of fractions obtained during the purification of recombinant clathrin expressed in insect cells. This example shows the results for recombinant full-length rat heavy (1675 HC) and the main proteolytic product of light chain LCa1 (rLCa1i*); details of the purification scheme are given in Materials and Methods. (A) Sample after cell lysis (lane 1); high-speed supernatant (lane 2) and pellet (lane 3) of the cell lysate; low-speed supernatant (lane 4) and pellet (lane 5) of sample in lane 2 after dialysis against low ionic strength solution; high-speed supernatant (lane 6) and pellet (lane 7) of sample in lane 4; high-speed supernatant (lane 8) and pellet (lane 9) of sample in lane 7 after addition of Tris solution (to depolymerize clathrin lattices). The samples correspond to equivalent fractions of the input (T), high-speed supernatant (S) and pellet (P) after centrifugation. (B) Fractionation by gel filtration of sample in lane 8. The figure shows fractions enriched in proteins eluting in the included volume; the elution time and profile of the first peak containing recombinant clathrin is the same as that of native triskelions (data not shown). (C) Elution profile of recombinant clathrin from an hydroxyapatite column loaded with the clathrin pool from B (underlined).
Similar expression yields were obtained with heavy chain corresponding to partial (1637 HC, 1643 HC, and 1661 HC) or complete (1630 HC) removal of the unstructured region on the C-terminal side of the tripod helix and with heavy chain constructs with internal deletions (1675ΔPIVYGQ HC, 1643ΔPIVYGQ HC, and 1675ΔQLMLTA HC) or amino acid substitutions (1643LML-AAA HC) in the unstructured region. Removal of the tripod (1596 HC) led to normal expression of the heavy chain but failure to occur in the high-speed pellet after the low salt concentration dialysis step; this observation suggests that the tripod region is required for lattice assembly, as inferred from the 8-Å cryo-electron microscopy (cryo-EM) map (Fotin et al., 2004b). The rat brain clathrin light chain a1 expressed in insect cells (rLCa1i) bound to heavy chain was subject to varied levels of proteolysis among different preparations, hence named here rLCa1i*. The limited proteolytic cleavage of rLCa1i, however, did not affect the outcome of the experiments described below.
In Vitro Reconstitution of Clathrin Triskelions
The complementary approach to generate clathrin is based in the purification of heavy chain triskelions expressed in insect cells followed by the addition of rat brain clathrin light chain a1 purified after their expression in E. coli (LCa1b). This procedure has two advantages. First, it substantially facilitates the expression of the heavy chain in insect cells; and second, it prevents light proteolytic degradation (Figure 3A). The in vitro assembly and uncoating properties of clathrin generated in this manner were indistinguishable from those of clathrin completely expressed in insect cells.
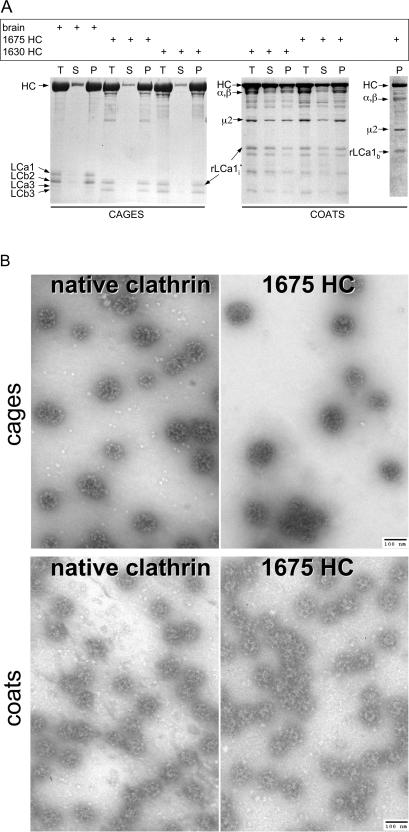
Figure 3.
Cage and coat assembly. (A) SDS-PAGE of cages and coats assembled as described in the text. The samples correspond to equivalent fractions of the input (T), high-speed supernatant (S), and pellet (P) after centrifugation of assemblies generated using native or recombinant clathrin (1675 HC and 1630 HC). Coats were assembled in the presence of AP-2. The bands corresponding to clathrin heavy chain (HC), native bovine brain light chains (LCa1, LCa3, LCb2, and LCb3), and recombinant rat light chain (rLCA1i and rLCA1b) are labeled; native bovine AP-2 large-chains (α/β) and AP-2 medium chain (μ2) are also labeled. The ς chain is not visible in the Figure. (B) Electron micrographs of fields of negatively stained clathrin cages or coats made from native or full-length (1675 HC) recombinant clathrin; coats were assembled in the presence of AP-2. The samples are from pellets obtained by high-speed centrifugation after resuspension in cage or coat buffer, respectively. Bar, 100 nm.
Assembly of Recombinant Triskelions into Cages and Coats
We first established that recombinant clathrin forms cages by comparing triskelions containing full-length recombinant heavy (1775 HC) and light chain (rLCa1i) with native clathrin. Samples of recombinant or native clathrin (∼1 mg/ml) were dialyzed overnight against cage assembly solution, and as expected most of the protein (∼90%) appeared in the pellet fractions after a high-speed centrifugation step (Figure 3A, cages). Cage formation was confirmed by negative stain electron microscopy (Figure 3B, cages); moreover, the images illustrate the similar appearance of cages assembled from recombinant or native clathrin. These observations suggest that the folding and properties of triskelions formed from recombinant and native clathrin are the same. We further confirmed that the recombinant clathrin is functional by showing that it forms coats in the presence of the heterotetrameric adaptor, AP-2. As expected for the assembly buffer we used (Gallusser and Kirchhausen, 1993) ∼50–70% of protein appears in the pellet fraction after high-speed centrifugation (Figure 3A, coats). Electron microscopy of negatively stained samples from the pellets confirmed formation of coats closely resembling those we have studied previously (mostly barrels; Gallusser and Kirchhausen, 1993; Smith et al., 1998; Fotin et al., 2004b) (Figure 3B, coats). We obtained similar results with all of the other heavy chain variants irrespective of their production method (Figure 3A; data not shown).
Hsc70 and Auxilin-dependent Disassembly of Coats Made with Recombinant Triskelions
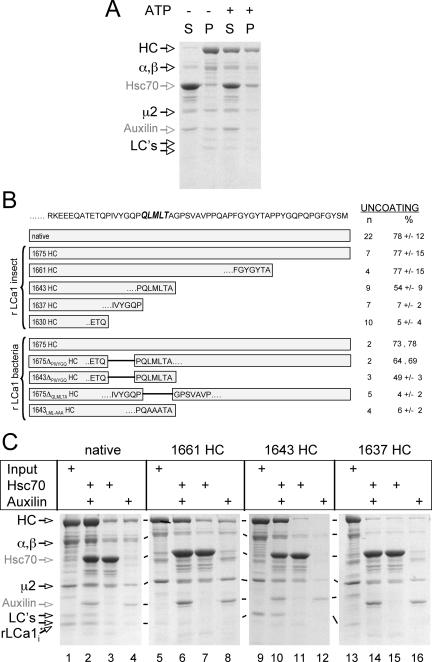
Using a centrifugation assay to follow the in vitro uncoating reaction, we reproduced earlier results showing the release of coat components in a reaction that depends on ATP, Hsc70 and a C-terminal fragment of auxilin containing the clathrin binding segment and the J-domain (auxilin [547-910]) (Ungewickell et al., 1997; Umeda et al., 2000). SDS-PAGE analysis of native clathrin coats subjected to the uncoating reaction showed ATP-dependent release into the supernatant fraction of proteins corresponding to native clathrin (HC and clathrin light chains LCa1 and LCb2 [LC's]) and AP-2 (α/β and μ2) (∼78% release; Figure 4, A and B). As expected, uncoating depends on the combined presence of Hsc70 and the auxilin fragment (Figure 4C, compare the release in lane 2 with the minimal release in lanes 3 and 4), and uncoating fails at 4°C (data not shown). We then used the same assay to test whether coats assembled with full-length recombinant triskelions (1675 HC) are also a substrate. We found that these coats undergo efficient release of coat components (∼77% release; Figure 4B), and we confirmed by electron microscopy that coats had indeed disappeared by the end of the reaction.
Figure 4.
Disassembly of clathrin coats. (A) Clathrin coats assembled in vitro with native clathrin and AP-2 were incubated with Hsc70 and Auxilin for 5 min at 37°C in the absence or presence of ATP, followed by high-speed centrifugation. The protein composition of the supernatants (S) and pellets (P) was analyzed by SDS-PAGE. HC and LCs, bovine brain clathrin heavy and light chains LCa1 and LCb2; α/β and μ2, components of the AP-2 complex. (B) The amino-acid sequence of the C-terminal segment of rat clathrin heavy chain is shown, and the proposed Hsc70 recognition sequence (QLMLT) is highlighted. The schematic representations indicate the truncated heavy chain constructs used in the uncoating experiments. In vitro-assembled coats were subjected to the uncoating reaction, and the extent of uncoating is expressed as percentage of coat proteins remaining in the supernatant of a high-speed centrifugation step as determined by SDS-PAGE analysis. n, number of independent experiments; data are presented as average ± SD. (C) Examples of supernatants analyzed by SDS-PAGE from uncoating reactions after high-speed centrifugation by using as substrate coats assembled with native bovine brain triskelions or with recombinant triskelions made of heavy (1661 HC, 1643 HC, and 1637 HC) and light chains (rLCa1i).
Identification of the Peptide Motif on the Heavy Chain Required for Uncoating
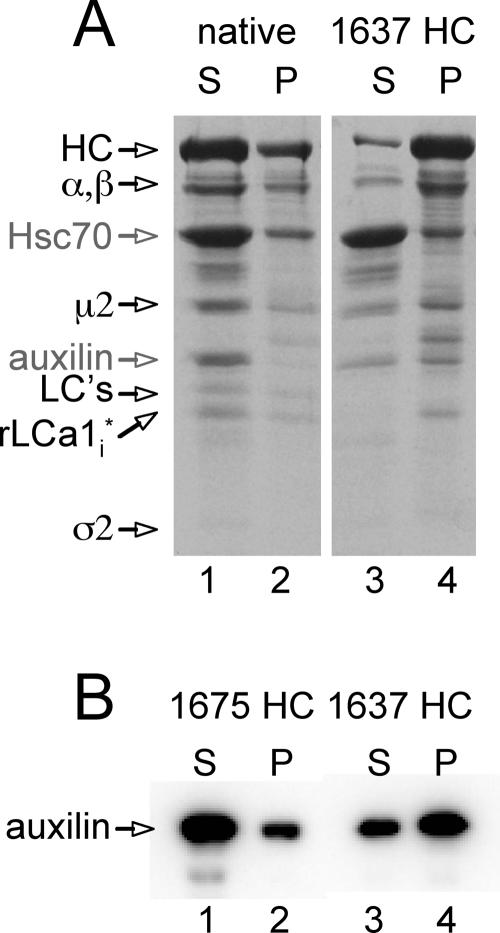
To test whether the QLMLT motif is required for the Hsc70/auxilin-dependent uncoating reaction, we prepared coats from clathrin heavy chain with different C-terminal truncations (summarized in Figure 4B). The efficiency of coat assembly and the appearance of the coats in electron micrographs were similar to those obtained using native clathrin (data not shown). Coats lacking approximately half of the unstructured segment, but still containing the QLMLT motif (1661 HC), are good substrates for the Hsc70/auxilin/ATP-catalyzed reaction, and they depolymerized as efficiently as coats assembled from native clathrin (∼77% release; Figure 4, B and C, lanes 6–8). Disassembly was confirmed by electron microscopy (Figure 5, 1661 HC). Coats assembled from clathrin truncated immediately after the QLMLT motif (1643 HC) showed partial uncoating (∼54% release; Figure 4, B and C, lanes 10–12). Coats assembled from clathrin lacking the QLMLT motif (1637 HC or 1630 HC) barely uncoated (<7% release; Figure 4, B and C, lanes 14–16). In this case, electron microscopy confirmed that the coats in the pellet after Hsc70/auxilin/ATP incubation were intact (Figure 5, 1637 HC). Further evidence for the important role of the QLMLT motif comes from the demonstration that exchange of LML with AAA or removal of the QLMLTA sequence but without deletion of the unstructured region completely prevents uncoating (∼4–6% release; Figure 4B, 1643LML-AAA HC and 1675ΔQLMLTA HC, respectively). In contrast, a small 6-residue shift of the motif toward the hub is tolerated (∼66% release; Figure 4B, 1675ΔPIVYGQ HC). Finally, a few residues immediately adjacent to the C terminus of the motif seem to be required, because their removal partially decreases uncoating efficiency (Figure 4B, 1643 HC and 1643ΔPIVYGQ HC). The inability to depolymerize cannot be attributed to a lack of auxilin binding to coats with truncated clathrin, because the auxilin fragment pelleted with 1637 HC coats, whereas a normal uncoating reaction leads to release of auxilin from its clathrin association and into the supernatant fraction, along with the other coat components (Figure 6). These data suggest that the postulated peptide motif containing the sequence QLMLT (or other related sequences with similar hydrophobic character) is likely to be the uncoating “substrate” recognized by Hsc70. The QLMLT motifs are located under the hub at the far end of the tripod legs (Fotin et al., 2004b), the central position within the triskelion where bound Hsc70 has been visualized by electron microscopy after the uncoating reaction (Heuser and Steer, 1989).
Figure 5.
Visualization of coat disassembly. Electron micrographs of negatively stained coats made from recombinant clathrin lacking part (1661 HC) or the complete C-terminal unstructured segment (1637 HC) and AP-2. The images are from pellets resuspended in coat buffer after high-speed centrifugation of samples subjected to the uncoating reaction in the presence of auxilin and ATP and the presence (top) or absence (bottom) of Hsc70. Bar, 100 nm.
Figure 6.
Binding of auxilin to clathrin coats. Coats assembled with native or recombinant full-length (1675 HC) or C-terminally truncated (1637 HC) clathrin were incubated with Hsc70, auxilin, and ATP and subjected to the uncoating reaction (see Materials and Methods). The products of the reaction were pelleted by high-speed centrifugation, and the protein composition of the supernatants and resuspended pellets was analyzed by SDS-PAGE and Coomassie Blue staining (A) or SDS-PAGE and Western blot (B). The auxilin fragment is soluble and occurs mainly in the supernatant fraction (lane 1) of the uncoated native and 1675 HC coats; in contrast, the auxilin fragment remains mainly in the pellet fraction (lane 4) of the recombinant 1637 HC coats that failed to uncoat: HC, clathrin heavy chain; 1675 HC, full-length recombinant clathrin heavy chain; 1637 HC, C-terminally truncated recombinant clathrin heavy chain; LCs, bovine brain light chains LCa1 and LCb2; rLCa1i, recombinant LCa1 expressed in insect cells; α/β, μ2, and ς2, components of the AP-2 complex; Auxilin, auxilin fragment (547-910).
Clathrin Assembly
Assembly of the C-terminally deleted recombinant clathrin trimers shows that the stability of a coat does not depend strongly on interactions of the 45-residue, nonhelical segment at the C terminus of the heavy chain. One model (the “ankle-brace”), suggested in our previous article (Fotin et al., 2004b) but now ruled out by this observation, was that this region is indeed important for coat integrity, e.g., through a contact with the ankle crossing, and that its capture by Hsc70 is part of the uncoating process. Thus, the interface between parallel distal and proximal segments along each edge of the lattice and perhaps the ankle crossing itself seem to be the principal assembly-promoting contacts. The distal-proximal contact, very much the more extensive of the two, may be the one that drives adaptor-free cage assembly at reduced pH.
Uncoating Mechanisms
Hsp70-like chaperones have an actin-like ATPase domain linked to a specialized, peptide-binding domain (Young et al., 2004). The peptide-binding site lies between two subdomains, a “base” and a “lid.” The latter closes down upon a suitably docked, target peptide. J-domain–containing cochaperones recruit Hsp70-like proteins in an ATP-bound conformation. In this state, the lid subdomain is open, and peptides can access the binding site. ATP hydrolysis, stimulated by the J-domain association and by peptide binding, leads to lid closure, tight peptide association, and release from the cochaperone. ADP-ATP nucleotide exchange permits peptide release and restarts the cycle. The QLMLT sequence shown here to be required for Hsc70-mediated uncoating is closely related to an “optimal” sequence for binding Hsc70, as selected from a phage display library (Takenaka et al., 1995). We conclude that the uncoating mechanism involves binding of this segment in the Hsc70 substrate groove, presumably accompanied by hydrolysis of ATP.
We can imagine two general types of uncoating mechanism, consistent with the structure of the clathrin coat and including engagement of the QLMLT segment by Hsc70:ATP. In one type, ATP hydrolysis and an accompanying conformational change in Hsc70 might cause the chaperone to “pull” on the heavy-chain terminus and to destabilize (e.g., by a torque-induced rotation) its contacts in the lattice. In the other type, Hsc70 could capture the QLMLT segment only during a transient fluctuation of a triskelion from its properly inserted position and would thus lower the barrier to full dissociation by interfering with a return to its equilibrium position. We expect that ongoing efforts to visualize uncoating intermediates by cryo-EM will enable us to distinguish between these models and to provide further mechanistic details.
ACKNOWLEDGMENTS
We are grateful to S. C. Harrison for helpful discussions throughout the project. This work has been supported by National Institutes of Health grant GM-036548 (to T.K.) T.B. is supported by a Human Frontier Science Program Fellowship.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-09-0870) on October 31, 2007.
REFERENCES
- Boll W., Ohno H., Songyang Z., Rapoport I., Cantley L. C., Bonifacino J. S., Kirchhausen T. Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 1996;15:5789–5795. [PMC free article] [PubMed] [Google Scholar]
- Chappell T. G., Welch W. J., Schlossman D. M., Palter K. B., Schlesinger M. J., Rothman J. E. Uncoating ATPase is a member of the 70 kilodalton family of stress proteins. Cell. 1986;45:3–13. doi: 10.1016/0092-8674(86)90532-5. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Boll W., Van Oijen A., Hariharan R., Chandran K., Nibert M. L., Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Fotin A., Cheng Y., Grigorieff N., Walz T., Harrison S. C., Kirchhausen T. Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating. Nature. 2004a;432:649–653. doi: 10.1038/nature03078. [DOI] [PubMed] [Google Scholar]
- Fotin A., Cheng Y., Sliz P., Grigorieff N., Harrison S. C., Kirchhausen T., Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004b;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- Fotin A., Kirchhausen T., Grigorieff N., Harrison S. C., Walz T., Cheng Y. Structure determination of clathrin coats to subnanometer resolution by single particle cryo-electron microscopy. J. Struct. Biol. 2006;156:453–460. doi: 10.1016/j.jsb.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall W. E., Higginbotham M. A., Chen C., Ingram M. F., Cyr D. M., Graham T. R. The auxilin-like phosphoprotein Swa2p is required for clathrin function in yeast. Curr. Biol. 2000;10:1349–1358. doi: 10.1016/s0960-9822(00)00771-5. [DOI] [PubMed] [Google Scholar]
- Gallusser A., Kirchhausen T. The beta 1 and beta 2 subunits of the AP complexes are the clathrin coat assembly components. EMBO J. 1993;12:5237–5244. doi: 10.1002/j.1460-2075.1993.tb06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene L. E., Eisenberg E. Dissociation of clathrin from coated vesicles by the uncoating ATPase. J. Biol. Chem. 1990;265:6682–6687. [PubMed] [Google Scholar]
- Greener T., Grant B., Zhang Y., Wu X., Greene L. E., Hirsh D., Eisenberg E. Caenorhabditis elegans auxilin: a J-domain protein essential for clathrin-mediated endocytosis in vivo. Nat. Cell Biol. 2001;3:215–219. doi: 10.1038/35055137. [DOI] [PubMed] [Google Scholar]
- Greener T., Zhao X., Nojima H., Eisenberg E., Greene L. E. Role of cyclin G-associated kinase in uncoating clathrin-coated vesicles from non-neuronal cells. J. Biol. Chem. 2000;275:1365–1370. doi: 10.1074/jbc.275.2.1365. [DOI] [PubMed] [Google Scholar]
- Gruschus J. M., Han C. J., Greener T., Ferretti J. A., Greene L. E., Eisenberg E. Structure of the functional fragment of auxilin required for catalytic uncoating of clathrin-coated vesicles. Biochemistry. 2004;43:3111–3119. doi: 10.1021/bi0354740. [DOI] [PubMed] [Google Scholar]
- Haynie D. T., Ponting C. P. The N-terminal domains of tensin and auxilin are phosphatase homologues. Protein Sci. 1996;5:2643–2646. doi: 10.1002/pro.5560051227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser J., Steer C. J. Trimeric binding of the 70-kD uncoating ATPase to the vertices of clathrin triskelia: a candidate intermediate in the vesicle uncoating reaction. J. Cell Biol. 1989;109:1457–1466. doi: 10.1083/jcb.109.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstein S. E., Ungewickell H., Ungewickell E. Mechanism of clathrin basket dissociation: separate functions of protein domains of the DnaJ homologue auxilin. J. Cell Biol. 1996;135:925–937. doi: 10.1083/jcb.135.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen J. H., Willingham M. C., Pastan I. H. Clathrin-coated vesicles: isolation, dissociation and factor-dependent reassociation of clathrin baskets. Cell. 1979;16:303–312. doi: 10.1016/0092-8674(79)90007-2. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Clathrin. Annu. Rev. Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T., Harrison S. C. Protein organization in clathrin trimers. Cell. 1981;23:755–761. doi: 10.1016/0092-8674(81)90439-6. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T., Harrison S. C., Chow E. P., Mattaliano R. J., Ramachandran K. L., Smart J., Brosius J. Clathrin heavy chain: molecular cloning and complete primary structure. Proc. Natl. Acad. Sci. USA. 1987a;84:8805–8809. doi: 10.1073/pnas.84.24.8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T., Scarmato P., Harrison S. C., Monroe J. J., Chow E. P., Mattaliano R. J., Ramachandran K. L., Smart J. E., Ahn A. H., Brosius J. Clathrin light chains LCA and LCB are similar, polymorphic and share repeated heptad motifs. Science. 1987b;236:320–324. doi: 10.1126/science.3563513. [DOI] [PubMed] [Google Scholar]
- Kruchten A. E., McNiven M. A. Dynamin as a mover and pincher during cell migration and invasion. J. Cell Sci. 2006;119:1683–1690. doi: 10.1242/jcs.02963. [DOI] [PubMed] [Google Scholar]
- Lee D. W., Wu X., Eisenberg E., Greene L. E. Recruitment dynamics of GAK and auxilin to clathrin-coated pits during endocytosis. J. Cell Sci. 2006;119:3502–3512. doi: 10.1242/jcs.03092. [DOI] [PubMed] [Google Scholar]
- Macia E., Ehrlich M., Massol R., Boucrot E., Brunner C., Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Massol R. H., Boll W., Griffin A. M., Kirchhausen T. A burst of auxilin recruitment determines the onset of clathrin-coated vesicle uncoating. Proc. Natl. Acad. Sci. USA. 2006;103:10265–10270. doi: 10.1073/pnas.0603369103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield C. J., Perrais D., Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Smith C. J., Roseman A. M., Harrison S. C., Kirchhausen T., Pearse B. M. Functional organization of clathrin in coats: combining electron cryomicroscopy and X-ray crystallography. Mol. Cell. 1999;3:761–770. doi: 10.1016/s1097-2765(01)80008-3. [DOI] [PubMed] [Google Scholar]
- Owen D. J., Collins B. M., Evans P. R. Adaptors for clathrin coats: structure and function. Annu. Rev. Cell Dev. Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Pishvaee B., Costaguta G., Yeung B. G., Ryazantsev S., Greener T., Greene L. E., Eisenberg E., McCaffery J. M., Payne G. S. A yeast DNA J protein required for uncoating of clathrin-coated vesicles in vivo. Nat. Cell Biol. 2000;2:958–963. doi: 10.1038/35046619. [DOI] [PubMed] [Google Scholar]
- Praefcke G. J., McMahon H. T. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Robinson M. S. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Scheele U., Kalthoff C., Ungewickell E. Multiple interactions of auxilin 1 with clathrin and the AP-2 adaptor complex. J. Biol. Chem. 2001;276:36131–36138. doi: 10.1074/jbc.M106511200. [DOI] [PubMed] [Google Scholar]
- Schmid S. L., Braell W. A., Rothman J. E. ATP catalyzes the sequestration of clathrin during enzymatic uncoating. J. Biol. Chem. 1985;260:10057–10062. [PubMed] [Google Scholar]
- Schmid S. L., Rothman J. E. Two classes of binding sites for uncoating protein in clathrin triskelions. J. Biol. Chem. 1985;260:10050–10056. [PubMed] [Google Scholar]
- Sever S. Dynamin and endocytosis. Curr. Opin. Cell Biol. 2002;14:463–467. doi: 10.1016/s0955-0674(02)00347-2. [DOI] [PubMed] [Google Scholar]
- Sever S., Skoch J., Bacskai B. J., Newmyer S. L. Assays and functional properties of auxilin-dynamin interactions. Methods Enzymol. 2005;404:570–585. doi: 10.1016/S0076-6879(05)04050-4. [DOI] [PubMed] [Google Scholar]
- Smith C. J., Grigorieff N., Pearse B. M. Clathrin coats at 21 Å resolution: a cellular assembly designed to recycle multiple membrane receptors. EMBO J. 1998;17:4943–4953. doi: 10.1093/emboj/17.17.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka I. M., Leung S. M., McAndrew S. J., Brown J. P., Hightower L. E. Hsc70-binding peptides selected from a phage display peptide library that resemble organellar targeting sequences. J. Biol. Chem. 1995;270:19839–19844. doi: 10.1074/jbc.270.34.19839. [DOI] [PubMed] [Google Scholar]
- Umeda A., Meyerholz A., Ungewickell E. Identification of the universal cofactor (auxilin 2) in clathrin coat dissociation. Eur. J. Cell Biol. 2000;79:336–342. doi: 10.1078/S0171-9335(04)70037-0. [DOI] [PubMed] [Google Scholar]
- Ungewickell E. The 70-kD mammalian heat shock proteins are structurally and functionally related to the uncoating protein that releases clathrin triskelia from coated vesicles. EMBO J. 1985;4:3385–3391. doi: 10.1002/j.1460-2075.1985.tb04094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E., Branton D. Assembly units of clathrin coats. Nature. 1981;289:420–422. doi: 10.1038/289420a0. [DOI] [PubMed] [Google Scholar]
- Ungewickell E., Ungewickell H., Holstein S. E. Functional interaction of the auxilin J domain with the nucleotide- and substrate-binding modules of Hsc70. J. Biol. Chem. 1997;272:19594–19600. doi: 10.1074/jbc.272.31.19594. [DOI] [PubMed] [Google Scholar]
- Ungewickell E., Ungewickell H., Holstein S. E., Lindner R., Prasad K., Barouch W., Martin B., Greene L. E., Eisenberg E. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- Vigers G. P., Crowther R. A., Pearse B. M. Location of the 100 kD-50 kD accessory proteins in clathrin coats. EMBO J. 1986a;5:2079–2085. doi: 10.1002/j.1460-2075.1986.tb04469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigers G. P., Crowther R. A., Pearse B. M. Three-dimensional structure of clathrin cages in ice. EMBO J. 1986b;5:529–534. doi: 10.1002/j.1460-2075.1986.tb04242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. C., Agashe V. R., Siegers K., Hartl F. U. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- Yu A., Rual J. F., Tamai K., Harada Y., Vidal M., He X., Kirchhausen T. Association of Dishevelled with the clathrin AP-2 adaptor is required for Frizzled endocytosis and planar cell polarity signaling. Dev. Cell. 2007;12:129–141. doi: 10.1016/j.devcel.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]